Abstract
Over the past two decades a growing body of evidence has demonstrated an important role of tight junction (TJ) proteins in the physiology and disease biology of gastrointestinal (GI) and liver disease. On one side, TJ proteins exert their functional role as integral proteins of TJs in forming barriers in the gut and the liver. Furthermore, TJ proteins can also be expressed outside TJs where they play important functional roles in signaling, trafficking and regulation of gene expression. A hallmark of TJ proteins in disease biology is their functional role in epithelial-to-mesenchymal transition. A causative role of TJ proteins has been established in the pathogenesis of colorectal cancer and gastric cancer. Among the best characterized roles of TJ proteins in liver disease biology is their function as cell entry receptors for hepatitis C virus – one of the most common causes of hepatocellular carcinoma. At the same time TJ proteins are emerging as targets for novel therapeutic approaches for GI and liver disease. Here we review our current knowledge of the role of TJ proteins in the pathogenesis of GI and liver disease biology and discuss their potential as therapeutic targets.
Keywords: tight junction, claudin-1, hepatitis C virus, hepatocellular carcinoma, colorectal cancer
Tight junctions (TJs) are intercellular adhesion complexes that are essential to the barrier function of epithelia and endothelia. They maintain cell polarity by limiting the movement of proteins within the plasma membrane and by regulating paracellular solute and water flux (for a recent review see[1]). Their functions are however not limited to these important structural gate and fence functions as TJs also act as signaling hubs[1, 2]. TJs are highly dynamic structures that constantly enable cells to adapt to their environment. Not surprisingly, perturbation of TJ protein expression or function and/or disruption of TJ integrity is associated with a variety of diseases, including skin, intestinal and lung diseases, and cancers[1, 3] (Table 1). Furthermore, pathogens have evolved strategies to overcome epithelial and endothelial barriers by using TJ components for their infection/invasion[1, 4]. TJs are composed of transmembrane proteins, including different claudins (CLDNs), tight junction-associated marvel proteins (TAMPs) such as occludin, junctional adhesion molecules (JAMs) as well as cytosolic proteins, which form what has been termed the junctional plaque and connect transmembrane components to the cytoskeleton[1] (Figure 1).
Table 1. Role of TJ proteins in disease.
Examples of TJ protein-disease associations and the underlying mechanisms are shown. CLDN: claudin, JAM: junctional adhesion molecule, TJ: tight junction, ZO: zona occludens
| TJ protein | Disease | TJ protein expression |
Described mechanism | References |
| JAM-A | Cancer | PI3K/MAPK signaling, Notch signaling, TGF-β1 signaling | [177, 178, 179, 180, 181, 182, 183, 184, 185] | |
| Brain | ↑ | |||
| Breast | ↑↓ | |||
| Gastric | ↑ | |||
| Lung | ↑ | |||
| Endometrial | ↓ | |||
| Pancreatic | ↓ | |||
| Hereditary diseases | [186] | |||
| Cystic fibrosis | ↓ | |||
| Viral infection | [187] | |||
| Retroviral infection (hydrocephalus, encephalitis) | ↑ | |||
| JAM-C | Cancer | LRP5/AKT/β-catenin/CCND1 signaling, PI3K/MAPK signaling | [188, 189, 190, 191, 192, 193, 194, 195] | |
| Fibrosarcoma | ↑ | |||
| Lung | ↑ | |||
| Melanoma | ↑ | |||
| Ovarian | ↑ | |||
| Coxsackie virus and adenovirus receptor (CAR) | Cancer | MyD88/IRAK-4/NF- B, ERK1/2 signaling, estrogen signaling | [196, 197, 198, 199, 200, 201, 202, 203, 204] | |
| Breast | ↑ | |||
| Endometrial | ↑ | |||
| Lung | ↑ | |||
| Oral | ↑ | |||
| Ovarian | ↑ | |||
| Thyroid | ↑ | |||
| CLDN1 | Cancer | Reactive oxygen species-mediated activation of heat shock factor 1 (HSF1) | [34, 78, 156, 157, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221] | |
| Breast | ↑↓ | |||
| Cervical | ↑ | |||
| Colorectal | ↑↓ | |||
| Gastric | ↑ | |||
| Liver | ↑ | |||
| Oral | ↑ | |||
| Ovarian | ↑ | |||
| Prostate | - | |||
| Thyroid neoplasma | ↑ | |||
| Hereditary disease | ||||
| Cystic fibrosis | - | [186] | ||
| CLDN2 | Cancer | EGFR/MEK/ERK signaling PI3K signaling | [212, 221, 222, 223, 224, 225, 226, 227, 228, 229] | |
| Breast | ↑↓ | |||
| Colorectal | ↑ | |||
| Lung | ↑ | |||
| Skin | ↑ | |||
| Prostate | ↓ | |||
| Inflammation | ||||
| Inflammatory bowel disease: Morbus Crohn | ↑ | [230, 231] | ||
| Collagenous colitis | ↑ | |||
| CLDN3 | Cancer | EGFR/MEK/ERK signaling PI3K/Akt signaling Wnt signaling Stat3 | [19, 217, 232, 233, 234, 235, 236, 237, 238] | |
| Breast | ↑ | |||
| Colorectal | ↑ | |||
| Endometrial | ↑↓ | |||
| Gastric | ↑ | |||
| Kidney | ↑ | |||
| Lung | ↑ | |||
| Ovarian | ↑ | |||
| Prostate | ||||
| Uterine | ↑ | |||
| Inflammation | ||||
| Inflammatory bowel disease: Morbus Crohn | ↓ | [230, 239] | ||
| Bacterial toxins | ||||
| Clostridium perfringens enterotoxin | ↓ | [240] | ||
| CLDN4 | Cancer | ERK signaling AMPK signaling | [19, 91, 217, 232, 235, 241, 242, 243] | |
| Breast | ↑ | |||
| Endometrial | ↑ | |||
| Gastric | ↑↓ | |||
| Kidney | ↑ | |||
| Lung | ↑ | |||
| Nasopharyngeal | ↑ | |||
| Ovarian | ↑ | |||
| Pancreatic | ↑ | |||
| Uterine | ↑ | |||
| Inflammation | ||||
| Collagenous colitis | ↓ | [231] | ||
| Hereditary diseases | [186] | |||
| Cystic fibrosis | ↓ | |||
| Bacterial toxins | ||||
| Clostridium perfringens enterotoxin | ↓ | [240] | ||
| CLDN7 | Cancer | ERK/MAPK signaling Wnt signaling Integrin/FAK signaling | [211, 217, 218, 232, 241, 243, 244] | |
| Breast | ↓ | |||
| Cervical | ↑ | |||
| Colon | ↑ | |||
| Gastric | ↑ | |||
| Liver | ↑ | |||
| Lung | ↑ | |||
| Nasopharyngeal | ↑ | |||
| Ovarian | ↑ | |||
| Pancreatic | ↑ | |||
| Prostate | ↑ | |||
| Thyroid neoplasma | ↑ | |||
| Inflammation | ||||
| Crohn's disease | - | [239] | ||
| Ulcerative colitis | ↓ | |||
| Celiac disease | - | |||
| CLDN11 | Cancer | TGF- , ERK, p38 signaling | [88] | |
| Gastric | ↑ | |||
| CLDN16 | Cancer | [245, 246, 247] | ||
| Breast | ↑ | |||
| Ovarian | ↑ | |||
| Renal | ↑ | |||
| Hereditary diseases | [248] | |||
| Familial hypomagnesemia | Mutation | |||
| CLDN20 | Cancer | [249] | ||
| Breast | ↑ | |||
| OCLN | Cancer | PI3K signaling MAPK signaling |
[218, 221] | |
| Thyroid neoplasma | Diverse expression | |||
| Infjammaflon | ↓ | [231, 239, 250] | ||
| Crohn's disease | ↓ | |||
| Ulcerative collitis | ↓ | |||
| Celiac Disease | ||||
| Hereditary diseases | [186] | |||
| Cystic fibrosis | ↑ | |||
| Vision loss | ||||
| Diabetic eye disease: diabetic retinopathy | ↑ | [251] | ||
| ZO-1 | Cancer | PI3K signaling MAPK signaling |
[207, 221] | |
| Breast | ↓ | |||
| Inflammation | ||||
| Inflammatory bowel disease: Morbus Crohn | ↓ | [230] | ||
| Vision loss | ||||
| Diabetic eye disease: diabetic retinopathy | ↑ | [251] |
Figure 1. Schematic representation of the expression and function of the major tight junction proteins addressed in this review.
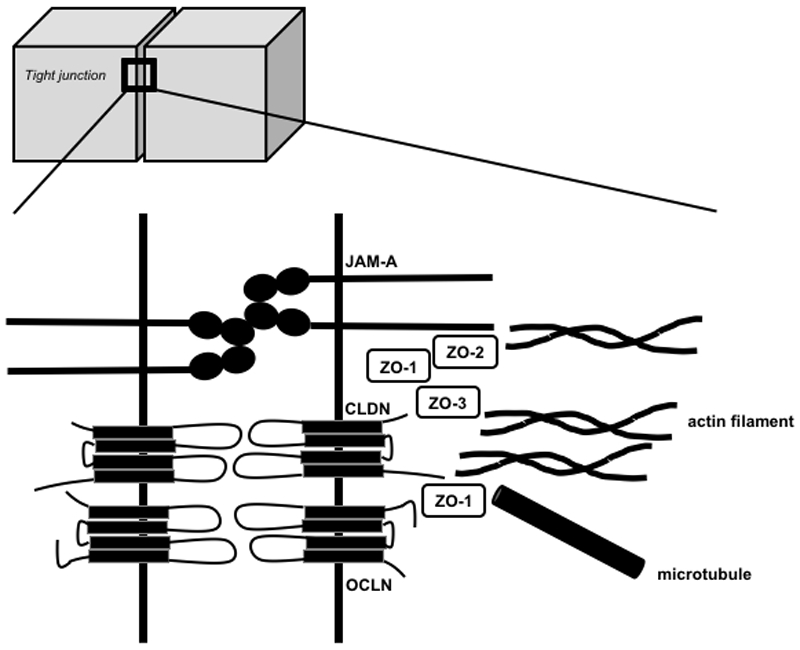
This simplified cartoon only displays the localization and interactions of the major tight junction (TJ) protein families that are addressed in this review. TJs are composed of transmembrane proteins, including different claudins (CLDNs), tight junction-associated marvel proteins (TAMPs, e.g. OCLN), junctional adhesion molecules (JAMs e.g. JAM-A) as well as cytosolic proteins (e.g. ZO-1, -2 and -3), which connect transmembrane components to the cytoskeleton (actin filaments, microtubules). For a more detailed description please refer to reference[1].
OCLN was the first identified integral membrane protein forming TJs. OCLN is a 65-kDa protein with 4 transmembrane domains. Posttranscriptional modification leads to several splice variants. OCLN contains a small intracellular loop and two extracellular loops (ECLs), ECL1 and ECL2, the latter being involved in homophilic interactions between OCLNs expressed on adjacent cells. The N- and C-terminal parts are both located within the cell. The C-terminal part is longer than the N-terminal part and its role in the modulation of TJ assembly, structure and function via posttranslational modifications of OCLN as well as for interaction with other TJ components and the cytoskeleton has been well studied[5] (Figure 1). OCLN contains a conserved four transmembrane marvel domain and is thus a member of the tight junction-associated marvel proteins (TAMPs) that also include tricellulin and marvelD3[6]. It has been shown that the three TAMPs have distinct but overlapping functions at the TJ. Tricellulin localizes at tricellular junctions formed by the corners of three epithelial cells while OCLN localizes at bicellular junctions. MarvelD3 has been reported to interact with tricellulin and OCLN, suggesting that it may be present at bi- and tricellular junctions[6].
The observation that TJs can form in the absence of OCLN[7] has led to the identification of CLDNs as integral TJ components. CLDNs form a family of 25-27 kDa proteins that in mammals comprises up to 27 members with high sequence homology. Like OCLN, CLDNs can also be subject to posttranslational and postranscriptional regulation. Their structure also resembles the one of OCLN, except for a shorter C-terminal part. The ECLs contribute through homophilic or heterophilic interactions with CLDNs or other integral membrane proteins located on adjacent cells to TJ formation[8]. The C-terminal part links the protein to intracellular TJ components and the actin cytoskeleton (Figure 1). Interestingly, CLDN expression patterns vary between different organs and cancers. CLDNs have thus been suggested as diagnostic markers and targets for cancer therapy[9, 10, 11].
Beside CLDNs and TAMPs that form TJ strands, additional transmembrane barrier proteins, including JAMs and related proteins, have been described (reviewed in[12, 13]. The best characterized JAM in the regulation of TJ barrier function is JAM-A, a member of the immunoglobulin (Ig) superfamily. The dimerization of two JAM-A molecules expressed on the same cell (cis-dimerization) contributes to the formation of a complex between transmembrane TJ proteins and cytoplasmic scaffold proteins[12] (Figure 1). Furthermore, JAM-A has been shown to act as a landmark for bicellular TJ formation[14] while lipolysis-stimulated lipoprotein receptor (LSR)/angulin-1, another member of the Ig superfamily of proteins, defines cell corners for tricellular TJ formation[15].
