Abstract
This protocol describes a culture system in which inner ear sensory tissue is produced from mouse embryonic stem cells under chemically defined conditions. This model is amenable to basic and translational investigations into inner ear biology and regeneration. In this protocol, mouse embryonic stem cells are aggregated in 96-well plates in medium containing extracellular matrix proteins to promote epithelialization. During the first 14 days a series of precisely timed protein and small molecule treatments sequentially induce epithelia that represent the mouse embryonic nonneural ectoderm, preplacodal ectoderm and otic vesicle epithelia. Ultimately, these tissues develop into cysts with a pseudostratified epithelium containing inner ear hair cells and supporting cells after 16–20 days. Concurrently, sensory-like neurons generate synapse-like structures with the derived hair cells. We have designated the stem cell-derived epithelia harboring hair cells, supporting cells and sensory-like neurons inner ear organoids. This method provides a reproducible and scalable means to generate inner ear sensory tissue in vitro.
INTRODUCTION
The proper development of organs requires the synchronized organization and coordinated differentiation of large cell populations. To date, a full appreciation of the molecular and cellular signaling mechanisms that govern cellular behaviors and cell fate decisions remains elusive. Culturing embryonic stem (ES) cells as a floating cell aggregate in three-dimensional (3D) culture has emerged as an effective method to generate complex organs in vitro; a controllable and readily accessible environment to investigate the mechanisms underlying organogenesis1,2. This approach has been used to generate mouse and human cortical tissue, neural retina, liver buds, spermatozoa, intestinal epithelium, anterior pituitary gland tissue, and inner ear sensory epithelium3–13. Moreover, in a 3D culture variant using tissue specific stem cells, the villi of the small intestine and liver tissue can be reconstituted from a single adult stem cell14,15. The key advantage of 3D culture over a monolayer (2D) culture is that the differentiating cells are free to self-organize into epithelia. In a 2D culture, by contrast, the cells adhere to a culture plate; thus, they are restricted to growing on a flat shape. The 3D environment allows cells to develop more naturally, as they would in the embryo.
In this protocol we describe a 3D culture method for the generation of inner ear sensory epithelia containing hair cells, which display some functional properties of native vestibular hair cells and are innervated by sensory-like neurons (Figure 1). To aid the reader with performing the method, we place special emphasis on the proper timing of treatments and the various morphological transformations that take place throughout the differentiation. Additionally, we provide instructions on how to prepare the derived tissue for immunohistochemical and electrophysiological analyses.
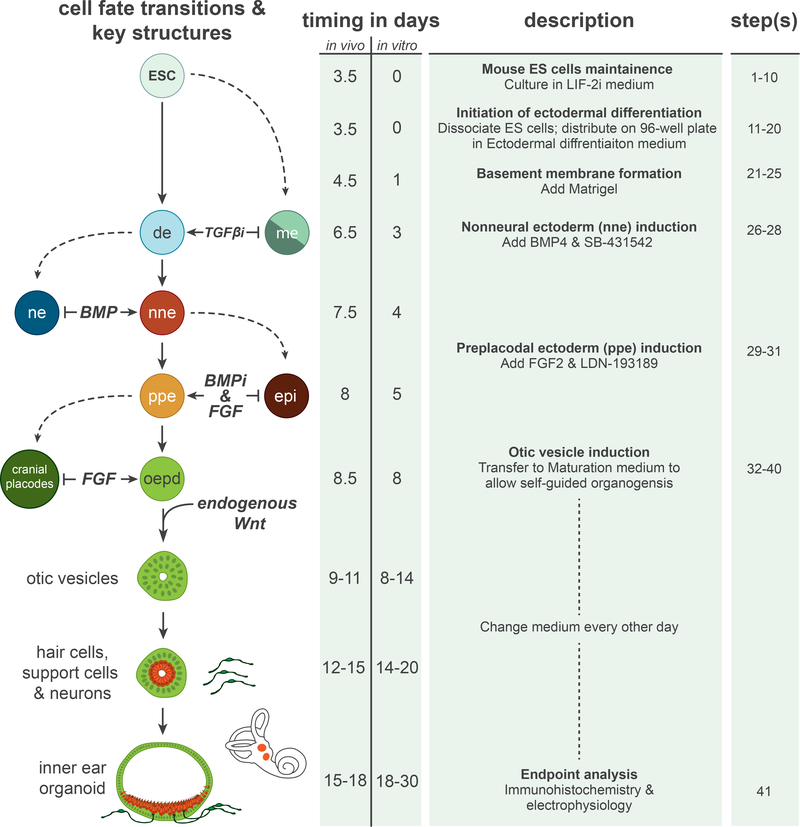
Figure 1: Overview of inner ear induction protocol.
A comparison of cell fate and morphological transitions that take place during in vivo and in vitro inner ear development. Note that the in vivo time points are approximate values derived from mouse developmental studies. Moreover, the cell fate transition model depicted in the left-hand column is a generalization of models derived from studies in various vertebrate species. The preplacodal fate, for instance, has not been as clearly defined in the mouse system as it has in the Xenopus, chick and zebrafish models. The right-hand column describes the experimental procedures and step numbers associated with each cell fate transition in the culture. ESC, embryonic stem cells; de, definitive ectoderm; nne, nonneural ectoderm; ne, neural ectoderm; me, mesendoderm; ppe, preplacodal ectoderm; epi, epidermis; oepd, otic-epibranchial placode domain.
Induction of the cranial placodes and inner ear during embryogenesis
The conditions described here for inner ear induction were selected on the basis of information gained from studies of mouse, chick, zebrafish, and Xenopus ear development. In general, vertebrate inner ear induction begins with the bifurcation of the definitive ectoderm into the nonneural and neural ectoderm. This occurs at approximately embryonic day (E) 6.5–7.5 in mice16–19. Studies in Xenopus, zebrafish and mouse ES cells have shown that bone morphogenetic protein (BMP) signaling is critical for ectodermal specification; high BMP signaling induces nonneural ectoderm in the lateral ectoderm, while low BMP signaling promotes neural plate formation at the midline20–25. At around E7.5–8, the preplacodal ectoderm develops in the head region of the embryo at the border between the nonneural and neural ectoderm, referred to as the preplacodal region (PPR). In vertebrates, attenuation of BMP signaling and concurrent fibroblast growth factor (FGF) signaling is necessary for PPR formation22,26–31. From approximately E8–9, varying inductive cues along the anterior-posterior axis act on the PPR to generate cranial placodes28,32,33. A defining characteristic of each placode—the adenohypophyseal (anterior pituitary gland), olfactory, lens, trigeminal, epibranchial and otic placodes—is a thicker pseudostratified morphology relative to the surrounding nonneural ectoderm28,32. Pax proteins can be used to distinguish placodes along the anterior-posterior axis: Pax6 is expressed in anterior placodes (adenohypophyseal, olfactory, and lens), Pax3 is expressed in the trigeminal placode, and Pax8 is expressed in posterior placodes (otic and epibranchial)34–36. The otic and epibranchial placodes arise from a broad Pax8+ region known as the otic-epibranchial placode domain (OEPD)36. In mice, the OEPD is specified by FGF signaling, attributable to Fgf3, Fgf8, and Fgf10 proteins emanating from the underlying mesenchyme and hindbrain region of the neural tube37,38. Further definition of the OEPD entails Wnt activity to specify the otic placode, whereas prolonged FGF signaling and additional BMP signaling specifies the epibranchial placodes39–41. Otic specification is denoted by an increase in expression of Pax2 protein39,42. From E8–8.5, the otic placode invaginates into the periotic mesenchyme to form the otic pit. The otic vesicle (also known as the otocyst) is formed when the otic pit pinches off from the surface ectoderm. The nonneural ectoderm surrounding the invaginating otic placode gives rise to epidermis on the surface of the embryo overlaying the development inner ear39.
The nascent otic vesicle is patterned into a nonsensory and prosensory region by various inductive cues from the surrounding mesenchyme and neural tube43. The prosensory region can be characterized by expression of Pax2, Pax8, Sox2, and Jag143,44. From E9–10, epithelial cells in the prosensory region either delaminate to become inner ear sensory neurons or remain in the epithelium to contribute to the sensory epithelia of the vestibular or cochlear organs45. During sensory epithelium development, Pax2 and Pax8 are downregulated, but Sox2 and Jag1 expression are maintained43. By E12.5 in the vestibule, the sensory epithelia contain supporting cells (CyclinD1+ Sox2+) and hair cells (Sox2+ Myo7a+ Brn3c+ Calretinin+)46–48.
Development of the protocol
Previous studies have shown that mouse pluripotent stem (PS) cells can be dissociated and aggregated into uniformly shaped spheroids in U-bottomed 96-well plates5–7,49. Culturing aggregates in a serum-free medium containing knockout serum replacement (KSR) promotes development of a neural epithelium. KSR is a defined medium-supplement optimized to grow pluripotent stem cells (Patent no. WO 98/30679). The efficient differentiation of mouse ES cells into neural ectoderm in medium containing KSR is thought to be due to the default pathway of neural ectodermal development49,50. KSR does not contain any factors important for mesendoderm (Activin/Nodal/BMP proteins) or nonneural (BMP proteins) induction. Modifying the signaling factors presented to the neural epithelium can induce specific regions of the forebrain, midbrain and hindbrain. Until recently, developing neuroepithelia within these cultures took the form of dispersed epithelial vesicles embedded in unorganized tissue. In a critical evolution of the method, Eiraku and colleagues demonstrated that extracellular matrix proteins (i.e. in the form of Matrigel) could be added to the culture medium to promote the development of a basement membrane on the surface of the aggregate5. With the structural support, the entire surface layer of cells self-organizes into a neural epithelium after 5 days in culture. These aggregates spontaneously developed into optic cups, which gave rise to complete neural retinas in about 18–20 days. This work provided proof-of-principle that organogenesis of complex sensory organs could occur in vitro.
The aforementioned studies were successful at deriving mature neural ectoderm tissues; however, the cranial placodes—including the otic placode—are derived from the nonneural ectoderm. Suga and coworkers recently demonstrated that by increasing the size of the aggregate, a nonneural ectoderm layer spontaneously arises on the surface of the cell aggregates leaving a neural layer underneath9. Mimicking in vivo nonneural induction, their data suggest that endogenous BMP signaling is likely mediating this process in 3D culture; however, the precise signaling mechanisms are unclear. Using a sonic hedgehog agonist they demonstrated sequential induction of adenohypophyseal placodes, vesicles, and functional anterior pituitary tissue from the nonneural epithelium. This result revealed that placode morphogenesis could be recapitulated in 3D culture. Specifically, placodal invagination and vesicle formation, two critical morphological steps in both anterior pituitary and inner ear development, were well preserved in the culture system.
