Abstract
Background
Genetic diversity is a characteristic trait of the hepatitis B virus (HBV) and has been associated with different clinical outcomes. In South Africa, HBV infection is a major public health concern. Most HBV infections are caused by genotype A strains. However rare cases of infection with HBV genotype D have been reported. The purpose of this study was to investigate the molecular characteristics of a rare HBV subgenotype D4 isolate.
Methods
The full-length genome of isolate ZADGM6964 was amplified in a one-step polymerase chain reaction. The amplified product was purified and cloned into a pGEM®-T Easy Vector System to investigate the genetic diversity of the viral quasi-populations. The primary isolate and clones were then directly sequenced and analysed using an array of bioinformatics software.
Results
Phylogenetic analysis showed that the primary isolate and cloned sequences formed a monophyletic cluster away from subgenotype D4 reference strains. Further recombination analysis revealed that isolate ZADGM6964 was in fact a D4/E recombinant strain with breakpoints identified within the X and overlapping pre-Core/Core open reading frames with a >70% bootstrap confidence level. The recombinant genotype D4/E was found to be unique from other D/E strains archived in the genetic database, GenBank.
Conclusion
This study represents the first ever report on the isolation and molecular characterization of an HBV D4/E recombinant strain in South Africa. The findings provide evidence of further HBV genetic diversity in South Africa than has been previously reported.
Keywords: Bioinformatics, Genetics, Virology
1. Introduction
Genetic diversity is a characteristic trait of the Hepatitis B virus (HBV). The HBV is a reverse transcribing DNA virus classified under the Hepadnaviridae family (McMahon, 2009). The viral reverse transcriptase lacks proofreading function and as such is prone to errors during replication (Kay and Zoulim, 2007). As genetic errors accumulate over time, multiple HBV genetic variants have evolved (Pourkarim et al., 2014; Kay and Zoulim, 2007; Norder et al., 2004). Eight genetically distinct HBV genotypes have been established and are designated A to H (Bell et al., 2016; Norder et al., 2004). Additionally, two putative HBV genotypes, I and J, are proposed (Yu et al., 2010; Tatematsu et al., 2009; Olinger et al., 2008). The HBV genotypes are further classified into subgenotypes; A1-A7, B1-B9, C1-C16, D1-D9, F1-F4 and I1-I2 (Pourkarim et al., 2014; Shi et al., 2013; Yousif and Kramvis, 2013).
The various HBV genotypes have distinct geographical distributions (Hwang and Cheung, 2011). Genotype A for example, has a more global distribution (Hwang and Cheung, 2011; Kramvis and Kew, 2007; Norder et al., 2004). Genotypes B and C are prevalent in Asia while Genotype D is found in the Mediterranean region, Eastern Europe and South America (Hwang and Cheung, 2011). Whereas genotype E is restricted to the African region, genotype F is found in Central and South America, genotype G in Europe and the Americas, and genotype H in Central America (Hwang and Cheung, 2011; Mulders et al., 2004; Odemuyiwa et al., 2001). Lastly, genotype I has been reported in North-Western China, Vietnam and Laos, while genotype J was isolated previously in a Japanese patient (Yu et al., 2010; Tatematsu et al., 2009; Olinger et al., 2008; Jutavijittum et al., 2007).
Aside from the HBV genotypes and subgenotypes, novel HBV variants arising from intergenotypic recombination events are increasingly being reported worldwide (Boyce et al., 2017; Araujo, 2015; Araujo et al., 2013; Martel et al., 2013; Osiowy et al., 2008; Owiredu et al., 2001; Su et al., 2014). These recombinant genotypes include A/B/C, A/C, A/D, A/E, A/G; A/C/G, B/C, C/D, C/F, C/J, D/E, D/F, and F/G (Araujo, 2015). Of the intergenotypic recombinant genotypes reported worldwide, genotypes B/C and C/D have been identified as the most predominant strains (Araujo, 2015).
Research interests in HBV genetic diversity lie in the evolutionary and clinical implications of the various genetic variants. This is because HBV is the cause of a potentially fatal form of viral hepatitis, known as hepatitis B (McMahon, 2009). To date, the World Health Organization (WHO) estimates that about 240 million people worldwide are living with chronic hepatitis B, accounting for 780 000 deaths annually (WHO, 2018). Furthermore, HBV genetic variants have been associated with differences in the progression of chronic hepatitis B, as well as response to antiviral and immuno-therapies (Shi, 2012). Ultimately, the evolution and expansion of HBV genetic variants may have critical implications for global efforts aimed at eliminating chronic hepatitis B by 2030 (WHO, 2016).
In South Africa, it is the genotype A which is found to predominantly circulate and has been associated with severe liver disease and rapid progression to hepatocellular carcinoma (Kew, 2008; Kramvis and Kew, 2007). Various studies have sufficiently addressed the molecular characteristics of genotype A as compared to the less common genotypes (B, C, D and E) found circulating in South Africa (Gededzha et al., 2016; Kramvis, 2008; Kimbi et al., 2004). These previous studies have found molecular variations unique to the genotype A strains. Additionally, intergenotypic recombinant events with the genotype A have also been identified in South Africa (Shi et al., 2012; Yang et al., 2006; Owiredu et al., 2001).
Despite the fact that the HBV genotype D is rarely isolated from the South African population, it remains the second most predominant HBV genotype in the country (Kew, 2008). Genotype D strains that have been reported to circulate in South Africa include subgenotype D1, D3, and D4, with D3 being the predominant strain (Gededzha et al., 2016; Amponsah-Dacosta et al., 2015; Mayaphi et al., 2013; Kew, 2008; Norder et al., 2004). The full-length genome of circulating subgenotype D4 strains in South Africa has not been sufficiently characterized. This study investigated the molecular characteristics of a rare HBV subgenotype D4 strain found circulating among the South African population.
2. Materials and methods
2.1. Study design
During a previous hepatitis B seroprevalence study (Amponsah-Dacosta et al., 2015), we identified a serum sample obtained from a 24-year-old male study patient from the North West province of South Africa. The serum sample was determined to be positive for the hepatitis B surface antigen (HBsAg) as well as IgM antibodies to the viral core antigen, but negative for antibodies to HBsAg. In addition to this, the patients' HBV viral load was 6.93 × 104 copies/ml, indicating active and potential acute HBV infection. By directly sequencing and analysing the surface (S) gene of the HBV strain from this study patient, it was determined that he was infected with an HBV subgenotype D4 which is uncommon in South Africa, leading to the interest for the current study. Ethics clearance, including consent to make use of the archived serum sample and to conduct this study, was granted by the University Research Ethics Committee (MREC/P/236/2014: PG and SMUREC/P/142/2015: PG).
