Abstract
Here we present APOBEC-Coupled Epigenetic Sequencing (ACE-Seq), a bisulfite-free method for localizing 5-hydroxymethylcytosine (5hmC) at single-base resolution with low DNA input. The method builds upon the observation that AID/APOBEC family DNA deaminase enzymes can potently discriminate between cytosine modification states, and exploits the non-destructive nature of enzymatic, rather than chemical, deamination. ACE-Seq yields high-confidence 5hmC profiles with at least 1000-fold less DNA input than conventional methods. Applying ACE-Seq to generate a base-resolution map of 5hmC in tissue-derived cortical excitatory neurons, we find that 5hmC is almost entirely confined to CG dinucleotides. The map permits cytosine, 5-methylcytosine (5mC) and 5hmC to be parsed and reveals genomic features that diverge from global patterns, including enhancers and imprinting control regions with high and low 5hmC/5mC ratios, respectively. Enzymatic deamination overcomes many challenges posed by bisulfite-based methods and expands the scope of epigenome profiling to include scarce samples and open new lines of inquiry regarding the role of cytosine modifications in genome biology.
Epigenetic modification of cytosine bases is crucial for proper regulation of gene expression in mammals1. Although 5-methylcytosine (5mC) is best characterized for its gene repressive roles, the types of known modifications greatly expanded with the identification of several oxidized forms of 5mC (ox-mCs) arising through the action of ten-eleven translocation (TET) family enzymes2–5. Ox-mCs serve as intermediates in active DNA demethylation, whereby repressive 5mC marks are erased; ox-mCs may also have independent epigenetic functions6. 5-hydroxymethylcytosine (5hmC) is by far the most abundant ox-mC, reaching levels as high as 1.8% of total cytosines in human neurons where 5mC comprises 4–5% of total C7. The highly oxidized bases 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) are also detected but are far less common: When quantified in parallel with 5hmC, 5fC was maximally detected at levels more than 3 orders of magnitude less (0.0007% of total cytosines in human neurons)7, while 5caC quantified around 5 ppm of total C in mouse embryonic stem cells (mESCs) and was undetectable in neurons3,8.
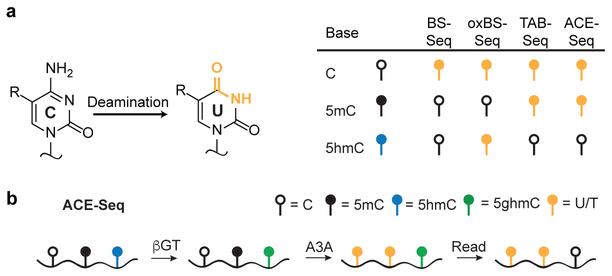
The most commonly-used approaches for localizing cytosine modifications rely upon differential chemical reactivity of cytosine variants in bisulfite sequencing (BS-Seq) (Fig. 1a)9. Incubation of DNA with bisulfite at extreme pH and elevated temperature promotes deamination of cytosine to uracil, whereas 5mC is largely unreacted (Supplementary Fig. 1). With the discovery of ox-mCs, the interpretation of BS-Seq became more complicated: Whereas 5fC and 5caC deaminate in BS-Seq, 5hmC forms a bulky adduct that is slow to deaminate, rendering 5hmC indistinguishable from 5mC10. In order to localize 5hmC specifically, several techniques have been advanced to change the bisulfite reactivities of 5mC versus 5hmC. In TET-assisted bisulfite sequencing (TAB-Seq)11, 5hmC bases are enzymatically modified by glucosylation (yielding 5ghmC), and 5mC is then selectively oxidized to 5caC in vitro by TET. Upon bisulfite treatment, all bases except the protected 5ghmC are deaminated. Alternatively, oxidative bisulfite sequencing (oxBS-Seq) employs selective oxidization of 5hmC to 5fC before bisulfite conversion12. Subtraction of oxBS-Seq from standard BS-Seq signals allows for indirect identification of 5hmC.
Figure 1. Reactivities of modified cytosines in sequencing approaches.
(a) Deamination underlies the differentiation of modified cytosines in current sequencing approaches. Standard BS-Seq converts cytosine to uracil and leaves 5mC and 5hmC unconverted, reading as C in sequencing. Modifications to 5mC and 5hmC in oxBS-Seq and TAB-Seq, when coupled to bisulfite, can differentiate these two bases. ACE-Seq uses enzymatic rather than bisulfite-mediated deamination to provide a comparable readout to TAB-Seq. (b) In the optimized ACE-Seq protocol, APOBEC3A catalyzes the enzymatic deamination of C and 5mC to completion, while 5hmC, which is highly resistant to deamination but is further protected by glucosylation, is localized by its non-conversion.
A key limitation of these methods is the use of bisulfite, as chemical deamination conditions can degrade as much as 99.9% of input genomic DNA (gDNA) 13. Thus, when samples of gDNA are limiting, experiments either provide limited coverage of the genome14, or methods using reduced representation or enrichment steps are performed15. Therefore, the characterization of specific primary cell types as well as rare cell populations undergoing dynamic epigenetic changes has been challenging, emphasizing the importance of methods that permit low amounts of starting DNA. For 5hmC detection, methods that avoid the use of bisulfite have been pursued, including nanopore, single-molecule real time (SMRT), and restriction enzyme-based approaches16–19. However, the limitations of each method have contributed to conflicting results, leaving, for example, the prevalence of 5hmC in non-CG sites a matter of debate20–23.
Recognizing the constraints of chemical deamination, we were drawn towards a natural analog of this reaction: enzymatic deamination by an AID/APOBEC family DNA deaminase. Here, we report the development, validation, and application of APOBEC-Coupled Epigenetic Sequencing (ACE-Seq). In our approach, deamination under near-physiological, nondestructive conditions permits single-base resolution 5hmC profiling with minimal DNA input.
RESULTS
Development of ACE-Seq.
Members of the AID/APOBEC family catalyze the deamination of cytosine to uracil in single-stranded DNA (ssDNA) and mediate critical functions in innate and adaptive immunity24. In prior work, we found that several family members can discriminate between cytosine modification states25; however, their overall poor catalytic activity prevented biotechnological applications. More recently, motivated by subsequent studies demonstrating that one human-specific family member, APOBEC3A (A3A), has high activity and a particular proficiency for 5mC deamination26,27, our attention turned to quantifying A3A’s activity on the full spectrum of natural cytosine modifications. We found that A3A indeed readily deaminates C and 5mC, but also discriminates potently against all three ox-mCs, with a ~5000-fold reduction in the 5hmC deamination rate relative to that of C28. This observation raised the prospect that A3A’s differential reactivity could be exploited to localize 5hmC in gDNA without the need for bisulfite.
We envisioned three potential barriers to using AID/APOBECs in sequencing. First, enzymatic deamination, like chemical deamination, requires ssDNA; unlike in BS-Seq, gDNA would need to be denatured under mild conditions that permit enzymatic activity in ACE-Seq. Second, different AID/APOBECs show distinctive sequence preferences, with 5’ bases influencing activity most strongly. A3A, for example, preferentially deaminates TTC, with reduced activity in disfavored contexts22. In ACE-Seq, sequence context preferences would have to be overcome. Third, ACE-Seq would require a sufficient window of enzymatic selectivity such that false-positive (C/5mC non-conversion) and false negative (5hmC conversion) readouts were minimized.