In our recent study3, we garnered greater control over the nonneural induction process by treating smaller, Matrigel-conditioned, aggregates with BMP4. The timing of BMP4 treatment (day 3) was critical; the surface layer of cells needed time to organize into an epithelium and commit to an ectodermal fate. If we waited until day 5, cells on the surface layer became committed to a neural epithelium fate that could not be reversed by BMP4 treatment. We also demonstrated that TGFß signaling must be inhibited for proper nonneural induction in 3D culture. We treated aggregates with a combination of BMP4 and a TGFß inhibitor, SB-431542 (SB). TGFß inhibition led to a reduction in the number of Brachyury+ mesendoderm-like cells following BMP4 treatment. This effect is consistent with the well-known role of TGFß signaling in mesoderm induction, as well as previous demonstrations that TGFß inhibition promotes ectoderm differentiation from ES cells51–53. From embryo studies, we knew that BMP signaling—although necessary for nonneural induction—needed to be attenuated for preplacodal induction22,28,32. By our estimation, 24–48 hours elapses between nonneural and preplacodal induction in the mouse embryo; thus, we added a BMP inhibitor, LDN-193189 (LDN), to the medium between days 4 and 5 after BMP/SB treatment. FGF signaling is also important for preplacodal induction. When we performed a combined treatment of FGF2 and LDN (FGF/LDN), the outer-epithelium developed a placode-like morphology and expressed several marker genes of the preplacodal ectoderm, including Six1, AP2, and Gata33.
At day 6 of differentiation, patches of cells in the outer epithelium began to express Pax8, a sign of OEPD development. This level of specificity was unexpected because BMP inhibition and FGF signaling are necessary for development of the entire PPR not just the OEPD28. One explanation may be that FGF2 and insulin found in the knockout serum replacement have a synergistic caudalizing effect on the tissue; this phenomenon was previously observed while deriving caudal portions of the brain in 3D culture54. We placed aggregates with Pax8-positive cells in a minimal medium known to support self-organizing tissues, to test if OEPD epithelium developed into otic vesicles. Under these conditions, the OEPD epithelium gave rise to vesicles containing Pax2+ Pax8+ Sox2+ and Jag1+ cells, reminiscent of prosensory cells in the otic vesicle44,55,56. By treating aggregates with a potent Wnt inhibitor we discovered that endogenous Wnt activity was involved in the process of vesicle production; Wnt inhibitor-treated aggregates produced few otic vesicles. With no additional treatments, aside from medium changes, cells in the epithelia of otic vesicles developed into hair cells and supporting cells. The derived hair cells expressed Myo7a, Brn3c, Calretinin, Sox2, and Pax2, denoting a vestibular hair cell identity47,48,57,58. This conclusion was further reinforced by the presence of pointed stereocilia bundles and elongated kinocilia with the length of greater than 10μm3,59. Moreover, the hair cells were spatially organized into clusters seen in the vestibular end organs, rather than in rows of hair cells in the organ of Corti of the cochlea. Quantitative analysis revealed that ~1500 hair cells developed in each cell aggregate by day 20 of differentiation.
We also consistently observed the development of sensory neuron-like cells that arose in discrete groups alongside the vestibular sensory epithelia. We observed ßIII-tubulin (TUJ1) and neurofilament-positive cells extending their processes toward hair cells and analysis of synaptic proteins confirmed the presence of synapse-associated proteins on the basal end of hair cells. CtBP2-positive puncta, whose number increased over time in culture, suggested the development of ribbon synapses in stem cell-derived hair cells. More extensive analysis, such as transmission electron microscopy and electrophysiological recording from button afferents, will be needed to confirm whether the synapse-like structures truly represent functional ribbon synapses.
Comparison with other methods
Several methods for generating inner ear cells from PS cells have been developed in recent years60–63. Of note, Oshima and colleagues provided the first demonstration that functional inner ear hair cells could be derived from ES and iPS cells61. The hair cells they derived had stereocilia and a kinocilium with similar architecture to native vestibular hair cells, expressed many hair cell-associated markers, and were shown to be mechanosensitive. They used FBS, insulin-like growth factor-1, Dkk1 (a Wnt signaling inhibitor) and SIS3 (a Smad3 inhibitor) followed by FGF2 treatment to stimulate otic progenitor development in stem cell aggregates. In order for these otic progenitors to develop into hair cell-like cells, however, they had to be plated on a mitotically inactivated layer of chicken utricle cells61. The need for exogenous tissue may limit the routine use of this method. Moreover, the efficiency of hair cell induction was low, accounting for only ~1–2% of the total population of cells. Additional methods utilized a monolayer culture to derive hair cells; thus, limiting the ability to compare the morphology of stem cell-derived and in vivo tissues. Moreover, stereocilia bundles with a kinocilium have not been observed in a completely monolayer format (i.e. without exogenous supporting/stromal cells). Notably, Ouji et al. recently reported two methods to produce Myo7a-positive cells from stem cells in a monolayer format62,64. They observed F-actin-positive protrusions from the cells reminiscent of hair bundles; however, individual stereocilia were not visible and they did not report whether a kinocilium was present. Consequently, it is unclear if the hair cell-like cells can attain the proper morphology under the conditions described by Ouji and collegues62,64. The method described here is advantageous because it does not require exogenous tissue, undefined media components, and generates inner ear sensory epithelia that appear to be structurally and physiologically comparable to native vestibular sensory epithelium. The average number of ~1,500 hair cells produced per aggregate—based on analysis of four separate experiments—is significant considering that each adult mouse inner ear contains ~30,000 hair cells (i.e. in both the vestibule and cochlea combined)65. Thus, ~20 aggregates provide a comparable number of hair cells to the entire inner ear.
Future applications
Investigating the signaling mechanisms and gene regulatory networks underlying nonneural, preplacodal and inner ear induction in mouse embryos is challenging; mouse embryos are inaccessible in vivo and cannot be maintained ex vivo for more than 2 days. Moreover, it is difficult to collect the large amount of embryonic tissue needed to perform biochemical analyses such as chromatin immunoprecipitation. The described 3D culture system addresses these issues because it is both accessible and the derived tissues are highly representative of their embryonic counterparts. We routinely perform experiments involving 200–400 aggregates, yet the method can be easily scaled to produce more aggregates. For larger scale experiments, a researcher should be able to simultaneously maintain 20 or more 96-wells plates totaling ~2000 aggregates. Further increases in scale may be attainable with automation. Besides studying early otic placode induction, this culture system can be used to evaluate hair cell and inner ear sensory neuron development, as well as hair cell-sensory neuron synaptogenesis. Additionally, recent advances in gene targeting technology may also be applied to this culture model to knockout and knock-in genes to study inner ear phenotypes66,67. The 20–30 day, all in vitro, format is a quick and cost effective alternative to generating an entire mouse line if inner ear analysis is all that is desired. The 3D culture system may also be suitable for mid- to high-throughput drug screening to uncover novel compounds with regenerative or toxic effects on inner ear sensory epithelia; however, several of the limitations discussed in the next section will need to be addressed to make the system more amenable to high-throughput applications. In addition, translation of our findings to human ES cells or iPS cells will be critical for potential applications in cellular therapies for vestibular disorders or hearing loss.
Limitations
Heterogeneity is both an advantage and a limitation of this 3D culture system. Unlike a monolayer format, cells in an aggregate receive varying concentrations of signaling cues depending on their spatial location within the aggregate. This often leads to unsynchronized differentiation of cells located at different depths of the aggregate; cells in the surface layer differentiate faster than those in the core. In our system, BMP4 signaling induces a nonneural ectoderm surface layer as well as Brachyury+ mesendodermal cells in the central core of the aggregate. BMP4 is known to induce mesendodermal cells from PS cells. Cells in the central core express the pluripotency gene Nanog through day 3 of differentiation; thus, BMP4 likely diffuses through the surface layer of the aggregates to act on these pluripotent cells. This cell layer gives rise to mesenchyme-like cells that eventually cover the surface of the aggregates and give rise to muscle, cartilage and adipose3. The presence of these cell types does not appear to encumber, and may play an essential role in, otic induction. Indeed, the periotic mesenchyme in vivo is an integral component of inner ear morphogenesis68. Due to this heterogeneity, however, the method would benefit from the use of florescent reporter PS cell lines to help identify key phenotypic changes. Specifically, PS cells containing fluorescently tagged marker genes for nonneural, preplacodal, otic prosensory cells, hair cells, supporting cells and inner ear neurons would be highly amenable to this method. Most notably, incorporation of reporter cell lines would facilitate florescence-activated cell sorting (FACS) analysis of key inner ear cell types. Another limitation is that the method only produces vestibular sensory organ-like structures. Further modifications to the protocol will be needed to investigate induction and differentiation of cochlear cell types.
Experimental design
ES cell culture in LIF-2i medium (prior to start of procedure).
We maintain our ES cells in Leukemia inhibitory factor (LIF)-2i medium because we have observed considerably less spontaneous differentiation under these conditions compared to fetal bovine serum or fetal calf serum (FBS/FCS)-based culture media. LIF-2i medium has been shown to restrict ES cells to a naïve state of pluripotency; therefore, it ensures a homogenous population of undifferentiated cells69. If acclimating ES cells grown under different conditions (e.g. in media containing FBS/FCS) to LIF-2i medium is required, we recommend passaging the cells 5–10 times prior to beginning a differentiation experiment to ensure that the cell population is homogeneous. We typically passage the cells 2–3 times after thawing before beginning a differentiation experiment. ES cells should be ~80% confluent before passaging or starting a differentiation experiment.
Nonneural and preplacedal induction (steps 1–31).