2.2. HBV full-length genome amplification
HBV DNA was extracted from 200μl of patient serum using the High Pure Viral Nucleic Acid Extraction Kit (Roche Diagnostics, Mannheim, Germany). A polymerase chain reaction (PCR) assay was used for the amplification of the HBV full-length genome using cycling conditions described previously by Günther et al. (1995). The assay was performed in a total volume of 50μL made-up of 25μL of 2X Kapa Taq Ready mix (Kapa Biosystems, Wilmington Massachusetts, USA), 15 μL of the DNA template, 8μL of PCR grade H2O, and 1μL each of 10μM primers P3 and P4 (Günther et al., 1995). Standard measures to avoid contamination were exercised, including making use of physically separated rooms for HBV DNA extraction, PCR master mix preparation, and loading of the template. Positive (ACCURUN® 325 HBV DNA positive control, SeraCare Life Sciences, USA; [initial viral load = 3.04 × 106 IU/ml]) and negative (nuclease-free PCR grade H2O) controls were included in all laboratory analysis as appropriate. The PCR amplified products were confirmed by a 2% agarose gel electrophoresis.
2.3. Cloning of the HBV full-length genome
The amplified HBV full-length genome was purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified PRC product was then cloned into a pGEM®-T Easy Vector System (Promega, Madison, USA) which was then cultured on Luria Broth agar plates (LB/ampicillin/IPTG/X-Gal) containing 100 μg/mL ampicillin as a selection antibiotic, 100mM IPTC (Isopropyl β-D-1-thiogalactopyranoside) to induce bacterial protein expression, and 100 mg/mL of X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) for blue/white bacterial colony screening. Only white colonies were sub-cultured in Luria Broth and the presence of the HBV full-length genome insert was confirmed using the one-step full-length HBV PCR described previously. Successfully cloned genomes, together with the primary isolate, were then purified and directly Sanger-sequenced using primers P1-P19 (Günther et al., 1995) on the ABI 3500XL Genetic Analyzer (ThermoFisher Scientific, Massachusetts, USA).
2.4. Sequence analysis and molecular characterization