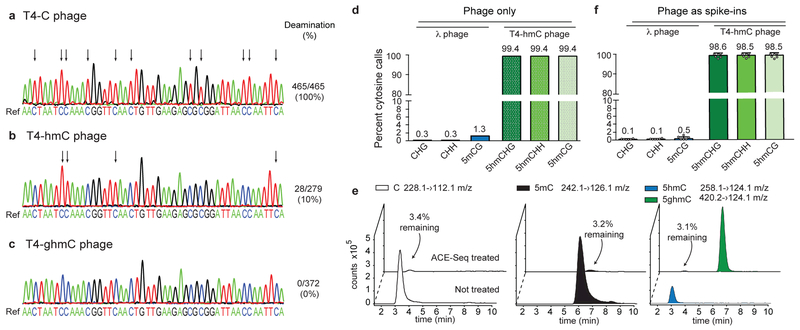
As a model system to develop ACE-Seq, we used phage gDNA, as these genomes offer known, homogenous modifications (Supplementary Fig. 2). The 169-kb T4 phage genome normally contains all cytosine bases replaced by 5ghmC (WT, here referred to as T4-ghmC). Established mutants in the 5ghmC pathway yield phage in which every encoded C is 5hmC (referred to as T4-hmC) or where all C bases are unmodified (referred to as T4-C) 29. To first evaluate conditions for ssDNA generation, 1 ng of gDNA from T4-C was subjected to brief heat denaturation, snap freezing, and subsequent incubation with excess A3A (5 µM). A target locus was then amplified and cloned, and individual clones were sequenced to guide method development. Some secondary structure elements proved resistant to deamination, but this could be overcome by denaturing in the presence of DMSO and performing A3A incubation under ramping temperature conditions (Supplementary Fig. 3). Efficient and complete deamination of the T4-C target locus was observed based on sequencing of five individual clones (Fig. 2a).
Figure 2. Development and validation of ACE-Seq.
(a-c) 1 ng of T4 phage genomic DNAs with homogeneous modifications (a: T4-C; b: T4-hmC; c: T4-ghmC) were heated, snap frozen, and incubated with A3A before amplification of a genomic segment, TA cloning, and Sanger sequencing of individual clones. Illustrative sequencing traces from individual clones are shown below the reference genome. Arrows denote deamination events (C>T transitions). Deamination events are quantified as the number of cytosines that were deaminated across the sum of all clones (93 cytosines per clone; T4-C 5 clones, T4-hmC 3 clones, T4-ghmC 4 clones). (d,f) Rates of non-conversion for enzymatically-methylated λ phage gDNA (5mCG, CH) and T4-hmC phage gDNA in ACE-Seq as determined by Illumina sequencing, using inputs of either (d) 1 ng of each alone or (f) 100 pg each as spike-ins averaged across six mammalian DNA samples (see Supplementary Table 1). Mean values listed above each bar, and error bars represent standard deviations. Individual data points are overlaid on the plot. (e) Representative LC-MS/MS traces of C, 5mC, 5hmC, and 5ghmC nucleosides after a 1:1 mix of methylated λ gDNA and T4-hmC gDNA was subjected to ACE-Seq treatment (compared to untreated control sample). Percentages denote amount detected after ACE-Seq treatment, averaged across three independent replicates.
When these conditions were next applied to T4-hmC, the majority of 5hmC bases were protected from deamination (Fig. 2b); however, some 5hmC deamination events were observed (~10% of cytosines across 3 separate clones). These events aligned with A3A preferences (i.e. TThmC sites), validating the prior observation that 5hmC can be deaminated by excess A3A, albeit inefficiently28. Having previously shown that AID/APOBECs discriminate against bulky 5-position modifications25,28, we considered whether further modification could protect 5hmC from deamination. Indeed, under conditions where some T4-hmC deamination occurs, we observed 0% deamination of T4-ghmC at any position across four clones sequenced (Fig. 2c). We therefore posited that in vitro glucosylation of 5hmC in gDNA could be used to prevent its sporadic deamination by A3A.
To simultaneously examine C, 5mC and 5hmC deamination, lambda (λ) phage gDNA (48.5-kb) that was enzymatically methylated at all CG sites (Supplementary Fig. 2) was pooled with T4-hmC. The pooled gDNA was sheared, and a low input sample (1 ng) was treated with T4 β−glucosyltransferase (βGT) to convert 5hmC in T4-hmC to 5ghmC, followed by incubation with excess A3A. The resulting products were analyzed using Illumina high-throughput sequencing after library preparation. Unbiased analysis of the λ phage genome showed robust C/5mC deamination: Non-conversion of C or 5mCG was detected for only ~6,800 of 2.4 million independently sequenced C sites and ~10,500 of ~800,000 5mCG sites, respectively (Fig. 2d, Supplementary Table 1). The C and 5mC non-conversion rates (0.3% and 1.3%, respectively) are similar or better than those observed with TAB-Seq (0.4%; 2.2%) on a comparable λ gDNA control11. In analyzing the enzymatically-glucosylated T4-hmC phage, ~99.4% of all 5hmC bases were called as C in sequencing (Fig. 2d), exceeding the sensitivity of TAB-Seq (84.4–92.0%)11,23,30. The ACE-Seq-treated phage gDNA was also digested to nucleosides and quantified via liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Efficient conversion of C and 5mC (~97% decrease in each signal) and protection of 5hmC were observed (Fig. 2e), and the necessity of βGT protection of 5hmC was also confirmed (Supplementary Fig. 4). Taken together, both sequencing- and LC-MS/MS-based approaches orthogonally confirm the robust conversion and protection efficiencies of ACE-Seq.
Thus, regarding the three barriers initially considered, in the optimized ACE-Seq protocol (Fig. 1b), a modified denaturation step permitted A3A to access its ssDNA substrate, and driving conditions with excess A3A allowed full C/5mC deamination in all sequence contexts. While these conditions resulted in some 5hmC deamination, these bases could be fully protected from deamination by glucosylation to generate a wide window for C/5mC versus 5hmC discrimination.
ACE-Seq is non-destructive.
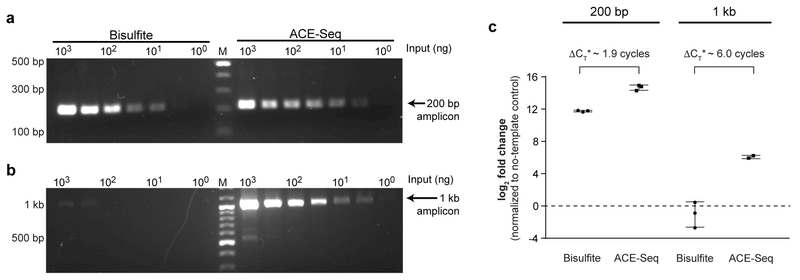
In typical bisulfite-based approaches, template gDNA damage limits the size of amplicons that can be characterized to those typically <300 bp31. To compare their impact, mouse embryonic stem cell (ESC) gDNA was treated with either BS-Seq or ACE-Seq conditions. After deamination, we attempted to amplify either short (200-bp) or long (1-kb) amplicons from a single target locus (Tbx5) using a fixed number of PCR cycles. For the short amplicon, although the ACE-Seq sample was more readily amplified, amplicons could be detected below 10 ng input gDNA with either condition (Fig. 3a). However, with the 1-kb locus, ACE-Seq amplicons were detectable with ~3-log lower DNA input, suggesting that BS-Seq had introduced substantial damage in the template (Fig. 3b). By contrast, 1-kb amplicons with ACE-Seq were detected at the same low input as the 200-bp amplicons, suggesting that gDNA is intact under ACE-Seq conditions. We validated this finding using quantitative PCR (Fig 3c; Supplementary Fig. 5), where BS-Seq shifts the threshold cycle number by >6.0 relative to ACE-Seq, suggesting a >64-fold decrease in intact template for the 1-kb amplicon.