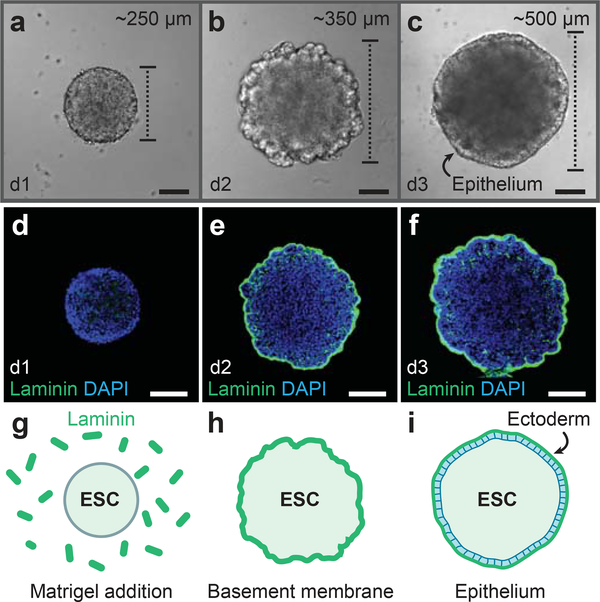
To start ectodermal differentiation, ES cells are dissociated and distributed, 3,000 cells per well, onto 96-well plates. The medium contains 1.5% knockout serum replacement which has been previously shown to be optimal for generating neural retinas5. This concentration also appears to be optimal for preplacodal induction, as higher concentrations seem to prohibit proper epithelium development. Matrigel, which contains extracellular matrix proteins such as Laminin and Entactin, should be added on day 1 at a final concentration of 2% (vol/vol; Fig. 2). We recommend using the growth factorreduced Matrigel. If a more defined medium composition is desired, a recent report suggests that a purified Laminin/Entactin complex can be used to replace Matrigel10. Likewise, we have used Geltrex (Gibco; cat. no. A1413202) with comparable results to Matrigel. With an optimal composition of extracellular matrix proteins, an epithelium should appear by day 3 (Fig. 2c), at which point BMP4 and the TGFß inhibitor SB-431542 (SB) are added to the culture medium.
Figure 2: Development of the definitive ectoderm epithelium.
a-c, DIC images of representative aggregates on days 1, 2, and 3 of 3D culture. d-i, Following Matrigel addition on day 1, Laminin is incorporated into a basement membrane (e, f, h, i) on the surface of the aggregate. An epithelium develops on the aggregate surface by day 3. See Table 1 for a list of antibodies used for characterization. ESC, embryonic stem cells. Scale bars, 100 μm
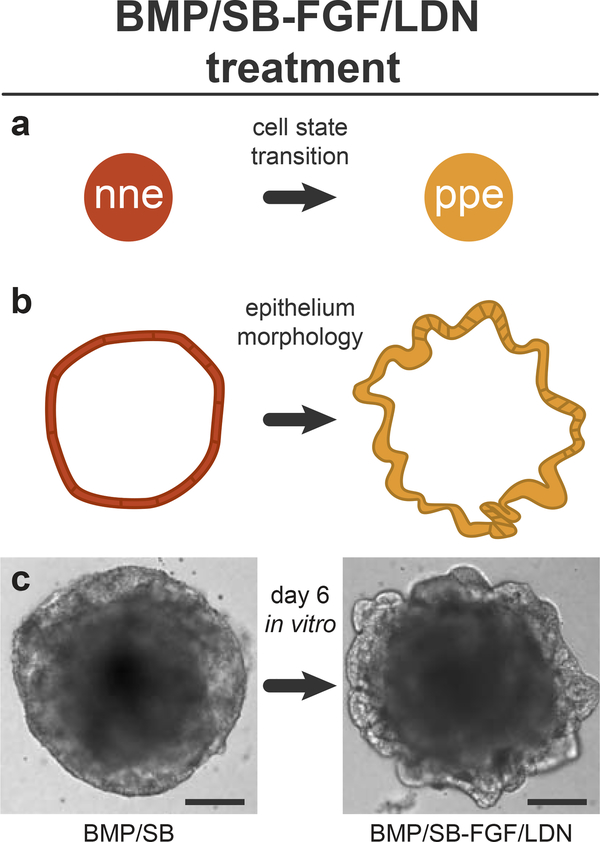
When attempting the protocol for the first time, we recommend performing several control treatments. At the least, set aside 8 aggregates (i.e. one vertical column) for each of these treatments: untreated control, BMP, SB, BMP/SB, BMP/SB-FGF, BMP/SB-LDN, and BMP/SB-FGF/LDN. We suspect that the optimal timing of treatments may vary slightly between ES cell lines. In our experience, the timing of FGF/LDN treatment is critical and should be varied between days 4 and 5 to optimize the culture. In our previous publication3, we used a powder form of LDN diluted in DMSO at a working concentration of 100 nM LDN; however, lot-to-lot variability was a recurrent issue. We now recommend using a pre-diluted version of LDN at a higher working concentration of 1 μM. The outer-epithelium of BMP/SB-FGF/LDN treated aggregates should thicken and express Pax8 by days 6–8 (Fig. 3 and 4).
Figure 3: Morphology changes following BMP/SB-FGF/LDN treatment.
a, FGF/LDN treatment induces preplacodal ectoderm from nonneural ectoderm. b, c, The morphology of the nonneural ectoderm layer on the surface of the aggregates thickens and ruffles in response to FGF/LDN treatment. Scale bars, 100 μm
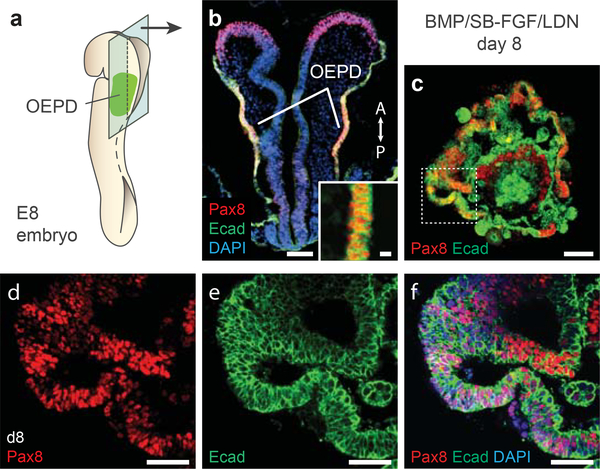
Figure 4: Aggregate organization on day 8 of differentiation.
a, b, The epithelium of the OEPD in an E8 mouse embryo and (c-f) the outer-epithelium of BMP/SB-FGF/LDN treated aggregates express Pax8 and Ecad. See Table 1 for a list of antibodies used for characterization. Panel b was previously published in Supplementary Figure 6 of Koehler et al. Nature 2013. All animal experiments were performed in accordance with the Indiana University Institutional Animal Care and Use Committee (IACUC) guidelines. Scale bars, 100 (b,c), 30 (d-f), and 10 (b-inset) μm.
Otic prosensory vesicle formation and hair cell induction (steps 32–29).
We describe two formats for long-term culture: 96- and 24-well formats. Although more labor-intensive, the 96-well format may be advantageous for analyses requiring the tracking of individual aggregates for the duration of the culture. To initiate self-organized development of otic vesicles, day 8 aggregates are transferred into a minimal medium containing DMEM/F12 and N2 supplement. The inner core of mesodermal and neural cells should migrate out to cover the surface of the aggregate (Fig. 5). Pax2+ Pax8+ vesicles should evaginate from the OEPD epithelium into this layer of cells between days 8–12 (Fig. 6a, b, d). By days 12–14, otic vesicles should express the prosensory markers Sox2 and Jag1 (Fig. 6c). Vesicles should also express Myo7a in a diffuse pattern similar to the developing otic vesicle in vivo3.
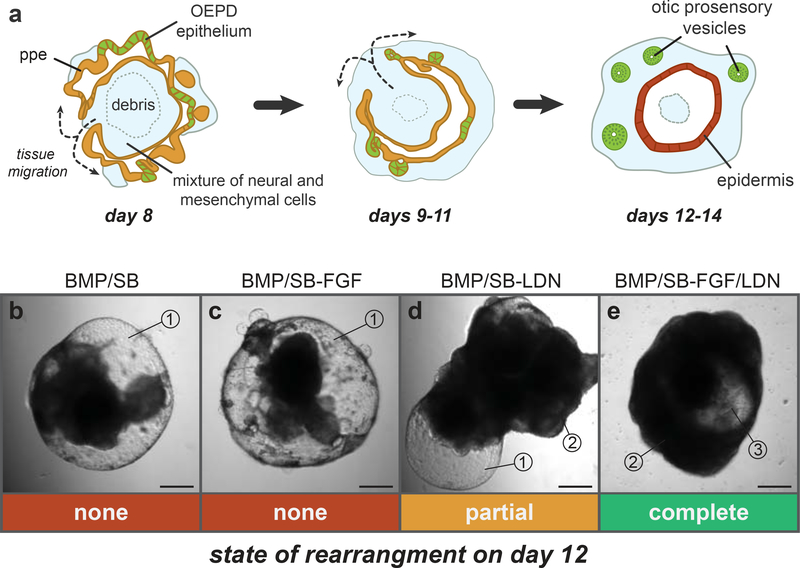
Figure 5: Cellular rearrangement between days 8 and 12 of differentiation.
a, Schematic of the stages of rearrangement during days 8, 9–11 and 12–14. b-e, Day 12 aggregates that received treatment with BMP/SB (b), BMP/SB-FGF (c), BMP/SB-LDN (d) or BMP/SB-FGF/LDN (e). The numbered indicators denote key characteristics to look for when assessing the state of cellular rearrangement: 1) An outer-layer of translucent epithelium indicates the development of epidermis and improper cellular rearrangement, 2) an outer-layer of opaque tissue indicates rearrangement, 3) the appearance of a translucent central core indicates the proper migration of tissue to the outer-surface of the aggregate. Nascent otic vesicles should be visible in translucent regions similar to #3. No rearrangement is seen in panels b and c, partial rearrangement is seen in panel d and complete rearrangement in panel e. Panels a-e were adapted from Supplementary Figure 9 of Koehler et al. Nature 2013. Scale bars, 250 μm.