The best studied cytoplasmic proteins of TJs are zona occludens (ZO) proteins. ZO-1, -2, and -3 can interact with each other as well as with several transmembrane proteins (OCLN, CLDNs, JAM-A) and F-actin (Figure 1). Cytoplasmic proteins of TJs have thus been suggested to act as scaffolds linking TJs to the actin cytoskeleton and microtubules[12].
Importantly, TJ proteins have been reported to be also localized at sites outside TJs (non-junctional expression). Indeed, CLDN, OCLN and ZO proteins can be expressed at the basolateral membrane, in the cytoplasm and/or the nucleus where they have important functions in addition to those observed in TJs. For TJ protein expressed at the basolateral membrane these non-canonical functions include endosomal trafficking, signaling and additional ion transport functions while TJ proteins in the nucleus have been shown to regulate gene transcription (reviewed in[16]). Noteworthy, non-junctional TJ proteins do not diffuse in a random manner throughout the membrane. Rather, by interacting with defined molecules within the membrane and/or their phosphorylation, non-junctional CLDNs have been shown to be stabilized in discrete domains within the basolateral membrane and contribute to cell adhesion through interactions with the extracellular matrix[17, 18]. Furthermore, CLDNs can regulate the expression/activity of matrix metalloproteases (MMPs) that contribute to matrix remodelling[19, 20, 21, 22]. These functions can contribute to epithelial-to-mesenchymal transition (EMT), a process by which polarized epithelial cells lose their contacts to neighbouring cells and enable them to migrate. EMT has been shown to play an important role in organogenesis (EMT type 1), homeostasis, inflammation and fibrosis (e.g. wound healing, fibrogenesis; EMT type 2) but can also promote tumorigenesis, invasion and metastasis (EMT type 3).
The role of TJ proteins in the physiology and disease biology of the GI system and the liver deserves special emphasis. The GI tract epithelium has to maintain a delicate however dynamic balance in allowing specific substances (food, ions and solutes) to pass through the epithelium while not allowing many others (e. g. pathogens) in order to maintain the delicate balance between immune tolerance and activation. These considerations are further enriched by the recent findings that a feed-forward loop may exist between the gut microbiota and mucosal barrier function in such regulatory schemes[23]. Moreover, studies have now also revealed non-canonical roles of specific TJ integral proteins in regulating cellular differentiation, proliferation and migration; cellular mechanisms implicated in normal repair/regeneration as well as the oncogenic growth during tumorigenesis[24, 25]. Accordingly, a causal role of TJ proteins in GI disease, including esophagitis, inflammatory bowel disease (IBD) and cancers has been demonstrated. Similar to the GI tract, the liver endothelial junctions are important for liver functions and TJ dysregulation has been observed in chronic liver disease and hepatocellular carcinoma (HCC). Interestingly, TJ proteins CLDN1 and OCLN have been shown to be essential entry factors for the hepatitis C virus (HCV)[26, 27]. Here we review our current knowledge of the role of TJ proteins in GI and liver disease and discuss their potential as therapeutic targets focussing on GI cancer and viral infection of the liver.
TJ proteins and the GI tract
The functional role of TJ proteins in the physiology of the GI system
The GI mucosal barrier plays an important role in the separation of the inside of the body from the outside environment. TJs are present on the apical end of the lateral membrane surface in epithelial cells and regulate paracellular transport and apicobasal cell polarity. The expression of different TJs in the gut varies according to the gut’s functional properties as well as localization in villus or crypt (small bowel vs colon). The proteins can be localized strictly at the apical cell-cell adhesion or extend to the lateral or basolateral surfaces[28, 29, 30, 31]. Moreover, expression and cellular distribution of the TJ proteins – such as the CLDN family of proteins – is associated with regulation of differentiation of the intestinal epithelium[32, 33, 34]: CLDN1 is mainly expressed at the apex of the epithelial cells with a reticular pattern in the colon. CLDN2 is expressed in both villus and crypt cells of the small intestine but restricted to undifferentiated crypt cells in the colon. CLDN3, -4, -7 and -8 are predominantly expressed in the distal parts (colon, sigmoid and rectum) of the GI tract while CLDN10 and -12 show an ubiquitous expression pattern throughout the GI tract.
Loss- and gain-of-function studies in mice have revealed specific roles for a number of CLDNs in the TJ barrier function, selective ion permeability, as well as their related pathological phenotypes. For instance, knockout of CLDN7 in mice has severe intestinal defects including mucosal ulcerations, epithelial cell sloughing and inflammation, which leads to the death of the mice[35, 36]. Similarly, constitutive overexpression of CLDN1 in the mouse gut epithelium (to mimic upregulated CLDN1 expression in colon cancer) demonstrated a key role of CLDN1 in normal colonic epithelial homeostasis by regulating Notch-signaling[21], while in combination with APC (adenomatous polyposis coli) mutation (APCmin mice) CLDN1 overexpression induced colon tumorigenesis[37]. Similarly, upregulated CLDN2 expression in the mouse gut epithelium demonstrated a critical and complex role of CLDN2 in intestinal homeostasis by regulating epithelial permeability, inflammation and proliferation[38, 39]. Moreover, CLDN8 contributed to regulation of paracellular Na+ permeability, protecting the leakage of Na+ into the intestinal lumen[40]. CLDN16 is responsible for the defective absorption of Ca2+ in the intestine causing primary hypercalciuria[41]. Furthermore, the interdependence between CLDN proteins in regulating intestinal homeostasis is well demonstrated by in vivo loss-of-function studies for CLDN15. CLDN15 knockout mice grow normally despite having a mega-intestine[42]. However, a double knockout of CLDN15 with CLDN2 chronically reduces the paracellular flow of Na+ from the intestinal submucosa into the lumen resulting in shunting of the nutrient absorption, malnourishment and death [43]. Of note, both CLDN2 and -15 are paracellular transporters of Na+[44]. Overall, these findings show a critical functional role of these proteins in regulating intestinal homeostasis.
Furthermore, OCLN and JAMs have been shown to contribute to regulate intestinal homeostasis. For example, mice lacking JAM-A display an alteration of intestinal homeostasis as shown by perturbed regulation of epithelial permeability, inflammation, and proliferation, and significant alteration in CLDN protein expression[45]. Next generation gene editing technology such as CRISPR and fluorescent gene-reporter tags will enable to better understand the details of the function of TJ proteins in regulating GI physiology.
Functional role of TJ proteins during GI neoplastic transformations and growth
During neoplastic transformation in GI cancer, the expression and localization of TJ proteins is perturbed by several mechanisms occuring at the transcriptional, translational and post-transcriptional level (Figure 2). Signaling mechanisms that are known to promote neoplastic growth and cancer malignancy, including receptor tyrosine kinase signaling, inflammatory signaling cascades and non-coding RNAs, have been shown to perturb TJ protein expression and function. Perturbed TJ protein expression or function alters downstream signaling that targets cellular pathways relevant for epithelial homeostasis, invasion, chronic inflammation and cancer (Zeb-1/E-cadherin, Wnt signaling, MMP9/Notch signaling and Src/PI3K/Akt signaling). Furthermore, disruption of TJs during infection or injury can result in increased permeability with translocation of bacteria and luminal antigen, which in turn increase IL-6/Stat3 signaling contributing to carcinogenesis (Figure 2). Delocalization of TJ proteins from their normal membrane-tethered expression appears to be common among inflammatory diseases and GI cancers. Aberrant cell signaling may contribute to this process. Notably, in Ras-overexpressing MDCK cells, CLDN1, OCLN, and ZO-1 were absent from the cell-cell contact sites however were present in the cell cytoplasm[46]. Inhibition of MEK1 activity recruited all three proteins to the cell membrane leading to a restoration of the TJ barrier function in these cells. In line with this, it has been shown that growth factor receptors, including EGF, HGF and IGF receptors, as well as proinflammatory and tumor promoting cytokines, including TNF-α, IFN-γ, IL-13, and IL-22, contribute to regulate CLDN expression[47, 48, 49, 50, 51]. It has also been reported that nonsteroidal anti-inflammatory drugs regulate CLDN expression in association with p38 MAPK activation in gastric epithelial cancer cells[52]. Furthermore, protein modifications by phosphorylation, sumoylation, palmitoylation[53, 54, 55] and endocytic recycling have emerged as potential mechanisms of regulating TJ protein function and expression [56, 57, 58].
Figure 2. Schematic representation of differential regulation of tight junction proteins and associated signaling in gastrointestinal cancer.

Signaling and molecular mechanisms that are known to promote neoplastic growth and cancer malignancy include the receptor tyrosine kinase signaling, inflammatory signaling cascades and non-coding RNAs that perturb tight junction (TJs) expression and function. TJ perturbation alters downstream signaling that target important cellular events in epithelial homeostasis, invasion, chronic inflammation and cancer (Zeb-1/E-cadherin, Wnt signaling, MMP9/Notch signaling and Src/PI3K/Akt signaling). Furthermore, disruption of TJs can result in increased permeability to promote translocation of bacteria and luminal antigens, which then activate IL-6/Stat3 signaling to induce carcinogenic processes.
Transcription factors known to be associated with cellular differentiation and EMT, including Snail, Cdx-2, HNF-α, and GATA-4[34, 59, 60], can bind to the promoter regions of specific CLDN genes to affect their expression in intestinal epithelial cells. In colon cancer cells, caudal homeobox proteins (Cdx1 & Cdx2) and GATA-4 in cooperation with the Wnt pathway are involved in CLDN1 promoter activation[34]. CLDN1 transcripts are regulated by Smad-4 (a known tumor suppressor) and HDAC inhibitors supporting a complex multiprotein regulatory scheme[61, 62]. Furthermore, transcription factor RUNX3, which is a gastric tumor suppressor, upregulates CLDN1 expression by binding to the promoter region of CLDN1 in gastric epithelial cells[63] and a similar regulatory scheme involving Cdx-proteins, GATA-4 and HNF- has been demonstrated for CLDN2 and -4. Additionally, various epigenetic regulatory mechanisms likely also contribute to the transcriptional regulation of CLDN expression (Table 2). Indeed, it has been shown that DNA hypermethylation associated with the downregulation of CLDN11 in gastric cancer cells[64] and CLDN7 in colon cancer cells[34]. Furthermore, loss of repressive histone methylations, including H3K27me3 and H4K20me3, is associated with the overexpression of CLDN4 in gastric cancer[65, 66].
Table 2. Perturbation of tight junction protein expression in gastrointestinal and liver disease.
Examples of perturbed CLDN and ZO protein expression in gastrointestinal and liver disease as well as the underlying mechanism are shown. CLDN: claudin, miR: micro- RNA, TJ: tight junction, ZO: zona occludens
| Disease/Cell type | TJ protein expression | Described mechanism | References |
|---|---|---|---|
| Gastric cancer | CLDN4↑ | Loss of repressive histone methylation (H3K27m3, H4K20m3) | [65, 66] |
| CLDN11↓ | DNA hypermethylation miR-421 | [64] [90] | |
| CLDN18↓ | miR-1303 | [92] | |
| Intestinal bowel disease-associated carcinoma | CLDN1↑ CLDN2↑ |
β-catenin activation | [39] |
| Colorectal cancer | CLDN1↑ | miR-155 Histone deacetylase-mediated binding of human antigen R and tristetraprolin to CLDN1 mRNA Increased Notch and Wnt signaling | [37, 59, 69, 70] |
| CLDN2↑ | Increased Notch and Wnt signaling | [37, 59] | |
| CLDN7↓ | DNA hypermethylation | [34] | |
| ZO-1 ↑ | β-catenin activation | [59] | |
| Hepatocellular carcinoma | CLDN1↑ | Reactive oxygen species-mediated activation of heat shock factor 1 (HSF1) | [156, 157] |
| CLDN11↓ | miR-99 | [153] |
Finally, microRNAs (miRNAs) post-transcriptionally regulate TJ formation and barrier function[67]. Indeed, different miRNAs have been shown to modulate CLDN1 expression (Table 2): targeting of CLDN1 mRNA by miR-29 has been shown to regulate intestinal permeability[68], while the regulation of CLDN1 mRNA by miR-155 plays an important role in promoting colorectal cancer (CRC) cell migration and invasion[69]. Moreover, the histone deacetylase has been shown to regulate CLDN1 mRNA stability in CRC cells through modulating the binding of the human antigen R and tristetraprolin to the 3′ UTR of CLDN1 mRNA[70].
TJ proteins and colorectal disease and cancer
Perturbation of the epithelial barrier function as well as TJ protein function and expression are hallmarks of GI disease including CRC. The breakdown of polarized epithelial barrier leads to the activation of specific signaling pathways as a response to injury. However, chronic activation of these signaling pathways can also promote cancer formation in premalignant epithelial tissues when TJs are chronically leaky. Furthermore, TJ proteins, especially CLDNs, have now been demonstrated to play an essential role in cell proliferation and neoplastic transformation during tumorigenic growth.