Figure 3. ACE-Seq is nondestructive.
Initial input levels of gDNA from mESCs were titrated from 1 µg to 1 ng, and the samples were treated with either BS-Seq or ACE-Seq protocols. Primers were designed to amplify either (a) a 200-bp amplicon or (b) a 1-kb amplicon from the Tbx5 genomic locus, using 35 cycles of PCR. Resulting amplicons were run on 1.5% agarose gels and stained with SybrSafe. Marker (M) is in the middle lane with bold bands at 1 kb and 500 bp. Bisulfite experiment was performed twice with similar results, and used to inform conditions for the ACE-Seq experiment. (c) The samples were additionally analyzed by qPCR. Plotted is the difference between threshold cycle (CT) in the absence of template (water-only control) versus reactions containing 1 µg of template. Unlike in (a,b) where input amount was normalized, the volume input was normalized with qPCR, resulting in 2-fold less BS-Seq input relative to ACE-Seq. Data are reported as collected, but ΔCT* denotes subtraction of 1 cycle from ACE-Seq measurements to account for a 2-fold difference in initial input due to dilution. qPCR data for other input concentrations of gDNA are reported in Supplementary Fig. 5. Individual triplicate data points are plotted, and error bars represent the standard deviation.
ACE-Seq detects true positive 5hmC bases.
We next characterized 5hmC at single-base resolution in mammalian cells. To highlight ACE-Seq’s utility on a specific tissue-derived cell subtype, we isolated gDNA from purified murine cortical excitatory neurons (NeuroD6/NEX+)32, which allowed comparison to gDNA from total mouse brain cortex previously characterized by TAB-Seq23. gDNA was also purified from wild-type and TET-triple knockout (TKO) mouse ESCs, the latter of which lacks authentic 5hmC. For each sample, gDNA was spiked with 0.5% of T4-hmC and CG-methylated λ phage gDNA as internal controls. Either 2 or 20 ng of gDNA was then subjected to the optimized ACE-Seq protocol and sequenced to an average depth of ~8–11x per strand, comparable to previous TAB-Seq experiments (Supplementary Table 1). Reads were aligned and complete strand-specific base-resolution 5hmC maps were established for each of the three samples (Fig. 4a). Analysis of the spike-in controls confirmed an average non-conversion rate of 5hmC (98.5%) versus C/5mC (0.1%/0.5%, respectively) (Fig. 2f), enabling robust 5hmC discrimination.
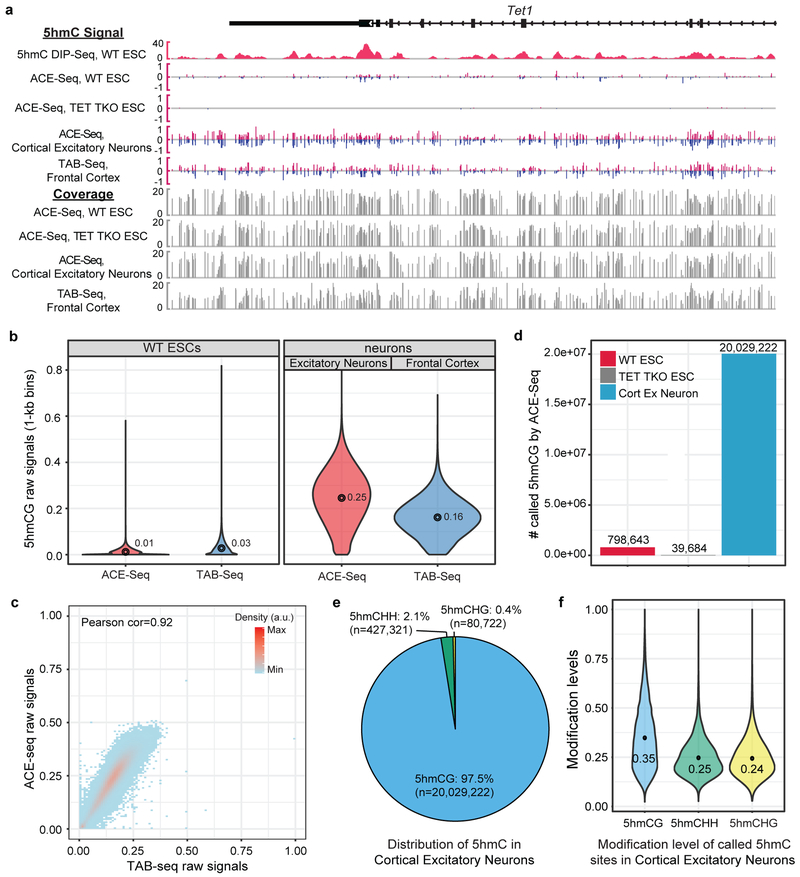
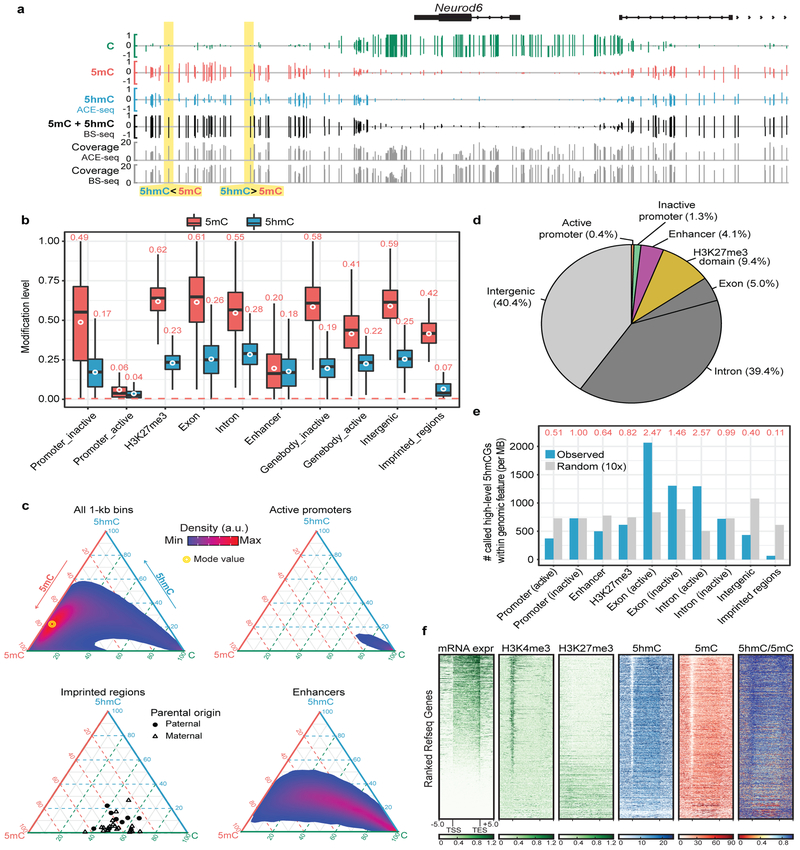
Figure 4. Generation of whole genome base-resolution maps of 5hmC using ACE-Seq.