Primary, as well as cloned HBV full-length genome sequences were analysed using various bioinformatics software programs. Briefly, the sequences were assembled, and base-calling performed using Chromas Pro software (Technelysium Pty. Ltd. South Brisbane, Australia). Edited sequences were then aligned and compared with a large dataset of HBV full-length genome reference strains retrieved from the genetic database, GenBank, using BioEdit version 7.0.5.3 (Hall, 1999). For the purpose of genotyping, sequences were first subjected to the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The evolutionary history of the sequences was then inferred by generating phylogenetic trees, using the Neighbour-Joining method set at 1000 bootstrap replications in Mega version 7 software program (Kumar et al., 2016).
Gene recombination analysis of the HBV full-length genome sequences was performed in Simplot version 3.5.1 (Lole et al., 1999). For prediction of phenotypic drug resistance and escape mutations within the polymerase (pol) and S open reading frames, respectively, sequences were submitted to the online algorithm, Geno-2-pheno HBV version 2.0 (http://hbv.geno2pheno.org/index.php). In addition to this, the pre-Core (preC), Core (C) and X gene open reading frames (ORFs) were manually investigated for genetic variations using the BioEdit software. The HBV full-length sequences generated in this study have been deposited in GenBank under the accession numbers [MH481858 – MH481862].
3. Results
3.1. Phylogenetic inference of HBV full-length genome sequences
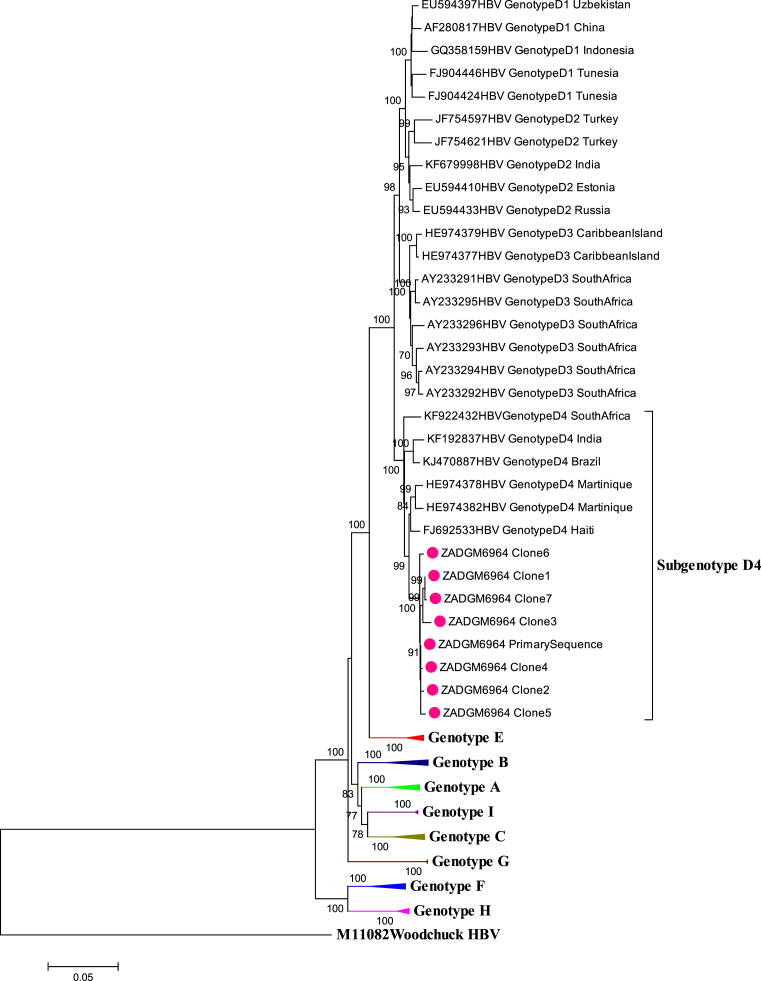
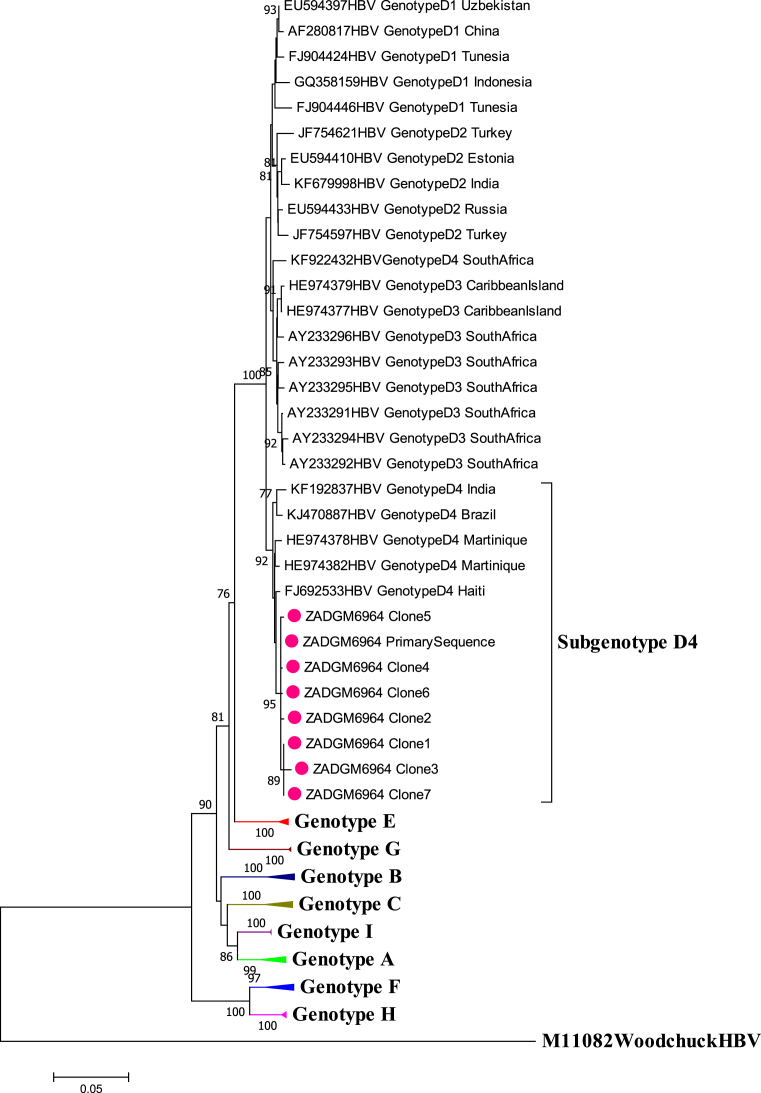
The primary HBV full-length genome of isolate ZADGM6964 was successfully amplified and sequenced. Seven clones of the primary isolate were also successfully isolated and sequenced. Phylogenetic analysis of the HBV full-length genome of the primary and cloned sequences showed that they clustered together with subgenotype D4 reference strains from Martinique, Haiti, Brazil, India and South Africa as shown in Fig. 1. It was observed however, that the primary and cloned sequences formed a monophyletic cluster away from the subgenotype D4 reference strains, prompting further investigation.
Fig. 1.
Phylogenetic inference of HBV full length genome sequences of isolate ZADGM6964. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
3.2. Identification of gene recombination events in HBV full-length genome sequences
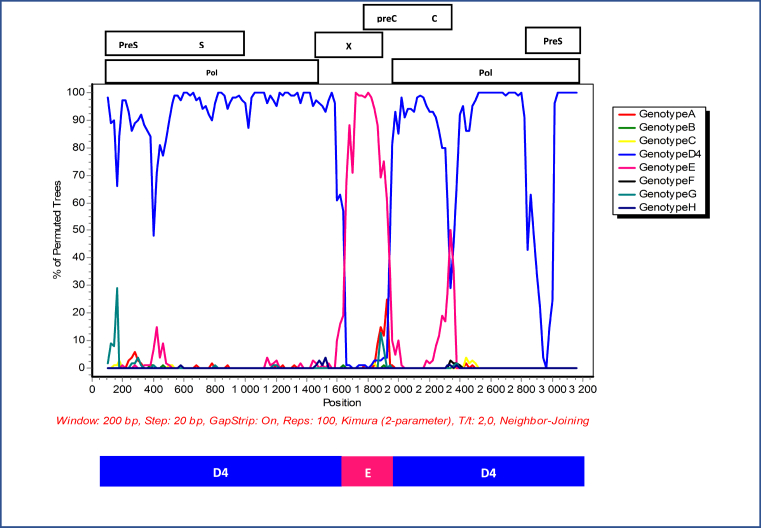
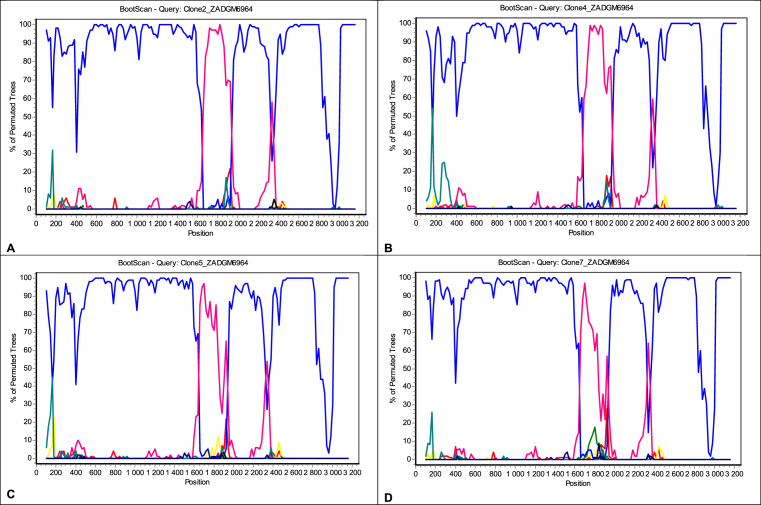