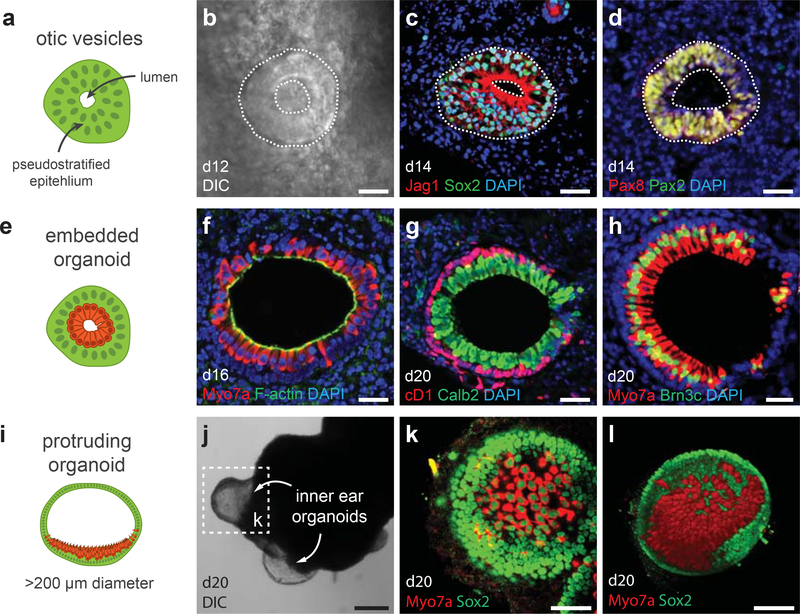
Figure 6: Hair cell induction and organoid types.
a, b, Otic vesicles can be seen using a conventional light microscope while aggregates are in culture. c, d, Otic vesicles express Jag1, Sox2, Pax2, and Pax8. e-l, There are two organoid variants: embedded (e-h) and protruding (i-l). The sensory epithelia of each inner ear organoids contain CyclinD1 (cD1)+ Sox2+ supporting cells and Myo7a+ Brn3c+ and Calretinin (Calb2)+ hair cells. i, j, Protruding organoids are typically >200 μm in diameter and are visible at low magnification or, occasionally, by the naked eye. See Table 1 for a list of antibodies used for characterization. Panels g and h were previously published in Supplementary Figure 13 of Koehler et al. Nature 2013. Scale bars, 250 (j), 100 (l), 50 (k), 25 (b-d) μm.
Hair cells, denoted by an elongated morphology and Myo7a, Brn3c, Sox2, Pax2, and Calretinin expression, should arise within the otic vesicle epithelia between days 14–16 (Fig. 6e-h). The luminal surface of the sensory epithelia should have strong F-actin bands indicative of cell-cell tight junctions (Fig. 6f). Supporting cell nuclei are positioned basal to the hair cell nuclei in the epithelium and can be identified by expression of Sox2 and CyclinD1 (Fig. 6g, k, l)3.
Inner ear organoid formation (step 40).
Between days 12–20, some vesicles become visible along the outer edge of the aggregates. We recommend monitoring these vesicles daily to see that they grow in size over time. By day 20–24, ~10–20% of aggregates should have produced vesicles that are 200–1500 μm in diameter, protrude from the aggregate surface, and contain sensory regions with numerous hair cells and nonsensory regions without hair cells (hereafter these structures will be referred to as inner ear organoids; Fig. 6i-l and 7). Protruding organoid formation appears to be highly sensitive to culture technique, proper manipulation and careful preparation of media throughout the protocol. Moreover, organoids are observed only on the outer surface of aggregates, suggesting that proper orientation of an otic vesicle is a necessary prerequisite to later organoid development. For instance, in the 24-well culture format, we have never seen organoid development at the interface of two conjoined aggregates; thus, we only culture 1–3 aggregates per well to maximize the surface area available for organoids to emerge. We recommend using protruding organoid development as a benchmark for proper execution of the protocol and health of the culture. We typically culture aggregates for no more than 30 days, however, longer culture is feasible. For long-term culture (>30 days), we recommend changing half of the medium daily and limiting the amount of time the plates remain outside of the incubator to ensure optimal health of the tissue.
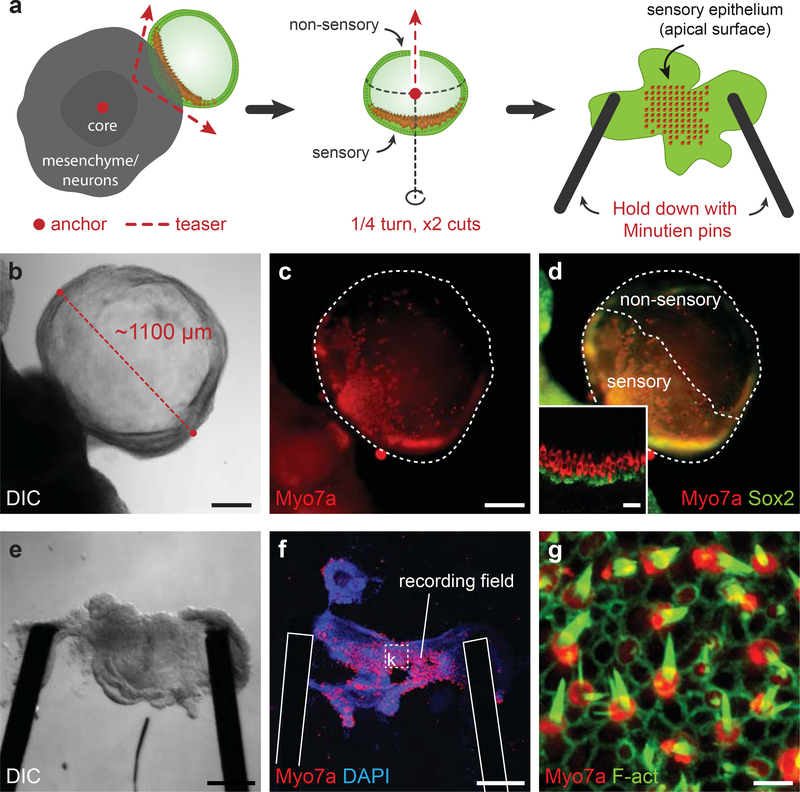
Figure 7: Inner ear organoid dissection for electrophysiological analysis.
a, A schematic of the organoid dissection method described in steps 62–72. The red dots and dotted lines with arrows refer the placement or motion that should be performed with two tungsten needles. b-d, Representative protruding organoid before dissection. e-g, Representative dissected sensory epithelium in an epithelium holder ready for experimentation. Post-recording immunostaining reveals the field of Myo7a+ hair cells with F-actin+ stereocilia from which electrophysiological recordings were taken. See Table 1 for a list of antibodies used for characterization. Panels b-g were previously published in Supplementary Figures 14 and 15 of Koehler et al. Nature 2013. Scale bars, 250 (b-f), 5 (g) μm.
Endpoint analysis (step 41).
We describe how to fix aggregates for immunohistochemical analysis (options A and B) and how to prepare samples for electrophysiological recordings (option C). We describe an approach for preparing aggregates for immunostaining in a cryosection (option A) and wholemount format (option B) that is suitable for fixing samples at any time point during the procedure. Due to their small size, it is important to carefully handle aggregates harvested between days 1–8 throughout the fixation and embedding process; aggregates are easily lost during this process. Imaging the aggregates in a wholemount format can be challenging due to the need to rotate the aggregate to reveal structures of interest. We have developed a custom imaging chamber system that is effective at preparing aggregates for imaging (Supplementary Fig. 1). Additionally, we describe an adapted version of a protocol used to isolate and prepare rodent utricles for electrophysiology experiments (option C). This technique can be used to isolate, dissect and position organoids for single-cell electrophysiological analyses (Fig. 7). For this method there is an optional dispase treatment that helps to break up the mesenchymal cells in the aggregate and greatly shortens the time it takes to liberate organoids from the aggregate. Dispase may disrupt certain structural components of the hair cell stereocilia; thus, depending on the endpoint analysis, it may be necessary to omit dispase treatment. Note that labeling the hair cells with a styryl dye such as FM1–43 or using a cell line with a hair cell-specific fluorescent reporter could improve the success of this dissection. Following dissection, we have cultured isolated epithelium for up to 5–6 hours while performing electrophysiology. If long-term isolated culture of epithelia is desired, based on previous studies of utricle explants, it is likely that dissected epithelia could be maintained for 10 or more days under the right culture conditions70.
MATERIALS
REAGENTS
Mouse ES cells acclimated to growth in LIF-2i medium and at 80% confluency. We recommend the use of any well-established mouse pluripotent stem cell line with this protocol. We primarily use a feeder-free ES cell line derived from R1 mice; however, this protocol has been tested with a mouse iPS cell line and several transgenic ES cell lines with comparable results.
GMEM (Gibco, cat. no. 11710–035)
Advanced DMEM/F12 (Gibco, cat. no. 12491–015)
Sodium Pyruvate, 100 mM (Stemcell Technologies, cat. no. 07000)
Non Essential Amino Acids, 10 mM (Stemcell Technologies, cat. no. 07600)
Penicillin-Streptomycin (Stemcell Technologies, cat. no. 07500)
Knockout Serum Replacement (KSR; Gibco, cat. no. 10828–010) CRITICAL KSR is light sensitive. Store in the dark at −20°C per the manufactures recommendation.
2-Mercaptoethanol (Gibco, cat. no. 21985–023)
Normocin (Invivogen, cat. no. ant-nr-1)
N2 Supplement (Gibco, cat. no. 17502–048)
GlutaMax (Gibco, cat. no. 35050–079)
Matrigel: We use the growth factor-reduced and LDEV-free Matrigel from BD Biosciences (cat. no. 354230). Each lot of Matrigel has a different total protein concentration, so we advise contacting BD Biosciences to order specific lots or lots with similar protein concentrations. We have used concentrations in the range of 8.7–9.2 mg/mL successfully with this protocol.
Recombinant BMP4 (Stemgent, cat. no. 03–0007)
FGF-2 (Peprotech, cat. no. 100–18B)
SB-431542 in solution (Stemgent, cat. no. 04–0010-05)
LDN-193189 in solution (Stemgent, cat. no. 04–0074-02) CRITICAL LDN-193189 is light sensitive. Store in the dark at −20°C per the manufactures recommendation.