(a) Snapshot of base-resolution 5hmC maps (ACE-Seq or TAB-Seq: red/blue for positive (Watson)/negative (Crick) strands, respectively) compared to DNA immunoprecipitation-based 5hmC map (DIP-Seq: pink) near the Tet1 locus (chr10:62,262,357–62,300,848; genome build: mm9). Only CGs sequenced to depth ≥2 are shown. Gray tracks denote sequencing coverage. All ACE-Seq data shown represent merged data sets from single experiments with 2 ng and 20 ng of input DNA (n = 2). (b) Violin plots comparing raw 5hmCG signals (fraction of C/(C+T)) within 1-kb genomic bins between ACE-Seq (red) or TAB-Seq (blue), with mean values above each plot. The width of the violin plot corresponds to the kernel probability density of the data at a given value, and the circle indicates the mean value. (c) Correlation density plot between ACE-Seq signals in neurons and TAB-Seq signals in frontal cortex (within 10-kb bins). a.u., arbitrary units. Correlation analysis was performed with 10-kb bins spanning the genome (n = 238,401). (d) Bar graph of statistically-significant 5hmCG sites (p value = 2.5e-4) in wild-type (WT) mouse ESCs, TET TKO mouse ESCs, and WT cortical excitatory neurons, with values listed above each bar. (e) Sequence context of statistically-significant 5hmC sites (in WT excitatory (ex) neurons) compared to the reference mouse genome (p value = 5e-8). (f) Violin plot of the distribution of modification levels of called 5hmCG, 5hmCHH, and 5hmCHG sites in WT ex neurons, with mean values are listed. The width of the violin plot corresponds to the kernel probability density of the data at a given value.
We first analyzed the raw 5hmC signal detectable in the CG context. 5hmCG is highly abundant in the cortical excitatory neurons (mean = 25%), and relatively lower in mESCs (mean = 1%), as anticipated (Fig. 4b). Given the high prevalence in neurons, we performed a pairwise comparison between the 5hmC signal from ACE-Seq in cortical excitatory neurons and that from TAB-Seq of the brain cortex and found a strong correlation (Fig. 4c, r=0.92), in agreement with the fact that the cortex contains ~85% excitatory neurons. Notably, only 2 or 20 ng of input gDNA was used in ACE-Seq in comparison to ~3 µg of cortical gDNA in TAB-Seq. A comparison between data collected using 2 ng versus 20 ng gDNA with ACE-Seq showed high correlation, indicating that ACE-Seq requires substantially less input DNA than bisulfite-based methods (Supplementary Fig. 6).
We next performed high-confidence calling of 5hmC sites in CGs. Using a p-value cut-off of 2.5e-4, we called 798,643 high-confidence 5hmCG sites in WT mESCs (Fig. 4d). By benchmarking ACE-Seq against TET TKO ESCs, our study offers the ability to empirically determine the false-discovery rate (FDR). Using the same statistical framework applied to WT ESCs, 39,684 false positive 5hmCG sites were detectable in TET TKO ESCs, resulting in an FDR of ~5.0% in these samples, identical to that estimated in TAB-Seq11. In neurons, where 5hmC is especially abundant, more than 20 million 5hmC sites were readily called, resulting in a calculated FDR of ~0.2%. Although detection of 5fC and 5caC is another theoretical source of false discovery—as A3A discriminates against all ox-mCs28—the 5hmC abundance significantly exceeds that of 5fC/5caC in all cell/tissue types (>100-fold in mESCs5; >1000-fold in mouse/human brain7,8), making 5fC/5caC’s contribution to the ACE-Seq signal negligible. Thus, ACE-Seq permits high-confidence 5hmC calling in both our ESC and neuronal samples.
5hmC in non-CG contexts is rare.
One area of disagreement in prior profiling has centered on the level and significance of 5hmC in non-CG (CH) contexts. When subtractive oxBS-Seq or a less-quantitative restriction enzyme-based method was applied to neuronal samples, 5hmCH levels were suggested to be relatively prevalent21,22. By contrast, TAB-Seq analysis of mouse cortical DNA indicates that 5hmC occurs almost exclusively in CG contexts (1.9% of 5hmC in CH contexts), with detectable but low levels in ESCs (0.11% in CH) 11,23. Using A3A provides an orthogonal approach to these methods and, unlike TET in TAB-Seq, which has a preference for oxidation of CGs33, has the added strength of being agnostic to the 3’ base28. Using a p-value cut-off of 5e-8, the FDR for 5hmCH detection in cortical excitatory neurons is 4.0%, obtained from analyzing the sequence-context-matched TET TKO samples. Using this statistical framework, ACE-Seq confirms that 5hmC is a rare modification in non-CG contexts, with only ~2.5% of 5hmC as 5hmCH (5hmCHH: 427,321 sites; 5hmCHG: 80,722 sites) in cortical excitatory neurons (Fig. 4e). Despite their rarity, the levels of 5hmC modification at called 5hmCH sites are largely comparable to those at 5hmCG sites (Fig. 4f). For mESCs, the direct comparison with TET TKO matched controls now allows us to state with statistical confidence that 5hmCH is not detectable in ESCs (that is, called 5hmCH in WT ESCs does not exceed that in TET TKO ESCs), at current sequencing depth.
5hmCG and 5mCG genomic distribution in excitatory neurons.
Having established the 5hmC landscape in cortical excitatory neurons, we next integrated our analysis with a neuronal subtype-matched BS-Seq dataset (Camk2a+)30 to uncover the true composition and genomic patterning of cytosine modifications in murine cortical excitatory neurons. By subtracting the ACE-Seq signals from those of BS-Seq, we constructed single-base resolution maps of C, 5mC, and 5hmC, revealing that sites that appear fully “methylated” in BS-Seq can vary substantially in terms of 5hmC and 5mC distribution (Fig. 5a, Supplementary Fig. 7). Across various classes of gene regulatory elements and genomic features, the levels of 5hmC are generally less than those of 5mC, and approximate 5hmC/5mC ratios remain nearly constant across many genomic features (5hmC/5mC ratio ~0.3–0.5, excluding outliers) (Fig. 5b).
Figure 5. Genomic distribution of 5hmC and 5mC in adult neurons.