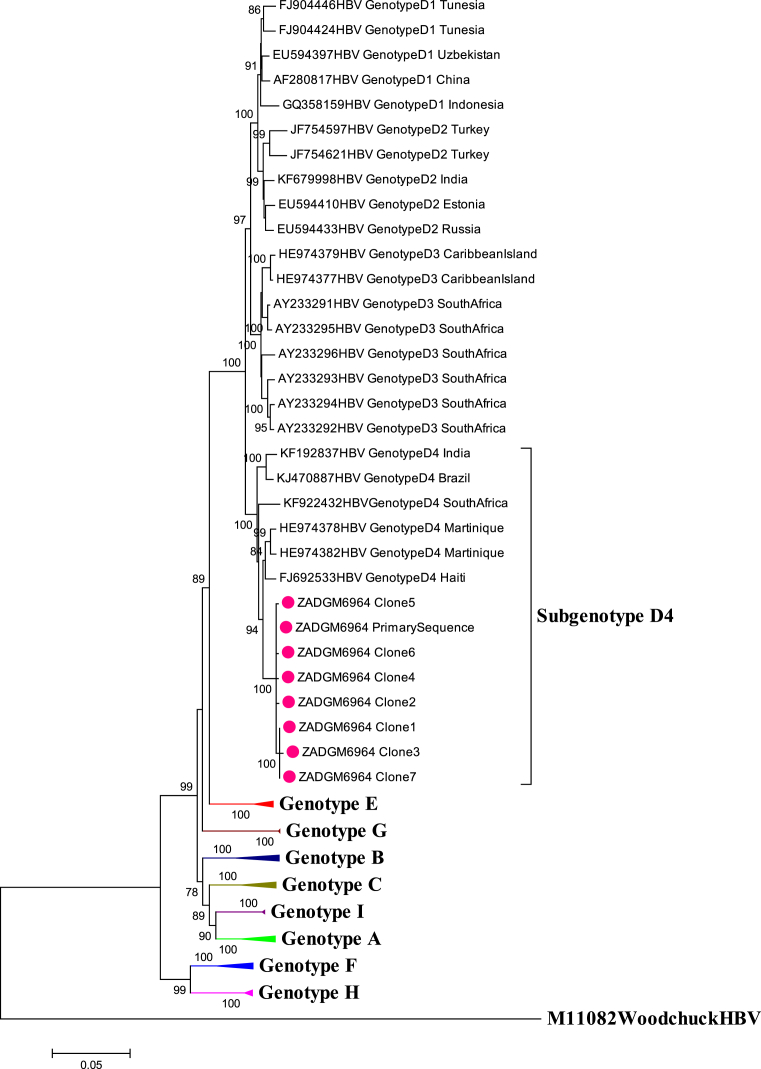
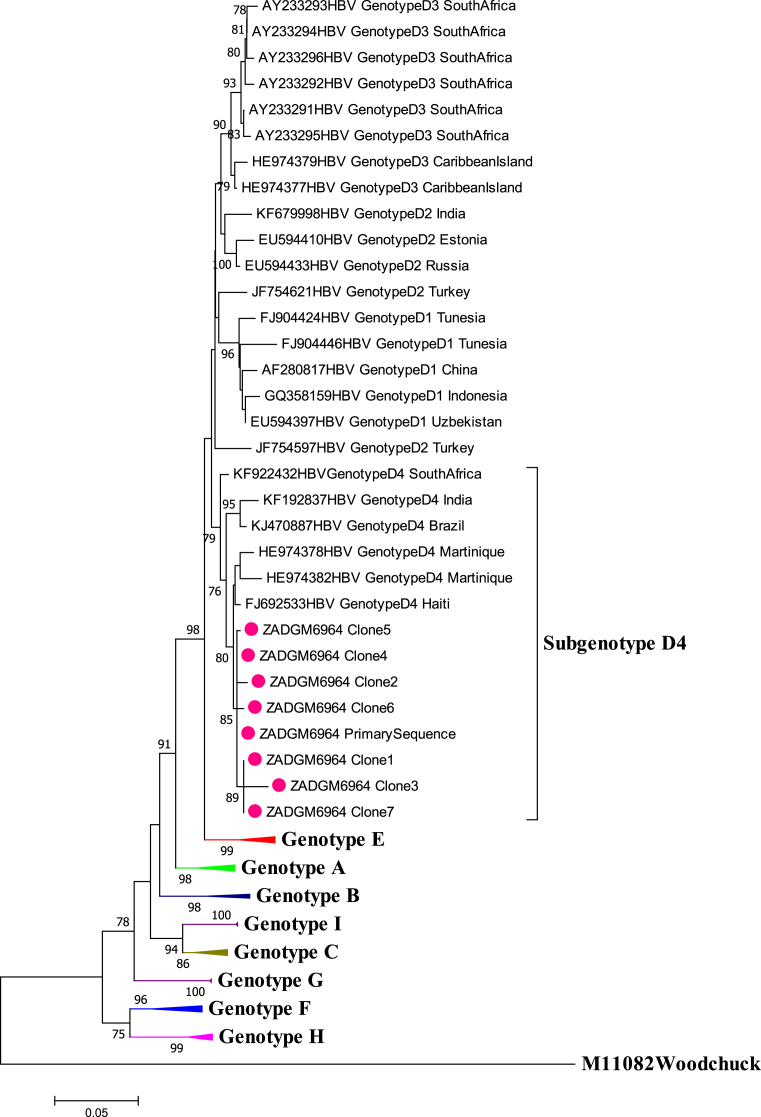
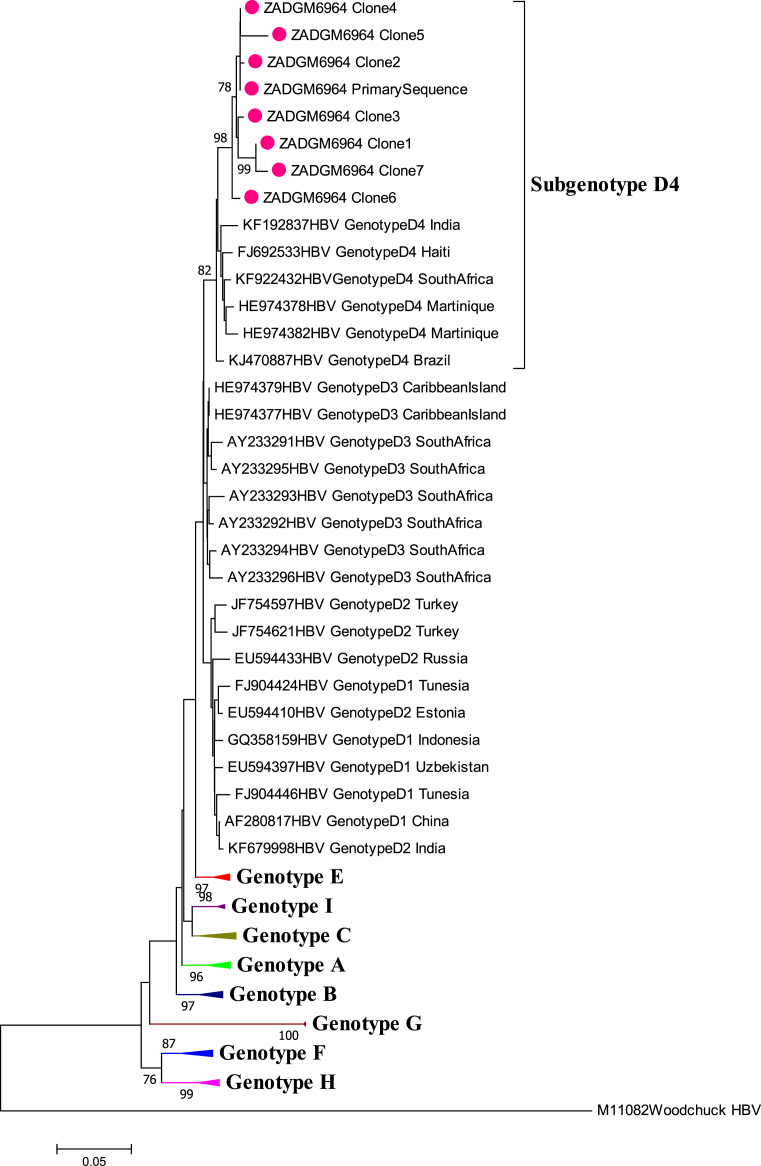
Gene recombination analysis revealed that the primary sequence was in fact a D4/E recombinant strain (Fig. 2). To confirm that the patient was infected with a pure D4/E recombinant strain and establish the sites where the gene recombination event occurred, the cloned sequences were also subjected to gene recombination analysis. The quasi-population of isolate ZADGM6964 had the same gene recombination pattern, suggesting infection by a pure D4/E recombinant strain (Fig. 3). There was no indication of dual infection with more than one HBV genotype. It was further observed that the recombination event occurred between nucleotide (nt) positions 1600–2000 within the X ORF and the pre-Core/Core (preC/C) overlap, with a >70% bootstrap confidence level based on the analysis performed in Simplot version 3.5.1. This observed recombination event was consistent in both the primary and cloned HBV full-length genome sequences (Figs. 2 and 3). It was also observed that the D4/E recombinant strain identified in this study was genetically unique from the HBV full-length genome sequences of other D/E recombinant strains available in GenBank (Fig. 4). In addition to this, each of the HBV ORFs of the primary and cloned sequences formed a monophyletic cluster away from reference sequences, with the X ORF showing higher diversity based on the topology of the branches (see Figs. 5, 6, 7, and 8).
Fig. 2.
Bootscan analysis of the primary HBV full length genome sequence of isolate ZADGM6964 generated in SimPlot v1.3. The X-axis represents the HBV nucleotide positions (nt) from the start of EcoR1 restriction site, while the Y-axis represents the percentage of permuted trees. Breakpoints were observed within the HBV X and preC/C overlap (nt1600 – 2000). The parental threshold of significance is set at 70% of the permutated trees. Blue peaks represent subgenotype D4 while fuchsia peaks represent genotype E.
Fig. 3.
Bootscan analysis of cloned HBV full length genome sequences of isolate ZADGM6964 generated in SimPlot v1.3. The quasi-population of isolate ZADGM6964 had the same gene recombination pattern, with breakpoints observed within the HBV X and preC//C overlap (nt1600 – 2000). The parental threshold of significance is set at 70% of the permutated trees. Blue peaks represent subgenotype D4 while fuchsia peaks represent genotype E. A = Clone 2, B = Clone 4, C = Clone 5, D = Clone 7.