Understanding how these complex signaling networks are altered in cancer cells represents a major challenge for the success of anti-cancer therapies. Upregulation or aberrant tissue expression of CLDNs may contribute to neoplasia by altering TJ structure and function or affecting cell signaling pathways. As stated above, the loss of cell polarity, due to TJ deregulation can abrogate the normal check-points. Moreover, studies linking EMT with the acquisition of stem cell characteristics have demonstrated important role of CLDNs in regulating the EMT process and cancer progression[16, 71, 72, 73]. In addition to regulating the barrier properties, TJs also serve as hubs for a multitude of signaling proteins including known tumor suppressor molecules like APC, PTEN (phosphatase and tensin homolog) and polarity proteins like Par-3[74, 75, 76]. Silencing of the expression and/or function of these proteins modulates CLDN expression and induces loss of polarity and EMT. Interestingly, genetic modulation of CLDN proteins in mice or cancer cells can similarly affect these signaling cascades, suggesting a feedback regulation.
Junctional proteins are known to play an important role or assist in cellular transformation when mislocalized from their normal membrane localization and could serve as oncogenic molecules. In this regard, the Wnt signaling pathway, essential for the differentiation of epithelial cells and imbalanced during intestinal epithelial oncogenic transformation is a major regulator of TJ protein expression[77]. For example, CLDN1 and CLDN2 proteins are known target genes of the Wnt/β-catenin signaling pathway with binding sites in the promoter of these genes[34, 59]. CLDN1 expression not only decreased significantly in response to the reduction of intracellular β-catenin by adenovirus-mediated transfer of wild-type APC into the APC-deficient colon cancer cells, but also two putative Tcf4 binding elements in the 5' flanking region of CLDN were confirmed to be responsible for activating its transcription[34]. Importantly, in the intestine CLDN1 is weakly expressed at the apical border of the lateral membrane of normal enterocytes but is strongly expressed at cell-cell boundaries as well as in the nucleus/cytoplasm of CRC cells. Many studies have demonstrated that the expression of CLDN1 at the mRNA and protein levels is increased in CRC tissue and correlates with tumor depth[78]. Additional studies using gene editing have further shown that an intricate interdependence between the Notch and Wnt-signaling upregulating CLDN1 expression to augment CRC progression[37] (Table 2). A role of the nuclear effectors of the Wnt signaling pathway is to bind directly to the CLDN2 promoter region and thereby enhance CLDN2 promoter activity. Also, a crosstalk between the Wnt signaling and Cdx related transcriptional activation machinery has been implicated in regulating CLDN2 promoter-activation[59]. Recent studies have also demonstrated that levels of CLDN1 and CLDN2 are elevated in IBD-associated carcinoma[39] (Table 2). Kinugasa et al.[79] demonstrated increased staining for CLDN1 in both high-grade dysplasia and ulcerative colitis (UC)-associated CRC when compared with normal or UC samples. CLDN1 overexpression modulates Notch-signaling in an MMP9-dependent manner to modulate barrier properties and immune homeostasis to promote susceptibility to inflammation-induced colitis and cancer[21]. Here it is worth noting that the outcome from a series of studies have now provided ample evidence for a role of deregulated CLDN1 expression in promoting invasive and metastatic abilities of the colon cancer cells. Notably, CLDN1 expression was sufficient to induce metastatic abilities in a colon cancer cell line that normally does not metastasize well in vivo. In contrast, stable genetic inhibition of CLDN1 in a poorly differentiated, highly metastatic and CLDN1 high colon cancer cell significantly inhibited its metastatic abilities in a splenic model of CRC metastasis[78]. An increase in CLDN2 expression also participates in promoting colorectal carcinogenesis potentially dependent on the EGFR/ERK1/2 signaling[80, 81]. Here, overexpression of CLDN2 in CLDN2 deficient CRC cells resulted in increased cell proliferation, anchorage-independent growth and tumor growth[81]. A similar effect of the Wnt-/ -catenin signaling upon gene expression of yet another component of the TJ complex, ZO-1, in human colonic cancer cell lines with low endogenous β-catenin has been reported suggesting potential contribution to the loss of epithelial polarization in neoplastic cells[59] (Table 2). Decreased ZO-1 expression was noted in the human digestive tract[82]. Using tissue biopsy samples, Mees et al.[83] also found that CRC in human exhibits significantly elevated expression levels of CLDN1 and -4 compared with normal mucosa. However, CLDN expression in colon cancer tissues is differential and downregulation of CLDN7 and -8 has been reported in colorectal adenoma samples compared with the normal intestinal tissues[84]. Collectively, these studies suggest that these proteins may serve as potential biomarkers for CRC progression and therapy resistance.
CLDNs and esophageal and gastric cancer
Similar to CRC, TJ proteins are also regulated aberrantly in the esophageal and gastric cancers and this abnormal expression correlates with specific clinicopathologic parameters. In the esophageal adenocarcinoma (EA), CLDN expression has been tested as potential biomarker for the transition of the Barrett’s esophagus to the EA. Indeed, CLDN2, -3, -4 and -7 are reported to have increased expression in EA compared to precancerous lesions and normal esophageal squamous mucosa[85, 86]. JAMs, which comprise the integral parts of TJs in the gastric epithelium, have been shown to promote proliferation, invasion, and inhibit apoptosis. JAM-B was upregulated significantly in tumor samples compared with adjacent normal tissues and was higher in high grade tumors than in the low grade and intermediate grade tumors[87].
An increased expression of CLDN2 is also associated with gastric cancer progression[88]. Additionally, expression of CLDN11 and -23 is downregulated in gastric cancer[89] and miR-421 was implicated in regulating CLDN11 expression to promote the proliferation, invasion and metastasis of gastric cancer cells[90] (Table 2). In contrast, CLDN23 positive expression was associated with poor prognostic outcomes of gastric cancer patients and may therefore serve as an independent predictor of patient survival. Similarly, upregulated expression of CLDN4 in gastric cancer was associated with cancer progression and poor prognosis[91]. Furthermore, CLDN4-expressing gastric adenocarcinoma AGS cells were found to have increased MMP2 and -9 expression, indicating that CLDN-mediated increased invasion may be mediated through the activation of MMPs[91]. Overall, these results suggest that CLDN4 overexpression may promote gastric cancer metastasis through the increased invasion of gastric cancer cells. Yet another CLDN family protein, CLDN18 is significantly downregulated in gastric cancer tissues and cell lines, which was associated with tumor size, location invasion, histologic type and tumor-node-metastasis stage. miR-1303 was demonstrated to have putative binding sites in CLDN18 mRNA 3'-UTR and visibly lower the expression of CLDN18[92] (Table 2). On the other hand, CLDN18.2 (isoform 2 of claudin-18) was retained on malignant transformation and was expressed in a significant proportion of primary gastric cancers and its metastases[9]. The expression of CLDN7 has also been reported to have the potential of serving as an independent indicator of the poor prognosis in gastric cancer[93].
Taken together, TJ proteins are tissue-specific regulators of the epithelial and endothelial barrier function and EMT, which cumulatively perturb the epithelial and immune homeostasis leading to carcinogenesis in CRC as well as gastric and esophageal cancer.
TJ proteins and the liver
The liver plays an essential role in homeostasis by its metabolic and storage functions. It is composed of different cell types, including two types of epithelial cells: hepatocytes and cholangiocytes. Liver epithelial junctions are important for liver function. Hepatocytes are liver parenchymal cells that exhibit a complex honeycomb morphology displaying at least two basolateral membranes (facing the sinusoidal blood) and two intercellular apical membranes (forming the bile canaliculi) separated by TJs. This peculiar architecture creates what has been termed the blood-biliary barrier[94] and enables hepatocytes to perform distinct secretory functions at the same time[95]. Hepatocytes produce and secrete bile into the bile canaliculi from where it is transported via intrahepatic and extrahepatic bile ducts, formed by cholangiocytes, to the gallbladder. Cholangiocytes contribute to modify the bile during its transport to the latter. Hepatocyte polarity and bile duct TJs play a major role in the liver[96, 97] and thus defects in hepatocyte and/or cholangiocyte TJ integrity can result in pathophysiological consequences.
Several lines of evidence indicate that disruption or loss-of-function of TJs contributes to the pathogenesis of cholestatic diseases including primary biliary cholangitis and primary sclerosing cholangitis[98, 99]. Interestingly, mutations in CLDN1 are associated with neonatal ichthyosis and sclerosing cholangitis (NISCH) syndrome where deficient CLDN1 expression may contribute to paracellular bile leakage through deficient TJs[100]. Mutations in ZO-2 have been described in familiar hypercholanemia[101]. The loss-of-function of TJ proteins is not lethal and the clinical manifestation is variable including very mild symptoms[102]. A comprehensive description of TJ protein alterations in biliary diseases is reviewed in reference[97].
Recent studies demonstrate that TJ protein expression is altered in HCC (primary liver cancer) and cholangiocarcinoma (biliary tract cancer). For example, CLDN1 has been shown to be up-regulated in advanced liver disease and HCC[103] and differential CLDN4 expression can help to distinguish these two forms of cancer at a molecular level[104]. Of note, TJ alteration in epithelia outside the liver can also contribute to liver disease. Indeed, dysfunction of the intestinal epithelial barrier - due to or unrelated to (aetiological factor(s) of) the underlying liver disease - has been associated with the pathogenesis of chronic liver disease and the development of complications in cirrhosis by favouring translocation of bacteria and bacterial products from the intestinal lumen into the systemic circulation[105].
CLDN1 and OCLN mediate hepatocyte entry of HCV
Ten years ago, expression cloning experiments uncovered CLDN1 to be required for HCV infection[26]. The role of OCLN in HCV infection was uncovered two years later using different approaches[27, 106, 107]. Subsequently several studies have characterized the underlying molecular mechanisms and highlighted the essential function played by these proteins in HCV entry and infection[4, 108, 109]. While over the past 20 years many host factors have been reported to contribute to the early steps of HCV infection[108, 110], CLDN1 and OCLN are regarded as two of the four major HCV host entry factors together with CD81[111] and scavenger receptor BI (SR-BI)[112] (Figure 3).
Figure 3. Functional roles of CLDN1 as hepatitis C virus entry factor.
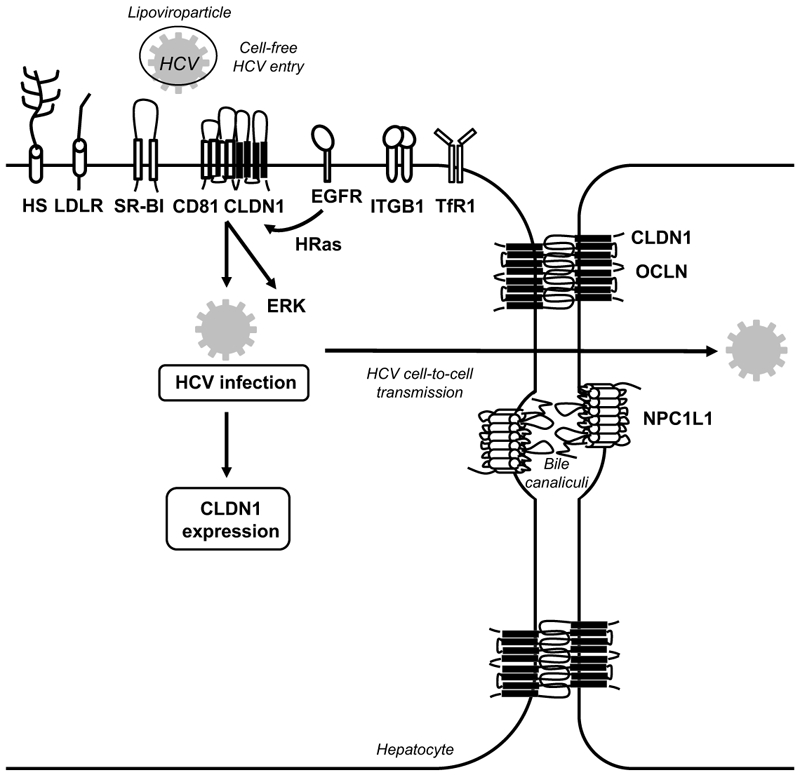
CLDN1 is one of the four main hepatitis C virus (HCV) host factors (i.e. SR-BI, CD81, CLDN1 and OCLN) essential for the early steps of HCV infection. Several other host factors (e.g. highly sulphated heparan sulfate (HS), low-density lipoprotein receptor (LDLR), epidermal growth factor receptor (EGFR), integrin beta 1 (ITGB1), transferrin receptor 1 (TfR1) and Niemann Pick C1 like 1 (NPC1L1)) contribute to viral binding and entry. EGFR-mediated signaling leads to the formation of a CD81-CLDN1 co-receptor complex that ultimately leads to viral internalization[116, 118]. HCV infection induces CLDN1-depend signaling via the ERK1/2 pathway[120]. HCV infection increases CLDN1 expression[122, 145]. TJ proteins involved in the HCV entry process are depicted in black, non-TJ host entry factors are depicted in white.