(a) Snapshot of base-resolution C (green), 5mC (red), and 5hmC (blue) maps near the Neurod6 gene locus (chr6:55,667,934–55,690,102; genome build: mm10). Only CGs sequenced to depth ≥2 are shown. Gray tracks denote sequencing coverage. All ACE-Seq data shown represent merged data sets from single experiments with 2 ng and 20 ng of input DNA (n = 2). (b) The modification levels of 5hmCG (blue) and 5mCG (red) for several classes of genomic elements. The red dashed line denotes the 5mCG non-conversion rate in ACE-Seq. Genic features were extracted from the UCSC Refseq Genes database, and imprinted regions were chosen from a prior study34. Transcriptional activity of promoters/gene bodies and the presence of H3K27me3-marked repressive domains reflect experimentally-determined results in Camk2a-positive cortical excitatory neurons from H3K4me3 and H3K27me3 ChIP-Seq experiments23. Enhancers (>1-kb from transcriptional start site (TSS)) are determined by ATAC-Seq experiments in Camk2a-positive cortical excitatory neurons23. The white circles denote the mean of 5mC or 5hmC levels across the indicated genomic elements, with mean values listed above each plot. The center line represents the median value, the box represents the interquartile range, and the whiskers represent the maximum and minimum values. (c) Ternary plots show the levels of C, 5mC, and 5hmC within 1-kb bins across the genome (all) or overlapping with representative genomic elements. (d) Pie chart shows the overlap of called 5hmCGs with genomic elements. Each called 5hmCG site is counted once: the overlap of a genomic element excludes all previously overlapped sites clockwise starting from active promoter. (e) The relative enrichment of 5hmCG (blue) and random sites (gray) at genomic elements (normalized to the coverage of the element type). ‘Random’ consists of 10 random samplings of genomic elements. Shown on the top are the ratios (red) between observed and random. (f) Heat-map representation of normalized RNA-Seq, H3K4me3 (ChIP-Seq), H3K27me3 (ChIP-Seq), 5hmC (ACE-Seq), 5mC (derived from BS-Seq and ACE-Seq), and 5hmC/5mC ratios within 33,136 mouse Refseq genes (gene length >200-bp). Genes were ranked by their expression levels in Camk2a-positive cortical excitatory neurons.
Our analysis revealed a few notable exceptions to the observation that 5hmC levels track with 5mC levels in most features. First, at promoter-distal enhancers (identified by ATAC-Seq), 5hmC levels are nearly equivalent to those of 5mC (5hmC/5mC ratio = 0.90), rather than being proportionally low as expected from the overall genomic trend (Fig. 5b). A compelling counterexample is provided by imprinting control regions (ICRs), which control parent-of-origin-dependent allelic-specific expression. Whole genome BS-Seq analysis previously identified differentially-methylated ICRs in the mouse brain34, but the contribution of 5hmC had not been defined. Focusing on 30 differentially-methylated regions (DMRs) in imprinting regions (including bona fide ICRs) (Supplementary Table 2), the average 5hmC level (6.7%) is well below the genomic baseline level, and even lower than expected relative to 5mC (5hmC/5mC ratio ~0.17) (Fig. 5b). The majority of imprinted regions follow this pattern, as 22 out of 30 imprinted regions show 5hmC levels <10%, and 5hmC/5mC ratios averaging 0.08 (Fig. 5c and Supplementary Fig. 8). These observations imply that TET binding or activity is negatively regulated at most imprinted regions to maintain allelic-specific 5mCG patterns in neurons.
To explore the intra-class heterogeneity, we analyzed tiled 1-kb genomic bins for C, 5mC, and 5hmC distribution and generated ternary plots that account for all three modification states (Fig. 5c). Across these bins, a wide distribution of states can be observed centered on the mode value of ~6% CG, ~72% 5mCG, and ~22% 5hmCG. While some genomic features—such as genic regions including exons—track well with the overall distribution, others show significant deviations (Supplementary Fig. 9). For example, regions overlapping with active promoters largely exhibit a homogeneous, hypomodified state, while enhancers display a “long tail” in the ternary plot, reflecting highly heterogeneous, partially-modified states of 5hmC and 5mC (Fig. 5c). These observations are notable in the context of the recent discovery that transcription factor (TF) binding can be differentially influenced by methylation of their target sequences35. The heterogeneity in 5hmC/5mC we observe may similarly influence TF activity in these regulatory regions.
Given the heterogeneity evident in the ternary plots, we next examined specific sites with high 5hmC levels. While the distribution of 5hmC in the ~20 million called sites varies as a function of genomic features and regulatory elements (Fig. 5d), the subset of CGs showing relatively high 5hmC modification levels (ACE-Seq signal >60%) is more enriched in transcriptionally-active genic regions (especially introns) and less enriched within intergenic regions (Supplementary Fig. 10). Consistent with this observation, high-level 5hmC sites are overrepresented in actively-transcribed exons (observed/random [o/r] = 2.47) and active introns (o/r = 2.57) compared to other genomic regions (Fig. 5e).
Finally, we rank-ordered the expression level of annotated genes and compared major histone modifications as well as our 5hmC and 5mC profiles within each gene locus (Fig. 5f). Notably, actively-transcribed genes, marked by higher H3K4me3 and lower H3K27me3 levels at promoters, also show higher intragenic 5hmC/5mC ratios compared with their inactive counterparts. These observations suggest that TET-mediated oxidation of 5mC within genic regions may be positively correlated with gene activity. Overall, ACE-Seq analysis thus provides a framework for deconvolution of BS-Seq data with high confidence to parse the roles of 5hmC and 5mC from an otherwise masked signal.
DISCUSSION
Recent work has demonstrated the potential for exploiting specific DNA-modifying enzymes to localize genomic features, as exemplified by the use of Tn5 transposase to localize open chromatin in ATAC-Seq and the monitoring of DNA polymerase kinetics in SMRT sequencing36,37. In the study of DNA cytosine modifications, although great gains have been made using bisulfite-based approaches, the excessive degradation of gDNA samples has remained the most significant limitation of chemical deamination. In this study, we took advantage of the substrate selectivity of DNA deaminases to devise an enzymatic method for base-resolution localization of 5hmC. The base-resolution maps of 5hmC generated by ACE-Seq in excitatory neurons correlate strongly with those from prior TAB-Seq studies on whole brain cortex, giving confidence to both techniques in that the orthogonal approaches generated similar output. The major advantage of ACE-Seq is that enzymatic deamination permits the generation of base-resolution 5hmC maps with up to 1000-fold less DNA input. The nondestructive nature of ACE-Seq was confirmed by generating both long and short amplicons from gDNA with equal efficiency after exposure of substrate gDNA to our procedure. Given this finding, ACE-Seq offers the potential to profile DNA from even more rare populations, such as those in early development or cell-free DNA samples, when coupled to advances that permit library construction from limited samples. Additionally, it may now be possible to examine read-lengths inaccessible to bisulfite-based methods such as multi-kb enhancer regions.
An added attribute of ACE-Seq is the robustness of discrimination of 5hmC from C and 5mC, which permits high-confidence profiling of 5hmC. The rate of C conversion is similar to bisulfite, while 5mC conversion and 5hmC non-conversion exceed those of established methods. The resulting statistical framework permits us to demonstrate confidently that 5hmC is a rare modification in non-CG contexts, a question of importance given the purported differences in 5hmC interactions with DNA binding partners such as MeCP2 in CG versus non-CG contexts38. Merging the base-resolution profiles from BS-Seq and ACE-Seq in purified excitatory neurons demonstrates the heterogeneity in 5hmC/5mC signals as a function of different genomic features and regulatory elements, where the functional impact of 5hmC enrichment and altered 5hmC/5mC ratios can now be better parsed. For example, in excitatory neurons, enhancers offer an example of a genomic feature with marked heterogeneity but overall high 5hmC/5mC ratios, whereas ICRs offer a notable contrast, as they are mainly comprised of 5hmC-depeleted regions with low 5hmC/5mC ratios. At present, bisulfite treatment remains useful for distinguishing C and 5mC; however, we envision that, building on this precedent, other schemes integrating APOBEC-mediated deamination could be optimized to discriminate between C and 5mC (Supplementary Fig. 11). Overall, ACE-Seq expands the repertoire of biotechnological approaches whereby exploiting nature’s toolbox of DNA-modifying enzymes can be used to great effect for characterizing and manipulating genomic DNA.