Neurobasal Medium (Gibco, cat. no. 21103–049)
B27 Supplement minus Vitamin A (Gibco, cat. no. 12587–010)
PD-0325901 in solution (Stemgent, cat. no. 04–0006-02)
CHIR99021 in solution (Stemgent, cat. no. 04–0004-02)
0.1% Gelatin (Stemcell Technologies, cat. no. 07903)
0.25% Trypsin-EDTA (Gibco, cat. no. 25200–072)
Phosphate Buffered Saline (PBS; Gibco, cat. no. 10010–023)
ESGRO® Leukemia Inhibitory Factor (LIF; Millipore, cat. no. ESG1106)
Dimethyl Sulfoxide (DMSO; Sigma, cat. no. D8418)
Paraformaldehyde (PFA, Electron Microscopy Sciences, cat. no. 15710)
MDX4–4210 platinum silicone elastomer A & B kit or equivalent silicone elastomer (Dow Corning)
Vacuum grease (Fisher Scientific, cat. no. S41718)
Normal goat serum (Vector Laboratories, cat. no. S-1000)
Urea (Sigma, cat. no. U5378)
Glycerol (Sigma, cat. no. G5516)
Triton X-100 (Sigma, cat. no. T8787)
Antibodies required for desired characterization (Table 1)
Table 1 -.
Antibodies needed for tissue characterization
| Useful marker for | Antibody | Host | Supplier | Catalog No. | Dilution |
|---|---|---|---|---|---|
| PSCa | Nanog | Rabbit | Abcam | AB80892 | 1:100 |
| MESb | Brachyury | Goat | Santa Cruz | SC17745 | 1:20 |
| NEd, MES | N-cadherin | Mouse | BD Biosciences | 610920 | 1:100 |
| NE | Sox1 | Rabbit | Cell Signaling | 4194 | 1:100 |
| PSC, DEc, NNEe, PPEf, EPIg, OTVh, SEi | E-cadherin | Mouse | BD Biosciences | 610181 | 1:250 |
| NNE, PPE | AP2 | Mouse | DSHB | 3B5 | 1:50 |
| PSC, DE, OTV, SE | Sox2 | Mouse | BD Biosciences | 561469 | 1:100 |
| NNE, PPE, OTV | GATA3 | Goat | R&D Systems | AF2605 | 1:50 |
| PPE | Six1 | Rabbit | Sigma | HPA001893 | 1:100 |
| OTV, NE | Pax8 | Rabbit | Abcam | AB97477 | 1:100 |
| OTV, HC (nuclei) | Pax2 | Rabbit | Invitrogen | 716000 | 1:100 |
| Anterior PPE, NE | Pax6 | Rabbit | Abcam | AB5790 | 1:200 |
| Intermediate PPE | Pax3 | Mouse | DSHB | N/A | 1:100 |
| EPI (nuclei) | p63 | Mouse | Santa Cruz | SC8431 | 1:50 |
| EPI (cytosol) | Cytokeratin-5 | Rabbit | Sigma | SAB4501651 | 1:100 |
| OTV, SE | p27kip1 | Mouse | BD Biosciences | 610241 | 1:100 |
| OTV, SE | Jagged-1 | Rabbit | LSBio | LSC138530 | 1:50 |
| Epithelium (Apical) | aPKC | Rabbit | Santa Cruz | SC208 | 1:50 |
| Epithelium (Basal) | Laminin-β1 | Rat | Abcam | AB44941 | 1:50 |
| HC (nuclei) | Brn3c | Mouse | Santa Cruz | SC81980 | 1:50 |
| HC (cytosol) | Myo7a | Rabbit | Proteus | 256790 | 1:100 |
| HC cuticle/kinocilia | α-Tubulin | Mouse | Sigma | T6793 | 1:100 |
| Neurons | ßlll-Tubulin (TuJ1) | Mouse | Covance | MMS-435P | 1:500 |
| SNk, HC | Calretinin (Calb2) | Mouse | Millipore | MAB1568 | 1:200 |
| SN | Brn3a | Mouse | Millipore | MAB1585 | 1:20 |
| SN | Islet1 (39.4D5) | Mouse | DSHB | 39.4D5 | 1:5 |
| SN | Neurofilament (L) | Rabbit | Millipore | AB1983 | 1:100 |
| Synapse | Synaptophysin | Rabbit | Invitrogen | 180130 | 1:100 |
| Synapse | Rab3 | Mouse | BD Biosciences | 610379 | 1:50 |
| Synapse | Snap25 | Mouse | BD Biosciences | 610366 | 1:100 |
| Ribbon Synapse | Ctbp2 | Mouse | BD Biosciences | 612044 | 1:50 |
PSC, pluripotent stem cell
MES, mesendoderm
DE, definitive ectoderm
NE, neural ectoderm
NNE, nonneural ectoderm
PPE, preplacodal ectoderm
EPI, epidermis
OTV, otic vesicle
SE, sensory epithelium
HC, hair cells
SN, sensory neurons
EQUIPMENT
Cell culture dish (60 mm; BD Falcon, cat. no. 353002)
Bacterial dish (100 mm; Fisher Scientific, cat. no. 0875712)
U-bottom 96-well plate (Lipidure-coat; Gel Company, cat. no. LCU96)
24-well plate (Lipidure-coat; Gel Company, cat. no. LCMD24)
Safe-Lock centrifuge tubes (1.5 and 2 mL; Eppendorf, cat. no. 022363204, 022363352)
Transfer pipets (Fisherbrand, cat. no. 13–711-7M)
Polystyrene round bottom centrifuge test tubes with cell strainer tops (5 mL; BD Falcon, cat. no. 352235)
Reagent Reservoirs (VistaLab, cat. no. 21–381-27F)
Tissue-Tek Cryomolds (Electron Microscopy Sciences, cat. no. 62534–10)
Olympus FV1000 multiphoton microscope or equivalent inverted confocal microscope equipped with a long-working distance objective (20–25X)
Silicone isolators with adhesive (20 mm diameter × 2.0 mm deep or equivalent; Electron Microscopy Sciences, cat. no. 70336–44)
Cover glasses (24 × 50–1 and 25 × 25 or equivalent; Fisher Scientific, cat. no. 12–544-14 and 12–548-C)
15 mm diameter round cover glasses (thickness no. 0; Assistent, cat. no. 01105509)
Dumont #5 forceps (Electron Microscopy Sciences, cat. no. 72701–1)
Tungsten needles (FST, cat. no. 10130–05, 10130–10)
Minutien Pins (FST, cat. no. 26001–10)
Biosafety cabinet
CO2 incubator
Hotplate
Water bath
Desiccator
Cryostat
REAGENT SETUP
Human recombinant BMP4 stock solution (100 ng/μL)
In the biosafety cabinet, add 100 μL of sterile 4 mM HCl to 10 μg of BMP4; vortex the solution and spin down in a tabletop centrifuge. Store BMP4 solution in 5 μL aliquots at −20°C for 6 months or at −80°C for 1 year.
Human recombinant FGF-2 stock solution (200 ng/μL)
In the biosafety cabinet, add 250 μL of sterile PBS or 5mM Tris (pH 7.6) to 50 μg of FGF-2; vortex the solution and spin down in a tabletop centrifuge. Store FGF-2 solution in 6 μL aliquots at −20°C for 6 months or at −80°C for 1 year.
LIF-2i ES cell maintenance medium
To prepare 100 mL basal LIF-2i medium, combine 48.75 mL DMEM/F12, 48.75 mL Neurobasal medium, 1 mL B27 supplement, 500 μL N2 supplement, and 1 mL GlutaMax in a sterile 250 mL bottle. This medium can be used for 3–4 weeks. Prepare complete LIF-2i medium immediately before use by adding 100 μL Penicillin-Streptomycin, 10 μL LIF, 1 μL PD-0325901, and 3 μL CHIR99021 to 10 mL of the basal LIF-2i medium. Complete LIF-2i medium can be used for up to 1 week at 4°C. CRITICAL STEP It is important to use B27 that does not contain Vitamin A for ES cell culture. Vitamin A can be converted into retinoic acid in culture and cause spontaneous neural induction. Serial passaging can eliminate any neuronal cells that may arise while adapting ES cells to LIF-2i medium.
Ectodermal differentiation medium
To prepare 100 mL ectodermal differentiation medium, combine 95.5 mL GMEM, 1.5 mL KSR, 1 mL sodium pyruvate, 1 mL non-essential amino acids, 1 mL Penicillin-Streptomycin, and 180 μL 2-Mercaptoethanol in a sterile 250 mL bottle. Complete ectodermal differentiation medium should be used for the duration of one experiment or 1 week at 4°C. Adjust the volume of medium made up according to the number of 96-well plates being seeded with ES cells. We typically make 40–50 mL for two 96-well plates and use the same medium for the Matrigel, BMP/SB and FGF/LDN treatments.
Maturation medium
To prepare 100 mL maturation medium, combine 97.9 mL Advanced DMEM/F12, 1 mL N2 supplement, 1 mL GlutaMax, 100 μL Normocin in a sterile 250 mL bottle. Complete maturation medium can be used for up to 2 weeks if stored at 4°C. CRITICAL STEP During the maturation phase of culture, Penicillin-Streptomycin should be replaced with Normocin which does not contain aminoglycosides. Aminoglycosides such as streptomycin are toxic to inner ear hair cells71. Ampicillin would be another suitable alternative.
ScaleA2 Solution
To prepare 1 L of ScaleA2 solution, dissolve 240.24 g (4 M final concentration) of urea powder in 750 mL of Milli-Q water. Add 1 mL of Triton X-100 (0.1% vol/vol final concentration) and 100 g of glycerol (10% wt/vol final concentration). Add Milli-Q water to a final of volume of 1 L. Store at room temperature (15–25°C) for at least 1 year.
Partially cured MDX4–4210 silicone elastomer
In a bacterial culture dish, stir together part A and B of the MDX4–4210 silicone elastomer at a 1:5 ratio using a handheld P1000 tip. The mixture should have a viscous paste-like consistency when properly mixed. Use the P1000 tip to trowel approximately 500 μL of the mixture into 1.5 mL Eppendorf tubes. Aliquots can be stored at −20°C indefinitely until needed.
EQUIPMENT SETUP
Wide-mouth P1000 tips
Use scissors to cut approximately 2 mm off the ends of P1000 tips. We typically cut the tips of an entire box of 96 tips and then autoclave the box to sterilize.