First binding studies indicated that CLDN1 was unable to bind the HCV envelope glycoprotein E2[26], suggesting that CLDN1 does not play the role of a primary receptor but rather of a co-receptor, which contributes to (a) step(s) subsequent to viral binding[26]. This was further confirmed in kinetic assays using anti-CLDN1 antibodies[113]. Several years later it was shown that in contrast to soluble E2, HCV E1E2 complexes can interact with the CLDN1 ECL1 and that this interaction is involved in viral fusion[114]. Based on the Coxsackie B virus cell entry model, it was suggested that HCV may first interact with host factors on the basolateral surface of hepatocytes and then move to the TJ co-entry factor(s), e.g. through CD81-lateral membrane movements[26, 115]. Elegant fluorescence resonance energy transfer studies showed that CLDN1 interacts with CD81 to promote viral internalization[116, 117, 118]. Interestingly, no cellular function for CD81-CLDN1 interaction has been reported so far and disruption of these complexes by defined anti-CLDN1 antibodies prevents HCV infection without affecting TJ integrity or any detectable adverse effect[113, 119, 120]. Indeed, several lines of evidence support a model in which the non-junctional form of CLDN1 rather than CLDN1 localized within TJ mediates HCV entry[117, 121]. The major pool of CLDN1 is expressed at TJs of hepatocytes and polarized hepatoma cells but a minor fraction is also located at the basal membranes of these cells[117, 122]. Of note, CD81-CLDN1 co-receptor association could only be detected at the basal membranes but not in TJ-associated pools of CLDN1 and CD81[117]. Furthermore, the ECL1 appears to be the critical part of the protein for HCV entry while the intracellular C-terminal part of CLDN1 that plays an important role for its interaction with intracellular TJ components is not required for this process[26, 121]. Interestingly, CLDN6 and CLDN9 - but not other members of the CLDN family of proteins - have been shown to be able to promote HCV entry into CLDN-deficient 293T-derived cell lines[123, 124]. This is most likely due to their ability to form co-receptor associations with CD81 like CLDN1 [118]. It is of interest to note that in experimental model systems using a liver tumor cell lines some HCV genotypes have been reported to be able to use either CLDN1 or CLDN6[125] through mutation in the HCV E1 envelope protein[126]. Whether CLDN6 or 9 can replace CLDN1 in the liver of HCV-infected patients remains questionable since CLDN6 and CLDN9 expression is very low or absent in human liver tissues[124, 125, 127]. Furthermore, treatment of HCV infection in human liver chimeric mice with a monoclonal CLDN1-specific antibody did not reveal any detectable escape (for a detailed review of the role of CLDN6 and CLDN9 in HCV entry please see[128]).
Like CLDN1, OCLN does not appear to play the role of a primary HCV attachment receptor but rather is required for late postbinding event(s) during the HCV entry process[107, 129] (Figure 3). Nevertheless, HCV might interact with OCLN during viral entry and/or in infected cells. Indeed, imaging studies evidenced a co-localization between OCLN and HCV E2 in the endoplasmic reticulum of hepatoma cells[130]. Furthermore, it was shown that an anti-E2 antibody could immunoprecipitate OCLN while GST-OCLN could pull down E2[129, 130]. The OCLN ECL2 appears to be important for this interaction with HCV E2 as well as for HCV entry[129]. However, the OCLN ECL2 was unable to pull down E2, suggesting that either this interaction might not be direct or not be visualized in the utilized experimental design[129]. Experiments using OCLN engineered to be recognized by anti-FLAG antibodies are in favour of a HCV-OCLN interaction as different clones displaying the FLAG epitope at different locations within ECL1 or ECL2 exhibited HCV isolate-dependent host factor activity[131]. Using kinetic assays in polarized cells, this study also showed that OCLN plays a role subsequent to SR-BI, CD81 and CLDN1[131]. These results were recently confirmed in kinetic assays using anti-OCLN mAbs directed against either the ECL1 or ECL2 of OCLN[132]. How and where HCV interacts with OCLN as well as what pool(s) of OCLN is(are) involved in this process remain to be further characterized. Although in liver sections OCLN has been located at apical surfaces of hepatocytes[117], a minor pool of this protein is expressed on the basolateral surface of hepatocytes. Indeed, OCLN is known to traffic through the basolateral membrane towards TJs[133] and its subcellular localization appears to be dependent on its phosphorylation status: phosphorylated forms of OCLN mainly are found in TJs of epithelial cells, while less phosphorylated forms are localized at the basolateral membrane and in the cytosol[134]. This is in line with a recent report showing that tumor-associated calcium signal transducer 2 (TACSTD2) regulates HCV entry by leading to the phosphorylation of CLDN1 and OCLN and regulates their subcellular localization[135]. The importance of the subcellular localization of OCLN for HCV entry is also underscored by the fact that only OCLN and its splice variant OCLN-ex7ext that both localize to the plasma membrane are able to promote HCV entry in contrast to other OCLN splice variants that exhibit an intracellular localization[136]. Of note, OCLN together with CD81 define the HCV species specificity as HCV non-permissive mouse cells acquire HCV-permissivity subsequent to human CD81 and OCLN expression both in vitro and in vivo[27, 137, 138, 139]. The species-specific determinants appear to be located within the second extracellular loop of OCLN[27, 139].
Beside cell-free HCV entry, CLDN1 and OCLN have also been shown to be important for HCV cell-to-cell transmission (Figure 3), a major mode of viral dissemination that enables the virus to avoid the host's immune surveillance and to establish chronic infection[140, 141, 142, 143]. The exact localization at the plasma membrane of this process as well as the form(s) of CLDN1 that contribute(s) to HCV cell-to-cell transmission remain unknown. The importance of CLDN1 and OCLN for the pathogenesis of HCV infection in vivo has been confirmed by observations of liver tissues from HCV-infected liver transplant patients. HCV recurrence was associated with an increase in CLDN1 and OCLN expression levels in hepatocytes over time after transplantation[144]. This is in line with findings indicating increased CLDN1 and OCLN expression levels in HCV-infected livers[122, 145, 146]. In contrast, in cell-based studies HCV infection was shown to downregulate CLDN1 and OCLN expression to prevent superinfection[106]. Differences in TJ protein expression upon HCV infection may thus exist depending on the analyzed samples.
CLDNs and HCC
The expression of several TJ proteins has been reported to be perturbed in liver tissue from HCC patients. Many studies have shown different expression levels of the individual CLDNs and OCLN and CLDN1 are being investigated as biomarkers for liver disease progression[103, 122, 147, 148, 149, 150, 151]. From these studies, it appears that expression of CLDNs is associated with more severe disease and/or bad prognosis in HCC patients (Table 2): epigenetic silencing of CLDN14 was significantly associated with advanced tumor state and tumor aggressiveness[152]; CLDN11 downregulation by miR-99 has been associated with metastasis of HCC[153]; and CLDN3 downregulation has been suggested to promote EMT via Wnt- -catenin signaling[154]. However, more data are needed to decipher the role of these TJ proteins in the pathogenesis of HCC. Several studies have shown an increase in CLDN1 expression on basolateral and apical hepatocyte membranes in cirrhotic livers and HCC compared to normal livers[103, 122, 149]. Interestingly, this increase was observed in tissues from both HCV-positive and -negative patients, although it appeared to be stronger in tissues from HCV-infected patients, as well as in HCC that developed on either cirrhotic or non-cirrhotic livers[103] and in paediatric HCC[149]. In advanced HCC down-regulation of CLDN1 has been observed[147, 151] which may correspond to the de-differentiation of cancer cells. Of note, a greater cytoplasmic localization of CLDN1 was observed in some HCC in line with reports indicating that CLDN localization has a causal role in cellular transformation[78, 155].
CLDN1 likely contributes to proliferation, motility and invasion by modulating cellular signaling. Overexpression of CLDN1 increases the migration and invasiveness of human hepatoma cells as well as normal liver cells through expression of MMP2 via the c-Abl-PKC pathway[20] (Figure 4). Furthermore, increased CLDN1 expression has been associated with mitochondrial dysfunction and invasiveness of hepatoma cells, and reactive oxygen species-mediated activation of heat shock factor 1 (HSF1) was demonstrated to increase CLDN1 expression in these cells[156, 157] (Table 2). These data are in line with reports indicating that CLDN1 enhances cell growth, migration and/or invasiveness of other cancer cell types such as oral squamous cell carcinoma cells, CRC cells, ovarian cancer-initiating cells or melanoma cells[69, 158, 159, 160, 161]. Taken together, these data suggest that by promoting cell migration and increasing the invasive behaviour of cancer cells, CLDN1 can contribute to cancer spread. Of note, CLDN1 has been shown to promote EMT in normal liver cells and HCC cells that thereby acquire an invasive phenotype[162]. This process is mediated by the c-Abl-Ras-Raf-1-ERK pathway and involves the transcription factors Slug and Zeb1[162] (Figure 4). Confirming the functional role of these signaling pathways, an CLDN1-specific antibody inhibits the HCV-induced increase in ERK1/2 phosphorylation in human liver tissue [120]. Further studies are needed to understand the detailed role of CLDNs in pathogenesis of liver disease and cancer.
Figure 4. Functional role of CLDN1 in signal transduction and EMT in liver disease.

In transformed liver cells, CLDN1 over-expression activates the c-Abl-PKC pathway to increase cellular migration and invasion via MMP2 activation[20] as well as the c-Abl-Ras-Raf-ERK pathway to promote EMT via the transcription factors Slug and Zeb1[162].
Targeting TJ proteins for therapeutic approaches in the gut and the liver
CLDNs as targets for CRC
While the field related to the role and regulation of TJ proteins in GI pathologies and oncogenic growth has taken a significant leap forward, therapeutic application of this knowledge is now emerging. Significant progress has been made at several fronts including the development of prognostic biomarkers, imaging and targeting. In this regard, CLDN proteins are currently investigated as potential biomarkers for disease progression and therapy resistance. A recent study has shown that serum levels of CLDN1 and CLDN7 may be a useful tool in the differential diagnosis of CRC[163]. Furthermore, a progressive increase in CLDN1 expression in colon cancer and the recent findings that infra-red imaging using CLDN1-targeted conjugated peptide can enhance the ability of conventional colonoscopy for detecting human colonic adenomas strongly supports the potential impact of CLDN1 as a biomarker[164]. CRC has been found to arise from missed polypoid and flat precancerous lesions which are more difficult to visualize by colonoscopy and the new CLDN1 targeted fluorescent peptides may be used to improve screening of high-risk patients with multiple polyps, inflammatory bowel disease, Lynch syndrome, or a family history of CRC.
Aiming to develop targeted therapies, several antibodies against the extracellular domain of CLDNs have been developed. Their therapeutic effects for cancer and metastasis are summarized in Table 3. Ideal monoclonal antibodies (IMAB) specific to the proteins expressed only on the tumor and hence avoiding potential off-target effects are actively being developed. Currently, monoclonal antibodies (mAbs) have been generated against CLDN1,-2, -3, -4, -6, and -18.2. The antibody against CLDN18.2 (claudiximab) is in clinical development for gastric cancer[9]. Interestingly, claudiximab significantly extends median survival when added to standard chemotherapy (13.2 vs 8.4 months) in patients with advanced gastric cancer[9]. Importantly, this target is not present in any healthy tissues except the lining of the stomach, thereby minimizing treatment side effects. In addition, recent studies have investigated the anti-tumor effect of anti-CLDN1 and anti-CLDN2 mAbs using cancer cell models[11, 165] (Table 3). Importantly, anti-CLDN mAbs have been shown to be safe and no relevant off-targets have been reported. Their marked therapeutic effects combined with excellent safety profiles are highly encouraging for their development in clinical applications.
Table 3. Preclinical and clinical development of antibodies directed against tight junction proteins.