Online Methods
Development of ACE-Seq protocol using T4 phage genomic DNA.
Wild-type human APOBEC3A was cloned, expressed, and purified as described previously, with additional details provided in Supplementary Protocol 128. For validation of ACE-Seq on T4 phage variants, genomic DNA was extracted from WT T4 phage (T4-ghmC, reference genome NCBI GenBank KJ477684.1), 147 T4 phage (T4-hmC; NCBI GenBank KJ477685.1), and GT7 T4 phage (T4-C, NCBI GenBank KJ477686.1). Modification content was validated by restriction digest and single-molecule PacBio sequencing in prior work29, as well as by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) in this study, using the parameters described below (Supplementary Fig. 2a). 1 ng of each gDNA sample in a volume of 5 µL was heated to 95 °C for 5 minutes in the presence or absence of 10% DMSO and immediately snap frozen in a dry ice/acetone bath. Then, the samples were incubated with 5 µM A3A at 37 ºC for 1 hr, 25 ºC for 1 hr, or under ramping temperature conditions from 4 ºC to 50 ºC over 2 hrs (Supplementary Fig. 3), in final buffer conditions containing 20 mM Tris, pH 6.5 and 0.1% Tween-20. A 550 bp amplicon was then PCR amplified from each reaction using Taq polymerase and primers optimized for bisulfite-converted DNA. The resulting amplicons were gel purified (Zymo Gel Extraction Kit) and TA cloned (Invitrogen TA Cloning Kit for Sequencing). Individual clones were Sanger sequenced and analyzed for C to T transitions across the amplicon.
Preparation of Phage Controls.
For whole-genome analyses of phage DNA samples, phage DNA was enzymatically methylated at CG sites by incubating with the CG methyltransferase M.SssI (ThermoFisher Scientific) and S-adenosylmethionine (SAM) at 37 °C. After four hours, additional enzyme and SAM were added to the reaction, and incubation continued for another four hours before purification (Zymo Genomic DNA Clean and Concentrator). Restriction digest with HpaII (NEB) as well as LC-MS/MS analysis (below) were used to assess methylation status (Supplementary Fig. 2b). The methylated phage DNA was combined in equal amounts with T4-hmC phage and together they were sheared to ~300 bp using a Covaris sonicator (20% duty factor, 200 cycles per burst, 150 s) and the fragment sizes were characterized using a Bioanalyzer.
Preparation of Mammalian Genomic DNA.
J1 WT ESCs were cultured in feeder-free gelatin-coated plates in Dulbecco’s Modified Eagle Medium (DMEM) (GIBCO) supplemented with 15% FBS (GIBCO), 2 mM L-glutamine (GIBCO), 0.1 mM 2-mercaptoethanol (Sigma), nonessential amino acids (GIBCO), and 1,000 units/mL LIF (Millipore, ESG1107), 3 µM of CHIR99021 (Stemgent) and 1 µM of PD032591 (Stemgent). The culture was passaged every 2–3 d using 0.05% Trypsin (GIBCO). TET triple knockout (TKO) mESCs were generated by as previously descibed39,40. Cortical excitatory neuronal nuclei (NeuroD6/NEX+) were purified from mouse brain as previously described32.
Optimized ACE-Seq protocol for whole genome sequencing.
Phage-only samples were analyzed with 1 ng each of pooled methylated phage and T4-hmC gDNA (Fig. 2d). For all mammalian DNA samples, a total of 2 or 20 ng of sheared gDNA (~300 bp) was analyzed, containing the sheared methylated phage and T4-hmC spike-in controls (0.5% total by mass). In a total volume of 5 µL, 1–20 ng of the gDNA mixture was glucosylated using UDP-glucose and T4 β-glucosyltransferase (βGT, NEB) at 37 ºC for 1 hr. 1 µL of DMSO was added and the sample was denatured at 95 ºC for 5 min and snap cooled by transfer to a PCR tube rack pre-incubated at −80 ºC. Before thawing, reaction buffer was overlaid to a final concentration of 20 mM MES pH 6.0 + 0.1 % Tween, and A3A was added to a final concentration of 5 μM in a total volume of 10 µL. The deamination reactions were incubated under linear ramping temperature conditions from 4–50 ºC over 2 hrs. After deamination, the samples were prepared for Illumina sequencing using the Accel Methyl-NGS kit (Swift Biosciences). The resulting ACE-Seq libraries were sequenced at 1.9 pM with single-end mode on a NextSeq 500 sequencer (Illumina) using the NextSeq 500/500 High Output kit v2 (150 cycles). A more detailed step-by-step protocol for ACE-Seq, with added rationale and discussion, is provided as Supplementary Protocol 2.
LC–MS/MS analysis of phage genome controls.
To determine the purity of the phage gDNA stocks and the efficiency of both the glucosylation and deamination steps in ACE-Seq, samples were analyzed by LC-MS/MS. For the phage validation experiments (Supplementary Fig. 2), untreated phage gDNA was used. For the experiments analyzing ACE-Seq efficiency (Fig. 2e; Supplementary Fig. 4), 15 ng of the phage-only samples (pooled samples with 7.5 ng each of methylated phage and T4-hmC gDNA) were treated using the ACE-Seq protocol, excluding either βGT or A3A, or excluding both βGT and A3A. The gDNA samples were degraded with 1 U DNA Degradase Plus (Zymo) in 10–15 µL at 37 °C for 4 hours. The nucleoside mixture was diluted ten-fold into 0.1% formic acid, and 2 ng was injected onto an Agilent 1200 Series HPLC with a 5 μm, 2.1 × 250 mm Supelcosil LC-18-S analytical column (Sigma) equilibrated to 45 °C in Buffer A (0.1% formic acid). The nucleosides were separated using a gradient of 0–10% Buffer B (0.1% formic acid, 30% (v/v) acetonitrile) over 8 min at a flow rate of 0.5 mL/min. Tandem MS/MS was performed in positive ion mode ESI on an Agilent 6460 triple-quadrupole mass spectrometer, with gas temperature of 225 °C, gas flow of 12 L/min, nebulizer at 35 psi, sheath gas temperature of 300 °C, sheath gas flow of 11 L/min, capillary voltage of 3,500 V, fragmentor voltage of 70 V, and delta EMV of +1,000 V. Collision energies were optimized to 10 V for C, U, T, and 5mC, and 25 V for 5hmC and 5ghmC. MRM mass transitions were C 228.1-›112.1 m/z, U 229.1-›113.0, T 243.1-›127.1, 5mC 242.1-›126.1; 5hmC 258.1-›124.1; and 5ghmC 420.2-›124.1. Standard curves were generated from standard nucleosides (Berry & Associates) ranging from 10 pmol to 2.4 fmol for all nucleosides, with the exception of 5ghmC for which a standard is not readily available. When possible, the sample peak areas were fit to the standard curve to determine amounts of each modified cytosine in the gDNA sample. To account for slight variations in the amount of material loaded, the calculated concentrations (or raw peak areas for 5ghmC) were normalized to that of G, allowing us to make comparisons between samples.