Fig. 4.
Comparison of the evolutionary history of recombinant genotype D4/E with selected D/E reference strains from GenBank. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. Where the subgenotype of D/E recombinant strains are known, these have been indicated. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
Fig. 5.
Phylogenetic inference of HBV Polymerase ORF sequences of isolate ZADGM6964. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
Fig. 6.
Phylogenetic inference of HBV Surface ORF sequences of isolate ZADGM6964. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
Fig. 7.
Phylogenetic inference of HBV preCore/Core ORF sequences of isolate ZADGM6964. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
Fig. 8.
Phylogenetic inference of HBV X ORF sequences of isolate ZADGM6964. The primary and cloned sequences are labelled with a dot while reference strains including the Woodchuck HBV outlier are represented by their accession numbers as they appear in GenBank. The percentage of replicate trees >75% in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branch nodes.
3.3. Genetic variability of HBV full-length genome sequences
Genetic variations were observed in all four ORFs of the HBV full-length genome sequences and these have been presented in Table 1. Classical HBV drug resistance-associated mutations and immune-, diagnostic- or vaccine-escape variations were not observed in the pol or S overlapping ORFs, respectively. However, stop codons were identified in the pol ORF of three of the seven cloned sequences leading to premature truncation of the polymerase protein and reverse transcriptase. Overall, the preC/C ORF remained relatively conserved, although stop codons were identified in one of the cloned sequences leading to premature truncation of the e antigen and core protein. In comparison, the X ORF within which the gene recombination event was observed appeared relatively variable for all study sequences. Variations such as the xQ146L, xH147A, xH148R, xA149P, xT150N, were unique to the X ORF of the primary and cloned sequences but not observed in subgenotype D4 reference strains retrieved from GenBank (Table 1).
Table 1.