The respective target, names of monoclonal antibodies (that have at least reached preclinical stage of development), clinical indication, and stage of development are shown. CLDN: claudin, HCV: hepatitis C virus, OCLN: occludin
| Targets | Monoclonal antibodies |
Clinical indication | Stage of development |
References |
|---|---|---|---|---|
| CLDN1 | OM-7D3-B3 and | HCV infection | Preclinical | [120] |
| H3L3 | [168] | |||
| 3A2 | HCV infection | Preclinical | [169] | |
| 6F6 | Colorectal cancer | Preclinical | [11] | |
| CLDN2 | xi-1A2 | Cancer | Preclinical | [165] |
| CLDN3 and CLDN4 | KM3907 | Cancer | Preclinical | [252] |
| 5A5 | Cancer | Preclinical | [253] | |
| CLDN4 | KM3900 | Pancreatic and ovarian cancers | Preclinical | [254] |
| CLDN6 | IMAB027 | Ovarian cancer | Phase I/II | NCT02054351 |
| CLDN18.2 | IMAB362 (claudiximab) | Gastroesophageal cancer | Phase II | NCT01630083 |
| OCLN | 1-3 and 37-5 | HCV infection | Preclinical | [132] |
CLDN1 and OCLN - targets for cure of HCV infection
CLDN1 was the first TJ protein to be explored as a therapeutic target for HCV infection using anti-CLDN1 antibodies directed against its extracellular domain(s) (Table 3). Such antibodies could be used to prevent liver graft infection in HCV-positive transplant recipients and as a promising alternative for patients who fail current anti-HCV therapies[166]. The first antibodies directed against human CLDN1 and blocking HCV infection were generated by genetic immunization in rats[113, 119]. They recognize a conformational-dependent epitope within the ECL1 and prevent CD81-CLDN1 co-receptor association at the basolateral membrane[113, 119]. They are characterized by pan-genotypic inhibition of the infection by all major HCV genotypes by blocking both cell-free virus entry and viral cell-to-cell transmission[19, 143]. Of note, studies in human liver-chimeric mice demonstrated that the lead antibody OM-7D3-B3 was not only able to prevent acute de novo infection with HCV (i.e. the anticipated effect of an entry inhibitor) but also to cure already established chronic HCV infection without detectable side or off-target effects[120]. These results highlighted the importance of viral dissemination for maintenance of chronic HCV infection. It is of interest to note that this antibody interfered with MAPK signalling suggesting an important role in this pathway potentially also contributing to its antiviral effect[120]. The therapeutic potential of this antibody is further underscored by the fact that it acts in synergy with HCV direct-acting antivirals (DAAs), the current state-of-the-art antiviral therapy, to clear viral infection and is also active on viral variants escaping DAAs[143, 167]. This antibody has recently been successfully humanized (IgG4) for further clinical development[168]. Subsequently, other CLDN1-specific antibodies inhibiting HCV infection have been reported: clones 3A2 and 7A5, generated in mice and recognizing the human CLDN1 ECL2 can prevent HCV infection of human liver-chimeric mice[169] while several antigen-binding fragments (Fab) and single chain antibody fragments selected using phage display were demonstrated to inhibit HCV infection in vitro when converted into human IgG1 or IgG4[170, 171]. Administration of CLDN1-specific antibodies has been shown to be very safe in various animal and human-cell based models without any adverse effects on the liver or other organs such as the gut or skin[120, 168, 172]. This is most likely to the mechanism of action of CLDN1-specific antibodies targeting the non-junctional expressed CLDN1 on the hepatocyte basolateral membrane without affecting TJ barrier function as shown in several TJ model systems[120, 168, 172].
Antibodies directed against OCLN have been more difficult to generate than anti-CLDN mAbs but very recently five mAbs with anti-HCV activity were described by two different groups[132, 173]. The mouse mAb (67-2) - raised against a linear peptide within OCLN ECL2 - was shown to recognize an epitope present in the ECL2 of both human and mouse OCLN[173]. Interestingly, this mAb hardly inhibited the infection of human hepatoma Huh7.5.1 cell monolayers with HCV when applied to the apical membrane of cells while it was able to efficiently prevent HCV infection when applied to the basolateral membrane of cells using a double-chamber culture system or a 3D culture model[173]. Of note, in line with the hypothesis that mAb 67-2 interacts with OCLN monomers expressed on the basolateral membrane, this mAb had no effect on TJ function in Eph4 cells[173]. Four rat mAbs - generated by genetic immunization and directed against either the ECL1 or ECL2 of OCLN – inhibited the entry of HCV into human hepatoma cells without affecting TJ barrier function of polarized cells. Since the mAb directed against ECL2 appeared to be more potent in inhibiting HCV infection than mAbs directed against ECL1 and in line with previous studies using OCLN mutants/chimeras having demonstrated the essential function of ECL2 for HCV infection[27, 129, 174], the authors hypothesized that the mAb directed against ECL2 may block an essential function of OCLN in the HCV entry process while mAbs directed against ECL1 may block HCV infection through steric hindrance. Two of those mAbs targeting either ECL1 or ECL2 (1-3 and 37-5) were shown to inhibit HCV infection in human liver chimeric mice without apparent side effects highlighting the possibility to target OCLN in vivo[132].
The positioning of CLDN1- and OCLN-specific antibodies in the widening arsenal of anti-HCV therapies is most likely for patients with multi-resistance to DAAs or in organ transplantation including HCV-positive donors where prevention of de novo infection may be preferable to cure of an established HCV infection. They may offer also perspectives to further shorten therapy regimens when combined with DAAs[175, 176].
Conclusions and future perspectives
Research in the last two decades has demonstrated an important role of TJ proteins in the physiology and disease biology in GI and liver disease. TJ proteins exert their functional role as integral proteins of TJs in forming barriers in the gut and the liver. Furthermore, TJ proteins are expressed non-junctionally where they play important roles in signaling, trafficking and regulation of gene expression outside the TJs. A hallmark of TJ proteins in disease biology is their role in EMT, which is relevant for organogenesis and differentiation (EMT type 1), inflammation and fibrosis (EMT type 2) and cancer metastasis/invasion (EMT type 3). A causative role of TJ proteins has been established in the pathogenesis of CRC and gastric cancer. Among the best characterized role of TJ proteins in liver disease biology is their function as cell entry receptors for HCV – one of the most common causes of HCC. At the same time TJ proteins are emerging as targets for novel therapeutic approaches for GI and liver disease: these include treatment of CRC and gastric cancer as well as antiviral therapy for chronic HCV infection complementing DAAs. Further studies are needed to study their role in chronic inflammation, fibrosis and their role as drivers for carcinogenesis. The understanding of these mechanisms offers new perspectives for novel therapeutic approaches for key unmet medical needs in the gut including CRC and gastric cancer as well as chronic liver disease and HCC.
Acknowledgements
The authors work is supported by ARC, Paris and Institut Hospitalo-Universitaire, Strasbourg (TheraHCC IHUARC IHU201301187), the European Union (ERC-AdG-HEPCIR, ERC-PoC-2016-PRELICAN, EU H2020-667273-HEPCAR, U Strasbourg Foundation HEPKIN), the National Institutes of Health (NCI 1R21CA209940-01A1, NIAID R03AI131066, NIAID 5U19AI123862-02), the Institut Universitaire de France (IUF), the IdEx Program of the University of Strasbourg, the Impulsion Program of the IDEXLYON, BX002086 (VA merit), CA216746 (NIH/NCI) and a pilot project award from Fred and Pamela Buffet Cancer Center, which is funded by a National Cancer Institute Cancer Center Support Grant under award number P30 CA036727. This work has been published under the framework of the LABEX ANR-10-LABX-0028_HEPSYS and benefits from funding from the state managed by the French National Research Agency as part of the Investments for the future program.
Abbreviations:
- APC
adenomatous polyposis coli
- CLDN
claudin
- CRC
colorectal cancer
- DAA
direct-acting antiviral
- EA
esophageal carcinoma
- ECL
extracellular loop
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- Fab
antigen-binding fragment
- GI
gastrointestinal
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- IBD
inflammatory bowel disease
- IgG
immunoglobulin G
- IMAB
ideal monoclonal antibody
- JAM
junctional adhesion molecule
- mAb
monoclonal antibody
- miR/miRNA
microRNA
- MMP
matrix metalloprotease
- TJ
tight junction
- OCLN
occludin
- SR-BI
scavenger receptor BI
- TAMP
tight junction-associated marvel proteins
- UC
ulcerative colitis
- ZO
zona occludens
Footnotes
Conflict of interests
TFB is a co-inventor of a patent/patent application of CLDN1-specific antibodies for prevention and treatment of HCV infection. TFB and MBZ are co-inventors on patent applications for anti-claudin 1 monoclonal antibodies for the prevention and treatment of liver disease and HCC.
References
- 1.Zihni C, Mills C, Matter K, et al. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol 2016;17:564–80. [DOI] [PubMed] [Google Scholar]
- 2.Singh AB, Uppada SB, Dhawan P. Claudin proteins, outside-in signaling, and carcinogenesis. Pflugers Arch 2017;469:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandner JM, Zorn-Kruppa M, Yoshida T, et al. Epidermal tight junctions in health and disease. Tissue Barriers 2015;3:e974451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeisel MB, Turek M, Baumert TF. Tight junctions and viral entry. Future Virology 2010;5:263–71. [Google Scholar]
- 5.Cummins PM. Occludin: one protein, many forms. Mol Cell Biol 2012;32:242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raleigh DR, Marchiando AM, Zhang Y, et al. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 2010;21:1200–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitou M, Furuse M, Sasaki H, et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell 2000;11:4131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers 2013;1:e25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. Journal of Hematology & Oncology 2017;10:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osanai M, Takasawa A, Murata M, et al. Claudins in cancer: bench to bedside. Pflugers Arch 2017;469:55–67. [DOI] [PubMed] [Google Scholar]
- 11.Cherradi S, Ayrolles-Torro A, Vezzo-Vie N, et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J Exp Clin Cancer Res 2017;36:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 2014;36:157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luissint AC, Nusrat A, Parkos CA. JAM-related proteins in mucosal homeostasis and inflammation. Semin Immunopathol 2014;36:211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severson EA, Parkos CA. Mechanisms of outside-in signaling at the tight junction by junctional adhesion molecule A. Ann N Y Acad Sci 2009;1165:10–8. [DOI] [PubMed] [Google Scholar]
- 15.Masuda S, Oda Y, Sasaki H, et al. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J Cell Sci 2011;124:548–55. [DOI] [PubMed] [Google Scholar]
- 16.Hagen SJ. Non-canonical functions of claudin proteins: Beyond the regulation of cell-cell adhesions. Tissue Barriers 2017;5:e1327839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu CJ, Mannan P, Lu M, et al. Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J Biol Chem 2013;288:12253–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Itallie CM, Tietgens AJ, LoGrande K, et al. Phosphorylation of claudin-2 on serine 208 promotes membrane retention and reduces trafficking to lysosomes. J Cell Sci 2012;125:4902–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005;65:7378–85. [DOI] [PubMed] [Google Scholar]
- 20.Yoon CH, Kim MJ, Park MJ, et al. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem 2010;285:226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pope JL, Bhat AA, Sharma A, et al. Claudin-1 Regulates Intestinal Epithelial Homeostasis through the Modulation of Notch Signaling. Gut 2014;63:622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamori H, Takino T, Kobayashi Y, et al. Claudin promotes activation of pro-matrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. J Biol Chem 2001;276:28204–11. [DOI] [PubMed] [Google Scholar]
- 23.Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012;491:254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol 2014;36:224–31. [DOI] [PubMed] [Google Scholar]
- 25.Salvador E, Burek M, Forster CY. Tight Junctions and the Tumor Microenvironment. Curr Pathobiol Rep 2016;4:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans MJ, von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 2007;446:801–5. [DOI] [PubMed] [Google Scholar]
- 27.Ploss A, Evans MJ, Gaysinskaya VA, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 2009;457:882–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev 2013;93:525–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Z, Ding L, Lu Q, et al. Claudins in intestines: Distribution and functional significance in health and diseases. Tissue Barriers 2013;1:e24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amasheh S, Fromm M, Gunzel D. Claudins of intestine and nephron – a correlation of molecular tight junction structure and barrier function. Acta Physiol (Oxf) 2011;201:133–40. [DOI] [PubMed] [Google Scholar]
- 31.Rahner C, Mitic LL, Anderson JM. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology 2001;120:411–22. [DOI] [PubMed] [Google Scholar]
- 32.Benoit YD, Pare F, Francoeur C, et al. Cooperation between HNF-1alpha, Cdx2, and GATA-4 in initiating an enterocytic differentiation program in a normal human intestinal epithelial progenitor cell line. Am J Physiol Gastrointest Liver Physiol 2010;298:G504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Escaffit F, Boudreau F, Beaulieu JF. Differential expression of claudin-2 along the human intestine: Implication of GATA-4 in the maintenance of claudin-2 in differentiating cells. J Cell Physiol 2005;203:15–26. [DOI] [PubMed] [Google Scholar]