Whole-aggregate imaging chamber
This is a modified version of a previously described imaging chamber72. Here we have optimized the chamber to allow for imaging through either side of the chamber (Suppl. Fig. 1). Remove the adhesive backing from a 1–2 mm deep silicone isolator and place firmly onto a 24 × 50 cover glass. Using forceps remove the protective cover from the top of the silicone isolator. The chamber is now ready to be loaded with a sample. After loading the sample, place a 25 × 25 cover glass on top of the silicone isolator to complete the chamber. If desired forceps can be used to separate the top cover glass in order to remove the sample. Chambers can be reused several times if the silicone isolator is gently cleaned with water between uses and kept free of debris. Multiple samples can be loaded into one chamber or multi-chamber silicone isolators can be used to separate samples.
Cover glass sensory epithelium holder
Cut one minutien pin in half to obtain two properly sized minutien pins for making the holder. Use a handheld P1000 tip to place a droplet of silicone elastomer onto a 15 mm cover glass in an off-center position. Use no. 5 forceps to embed one end of the minutien pins into the silicone droplet so that the other ends converge on the center of the cover glass. Position the pins so that 500–1500 μm gap is present between each end; the epithelium will be held in place between these two ends. Epithelia sizes vary, so we recommend making a variety of epithelium holders with different sized gaps. Use forceps to flatten the pins against the cover glass. Place the epithelium holder directly on the surface of a >100°C hotplate for 5–10 minutes to cure the silicone elastomer. Holders can be stored at room temperature in a bacterial culture dish indefinitely. Before use, place a droplet of 10% Matrigel in DMEM/F12 on the holder and incubate at 37°C for 1-hour.
PROCEDURE
ES cell maintenance and passaging
TIMING 30 min
In the biosafety cabinet, coat a 60 mm culture dish with 2 mL of 0.1% gelatin. Leave for 20 minutes and in the mean time proceed to step 2.
Warm at least 1 mL of 0.25% Trypsin-EDTA and 20 mL of LIF-2i medium in a 37°C water bath.
To begin dissociation of the 80% confluent ES cells, aspirate the spent LIF-2i medium and wash once with PBS.
Add 400 μL of 0.25% Trypsin-EDTA and incubate at RT for ~1–2 min. After 30 seconds, shake the plate horizontally to break up the cells. Under a microscope, confirm that the cells have rounded and are detached from the surface of the plate.
Collect the dissociated cells into 1 mL of LIF-2i medium and transfer the cells to a 2 mL microcentrifuge tube.
Triturate the cells using a P1000 tip to break up clumps into single-cells. CRITICAL STEP Care should be taken to not introduce air bubbles into the medium.
Centrifuge for 2–5 minutes at 200 g.
While the cell suspension is in the centrifuge, aspirate the 0.1% gelatin from the 60 mm plate from step 1. Leave the plate for ~5 minutes to dry.
Discard the supernatant and resuspend the cells from step 7 in 1 mL LIF-2i Medium.
Plate the cells on the dried gelation coated plate (from step 8). Select plating density based on when you next wish to split the cells. Density should be between 1:10 (for next passage in ~2 days) and 1:50 (for next passage in ~4 days). Incubate cells at 37°C in 5.0% CO2 until ready for passage.
TROUBLESHOOTING
ES cell differentiation: Initiating ectodermal induction (two 96-well plates)
TIMING 45 min
-
11
When cells are 80% confluent, aspirate the LIF-2i medium and wash three times with PBS at RT. CRITICAL STEP It is important to remove all trace of LIF-2i medium, as signaling factors within the LIF-2i can influence differentiation.
-
12
Add 400 μL of 0.25% Trypsin-EDTA and incubate at RT or 37°C for ~1–2 min. After 30 seconds, shake the plate horizontally to break up the cells. Under a microscope, confirm that the cells have rounded and are detached from the surface of the plate.
-
13
Collect the dissociated cells into 1 mL of Ectodermal differentiation medium and transfer them into a 2 mL microcentrifuge tube.
-
14
Break the cell clumps into single-cells by pipetting with a P1000 tip. Pellet the cells, by centrifugation at 200 g for 2–5 minutes.
-
15
Completely remove the supernatant and resuspend the cell pellet in 1 mL Ectodermal differentiation medium.
-
16
Forcefully pipet 1 mL of fresh Ectodermal differentiation medium through a cell-strainer-top test tube to prime the strainer. Pipet the 1 mL of ES cell suspension drop-wise onto the cell strainer. Next, pipet 1 mL of fresh Ectodermal differentiation medium drop-wise onto the cell strainer. There should be 3 mL in the test tube. CRITICAL STEP This step is important for ensuring that the cells are completely dissociated into single cells.
-
17
Mix the cell suspension by pipetting with a P1000 tip and determine the concentration of cells using a hemocytometer.
-
18
Dilute the appropriate volume of cell suspension in 22 mL of fresh Ectodermal differentiation medium to acquire a final concentration of 30,000 cells per mL (i.e. 6.6×105 total cells). For example, if the cell suspension contains 1×106 cells per mL, dilute 0.66 mL of cell suspension (6.6×105 divided by 1×106) in 21.34 mL of Ectodermal differentiation medium. Invert several times to mix.
-
19
Pour the cell suspension into a reservoir and aliquot 100 μL of cell suspension into each well of two 96-well plates using a multichannel pipette.
-
20
Place the plates in a 37°C incubator with 5.0% CO2 for 24 hours.
Differentiation day 1: Addition of Matrigel
TIMING 30 min
-
21
Mix 10.56 mL of Ectodermal differentiation medium with 440 μL of Matrigel to make 11 mL of complete medium. CRITICAL STEP Keep the medium and Matrigel on ice or in a 4°C refrigerator immediately prior to mixing. Matrigel will become gelatinous at temperatures above 15°C. It is important to work quickly and hold the medium up to a light to see that the Matrigel is properly mixed.
-
22
Warm Ectodermal differentiation medium containing Matrigel in 37°C water bath.
-
23
Holding the multichannel pipette at an angle, carefully remove 50 μL of media from each well of the plate from step 20 so that the cell aggregate at the bottom of the well remains undisturbed.
-
24
Add 50 μL of the Ectodermal differentiation medium containing Matrigel to each well. Note that the final concentration of Matrigel is 2% v/v.
-
25
Mix by pipetting 4–6 times. Return plates to the incubator for 48 hours.
Differentiation day 3: Addition of BMP4 and SB-431542 (BMP/SB)
TIMING 30 min
-
26
Prepare an appropriate amount of Ectodermal differentiation medium for each experimental condition. Typically 6 mL is sufficient for two 96-well plates.
-
27
Add BMP4 and SB-431542 to the Ectodermal differentiation medium at a 5X concentration. For 6 mL, add 3 μL of BMP4 and 3 μL of SB-431542 (i.e. 0.5 μL per 1 mL).
-
28
Add 25 μL of Ectodermal differentiation medium containing BMP/SB to each well of the plate from step 26. Each well now contains 125 μL of medium with a final concentration of 10 ng/mL BMP4 and 1 μM SB-431542. Incubate cells for a further 24–48 hours.
Differentiation day 4–5: Addition of FGF-2 and LDN-193189 (FGF/LDN)
TIMING 30 min
-
29
Prepare an appropriate amount of Ectodermal differentiation medium for each experimental condition. Typically 6 mL is sufficient for two 96-well plates.
-
30
Add FGF-2 and LDN-193189 to the Ectodermal differentiation medium at a 6X concentration. For 6 mL, add 4.5 μL of FGF-2 (i.e. 0.75 μL per 1 mL) and 3.6 μL of LDN-193189 (i.e. 0.6 μL per 1 mL).
-
31
Add 25 μL of Ectodermal differentiation medium containing FGF/LDN to each well of the plates from step 28. Each well now contains 150 μL of medium with a final concentration of 25 ng/mL FGF-2 and 1 μM LDN-193189. Continue to incubate cells for 3–4 days.
Differentiation day 8: Transition to long-term culture
TIMING 45 min
-
32
Using a P1000 pipette tip, transfer the aggregates from each well into a 15 mL conical tube. Use different 15 mL tubes for different conditions if necessary.
-
33
Allow the aggregates to settle at the bottom of the tube for 1 minute and then carefully aspirate the media.
-
34
Add at least 5 mL pre-warmed DMEM/F12 to wash the aggregates.
-
35
Repeat steps 33 and 34 at least 3 times.
-
36
Remove excess DMEM/F12.
-
37
Re-suspend the aggregates in complete Maturation medium containing 1% Matrigel. Transfer the aggregates to a 100 mm bacterial dish using a wide-mouth P1000. Add an appropriate volume of additional Maturation medium to allow 1 mL containing 1–3 aggregates to be distributed into individual wells of a 24-well plate. CRITICAL STEP Use an additional 1–2 mL of Maturation medium to account for medium spreading out across the surface of the plate.
-
38
Using a wide-mouth P1000 tip, pipet 1–3 aggregates in 1 mL into wells of a 24-well plate. Aggregates in the bacterial plate should be visible without a microscope.
Alternatively, single aggregates can be transferred to a new 96-well plate in 150–200 μL of Maturation medium containing Matrigel.
-
39
Incubate cells for a further 22 days (24 well plates) or 12 days (96 well plates). For 24-well plates, beginning on day 10, replace half of the medium with Maturation medium that does not contain Matrigel every other day (i.e. day 12, 14, 16, etc.). Half of the medium should be replaced in 96-well plates every day beginning on day 9.
-
40
Under these conditions, organoids can be cultured for up to 30 days (24 well plates) or 20 days (96 well plates). Longer culture durations will likely require different media compositions or culture formats.
TROUBLESHOOTING
End point analysis
41 Aggregates can be fixed at any point during the procedure for imaging as a cryosection (option A) or as a whole mount (option B). Option C describes how to prepare sensory epithelia for electrophysiological recording, which can be performed following the appearance of sensory epithelia with hair cells (days 16–30).
A). Preparation of aggregates for imaging – cryosections
TIMING 1 week
Pipet aggregates into a 2 mL tube using a P1000 tip (during days 1–6 or a wide-mouth P1000 (after day 6).
Aspirate the medium and immediately add 4% PFA in PBS to the aggregates.
Incubate the aggregates in 4% PFA overnight at 4°C to fix the tissue.
Wash the aggregates at least 3 times with PBS. PAUSE POINT Fixed aggregates can be stored for several months in PBS at 4°C. For long-term storage, we recommend storing the aggregates in PBS containing 0.02% sodium azide to discourage bacterial growth.
Incubate the aggregates in a series of sucrose-PBS solutions (i.e. 10% sucrose, 20% sucrose, and 30% sucrose) for 30 minutes each with rocking. PAUSE POINT Sucrose treated aggregates can be stored at 4°C for up to 1 week.