Genetic diversity of the quasi-population of isolate ZADGM6469.
| Open Reading Frame | Sequence | Genetic Variations | Reported Significance |
|---|---|---|---|
| Polymerase | Primary Sequence | No drug resistance mutations. Premature truncation of the polymerase protein including the reverse transcriptase expressed by Clones 2, 3 and 6; associated with potential impaired viral replication. |
|
| Clone 1 | M1T, R223K, T372I | ||
| Clone 2 | E24G, I193T, R196*, D366G | ||
| Clone 3 | M1T, R233K, F244L, V246E, P248L, S251*, T372I | ||
| Clone 4 | F665L | ||
| Clone 5 | P812L | ||
| Clone 6 | N400H, W578* | ||
| Clone 7 | M1T, R223K, T372I | ||
| Surface | Primary Sequence | No immune-, diagnostic- or vaccine- escape mutations. | |
| Clone 1 | D43N, P192S | ||
| Clone 2 | F13L, S85F, T186A | ||
| Clone 3 | D43N, L64N, W66R, P68S, Q71K, G22A, P192S | ||
| Clone 4 | |||
| Clone 5 | |||
| Clone 6 | |||
| Clone 7 | D43N, P192S | ||
| X | Primary Sequence | Q146L, H147A, H148R, A149P, T150N | High rate of variability potentially resulting from gene recombination event. |
| Clone 1 | D119A, W120S, H145R, T150N | ||
| Clone 2 | R138S, Q146L, H147A, H148R, A149P, T150N | ||
| Clone 3 | Deletion at aa144-146, A149P, T150N | ||
| Clone 4 | K130R, G136S, R138S, Q146L, H147A, H148R, A149R, T150N | ||
| Clone 5 | F112L, K140R, V142C, insertion at aa143, Q146L, H147A, H148R, A149P, T150N | ||
| Clone 6 | V92F, I127N, V133E, R138S, A149P | ||
| Clone 7 | D119A, W120S, L123S, K130T, Deletion at aa133-142, H145R, Deletion at aa146-149, T150 | ||
| PreCore/Core | Primary Sequence | Premature truncation of e antigen and core protein may lead to potential immune and diagnostic escape in Clone 3. | |
| Clone 1 | C149A | ||
| Clone 2 | F113G, E117G | ||
| Clone 3 | D40E, L42*, P45R, H52G, L76*, V85L, H104Q, C149A | ||
| Clone 4 | |||
| Clone 5 | S87N | ||
| Clone 6 | I3S, V13M | ||
| Clone 7 | C149A |
Note: Amino acid variations are listed as they occur in the complete ORFs of the HBV genome.
The * denotes a stop codon. aa refers to amino acid.
4. Discussion
If the universal goal of eliminating hepatitis B by the year 2030 is to be achieved, it is important to appreciate that the evolving genetic diversity of HBV may have implications for the expansion of the epidemic as well as diagnosis, vaccine efficacy, treatment response and disease progression (Castelhanoa et al., 2017; WHO, 2016; Kramvis et al., 2002; Kao et al., 2000). It is therefore important to investigate the molecular characteristics of the viral quasi-population of suspected recombinant strains. Historically, the HBV genotype A has been the dominant genotype driving the high burden of hepatitis B in South Africa (Kramvis et al., 2005). As international migration has increased over the years, there has been an observed increase in the variability of circulating strains in the country (Kramvis, 2008; Kramvis and Kew, 2007). With other minor HBV genotypes like genotypes D and E co-circulating in South Africa, the potential for recombination between these genotypes is substantial.
Intergenotypic recombination between the HBV genotypes D and E are not novel, but have been reported previously in Ethiopia, Gabon, Ghana, Kenya, Niger and Sudan (Boyce et al., 2017; Brah et al., 2016; Ochwoto et al., 2016; Yousif et al., 2013; Mahgoub et al., 2011; Chekaraou et al., 2010; Garmiri et al., 2009). In most of these countries, genotypes D and E are often found to be co-circulating (Ochwoto et al., 2016; Garmiri et al., 2009; Kramvis, 2008; Kramvis and Kew, 2007; Norder et al., 2004). In South Africa however, the D/E recombinant strain has not been reported previously. Instead, intergenotypic recombination between the more predominant genotypes A/D, as well as A/C and A/B/C have been reported to circulate in South Africa (Shi et al., 2012; Yang et al., 2006; Owiredu et al., 2001). According to Araujo (2015), intergenotypic recombinant strains typically share similar circulation patterns with their parental genotypes. This could potentially explain why the intergenotypic recombinant strains that have so far been reported in South Africa have been associated with the more predominant genotype A.
Phylogenetic analysis showed that while the HBV full-length primary and cloned sequences clustered with HBV subgenotype D4 reference strains, they formed a separate monophyletic cluster. Additionally, when compared to previously identified HBV D/E recombinant strains, the strain isolated in this study was found to be unique. This distinction between the HBV D/E recombinant strain identified in this study compared to those reported previously could be attributed to the parental genotype D strain. While the parental strain in this study was a subgenotype D4, those from Ethiopia and Sudan for example are recorded as subgenotype D7 in GenBank. Further analysis revealed that each of the HBV ORFs of the primary and cloned sequences formed a monophyletic cluster away from reference strains. The pol, S and the relatively conserved preC/C ORFs clustered in a similar way showing genetic relatedness among the study sequences and with subgenotype D4 reference sequences. This is likely due to the pure genome sequence of these ORFs. The X ORF cluster however, showed a higher diversity indicated by the topology of the branches, possibly a result of the gene recombination event occurring within this region of the genome.
The previously identified HBV D/E recombinant strains share similarities in the sites where the intergenotypic recombination events occur, typically corresponding with the pol, X and preC/C ORFs (Boyce et al., 2017). In Sudan, Mahgoub et al. (2011) reported gene recombination between genotypes D and E with breakpoints occurring in the preS1, X and preC/C gene regions. In agreement with our findings, a study conducted in Niger identified gene recombination events in the X and preC/C ORFs (Chekaraou et al., 2010). It was important to find that the quasi-population of the primary isolate had the same recombination pattern, suggesting that the study patient was infected with an HBV D4/E recombinant strain as opposed to co-infection with pure HBV genotypes D and E parental strains. This is consistent with the recombinant genotype D/E infections reported in a previous study (Mahgoub et al., 2011).
It is worth noting that HBV gene recombination events are not randomly distributed across the genome. Instead, nt1700–2000 and nt2100–2300 are favoured sites for gene recombination compared to other regions of the genome (Araujo, 2015). Accordingly, previous studies have identified breakpoints in HBV D/E recombinant strains which span across these specified favoured sites within the genome (Boyce et al., 2017; Mahgoub et al., 2011; Chekaraou et al., 2010). Consistent with these previous reports, breakpoints were observed within nt1600 and 2000 in both the primary and cloned sequences in this study.