- 34.Bhat AA, Sharma A, Pope J, et al. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PLoS One 2012;7:e37174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka H, Takechi M, Kiyonari H, et al. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut 2015;64:1529–38. [DOI] [PubMed] [Google Scholar]
- 36.Ding L, Lu Z, Foreman O, et al. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 2012;142:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pope JL, Ahmad R, Bhat AA, et al. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Molecular Cancer 2014;13:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad R, Chaturvedi R, Olivares-Villagomez D, et al. Targeted colonic claudin-2 expression renders resistance to epithelial injury, induces immune suppression, and protects from colitis. Mucosal Immunol 2014;7:1340–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber CR, Nalle SC, Tretiakova M, et al. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest 2008;88:1110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu ASL, Enck AH, Lencer WI, et al. Claudin-8 Expression in Madin-Darby Canine Kidney Cells Augments the Paracellular Barrier to Cation Permeation. Journal of Biological Chemistry 2003;278:17350–9. [DOI] [PubMed] [Google Scholar]
- 41.Claverie-Martin F Familial hypomagnesaemia with hypercalciuria and nephrocalcinosis: clinical and molecular characteristics. Clinical Kidney Journal 2015;8:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura A, Kitano Y, Hata M, et al. Megaintestine in Claudin-15–Deficient Mice. Gastroenterology 2008;134:523–34.e3. [DOI] [PubMed] [Google Scholar]
- 43.Wada M, Tamura A, Takahashi N, et al. Loss of Claudins 2 and 15 From Mice Causes Defects in Paracellular Na+ Flow and Nutrient Transport in Gut and Leads to Death from Malnutrition. Gastroenterology 2013;144:369–80. [DOI] [PubMed] [Google Scholar]
- 44.Van Itallie CM, Holmes J, Bridges A, et al. The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. Journal of Cell Science 2008;121:298–305. [DOI] [PubMed] [Google Scholar]
- 45.Laukoetter MG, Nava P, Lee WY, et al. JAM-A regulates permeability and inflammation in the intestine in vivo. The Journal of Experimental Medicine 2007;204:3067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y-h, Lu Q, Schneeberger EE, et al. Restoration of Tight Junction Structure and Barrier Function by Down-Regulation of the Mitogen-activated Protein Kinase Pathway in Ras-transformed Madin-Darby Canine Kidney Cells. Molecular Biology of the Cell 2000;11:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh AB, Harris RC. Epidermal Growth Factor Receptor Activation Differentially Regulates Claudin Expression and Enhances Transepithelial Resistance in Madin-Darby Canine Kidney Cells. Journal of Biological Chemistry 2004;279:3543–52. [DOI] [PubMed] [Google Scholar]
- 48.Twiss F, Oldenkamp M, Hiemstra A, et al. HGF signaling regulates Claudin-3 dynamics through its C-terminal tyrosine residues. Tissue Barriers 2013;1:e27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong CX, Zhao W, Solomon C, et al. The Intestinal Epithelial Insulin-Like Growth Factor-1 Receptor Links Glucagon-Like Peptide-2 Action to Gut Barrier Function. Endocrinology 2014;155:370–9. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Mumm JB, Herbst R, et al. IL-22 Increases Permeability of Intestinal Epithelial Tight Junctions by Enhancing Claudin-2 Expression. The Journal of Immunology 2017. [DOI] [PubMed] [Google Scholar]
- 51.Leppkes M, Roulis M, Neurath MF, et al. Pleiotropic functions of TNF-α in the regulation of the intestinal epithelial response to inflammation. International Immunology 2014;26:509–15. [DOI] [PubMed] [Google Scholar]
- 52.Huang X-z, Chen Y, Wu J, et al. Aspirin and non-steroidal anti-inflammatory drugs use reduce gastric cancer risk: A dose-response meta-analysis. Oncotarget 2017;8:4781–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Li Y-X, Chen M-H, et al. Changes in the phosphorylation of claudins during the course of experimental colitis. International Journal of Clinical and Experimental Pathology 2015;8:12225–33. [PMC free article] [PubMed] [Google Scholar]
- 54.Itallie CMV, Mitic LL, Anderson JM. SUMOylation of claudin-2. Annals of the New York Academy of Sciences 2012;1258:60–4. [DOI] [PubMed] [Google Scholar]
- 55.Heiler S, Mu W, Zöller M, et al. The importance of claudin-7 palmitoylation on membrane subdomain localization and metastasis-promoting activities. Cell Communication and Signaling : CCS 2015;13:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Utech M, Mennigen R, Bruewer M. Endocytosis and Recycling of Tight Junction Proteins in Inflammation. Journal of Biomedicine and Biotechnology 2010;2010:484987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dukes JD, Whitley P, Chalmers AD. The PIKfyve Inhibitor YM201636 Blocks the Continuous Recycling of the Tight Junction Proteins Claudin-1 and Claudin-2 in MDCK cells. PLOS ONE 2012;7:e28659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatovic S, Johnson A, Sladojevic N, et al. Endocytosis of tight junction proteins and the regulation of degradation and recycling: Endocytic sorting of tight junction proteins, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mankertz J, Hillenbrand B, Tavalali S, et al. Functional crosstalk between Wnt signaling and Cdx-related transcriptional activation in the regulation of the claudin-2 promoter activity. Biochemical and Biophysical Research Communications 2004;314:1001–7. [DOI] [PubMed] [Google Scholar]
- 60.Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. Journal of Cell Science 2004;117:1675–85. [DOI] [PubMed] [Google Scholar]
- 61.Shiou S-R, Singh AB, Moorthy K, et al. Smad4 Regulates Claudin-1 Expression in a Transforming Growth Factory-β–Independent Manner in Colon Cancer Cells. Cancer Research 2007;67:1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishnan M, Singh AB, Smith JJ, et al. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene 2010;29:305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang TL, Ito K, Ko TK, et al. Claudin-1 Has Tumor Suppressive Activity and Is a Direct Target of RUNX3 in Gastric Epithelial Cells. Gastroenterology 2010;138:255–65.e3. [DOI] [PubMed] [Google Scholar]
- 64.Agarwal R, Mori Y, Cheng Y, et al. Silencing of Claudin-11 Is Associated with Increased Invasiveness of Gastric Cancer Cells. PLoS ONE 2009;4:e8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwon MJ, Kim S-H, Jeong HM, et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Laboratory Investigation 2011;91:1652. [DOI] [PubMed] [Google Scholar]
- 66.Kwon MJ, Kim S-S, Choi Y-L, et al. Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis 2010;31:974–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cichon C, Sabharwal H, Rüter C, et al. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2014;2:e944446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Q, Costinean S, Croce CM, et al. microRNA 29 Targets NKRF and Claudin 1 to Increase Intestinal Permeability, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang GJ, Xiao HX, Tian HP, et al. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med 2013;31:1375–80. [DOI] [PubMed] [Google Scholar]
- 70.Sharma A, Bhat AA, Krishnan M, et al. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis 2013;34:2610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jordan NV, Johnson GL, Abell AN. Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell Cycle 2011;10:2865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon MJ. Emerging Roles of Claudins in Human Cancer. International Journal of Molecular Sciences 2013;14:18148–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Seminars in Cell & Developmental Biology 2015;42:58–65. [DOI] [PubMed] [Google Scholar]
- 74.Breitman M, Zilberberg A, Caspi M, et al. The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. Biochimica et Biophysica Acta (BBA) – Molecular Cell Research 2008;1783:1792–802. [DOI] [PubMed] [Google Scholar]
- 75.Berglund F, Weerasinghe NR, Davidson L, et al. Disruption of epithelial architecture caused by loss of PTEN or by oncogenic mutant p110α/PIK3CA but not by HER2 or mutant AKT1. Oncogene 2013;32:4417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nature Cell Biology 2005;7:262. [DOI] [PubMed] [Google Scholar]
- 77.Fevr T, Robine S, Louvard D, et al. Wnt/β-Catenin Is Essential for Intestinal Homeostasis and Maintenance of Intestinal Stem Cells. Molecular and Cellular Biology 2007;27:7551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dhawan P, Singh AB, Deane NG, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 2005;115:1765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinugasa T, Akagi Y, Yoshida T, et al. Increased Claudin-1 Protein Expression Contributes to Tumorigenesis in Ulcerative Colitis-associated Colorectal Cancer. Anticancer Research 2010;30:3181–6. [PubMed] [Google Scholar]
- 80.Ahmad R, Kumar B, Pan K, et al. HDAC-4 regulates claudin-2 expression in EGFR-ERK1/2 dependent manner to regulate colonic epithelial cell differentiation. Oncotarget 2017;8:87718–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dhawan P, Ahmad R, Chaturvedi R, et al. Claudin-2 Expression Increases Tumorigenicity of Colon Cancer Cells: Role of Epidermal Growth Factor Receptor Activation. Oncogene 2011;30:3234–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawauchiya T, Takumi R, Kudo Y, et al. Correlation between the destruction of tight junction by patulin treatment and increase of phosphorylation of ZO-1 in Caco-2 human colon cancer cells. Toxicology Letters 2011;205:196–202. [DOI] [PubMed] [Google Scholar]
- 83.Mees ST, Mennigen R, Spieker T, et al. Expression of tight and adherens junction proteins in ulcerative colitis associated colorectal carcinoma: upregulation of claudin-1, claudin-3, claudin-4, and beta-catenin. Int J Colorectal Dis 2009;24:361–8. [DOI] [PubMed] [Google Scholar]
- 84.Gröne J, Weber B, Staub E, et al. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. International Journal of Colorectal Disease 2007;22:651–9. [DOI] [PubMed] [Google Scholar]
- 85.Abu-Farsakh S, Wu T, Lalonde A, et al. High expression of Claudin-2 in esophageal carcinoma and precancerous lesions is significantly associated with the bile salt receptors VDR and TGR5. BMC Gastroenterology 2017;17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takala H, Saarinio J, Wiik HI, et al. Claudins 1, 3, 4, 5 and 7 in esophageal cancer: loss of claudin 3 and 4 expression is associated with metastatic behavior. APMIS 2007;115:838–47. [DOI] [PubMed] [Google Scholar]
- 87.Zhao H, Yu H, Martin TA, et al. The role of JAM-B in cancer and cancer metastasis (Review). Oncology Reports 2016;36:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin Z, Zhang X, Liu Z, et al. The distinct expression patterns of claudin-2, -6, and –11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagnostic Pathology 2013;8:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu Y, Jing J, Sun L, et al. Expression of claudin-11, -23 in different gastric tissues and its relationship with the risk and prognosis of gastric cancer. PLOS ONE 2017;12:e0174476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang P, Zhang M, Liu X, et al. MicroRNA-421 promotes the proliferation and metastasis of gastric cancer cells by targeting claudin-11. Experimental and Therapeutic Medicine 2017;14:2625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang T-L, Changchien T-T, Wang C-C, et al. Claudin-4 expression in gastric cancer cells enhances the invasion and is associated with the increased level of matrix metalloproteinase-2 and -9 expression. Oncology Letters 2014;8:1367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang S-j, Feng J-f, Wang L, et al. miR-1303 Targets Claudin-18 Gene to Modulate Proliferation and Invasion of Gastric Cancer Cells. Digestive Diseases and Sciences 2014;59:1754–63. [DOI] [PubMed] [Google Scholar]
- 93.Johnson AH, Frierson HF, Zaika A, et al. Expression of Tight-Junction Protein Claudin-7 Is an Early Event in Gastric Tumorigenesis. The American Journal of Pathology 2005;167:577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kojima T, Yamamoto T, Murata M, et al. Regulation of the blood-biliary barrier: interaction between gap and tight junctions in hepatocytes. Med Electron Microsc 2003;36:157–64. [DOI] [PubMed] [Google Scholar]
- 95.Wang L, Boyer JL. The maintenance and generation of membrane polarity in hepatocytes. Hepatology 2004;39:892–9. [DOI] [PubMed] [Google Scholar]
- 96.Gissen P, Arias IM. Structural and functional hepatocyte polarity and liver disease. J Hepatol 2015;63:1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rao RK, Samak G. Bile duct epithelial tight junctions and barrier function. Tissue Barriers 2013;1:e25718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiesner RH. Current concepts in primary sclerosing cholangitis. Mayo Clin Proc 1994;69:969–82. [DOI] [PubMed] [Google Scholar]
- 99.Nakanuma Y, Tsuneyama K, Gershwin ME, et al. Pathology and immunopathology of primary biliary cirrhosis with emphasis on bile duct lesions: recent progress. Semin Liver Dis 1995;15:313–28. [DOI] [PubMed] [Google Scholar]
- 100.Grosse B, Cassio D, Yousef N, et al. Claudin-1 involved in neonatal ichthyosis sclerosing cholangitis syndrome regulates hepatic paracellular permeability. Hepatology 2012;55:1249–59. [DOI] [PubMed] [Google Scholar]
- 101.Carlton VE, Harris BZ, Puffenberger EG, et al. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet 2003;34:91–6. [DOI] [PubMed] [Google Scholar]
- 102.Szepetowski S, Lacoste C, Mallet S, et al. [NISCH syndrome, a rare cause of neonatal cholestasis: A case report]. Arch Pediatr 2017;24:1228–34. [DOI] [PubMed] [Google Scholar]