Input DNA comparison of ACE-Seq and BS-Seq by fixed-cycle PCR or by quantitative PCR (qPCR).
Half log dilutions of WT ESC DNA (from 1 µg to 1 ng) were subjected to standard bisulfite treatment (Qiagen EpiTect Bisulfite Kit) or to ACE-Seq, following the optimized protocol with two exceptions: first, the samples contained WT ESC gDNA that was not sheared and did not contain phage spike-in DNA, and, second, the samples were not subjected to library preparation after the deamination reactions. For fixed-cycle PCR analysis, after the deamination procedure, 0.5 or 1 µL of each reaction (normalized to contain an equivalent amount of starting template DNA) was used to seed two different PCR reactions: Either short (200-bp) and long (1-kb) fragments were amplified from the Tbx5 genomic region using primers designed with a bisulfite-optimized algorithm (MethPrimer) and KAPA Hifi HotStart Uracil+ ReadyMix (KAPA Biosystems). After 35 cycles of PCR, resulting products were visualized on a 1.5% agarose gel stained with SybrSafe (Fig. 3a,b). For qPCR analysis (Fig. 3c; Supplementary Fig. 5), 0.5 µL of the treated samples were combined with 500 nM each of the forward and reverse primers, to amplify either the 200-bp amplicon or the 1-kb amplicon, and amplified using the KAPA SYBR Fast Rox low qPCR Mastermix kit (KAPA Biosystems). For the 200-bp amplicon, a two-step PCR protocol was used in which the samples were initially denatured at 95 °C for 3 minutes, and then cycled between 95 °C (15 seconds) and 63 °C (20 seconds) for a total of 35 cycles. For the 1-kb amplicon, a two-step PCR protocol was used in which the samples were initially denatured at 95 °C for 5 minutes, and then cycled between 95 °C (30 seconds) and 66 °C (90 seconds) for a total of 41 cycles. Calculated CT values for each sample were normalized to the CT values calculated in the no template controls (resulting from primer dimer signal), and one cycle was subtracted from ACE-Seq measurements to account for differences in gDNA input associated with sample dilution. Resulting qPCR products were run on 1% agarose gels and stained with SybrSafe to confirm specific amplification (Supplementary Fig. 5); notably, the background signals from the “no template” and bisulfite-treated samples for the 1-kb amplicon were exclusively from primer dimer amplification and were not specific to the desired 1-kb product.
Data processing for whole-genome ACE-Seq.
Sequencing reads were processed as previously reported41. Briefly, raw reads were trimmed for low-quality bases and adaptor sequences using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), and the data quality was examined with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed reads were mapped against the reference genomes with Bismark (v0.14.3) 42. PCR duplicates were removed using the Picard program (http://picard.sourceforge.net/). To eliminate reads from strands not deaminated by A3A, reads with three or more consecutive non-converted cytosines in the CH context were removed. Raw signals were calculated as % of C/(C+T) at each site. Statistics of all genome-scale sequencing libraries are summarized in Supplementary Table 1.
Statistical calling of 5hmC and assessing FDR of whole-genome ACE-Seq.
For each cytosine within CG dinucleotides, we counted the number of “C” bases from ACE-Seq reads as 5hmC (denoted NC) and the number of “T” bases as methylated or unmodified cytosines (denoted NT). For statistical calling, we used the binomial distribution [N as the sequencing coverage (NT + NC) and p as the error rate of A3A deamination (0.47%, averaged non-conversion rate for 5mCG in spiked-in λ phage DNA, from six independent measurements)] to assess the probability of observing NT or greater by chance (Fig. 4e). To estimate empirical FDR of calling 5hmC-modified CGs, we repeated the steps above on ACE-Seq signals of a negative control sample (TET TKO ESCs) in which all authentic ox-mCs are absent. The FDR for a given p-value cutoff of the binomial distribution is the number of called CGs in negative controls divided by the number detected in the sample. For estimating the contribution of 5fC/5caC to ACE-Seq signals, we used the global modification levels of 5hmC (~5,000 ppm), 5fC (~10 ppm) and 5caC (undetectable) measured by the quantitative mass spectrometry analysis of adult mouse brain8,8,43. This suggests that the false positive signal derived from 5fC/5caC is at most 0.2%. For calling 5hmC in non-CG contexts (CHH or CHG), a sequence-context-matched error rate of A3A deamination (0.10%) was used to calculate empirical FDR (Fig. 4e). We restricted our statistical analysis to CG, CHH, or CHG sites covered by at least five reads per strand.
Pairwise Comparisons with ACE-Seq.
For pairwise comparison between ACE-Seq and TAB-Seq in neurons, the raw 5hmCG signals were calculated within tiled 10-kb genomic bins (Fig. 4b). For pairwise comparison between ACE-Seq performed with 2 ng versus 20 ng of gDNA, the raw 5hmCG signals were similarly calculated within tiled 10-kb genomic bins (Supplementary Fig. 6). Pearson correlation coefficients were calculated using R function cor.
Calculating the true level of 5mCGs by combining ACE-Seq with BS-Seq.
For each CG site, the levels of 5mC and 5hmC were estimated using the MLML tool44. This approach arrives at maximum likelihood estimates for the 5mC and 5hmC levels by combining data from ACE-Seq and BS-Seq (See “Published data sets” below). Only CG sites with 0 conflicts were considered for further analysis. From the MLML output, the level of unmodified CG was estimated by [100% − (abundance of 5hmC + 5mC)]. The results were further filtered, such that CG, 5mCG, and 5hmCG levels were nonnegative. For generating ternary plots (Fig. 5; Supplementary Fig. 8), levels of CG, 5mCG, and 5hmCG (as percentage of the sum of [CG + 5mCG + 5hmCG]) were calculated within tiled 1-kb genomic bins across the genome.
Quantifying enrichment of called 5hmCGs at genomic elements.
To calculate the enrichment of statistically-confident 5hmCGs at a set of genomic elements (Fig. 5e, Supplementary Fig. 10), we counted the number of overlapping 5hmCGs and divided by the average of 10 random samplings of called 5hmCGs. The sampling involves the shuffling of genomic elements within the same chromosome. We then normalized this enrichment value by the genomic span of the corresponding set of genomic elements.
Genome browser visualization.
We used Integrative Genomics Viewer (IGV, v2.3.91) 45 to visualize ACE-Seq signals using mm9 (Fig. 4a) or mm10 (Fig. 5a; Supplementary Fig. 7) Refseq transcript annotation as reference. For ACE-Seq, TAB-Seq, and BS-Seq datasets, modified cytosines are indicated by upward (plus strand) and downward (minus strand) ticks, with the height of each tick representing the fraction of modification at the site ranging from 0 to 1. For RNA-Seq, ATAC-Seq, and ChIP-Seq data, read density was normalized to 10 million reads.
Analysis of 5hmCGs and 5mCGs within imprinted regions.