The findings of this study should be carefully considered in light of some limitations, including the incomplete clinical records of the study patient (such as liver function test results) and the modest sample size. As the HBV D4/E recombinant strain is rare to South Africa and was a unique finding, it was not possible to genetically characterize more than one primary isolate. Additionally, the clinical implications of infection with an HBV D4/E recombinant strain could not be established within the scope of this study based on the cross-sectional design of the parental seroprevalence study and the unavailability of the study patients' clinical data. Despite this, mutational analysis showed the absence of drug resistance and vaccine-escape variants. Premature truncation of HBV reverse transcriptase, e antigen and core proteins, was observed within the quasi-population of isolate ZADGM6964. While truncated reverse transcriptase has been previously associated with replication deficient viruses, truncated e antigen and core proteins have been linked with reduced expression of the e antigen, leading to potential immune- and diagnostic-escape (Tong et al., 2013).
5. Conclusion
This study represents the first ever report on the isolation and molecular characterization of an HBV D4/E recombinant strain in South Africa. While the HBV subgenotype D4 has been isolated previously in South Africa, the full-length genome has not been adequately characterized. It is possible that previously reported subgenotype D4 strains circulating in South Africa are not pure D4 but D4/E recombinants, although this would require further investigation. The findings from this study provide evidence of further HBV genetic diversity in South Africa than has been previously reported. The study recommends continuous surveillance of HBV genetic diversity by full-length genome sequencing to document and follow-up on infections with unique genetic strains. The clinical implications of infection with these unique strains also need to be investigated.
Declarations
Author contribution statement
Edina Amponsah-Dacosta, Andrew M. Musyoki: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Mmatsatsi K. Matlou, Lucinda R. Gaelejwe: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
J. Nare Rakgole: Analyzed and interpreted the data; Wrote the paper.
Selokela G. Selabe: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by grants from the Poliomyelitis Research Foundation (PRF) [grant numbers: 11/74 and 15/37] and the National Health Laboratory Service (NHLS) [grant number: GRANT004_94329].
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GenBank under the accession numbers MH481858 – MH481862.
References
- Amponsah-Dacosta E., Lebelo R.L., Rakgole J.N., Selabe S.G., Gededzha M.P., Mayaphi S.H., Powell E.A., Blackard J.T., Mphahlele M.J. Hepatitis B virus infection in post-vaccination South Africa: occult HBV infection and circulating surface gene variants. J. Clin. Virol. 2015;63:12–17. doi: 10.1016/j.jcv.2014.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo N.M., Araujo O.C., Silva E.M., Villela-Nogueira C.A., Nabuco L.C., Parana R., Bessone F., Gomes S.A., Trepo C., Kay A. Identification of novel recombinants of hepatitis B virus genotypes F and G in human immunodeficiency virus-positive patients from Argentina and Brazil. J. Gen. Virol. 2013;94:150–158. doi: 10.1099/vir.0.047324-0. [DOI] [PubMed] [Google Scholar]
- Araujo N.M. Hepatitis B virus intergenotypic recombinants worldwide: an overview. Infect. Genet. Evol. 2015;36:500–510. doi: 10.1016/j.meegid.2015.08.024. [DOI] [PubMed] [Google Scholar]
- Bell T.G., Yousif M., Kramvis A. Bioinformatic curation and alignment of genotyped hepatitis B virus (HBV) sequence data from the GenBank public database. SpringerPlus. 2016;5:1896. doi: 10.1186/s40064-016-3312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce C.L., Ganova-Raeva L., Archampong T.N.A., Lartey M., Sagoe K.W., Obo-Akwa A., Kenu E., Kwara A., Blackard J.T. Identification and comparative analysis of hepatitis B virus genotype D/E recombinants in Africa. Virus Gene. 2017;53:538–547. doi: 10.1007/s11262-017-1469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brah S., Moussa S., Inoua A., Alhousseini D.M., Daou M., Madougou B., Romera M.H., Hamadou A., Adehossi E., Parola P., Colson P. Molecular characterization of hepatitis B virus from chronically-infected patients in Niamey, Niger. Int. J. Infect. Dis. 2016;45:18–23. doi: 10.1016/j.ijid.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Castelhanoa N., Araujoc N.M., Arenas M. Heterogeneous recombination among Hepatitis B virus genotypes. Infect. Genet. Evol. 2017;54:486–490. doi: 10.1016/j.meegid.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Chekaraou M.A., Brichler S., Mansour W., Gal F.L., Garba A., Dény P., Gordien E. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J. Gen. Virol. 2010;91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
- Garmiri P., Loua A., Haba N., Candotti D., Allain J.P. Deletions and recombinations in the core region of hepatitis B virus genotype E strains from asymptomatic blood donors in Guinea, west Africa. J. Gen. Virol. 2009;90:2442–2451. doi: 10.1099/vir.0.012013-0. [DOI] [PubMed] [Google Scholar]
- Gededzha M.P., Muzeze M., Burnett R.J., Amponsah-Dacosta E., Mphahlele M.J., Selabe S.G. Complete genome analysis of hepatitis B virus in human immunodeficiency virus infected and uninfected South Africans. J. Med. Virol. 2016;88:1560–1566. doi: 10.1002/jmv.24502. [DOI] [PubMed] [Google Scholar]
- Günther S., Li B.C., Miska S., Krüger D.H., Meisel H., Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hwang E.W., Cheung R. Global epidemiology of HBV infection. N. Am. J. Med. Sci. 2011;4:7–10. [Google Scholar]
- Jutavijittum P., Yousukh A., Samountry B., Samountry K., Ounavong A., Thammavong T., Keokhamphue J., Toriyama K. Seroprevalence of hepatitis B and C virus infections among Lao blood donors. Southeast Asian J. Trop. Med. Publ. Health. 2007;38:674–679. [PubMed] [Google Scholar]