- 103.Holczbauer A, Gyongyosi B, Lotz G, et al. Increased expression of claudin-1 and claudin-7 in liver cirrhosis and hepatocellular carcinoma. Pathol Oncol Res 2014;20:493–502. [DOI] [PubMed] [Google Scholar]
- 104.Lodi C, Szabo E, Holczbauer A, et al. Claudin-4 differentiates biliary tract cancers from hepatocellular carcinomas. Mod Pathol 2006;19:460–9. [DOI] [PubMed] [Google Scholar]
- 105.Pijls KE, Jonkers DM, Elamin EE, et al. Intestinal epithelial barrier function in liver cirrhosis: an extensive review of the literature. Liver Int 2013;33:1457–69. [DOI] [PubMed] [Google Scholar]
- 106.Liu S, Yang W, Shen L, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol 2009;83:2011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benedicto I, Molina-Jimenez F, Bartosch B, et al. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J Virol 2009;83:8012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol 2013;369:87–112. [DOI] [PubMed] [Google Scholar]
- 109.Colpitts CC, Baumert TF. Claudins in viral infection: from entry to spread. Pflugers Arch 2017;469:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Douam F, Lavillette D, Cosset FL. The mechanism of HCV entry into host cells. Prog Mol Biol Transl Sci 2015;129:63–107. [DOI] [PubMed] [Google Scholar]
- 111.Pileri P, Uematsu Y, Campagnoli S, et al. Binding of hepatitis C virus to CD81. Science 1998;282:938–41. [DOI] [PubMed] [Google Scholar]
- 112.Scarselli E, Ansuini H, Cerino R, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 2002;21:5017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krieger SE, Zeisel MB, Davis C, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 2010;51:1144–57. [DOI] [PubMed] [Google Scholar]
- 114.Douam F, Dao Thi VL, Maurin G, et al. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 2014;59:776–88. [DOI] [PubMed] [Google Scholar]
- 115.Brazzoli M, Bianchi A, Filippini S, et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol 2008;82:8316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harris HJ, Farquhar MJ, Mee CJ, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol 2008;82:5007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mee CJ, Harris HJ, Farquhar MJ, et al. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol 2009;83:6211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harris HJ, Davis C, Mullins JG, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 2010;285:21092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fofana I, Krieger SE, Grunert F, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 2010;39:953–64. [DOI] [PubMed] [Google Scholar]
- 120.Mailly L, Xiao F, Lupberger J, et al. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat Biotechnol 2015;33:549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cukierman L, Meertens L, Bertaux C, et al. Residues in a highly conserved claudin-1 motif are required for hepatitis C virus entry and mediate the formation of cell-cell contacts. J Virol 2009;83:5477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reynolds GM, Harris HJ, Jennings A, et al. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology 2008;47:418–27. [DOI] [PubMed] [Google Scholar]
- 123.Zheng A, Yuan F, Li Y, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol 2007;81:12465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meertens L, Bertaux C, Cukierman L, et al. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol 2008;82:3555–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haid S, Grethe C, Dill MT, et al. Isolate-dependent use of claudins for cell entry by hepatitis C virus. Hepatology 2014;59:24–34. [DOI] [PubMed] [Google Scholar]
- 126.Hopcraft SE, Evans MJ. Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins. Hepatology 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fofana I, Zona L, Thumann C, et al. Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J Virol 2013;87:10405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tawar RG, Colpitts CC, Lupberger J, et al. Claudins and pathogenesis of viral infection. Semin Cell Dev Biol 2015;42:39–46. [DOI] [PubMed] [Google Scholar]
- 129.Liu S, Kuo W, Yang W, et al. The second extracellular loop dictates Occludin-mediated HCV entry. Virology 2010;407:160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Benedicto I, Molina-Jimenez F, Barreiro O, et al. Hepatitis C virus envelope components alter localization of hepatocyte tight junction-associated proteins and promote occludin retention in the endoplasmic reticulum. Hepatology 2008;48:1044–53. [DOI] [PubMed] [Google Scholar]
- 131.Sourisseau M, Michta ML, Zony C, et al. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog 2013;9:e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shimizu Y, Shirasago Y, Kondoh M, et al. Monoclonal antibodies against occludin completely prevented hepatitis C virus infection in a mouse model. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matter K, Balda MS. Biogenesis of tight junctions: the C-terminal domain of occludin mediates basolateral targeting. J Cell Sci 1998;111 (Pt 4):511–9. [DOI] [PubMed] [Google Scholar]
- 134.Saitou M, Ando-Akatsuka Y, Itoh M, et al. Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J Cell Biol 1997;73:222–31. [PubMed] [Google Scholar]
- 135.Sekhar V, Pollicino T, Diaz G, et al. Infection with hepatitis C virus depends on TACSTD2, a regulator of claudin-1 and occludin highly downregulated in hepatocellular carcinoma. PLoS Pathog 2018;14:e1006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kohaar I, Ploss A, Korol E, et al. Splicing diversity of the human OCLN gene and its biological significance for hepatitis C virus entry. J Virol 2010;84:6987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 2011;474:208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dorner M, Horwitz JA, Donovan BM, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature 2013;501:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ding Q, von Schaewen M, Hrebikova G, et al. Mice Expressing Minimally Humanized CD81 and Occludin Genes Support Hepatitis C Virus Uptake In Vivo. J Virol 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Timpe JM, Stamataki Z, Jennings A, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 2008;47:17–24. [DOI] [PubMed] [Google Scholar]
- 141.Witteveldt J, Evans MJ, Bitzegeio J, et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol 2009;90:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brimacombe CL, Grove J, Meredith LW, et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol 2011;85:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiao F, Fofana I, Heydmann L, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog 2014;10:e1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mensa L, Crespo G, Gastinger MJ, et al. Hepatitis C virus receptors claudin-1 and occludin after liver transplantation and influence on early viral kinetics. Hepatology 2011;53:1436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zadori G, Gelley F, Torzsok P, et al. Examination of claudin-1 expression in patients undergoing liver transplantation owing to hepatitis C virus cirrhosis. Transplant Proc 2011;43:1267–71. [DOI] [PubMed] [Google Scholar]
- 146.Nakamuta M, Fujino T, Yada R, et al. Expression profiles of genes associated with viral entry in HCV-infected human liver. J Med Virol 2011;83:921–7. [DOI] [PubMed] [Google Scholar]
- 147.Higashi Y, Suzuki S, Sakaguchi T, et al. Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res 2007;139:68–76. [DOI] [PubMed] [Google Scholar]
- 148.Orban E, Szabo E, Lotz G, et al. Different expression of occludin and ZO-1 in primary and metastatic liver tumors. Pathol Oncol Res 2008;14:299–306. [DOI] [PubMed] [Google Scholar]
- 149.Zhou S, Parham DM, Yung E, et al. Quantification of glypican 3, beta-catenin and claudin-1 protein expression in hepatoblastoma and paediatric hepatocellular carcinoma by colour deconvolution. Histopathology 2015;67:905–13. [DOI] [PubMed] [Google Scholar]
- 150.Bouchagier KA, Assimakopoulos SF, Karavias DD, et al. Expression of claudins-1, -4, -5, -7 and occludin in hepatocellular carcinoma and their relation with classic clinicopathological features and patients' survival. In Vivo 2014;28:315–26. [PubMed] [Google Scholar]
- 151.Chen YJ, You ML, Chong QY, et al. Autocrine Human Growth Hormone Promotes Invasive and Cancer Stem Cell-Like Behavior of Hepatocellular Carcinoma Cells by STAT3 Dependent Inhibition of CLAUDIN-1 Expression. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li CP, Cai MY, Jiang LJ, et al. CLDN14 is epigenetically silenced by EZH2-mediated H3K27ME3 and is a novel prognostic biomarker in hepatocellular carcinoma. Carcinogenesis 2016;37:557–66. [DOI] [PubMed] [Google Scholar]
- 153.Yang J, Liu X, Yuan X, et al. miR-99b promotes metastasis of hepatocellular carcinoma through inhibition of claudin 11 expression and may serve as a prognostic marker. Oncol Rep 2015;34:1415–23. [DOI] [PubMed] [Google Scholar]
- 154.Jiang L, Yang YD, Fu L, et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget 2014;5:7663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bhat AA, Ahmad R, Uppada SB, et al. Claudin-1 promotes TNF-alpha-induced epithelial-mesenchymal transition and migration in colorectal adenocarcinoma cells. Exp Cell Res 2016;349:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim JH, Kim EL, Lee YK, et al. Decreased lactate dehydrogenase B expression enhances claudin 1-mediated hepatoma cell invasiveness via mitochondrial defects. Exp Cell Res 2011;317:1108–18. [DOI] [PubMed] [Google Scholar]
- 157.Lee JH, Lee YK, Lim JJ, et al. Mitochondrial Respiratory Dysfunction Induces Claudin-1 Expression via Reactive Oxygen Species-mediated Heat Shock Factor 1 Activation, Leading to Hepatoma Cell Invasiveness. J Biol Chem 2015;290:21421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oku N, Sasabe E, Ueta E, et al. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res 2006;66:5251–7. [DOI] [PubMed] [Google Scholar]
- 159.Dos Reis PP, Bharadwaj RR, Machado J, et al. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 2008;113:3169–80. [DOI] [PubMed] [Google Scholar]
- 160.Babkair H, Yamazaki M, Uddin MS, et al. Aberrant expression of the tight junction molecules claudin-1 and zonula occludens-1 mediates cell growth and invasion in oral squamous cell carcinoma. Hum Pathol 2016;57:51–60. [DOI] [PubMed] [Google Scholar]
- 161.Leotlela PD, Wade MS, Duray PH, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene 2007;26:3846–56. [DOI] [PubMed] [Google Scholar]
- 162.Suh Y, Yoon CH, Kim RK, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013;32:4873–82. [DOI] [PubMed] [Google Scholar]
- 163.Karabulut M, Alis H, Bas K, et al. Clinical significance of serum claudin-1 and claudin-7 levels in patients with colorectal cancer. Molecular and Clinical Oncology 2015;3:1255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rabinsky EF, Joshi BP, Pant A, et al. Overexpressed Claudin-1 Can Be Visualized Endoscopically in Colonic Adenomas In Vivo. Cellular and molecular gastroenterology and hepatology 2016;2:222–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hashimoto Y, Hata T, Tada M, et al. Safety evaluation of a human chimeric monoclonal antibody that recognizes the extracellular loop domain of claudin-2. European Journal of Pharmaceutical Sciences 2018;117:161–7. [DOI] [PubMed] [Google Scholar]
- 166.Zeisel MB, Crouchet E, Baumert TF, et al. Host-Targeting Agents to Prevent and Cure Hepatitis C Virus Infection. Viruses 2015;7:5659–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xiao F, Fofana I, Thumann C, et al. Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut 2015;64:483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Colpitts CC, Tawar RG, Mailly L, et al. Humanisation of a claudin-1-specific monoclonal antibody for clinical prevention and cure of HCV infection without escape. Gut 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fukasawa M, Nagase S, Shirasago Y, et al. Monoclonal antibodies against extracellular domains of claudin-1 block hepatitis C virus infection in a mouse model. J Virol 2015;89:4866–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hotzel I, Chiang V, Diao J, et al. Efficient production of antibodies against a mammalian integral membrane protein by phage display. Protein Eng Des Sel 2011;24:679–89. [DOI] [PubMed] [Google Scholar]
- 171.Paciello R, Urbanowicz RA, Riccio G, et al. Novel human anti-claudin 1 mAbs inhibit hepatitis C virus infection and may synergize with anti-SRB1 mAb. J Gen Virol 2016;97:82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fofana I, Fafi-Kremer S, Carolla P, et al. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology 2012;143:223–33 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Okai K, Ichikawa-Tomikawa N, Saito AC, et al. A novel occludin-targeting monoclonal antibody prevents hepatitis C virus infection in vitro. Oncotarget 2018;9:16588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Michta ML, Hopcraft SE, Narbus CM, et al. Species-specific regions of occludin required by hepatitis C virus for cell entry. J Virol 2010;84:11696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Felmlee DJ, Coilly A, Chung RT, et al. New perspectives for preventing hepatitis C virus liver graft infection. Lancet Infect Dis 2016;16:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Colpitts CC, Chung RT, Baumert TF. Entry Inhibitors: A Perspective for Prevention of Hepatitis C Virus Infection in Organ Transplantation. ACS Infect Dis 2017;3:620–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rosager AM, Sorensen MD, Dahlrot RH, et al. Expression and prognostic value of JAM-A in gliomas. J Neurooncol 2017;135:107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.McSherry EA, McGee SF, Jirstrom K, et al. JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer 2009;125:1343–51. [DOI] [PubMed] [Google Scholar]
- 179.Brennan K, McSherry EA, Hudson L, et al. Junctional adhesion molecule-A is co-expressed with HER2 in breast tumors and acts as a novel regulator of HER2 protein degradation and signaling. Oncogene 2013;32:2799–804. [DOI] [PubMed] [Google Scholar]