For Fig. 5 and Supplementary Fig. 9, we analyzed 5hmCG and 5mCG modification levels within 30 of 32 well-established parental origin-dependent allele specific regions34 (see Supplementary Table 2). The Dlk1-Gtl2 IG and Igf2r loci were excluded due to aberrantly high 5mC levels and low coverage, respectively. The mean 5hmC levels were calculated for each imprinted region using the base-resolution ACE-Seq dataset of cortical excitatory neurons generated in this study. The true 5mC levels were then estimated via subtraction of the ACE-Seq signal from previously-published bisulfite-sequencing data in mouse brain34. For imprinted regions (n=22) with less than 10% mean 5hmC levels, we denote them as 5hmC-low imprinted regions in Supplementary Fig. 9. Those (n=8) with >10% mean 5hmC levels were denoted as 5hmC-high imprinted regions.
Statistical analysis.
No statistical methods were used to predetermine sample size for any experiments. All group results are expressed as mean +/− standard deviation unless otherwise stated. Specific p-values used for calling modified cytosine bases are explicitly stated in the text and figure legends. Each figure legend explicitly states the number of independent experiments. Statistical analyses for graphs were performed in GraphPad PRISM 7.
Published data sets.
For Fig. 4a-c, we used the following published data sets: 5hmC DIP-Seq in mouse ESCs46, TAB-Seq in mouse ESCs11, TAB-Seq in mouse PFC23. For Fig. 5 and Supplementary Fig. 7, we used whole-genome BS-Seq, RNA-Seq, ATAC-Seq, and ChIP-Seq for H3K4me1, H3K4me3, H3K27me3, and H3K27ac in purified mouse cortical excitatory neurons30.
Life Science Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability.
All sequencing data associated with this study have been deposited to Gene Expression Omnibus (GEO) under the accession number GSE116016.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Zhaolan Zhou, Maria Fasolino, Alexandra Bryson, and Jennifer Myers SanMiguel for useful discussion and reagents. This work was supported by the National Institutes of Health through R21-HG009545 (to R.M.K.) and by the Penn Epigenetics Institute. Additional support included R00-HG007982 (to H.W.), DP2-HL142044 (to H.W.), and R01-GM118501 (to R.M.K.). E.K.S. and J.E.D. are NSF Graduate Research Fellows. J.E.D. and E.B.F. were supported on NIH training grant T32-GM07229, and M.Y.L. by F30-CA196097.
Footnotes
AUTHOR CONTRIBUTIONS
E.K.S., C.S.N., R.M.K., and H.W. conceived of and developed the ACE-Seq approach. E.K.S. conducted all experiments, with assistance from J.E.D., M.Y.L., E.B.F., P.H., and Y.H. F.D.B. contributed to phage experiment design. H.W. performed computational analysis. E.K.S., H.W., and R.M.K. analyzed the results and wrote the manuscript, with contributions from all authors.
COMPETING FINANCIAL INTERESTS
Aspects of the ACE-Seq protocol have been non-exclusively licensed.
REFERENCES
- 1.Schubeler D Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L & Rao A Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C & Zhang Y Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C & Xu GL Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C & Carell T The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angew. Chem. Int. Ed Engl 50, 7008–7012 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Kohli RM & Zhang Y TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner M, Steinbacher J, Kraus TF, Michalakis S, Hackner B, Pfaffeneder T, Perera A, Muller M, Giese A, Kretzschmar HA & Carell T Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew. Chem. Int. Ed Engl 54, 12511–12514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, Murrell A & Balasubramanian S 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol 11, 555–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H & Zhang Y Charting oxidized methylcytosines at base resolution. Nat. Struct. Mol. Biol 22, 656–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR & Rao A The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B & He C Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W & Balasubramanian S Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K & Okamoto A Degradation of DNA by bisulfite treatment. Bioorg. Med. Chem. Lett 17, 1912–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, Bui B, Sejnowski TJ, Harkins TT, Mukamel EA, Behrens MM & Ecker JR Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark SJ, Lee HJ, Smallwood SA, Kelsey G & Reik W Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol 17, 72–016–0944–x (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booth MJ, Raiber EA & Balasubramanian S Chemical methods for decoding cytosine modifications in DNA. Chem. Rev 115, 2240–2254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahid OK, Zhao BS, He C & Hall AR Quantifying mammalian genomic DNA hydroxymethylcytosine content using solid-state nanopores. Sci. Rep 6, 29565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chavez L, Huang Y, Luong K, Agarwal S, Iyer LM, Pastor WA, Hench VK, Frazier-Bowers SA, Korol E, Liu S, Tahiliani M, Wang Y, Clark TA, Korlach J, Pukkila PJ, Aravind L & Rao A Simultaneous sequencing of oxidized methylcytosines produced by TET/JBP dioxygenases in Coprinopsis cinerea. Proc. Natl. Acad. Sci. U. S. A 111, E5149–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, Wang H, Sun D, Cheng X, Zhu Z, Pradhan S & Zheng Y High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell. Rep 3, 567–576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, Dai N, Borgaro JG, Quimby A, Sun D, Correa IR Jr, Zheng Y, Zhu Z & Guan S A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell 57, 750–761 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Mellen M, Ayata P & Heintz N 5-Hydroxymethylcytosine Accumulation in Postmitotic Neurons Results in Functional Demethylation of Expressed Genes. Proc. Natl. Acad. Sci. U. S. A 114, E7812–E7821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross JA, Pacis A, Chen GG, Barreiro LB, Ernst C & Turecki G Characterizing 5-hydroxymethylcytosine in human prefrontal cortex at single base resolution. BMC Genomics 16, 672–015- 1875–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM & Ecker JR Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siriwardena SU, Chen K & Bhagwat AS Functions and Malfunctions of Mammalian DNA-Cytosine Deaminases. Chem. Rev 116, 12688–12710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y & Kohli RM AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol 8, 751–758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wijesinghe P & Bhagwat AS Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res 40, 9206–9217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL & Harris RS Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem 287, 34801–34808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schutsky EK, Nabel CS, Davis AKF, DeNizio JE & Kohli RM APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Res 45, 7655–7665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA & Bushman FD Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9. MBio 6, e00648–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR & Nathans J Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR & Clark SJ Identification and resolution of artifacts in bisulfite sequencing. Methods 27, 101–107 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Johnson BS, Zhao YT, Fasolino M, Lamonica JM, Kim YJ, Georgakilas G, Wood KH, Bu D, Cui Y, Goffin D, Vahedi G, Kim TH & Zhou Z Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat. Med 23, 1203–1214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P & Xu Y Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL & Ren B Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schubeler D, Vinson C & Taipale J Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, 10.1126/science.aaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J & Turner S Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH & Greenberg ME Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F & Jaenisch R One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu F, Liu Y, Jiang L, Yamaguchi S & Zhang Y Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28, 2103–2119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Wu X, Shen L & Zhang Y Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol 32, 1231–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A & Balasubramanian S 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem 6, 1049–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu J, Zhou M, Song Q, Hong EE & Smith AD MLML: consistent simultaneous estimates of DNA methylation and hydroxymethylation. Bioinformatics 29, 2645–2646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorvaldsdottir H, Robinson JT & Mesirov JP Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K & Zhang Y Genome-wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell 153, 692–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data associated with this study have been deposited to Gene Expression Omnibus (GEO) under the accession number GSE116016.