Pipet the aggregates into a cryomold and carefully remove the sucrose solution using a P20 pipette. Position the aggregates in the center of the cryomold.
Slowly add tissue-freezing medium along the outer-walls of the cryomold. Do not allow the aggregates to float away from the bottom of the cryomold.
Place the cryomolds in a vacuum desiccator for 30 minutes to remove bubbles from the tissue-freezing medium.
Freeze the samples on dry ice for 30 minutes or until the tissue-freezing medium is opaque. PAUSE POINT Frozen tissue blocks can be stored at −80°C in a sealed container or ziplocked bags for several years.
Cryosection tissue blocks into 10–15 μm thick sections on a cryostat.
Perform immunohistochemistry according to a standard protocol using the antibodies in table
TROUBLESHOOTING
B). Preparation of aggregates for imaging – wholemount
TIMING 2 week
Perform steps 41–44.
Incubate the aggregates in PBS containing 10% normal goat serum, and 0.1% Triton X-100 at room temperature overnight on shaker. CRITICAL STEP Goat serum can be replaced with bovine serum albumin or normal horse serum depending on the antibodies being used.
Remove the blocking solution and suspend the aggregates in PBS containing 3% normal goat serum, 0.1% Triton X-100, and primary antibodies. Incubate at room temperature for 2–3 days on a shaker.
Wash 3 times with PBS containing 0.1% Triton X-100 for 1 hour each at room temperature on a shaker.
Incubate the aggregates in PBS containing 3% normal goat serum, 0.1% Triton X-100, and secondary antibodies at room temperature for 2–3 days on a shaker. CRITICAL STEP Wrap the samples in foil to protect the fluorophores from photobleaching during this and subsequent steps.
Repeat step 55.
Incubate aggregates in scaleA2 solution for 4–5 days prior to imaging. CRITICAL STEP ScaleA2 solution will cause slight enlargement of the tissue.
To prepare for imaging, transfer ~250–500 μL (depending on the volume of the chamber) of scaleA2 solution containing an aggregate to an imaging chamber.
Affix the top coverslip slowly. Some liquid will escape. A small air bubble will not be detrimental to later imaging.
While imaging, flip the imaging chamber to image the specimen from multiple orientations.
C). Preparation of sensory epithelia for electrophysiological recordings
TIMING 1–2 hours
Between days 20–30, identify an aggregate that has a protruding organoid with a long-axis diameter greater than 750 μm.
Under a dissecting scope, use two tungsten needles to tease the organoid out of the cell aggregate. Attempt to clean mesenchymal and neuronal cells from the surface of the organoid using the tungsten needles. Try not to puncture the organoid while dissecting. CRITICAL STEP It is difficult to assess the morphology of the portion of the organoid embedded in the aggregate. As this region most often harbors hair cells, extreme care should be taken not to destroy this portion of epithelium. Begin dissecting from the center of the aggregate out toward the surface. Puncture and anchor the center of the aggregate with one needle while teasing outward with the other (Fig. 7a, left).
Use an inverted light-microscope to examine the organoid. Note the apparent thickness of the epithelium from multiple orientations. Regions of the epithelium containing hair cells appear thicker (~30 μm) than regions devoid of hair cells (~15 μm).
Back under the dissecting scope, use the tungsten needles to puncture the organoid in a region not containing hair cells. Use one needle to anchor the tissue while pulling away rapidly with the other to create incisions in the epithelium. Attempt to make two incisions in an X-pattern opposite the region that likely contains hair cells (Fig. 7a, middle).
Use the tungsten needles to flatten the epithelium in a four-leafed clover pattern. The inside surface of the organoid (i.e. the apical end of the hair cells) should be facing up.
Submerge an epithelium holder in the medium near the flattened epithelium.
Use a pipet tip to gently position the flatten epithelium onto the center of the epithelium holder.
Use fine forceps to pull up one minutien pin and a tungsten needle to position the epithelium underneath (Fig. 7a, right).
With one end of the epithelium secured, repeat viii for the other end.
Lightly blow medium across the secured epithelium with a P200 pipette to test the stability of the epithelium preparation.
The epithelium is now ready to be transferred to an electrophysiology rig for further experimentation.
TIMING
Steps 1–10, Maintenance and passaging ES cells: 30 min
Steps 11–20, ES cell differentiation: Initiating ectodermal induction (two 96-well plates): 45 min
Steps 21–25, Differentiation day 1: Addition of Matrigel: 30 min
Steps 26–28, Differentiation day 3: Addition of BMP4 and SB-431542 (BMP/SB): 30 min
Steps 29–31, Differentiation day 4–5: Addition of FGF-2 and LDN-193189 (FGF/LDN): 30 min
Steps 32–40, Differentiation day 8: Transition to long-term culture: 45 min
Steps 1–40, Total time to generate inner ear organoids: 20–30 days
Step 41 option A, Preparation of aggregates for imaging - cryosections: 1 week
Step 41 option B, Preparation of aggregates for imaging - wholemount: 1 week
Step 41 option C, Preparation of sensory epithelia for electrophysiological recordings: 1–2 hours
TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2 -.
Troubleshooting
| Steps | Problem | Possible Reason | Solution |
|---|---|---|---|
| 1–10 | ES cells do not attach to the plate | Insufficient gelatin coating | Re-plate the cells on a new plate with fresh gelatin coating |
| Cells are not yet adapted to LIF-2i medium | Perform several passages | ||
| 11–28 | Epithelium does not form or thicken over time in control samples | Ectodermal medium was not made properly | Check reagent expiration dates; Make new Ectodermal medium |
| Matrigel was not mixed into medium properly | Make sure that Matrigel is added to Ectoderm differentiation medium during preparation at 4°C. | ||
| 29–31 | Epithelium does not thicken and/or ruffle | One of the small molecules or proteins has expired or is not present at an optimal concentration | Test each treatment separately and replace small molecule or protein stocks |
| Matrigel concentration is not optimal | Reduce/Increase Matrigel concentration | ||
| 29–31 | The aggregates themselves or the medium color in wells on the edge of the plate look different from the rest of the plate | Disproportionate evaporative loss in the edge wells | Check that incubator is properly humidified |
| Only analyze aggregates from interior wells | |||
| 32–40 | Cellular rearrangement does not occur during days 8–12 | Cell aggregates were transferred to floating culture too early/late | Transfer aggregates at a different time point between days 7–8.5 |
| 32–40 | No Pax2, Pax8, Sox2, Ecad+ vesicles develop by day 12 | Improper cellular rearrangement | Transfer aggregates at a different time point between days 7–8.5 |
| Carry-over of signaling molecules/proteins from day 0–8 culture | Wash aggregates with DMEM/F12 at least 3 times prior culture in maturation medium | ||
| Check that proper morphology is observed during days 0–8 | |||
| Aggregates are damaged during transfer to maturation medium | Use wide-mouth or cut P1000 tips or a sterile transfer pipet to manipulate cell aggregates | ||
| 29–40 | Excessive development of beating myocytes, cartilaginous masses and adipose apparent between days 14–20 | LDN-193189 concentration was suboptimal; insufficient suppression of mesenchyme development | Increase the concentration of LDN-193189 (1 μM should be sufficient) |
| 32–40 | Large translucent organoids do not develop by day 18–20 | Too many aggregates were cultured together | Reduce the number of aggregates in each well on 24-well plates |
| Matrigel concentration is too high during floating culture | Reduce Matrigel concentration to 0–0.5% at the start of floating culture | ||
| 41–51 | No hair cells observed in cyrosectioned organoids | Incomplete analysis | Ensure that the entire organoid is analyzed; cyrosections through nonsensory regions give the appearance of no hair cells |
ANTICIPATED RESULTS
For reproducible results we have found that it is absolutely critical to use fresh medium and well-preserved small molecule and recombinant protein stocks for each experiment. In addition, to reduce the time for performing each manipulation of the aggregates, we advise only running 2–3 96-well plates per experiment until the protocol is mastered. With experience, the various morphological changes can be readily identified under a light microscope.
With proper dissociation, counting, and pipetting we have found that cell aggregates are 250–270 μm in diameter 24 hours after plating (i.e. at the time of Matrigel addition). As a quality control measure, we check to see that there is minimal debris floating around the aggregates (i.e. denoting cellular death) and that the average aggregate diameter across all 96-well plates has a standard deviation of 5 μm or less (Fig. 2a). Following another 48 hours of culture (d3), the aggregates should roughly double in size to 480–500 μm in diameter and a translucent epithelium should be visible on the surface of the aggregate (Fig. 2c). Our previous data suggest that this epithelium is representative of the definitive ectoderm because it expresses E-cadherin and does not express the neural marker Sox1, the mesendodermal marker Brachyury or the pluripotency marker Nanog3. Following BMP/SB treatment on day 3, AP2 expression can be seen in the outer-epithelium beginning on day 4. If the FGF/LDN treatment timing is optimal, on day 6 of differentiation, the outer-epithelium should appear to thicken (~30 μm apparent thickness) relative to the epithelium of aggregates treated with BMP/SB alone (~15 μm apparent thickness). We also observe a distinctive ruffling of the outer-epithelium (Fig. 3). Over the next 24–36 hours these morphological characteristics become more apparent and should be quite obvious if the experiment was performed correctly. Between days 6–8, patches of Pax8+ E-cadherin (Ecad)+ epithelium are visible in the outer-epithelium indicating the development of OEPD tissue (Fig 4).
Upon transfer to maturation medium (d8–11), the aggregates lose discernable morphological features as the inner core cells migrate to the surface and cover the outer-epithelium (Fig. 5a). By day 12, however, in properly treated and manipulated aggregates, the core of the aggregate should become partially translucent, indicating the evacuation of the inner core cells; the size of each aggregate should be ~1.5–2 mm. It is common for aggregates to conjoin to one another after day 12 of the culture if not cultured individually. Also, by day 12, vesicles should be visible by looking alone the edges of the aggregates or at the translucent central region of the aggregate (Fig. 6a-d). To have confidence that an experiment was performed correctly, we recommend checking to see that at least 10% of the aggregates have vesicles overtly visible using a light microscope by day 12–14. Immunohistochemistry should be performed to assess the true number of vesicles generated, which should be in the range of 15–25 vesicles per aggregate. On day 12, vesicles display a wide range of intraluminal diameters from 1–100 μm. Two organoid variants should develop during days 14–30: embedded and protruding (Fig. 6e-l, 7b-d). The majority organoids containing hair cells will remain embedded in the tissue for the duration of the culture. These are difficult to see with the naked eye; however, protruding organoids are readily apparent. In total, we normally see that 3–6 organoids develop from the original group of vesicles per aggregate by day 20. Protruding organoids should be visible in ~15% of the aggregates.