- 180.McSherry EA, Brennan K, Hudson L, et al. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res 2011;13:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gotte M, Mohr C, Koo CY, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 2010;29:6569–80. [DOI] [PubMed] [Google Scholar]
- 182.Goetsch L, Haeuw JF, Beau-Larvor C, et al. A novel role for junctional adhesion molecule-A in tumor proliferation: modulation by an anti-JAM-A monoclonal antibody. Int J Cancer 2013;132:1463–74. [DOI] [PubMed] [Google Scholar]
- 183.Ikeo K, Oshima T, Shan J, et al. Junctional adhesion molecule-A promotes proliferation and inhibits apoptosis of gastric cancer. Hepatogastroenterology 2015;62:540–5. [PubMed] [Google Scholar]
- 184.Zhang M, Luo W, Huang B, et al. Overexpression of JAM-A in non-small cell lung cancer correlates with tumor progression. PLoS One 2013;8:e79173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhao C, Lu F, Chen H, et al. Dysregulation of JAM-A plays an important role in human tumor progression. Int J Clin Exp Pathol 2014;7:7242–8. [PMC free article] [PubMed] [Google Scholar]
- 186.Coyne CB, Vanhook MK, Gambling TM, et al. Regulation of airway tight junctions by proinflammatory cytokines. Mol Biol Cell 2002;13:3218–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Williams DW, Anastos K, Morgello S, et al. JAM-A and ALCAM are therapeutic targets to inhibit diapedesis across the BBB of CD14+CD16+ monocytes in HIV-infected individuals. J Leukoc Biol 2015;97:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fuse C, Ishida Y, Hikita T, et al. Junctional adhesion molecule-C promotes metastatic potential of HT1080 human fibrosarcoma. J Biol Chem 2007;282:8276–83. [DOI] [PubMed] [Google Scholar]
- 189.Hao S, Yang Y, Liu Y, et al. JAM-C promotes lymphangiogenesis and nodal metastasis in non-small cell lung cancer. Tumour Biol 2014;35:5675–87. [DOI] [PubMed] [Google Scholar]
- 190.Arcangeli ML, Frontera V, Bardin F, et al. The Junctional Adhesion Molecule-B regulates JAM-C-dependent melanoma cell metastasis. FEBS Lett 2012;586:4046–51. [DOI] [PubMed] [Google Scholar]
- 191.Ghislin S, Obino D, Middendorp S, et al. Junctional adhesion molecules are required for melanoma cell lines transendothelial migration in vitro. Pigment Cell Melanoma Res 2011;24:504–11. [DOI] [PubMed] [Google Scholar]
- 192.Langer HF, Orlova VV, Xie C, et al. A novel function of junctional adhesion molecule- C in mediating melanoma cell metastasis. Cancer Res 2011;71:4096–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leinster DA, Colom B, Whiteford JR, et al. Endothelial cell junctional adhesion molecule C plays a key role in the development of tumors in a murine model of ovarian cancer. FASEB J 2013;27:4244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang Y, Xia F, Liu X, et al. JAM3 maintains leukemia-initiating cell self-renewal through LRP5/AKT/beta-catenin/CCND1 signaling. J Clin Invest 2018;128:1737–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ebnet K Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol Rev 2017;97:1529–54. [DOI] [PubMed] [Google Scholar]
- 196.Martin TA, Watkins G, Jiang WG. The Coxsackie-adenovirus receptor has elevated expression in human breast cancer. Clin Exp Med 2005;5:122–8. [DOI] [PubMed] [Google Scholar]
- 197.Bruning A, Stickeler E, Diederich D, et al. Coxsackie and adenovirus receptor promotes adenocarcinoma cell survival and is expressionally activated after transition from preneoplastic precursor lesions to invasive adenocarcinomas. Clin Cancer Res 2005;11:4316–20. [DOI] [PubMed] [Google Scholar]
- 198.Giaginis CT, Zarros AC, Papaefthymiou MA, et al. Coxsackievirus and adenovirus receptor expression in human endometrial adenocarcinoma: possible clinical implications. World J Surg Oncol 2008;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen Z, Wang Q, Sun J, et al. Expression of the coxsackie and adenovirus receptor in human lung cancers. Tumour Biol 2013;34:17–24. [DOI] [PubMed] [Google Scholar]
- 200.Saito K, Sakaguchi M, Iioka H, et al. Coxsackie and adenovirus receptor is a critical regulator for the survival and growth of oral squamous carcinoma cells. Oncogene 2014;33:1274–86. [DOI] [PubMed] [Google Scholar]
- 201.Reimer D, Steppan I, Wiedemair A, et al. Soluble isoforms but not the transmembrane form of coxsackie-adenovirus receptor are of clinical relevance in epithelial ovarian cancer. Int J Cancer 2007;120:2568–75. [DOI] [PubMed] [Google Scholar]
- 202.Giaginis C, Zarros A, Alexandrou P, et al. Evaluation of coxsackievirus and adenovirus receptor expression in human benign and malignant thyroid lesions. APMIS 2010;118:210–21. [DOI] [PubMed] [Google Scholar]
- 203.Vindrieux D, Le Corre L, Hsieh JT, et al. Coxsackie and adenovirus receptor is a target and a mediator of estrogen action in breast cancer. Endocr Relat Cancer 2011;18:311–21. [DOI] [PubMed] [Google Scholar]
- 204.Maekawa Y, Ouzounian M, Opavsky MA, et al. Connecting the missing link between dilated cardiomyopathy and viral myocarditis: virus, cytoskeleton, and innate immunity. Circulation 2007;115:5–8. [DOI] [PubMed] [Google Scholar]
- 205.Leech AO, Cruz RG, Hill AD, et al. Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann Transl Med 2015;3:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Oliveira SS, Morgado-Diaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci 2007;64:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Itoh M, Bissell MJ. The organization of tight junctions in epithelia: implications for mammary gland biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia 2003;8:449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Akasaka H, Sato F, Morohashi S, et al. Anti-apoptotic effect of claudin-1 in tamoxifen-treated human breast cancer MCF-7 cells. BMC Cancer 2010;10:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu Y, Wang L, Lin XY, et al. Anti-apoptotic effect of claudin-1 on TNF-alpha-induced apoptosis in human breast cancer MCF-7 cells. Tumour Biol 2012;33:2307–15. [DOI] [PubMed] [Google Scholar]
- 210.Achari C, Winslow S, Larsson C. Down Regulation of CLDND1 Induces Apoptosis in Breast Cancer Cells. PLoS One 2015;10:e0130300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lee JW, Lee SJ, Seo J, et al. Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol 2005;97:53–9. [DOI] [PubMed] [Google Scholar]
- 212.Kinugasa T, Huo Q, Higashi D, et al. Selective up-regulation of claudin-1 and claudin- 2 in colorectal cancer. Anticancer Res 2007;27:3729–34. [PubMed] [Google Scholar]
- 213.Caruso M, Fung KY, Moore J, et al. Claudin-1 Expression Is Elevated in Colorectal Cancer Precursor Lesions Harboring the BRAF V600E Mutation. Transl Oncol 2014;7:456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huo Q, Kinugasa T, Wang L, et al. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Res 2009;29:851–7. [PubMed] [Google Scholar]
- 215.Resnick MB, Konkin T, Routhier J, et al. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol 2005;18:511–8. [DOI] [PubMed] [Google Scholar]
- 216.Singh AB, Sharma A, Dhawan P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis 2012;33:2538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sheehan GM, Kallakury BV, Sheehan CE, et al. Loss of claudins-1 and -7 and expression of claudins-3 and -4 correlate with prognostic variables in prostatic adenocarcinomas. Hum Pathol 2007;38:564–9. [DOI] [PubMed] [Google Scholar]
- 218.Tzelepi VN, Tsamandas AC, Vlotinou HD, et al. Tight junctions in thyroid carcinogenesis: diverse expression of claudin-1, claudin-4, claudin-7 and occludin in thyroid neoplasms. Mod Pathol 2008;21:22–30. [DOI] [PubMed] [Google Scholar]
- 219.Lv J, Sun B, Mai Z, et al. CLDN-1 promoted the epithelial to migration and mesenchymal transition (EMT) in human bronchial epithelial cells via Notch pathway. Mol Cell Biochem 2017;432:91–8. [DOI] [PubMed] [Google Scholar]
- 220.Miwa N, Furuse M, Tsukita S, et al. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res 2001;12:469–76. [DOI] [PubMed] [Google Scholar]
- 221.Gonzalez-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta 2008;1778:729–56. [DOI] [PubMed] [Google Scholar]
- 222.Tabaries S, Dong Z, Annis MG, et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011;30:1318–28. [DOI] [PubMed] [Google Scholar]
- 223.Kimbung S, Kovacs A, Bendahl PO, et al. Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol Oncol 2014;8:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tabaries S, Annis MG, Hsu BE, et al. Lyn modulates Claudin-2 expression and is a therapeutic target for breast cancer liver metastasis. Oncotarget 2015;6:9476–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tabaries S, Dupuy F, Dong Z, et al. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol Cell Biol 2012;32:2979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Buchert M, Papin M, Bonnans C, et al. Symplekin promotes tumorigenicity by upregulating claudin-2 expression. Proc Natl Acad Sci U S A 2010;107:2628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ikari A, Sato T, Watanabe R, et al. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim Biophys Acta 2012;1823:1110–8. [DOI] [PubMed] [Google Scholar]
- 228.Ikari A, Watanabe R, Sato T, et al. Nuclear distribution of claudin-2 increases cell proliferation in human lung adenocarcinoma cells. Biochim Biophys Acta 2014;1843:2079–88. [DOI] [PubMed] [Google Scholar]
- 229.Hintsala HR, Siponen M, Haapasaari KM, et al. Claudins 1, 2, 3, 4, 5 and 7 in solar keratosis and squamocellular carcinoma of the skin. Int J Clin Exp Pathol 2013;6:2855–63. [PMC free article] [PubMed] [Google Scholar]
- 230.Zeissig S, Burgel N, Gunzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut 2007;56:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Burgel N, Bojarski C, Mankertz J, et al. Mechanisms of diarrhea in collagenous colitis. Gastroenterology 2002;123:433–43. [DOI] [PubMed] [Google Scholar]
- 232.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 2006;6:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kolokytha P, Yiannou P, Keramopoulos D, et al. Claudin-3 and claudin-4: distinct prognostic significance in triple-negative and luminal breast cancer. Appl Immunohistochem Mol Morphol 2014;22:125–31. [DOI] [PubMed] [Google Scholar]
- 234.de Souza WF, Fortunato-Miranda N, Robbs BK, et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS One 2013;8:e74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Konecny GE, Agarwal R, Keeney GA, et al. Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol Oncol 2008;109:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Santin AD, Bellone S, Marizzoni M, et al. Overexpression of claudin-3 and claudin-4 receptors in uterine serous papillary carcinoma: novel targets for a type-specific therapy using Clostridium perfringens enterotoxin (CPE). Cancer 2007;109:1312–22. [DOI] [PubMed] [Google Scholar]
- 237.Satake S, Semba S, Matsuda Y, et al. Cdx2 transcription factor regulates claudin-3 and claudin-4 expression during intestinal differentiation of gastric carcinoma. Pathol Int 2008;58:156–63. [DOI] [PubMed] [Google Scholar]
- 238.Choi YL, Kim J, Kwon MJ, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol 2007;22:1185–95. [DOI] [PubMed] [Google Scholar]
- 239.Luettig J, Rosenthal R, Barmeyer C, et al. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers 2015;3:e977176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.McClane BA. The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 2001;39:1781–91. [DOI] [PubMed] [Google Scholar]
- 241.Suren D, Yildirim M, Kaya V, et al. Expression patterns of claudins 1, 4, and 7 and their prognostic significance in nasopharyngeal carcinoma. J BUON 2015;20:212–7. [PubMed] [Google Scholar]
- 242.Pan XY, Wang B, Che YC, et al. Expression of claudin-3 and claudin-4 in normal, hyperplastic, and malignant endometrial tissue. Int J Gynecol Cancer 2007;17:233–41. [DOI] [PubMed] [Google Scholar]
- 243.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol 2010;2010:541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Philip R, Heiler S, Mu W, et al. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget 2015;6:2046–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kuo SJ, Chien SY, Lin C, et al. Significant elevation of CLDN16 and HAPLN3 gene expression in human breast cancer. Oncol Rep 2010;24:759–66. [DOI] [PubMed] [Google Scholar]
- 246.Rangel LB, Sherman-Baust CA, Wernyj RP, et al. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene 2003;22:7225–32. [DOI] [PubMed] [Google Scholar]
- 247.Men W, Martin TA, Ruge F, et al. Expression of claudins in human clear cell renal cell carcinoma. Cancer Genomics Proteomics 2015;12:1–8. [PubMed] [Google Scholar]
- 248.Simon DB, Lu Y, Choate KA, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999;285:103–6. [DOI] [PubMed] [Google Scholar]
- 249.Martin TA, Lane J, Ozupek H, et al. Claudin-20 promotes an aggressive phenotype in human breast cancer cells. Tissue Barriers 2013;1:e26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Forster C, Kahles T, Kietz S, et al. Dexamethasone induces the expression of metalloproteinase inhibitor TIMP-1 in the murine cerebral vascular endothelial cell line cEND. J Physiol 2007;580:937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Felinski EA, Antonetti DA. Glucocorticoid regulation of endothelial cell tight junction gene expression: novel treatments for diabetic retinopathy. Curr Eye Res 2005;30:949–57. [DOI] [PubMed] [Google Scholar]
- 252.Kato-Nakano M, Suzuki M, Kawamoto S, et al. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res 2010;30:4555–62. [PubMed] [Google Scholar]
- 253.Li X, Iida M, Tada M, et al. Development of an anti-claudin-3 and -4 bispecific monoclonal antibody for cancer diagnosis and therapy. J Pharmacol Exp Ther 2014;351:206–13. [DOI] [PubMed] [Google Scholar]
- 254.Suzuki M, Kato-Nakano M, Kawamoto S, et al. Therapeutic antitumor efficacy of monoclonal antibody against Claudin-4 for pancreatic and ovarian cancers. Cancer Sci 2009;100:1623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]