Supplementary Material
Supplementary Figure 1: Aggregate imaging chamber. a, Typical imaging chamber constructed from two cover glasses and a silicone isolator. One aggregate (sample) is loaded in the chamber. Note the air bubble that forms during the loading process. b, Projection drawing of an aggregate imaging chamber. Note that this chamber design is an example and can be easily customized using different sized cover glasses or silicone isolators.
Acknowledgments
The authors would like to thank Andrew Mikosz, Dharmeshkumar Patel and Sreeparna Majumdar for their technical assistance and comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants RC1DC010706, R21DC012617, R01DC013294 and an Indiana University School of Medicine Biomedical Research Grant (to E.H.). K.R.K. was supported by a Paul and Carole Stark Neurosciences Fellowship and an Indiana Clinical and Translational Science Institute Predoctoral Fellowship (NIH TL1RR025759).
Footnotes
Competing financial interests The authors declare no competing financial interests.
REFERENCES
- 1.Sasai Y Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–326 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Sasai Y, Eiraku M & Suga H In vitro organogenesis in three dimensions: self-organising stem cells. Development (Cambridge, England) 139, 4111–4121 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Koehler KR, Mikosz AM, Molosh AI, Patel D & Hashino E Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Cordero A et al. Photoreceptor precursors derived from three-dimensional embryonic stem cell cultures integrate and mature within adult degenerate retina. Nature Biotechnology 31, 741–747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiraku M et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Eiraku M et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Eiraku M & Sasai Y Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nature Protocols 7, 69–79 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Nakano T et al. Self-Formation of Optic Cups and Storable Stratified Neural Retina from Human ESCs. Cell Stem Cell 10, 771–785 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Suga H et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature 480, 57–62 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Nasu M et al. Robust Formation and Maintenance of Continuous Stratified Cortical Neuroepithelium by Laminin-Containing Matrix in Mouse ES Cell Culture. PLoS ONE 7, e53024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spence JR et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCracken KW, Howell JC, Wells JM & Spence JR Generating human intestinal tissue from pluripotent stem cells in vitro. Nature Protocols 6, 1920–1928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Huch M et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L et al. Location of transient ectodermal progenitor potential in mouse development. Development 140, 4533–4543 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Cajal M et al. Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo. Development 139, 423–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beddington SP An autoradiographic analysis of the potency of embryonic ectoderm in the 8th day postimplantation mouse embryo. J Embryol Exp Morphol 64, 87–104 (1981). [PubMed] [Google Scholar]
- 19.Beddington RS An autoradiographic analysis of tissue potency in different regions of the embryonic ectoderm during gastrulation in the mouse. J Embryol Exp Morphol 69, 265–285 (1982). [PubMed] [Google Scholar]
- 20.Wilson PA & Hemmati-Brivanlou A Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376, 331–333 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Wilson PA, Lagna G, Suzuki A & Hemmati-Brivanlou A Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development (Cambridge, England) 124, 3177–3184 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Kwon H-J, Bhat N, Sweet EM, Cornell RA & Riley BB Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS genetics 6, e1001133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barth KA et al. Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development (Cambridge, England) 126, 4977–4987 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Neave B, Holder N & Patient R A graded response to BMP-4 spatially coordinates patterning of the mesoderm and ectoderm in the zebrafish. Mechanisms of development 62, 183–195 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Harvey NT et al. Response to BMP4 signalling during ES cell differentiation defines intermediates of the ectoderm lineage. Journal of cell science 123, 1796–1804 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Pieper M, Ahrens K, Rink E, Peter A & Schlosser G Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development (Cambridge, England) 139, 1175–1187 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Reichert S, Randall RA & Hill CS A BMP regulatory network controls ectodermal cell fate decisions at the neural plate border. Development (Cambridge, England) 140, 4435–4444 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Grocott T, Tambalo M & Streit A The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Developmental biology 370, 3–23 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Ahrens K & Schlosser G Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Developmental biology 288, 40–59 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Brugmann SA, Pandur PD, Kenyon KL, Pignoni F & Moody SA Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development (Cambridge, England) 131, 5871–5881 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Litsiou A, Hanson S & Streit A A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development (Cambridge, England) 132, 4051–4062 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Schlosser G Induction and specification of cranial placodes. Developmental biology 294, 303–351 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Patthey C & Gunhaga L Specification and regionalisation of the neural plate border. The European journal of neuroscience 34, 1516–1528 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya S & Bronner-Fraser M Competence, specification and commitment to an olfactory placode fate. Development (Cambridge, England) 135, 4165–4177 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Baker CV, Stark MR, Marcelle C & Bronner-Fraser M Competence, specification and induction of Pax-3 in the trigeminal placode. Development (Cambridge, England) 126, 147–156 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Ladher RK, O’Neill P & Begbie J From shared lineage to distinct functions: the development of the inner ear and epibranchial placodes. Development (Cambridge, England) 137, 1777–1785 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Wright TJ & Mansour SL Fgf3 and Fgf10 are required for mouse otic placode induction. Development (Cambridge, England) 130, 3379–3390 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Urness LD, Paxton CN, Wang X, Schoenwolf GC & Mansour SL FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Developmental biology 340, 595–604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohyama T, Mohamed OA, Taketo MM, Dufort D & Groves AK Wnt signals mediate a fate decision between otic placode and epidermis. Development (Cambridge, England) 133, 865–875 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Riccomagno MM, Takada S & Epstein DJ Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes & Development 19, 1612–1623 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freter S, Muta Y, Mak S-S, Rinkwitz S & Ladher RK Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Development (Cambridge, England) 135, 3415–3424 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Groves AK & Bronner-Fraser M Competence, specification and commitment in otic placode induction. Development (Cambridge, England) 127, 3489–3499 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Groves AK & Fekete DM Shaping sound in space: the regulation of inner ear patterning. Development (Cambridge, England) 139, 245–257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neves J, Parada C, Chamizo M & Giraldez F Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: a mechanism for sensory organ specification. Development (Cambridge, England) 138, 735–744 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Appler JM & Goodrich LV Connecting the ear to the brain: Molecular mechanisms of auditory circuit assembly. Progress in neurobiology 93, 488–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laine H, Sulg M, Kirjavainen A & Pirvola U Cell cycle regulation in the inner ear sensory epithelia: role of cyclin D1 and cyclin-dependent kinase inhibitors. Developmental biology 337, 134–146 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Oesterle EC, Campbell S, Taylor RR, Forge A & Hume CR Sox2 and Jagged1 Expression in Normal and Drug-Damaged Adult Mouse Inner Ear. Journal of the Association for Research in Otolaryngology : JARO 9, 65–89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang M, Gao WQ, Hasson T & Shin JJ Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development (Cambridge, England) 125, 3935–3946 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Watanabe K et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nature Neuroscience 8, 288–296 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Muñoz-Sanjuán I & Brivanlou AH Neural induction, the default model and embryonic stem cells. Nature Reviews Neuroscience 3, 271–280 (2002). [DOI] [PubMed] [Google Scholar]
- 51.James D, Levine AJ, Besser D & Hemmati-Brivanlou A TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development (Cambridge, England) 132, 1273–1282 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Camus A, Perea-Gomez A, Moreau A & Collignon J Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Developmental biology 295, 743–755 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnology 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muguruma K et al. Ontogeny-recapitulating generation and tissue integration of ES cell-derived Purkinje cells. Nature Neuroscience 13, 1171–1180 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Bouchard M, de Caprona D, Busslinger M, Xu P & Fritzsch B Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC developmental biology 10, 89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christophorou NAD, Mende M, Lleras-Forero L, Grocott T & Streit A Pax2 coordinates epithelial morphogenesis and cell fate in the inner ear. Developmental biology 345, 180–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warchol ME & Richardson GP Expression of the Pax2 transcription factor is associated with vestibular phenotype in the avian inner ear. Developmental neurobiology 69, 191–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lysakowski A et al. Molecular microdomains in a sensory terminal, the vestibular calyx ending. Journal of Neuroscience 31, 10101–10114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li A, Xue J & Peterson EH Architecture of the mouse utricle: macular organization and hair bundle heights. Journal of neurophysiology 99, 718–733 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Li H, Roblin G, Liu H & Heller S Generation of hair cells by stepwise differentiation of embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 100, 13495–13500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oshima K et al. Mechanosensitive Hair Cell-like Cells from Embryonic and Induced Pluripotent Stem Cells. Cell 141, 704–716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ouji Y, Ishizaka S, Nakamura-Uchiyama F & Yoshikawa M In vitro differentiation of mouse embryonic stem cells into inner ear hair cell-like cells using stromal cell conditioned medium. Cell Death and Disease 3, e314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen W et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature 490, 278–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ouji Y, Ishizaka S, Nakamura-Uchiyama F, Wanaka A & Yoshikawa M Induction of inner ear hair cell-like cells from Math1-transfected mouse ES cells. Cell Death and Disease 4, e700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brigande JV & Heller S Quo vadis, hair cell regeneration? Nature Neuroscience 12, 679–685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding Q et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12, 238–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Q et al. Enhanced Efficiency of Human Pluripotent Stem Cell Genome Editing through Replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trowe M-O et al. Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Developmental biology 342, 51–62 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Ying Q-L et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaboyard S et al. Three-dimensional culture of newborn rat utricle using an extracellular matrix promotes formation of a cyst. Neuroscience 133, 253–265 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Lerner SA, Matz GJ & Hawkins JE Aminoglycoside ototoxicity. (Little Brown and Company, 1981). [Google Scholar]
- 72.Yokomizo T et al. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nature Protocols 7, 421–431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Aggregate imaging chamber. a, Typical imaging chamber constructed from two cover glasses and a silicone isolator. One aggregate (sample) is loaded in the chamber. Note the air bubble that forms during the loading process. b, Projection drawing of an aggregate imaging chamber. Note that this chamber design is an example and can be easily customized using different sized cover glasses or silicone isolators.