- Kao J.H., Wu N.H., Chen P.J., Lai M.Y., Chen D.S. Hepatitis B genotypes and the response of interferon therapy. J. Hepatol. 2000;33:998–1002. doi: 10.1016/s0168-8278(00)80135-x. [DOI] [PubMed] [Google Scholar]
- Kay A., Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Kew M.C. Hepatitis B virus the burden of the disease in South Africa. South Afr. J. Epidemiol. Infect. 2008;23:4–8. [Google Scholar]
- Kimbi G.C., Kramvis A., Kew M.C. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J. Gen. Virol. 2004;85:1211–1220. doi: 10.1099/vir.0.19749-0. [DOI] [PubMed] [Google Scholar]
- Kramvis A., Kew M., Franҫois G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- Kramvis A., Kew M.C. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol. Res. 2007;37:S9–S19. doi: 10.1111/j.1872-034X.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- Kramvis A., Wietzmann L., Owiredu W.K.B.A., Kew M.C. Analysis of the complete genome of subgroup A hepatitis B virus isolates from South Africa. J. Gen. Virol. 2002;83:835–839. doi: 10.1099/0022-1317-83-4-835. [DOI] [PubMed] [Google Scholar]
- Kramvis A. Molecular characterization of the genotypes and mutants of Hepatitis B Virus from South Africa. South. Afr. J. Epidemiol. Infect. 2008;23:29–32. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahgoub S., Candotti D., El Ekiaby M., Allain J.P. Hepatitis B virus (HBV) infection and recombination between HBV genotypes D and E in asymptomatic blood donors from Khartoum, Sudan. J. Clin. Microbiol. 2011;49:289–306. doi: 10.1128/JCM.00867-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel N., Gomes S.A., Chemin I., Trepo C., Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J. Virol Methods. 2013;193:653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- Mayaphi S.H., Martin D.J., Mphahlele M.J., Blackard J.T., Bowyer S.M. Variability of the preC/C region of hepatitis B virus genotype A from a South African cohort predominantly infected with HIV. J. Med. Virol. 2013;85:1883–1892. doi: 10.1002/jmv.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon B.J. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45–S55. doi: 10.1002/hep.22898. [DOI] [PubMed] [Google Scholar]
- Mulders M.N., Venard V., Njayou M., Njayou M., Edorh A.P., Oyefolu A.O.B., Kehinde M.O., Tamfum J.M., Nebie Y.K., Maiga I., Ammerlaan W., Fack F., Omilabu S.A., le Faou A., Muller C.P. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J. Infect. Dis. 2004;190:400–408. doi: 10.1086/421502. [DOI] [PubMed] [Google Scholar]
- Norder H., Courouce A.M., Coursaget P., Echevarria J.M., Lee S.D., Mushahwar I.K., Robertson B.H., Locarnini S., Magnius L.O. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- Ochwoto M., Kimotho J.H., Oyugi J., Okoth F., Kioko H., Mining S., Budambula N.L., Giles E., Andonov A., Songok E., Osiowy C. Hepatitis B infection is highly prevalent among patients presenting with jaundice in Kenya. BMC Infect. Dis. 2016;16:101. doi: 10.1186/s12879-016-1409-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odemuyiwa S.O., Mulders M.N., Oyedele O.I., Ola S.O., Odaibo G.N., Olaleye D.O., Muller C.P. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J. Med. Virol. 2001;65:463–469. [PubMed] [Google Scholar]
- Olinger C.M., Jutavijittum P., Hubschen J.M., Yousukh A., Samountry B., Thammavong T., Toriyama K., Muller C.P. Possible new hepatitis B virus genotype, southeast Asia. Emerg. Infect. Dis. 2008;14:1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiowy C., Gordon D., Borlang J., Giles E., Villeneuve J.P. Hepatitis B virus genotype G epidemiology and co-infection with genotype A in Canada. J. Gen. Virol. 2008;89:3009–3015. doi: 10.1099/vir.0.2008/005124-0. [DOI] [PubMed] [Google Scholar]
- Owiredu W.K., Kramvis A., Kew M.C. Hepatitis B virus DNA in serum of healthy black African adults positive for hepatitis B surface antibody alone; possible association with recombination between genotypes A and D. J. Med. Virol. 2001;64:441–454. doi: 10.1002/jmv.1070. [DOI] [PubMed] [Google Scholar]
- Pourkarim M.R., Amini-Bavil-Olyaee S., Kurbanov F., van Ranst M., Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J. Gastroenterol. 2014;20:7152–7168. doi: 10.3748/wjg.v20.i23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Carr M.J., Dunford L., Zhu C., Hall W.W., Higgins D.G. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology. 2012;427:51–59. doi: 10.1016/j.virol.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Shi W., Zhang Z., Ling C., Zheng W., Zhu C., Carr M.J., Higgins D.G. Hepatitis B virus subgenotyping: history, effects of recombination, misclassifications, and corrections. Infect. Genet. Evol. 2013;16:355–361. doi: 10.1016/j.meegid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Shi Y.H. Correlation between hepatitis B virus genotypes and clinical outcomes. Jpn. J. Infect. Dis. 2012;65:476–482. doi: 10.7883/yoken.65.476. [DOI] [PubMed] [Google Scholar]
- Su H., Liu Y., Xu Z., Cheng S., Ye H., Xu Q., Liu Q., Tan S., Xu D., Liu Y. A novel complex A/C/G intergenotypic recombinant of hepatitis B virus isolated in southern China. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Tanaka Y., Kurbanov F., Sugauchi F., Mano S., Maeshiro T., Nakayoshi T., Wakuta M., Miyakawa Y., Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype. J. Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li J., Wands J.R., Wen Y. Hepatitis B virus genetic variants: biological properties and clinical implications. Emerg. Microb. Infect. 2013;2 doi: 10.1038/emi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2016. Draft Global Health Sector Strategies.http://apps.who.int/gb/ebwha/pdf_files/WHA69/A69_32-en.pdf?ua=1 Viral hepatitis, 2016-2021. Available at: [accessed August 2018] [Google Scholar]
- World Health Organization . 2018. Hepatitis B.http://www.who.int/immunization/diseases/hepatitisB/en/ Available at: [accessed August 2018] [Google Scholar]
- Yang J., Xing K., Deng R., Wang J., Wang X. Identification of hepatitis B virus putative intergenotype recombinants by using fragment typing. J. Gen. Virol. 2006;87:2203–2215. doi: 10.1099/vir.0.81752-0. [DOI] [PubMed] [Google Scholar]
- Yousif M., Kramvis A. Genotype D of hepatitis B virus and its subgenotypes: an update. Hepatol. Res. 2013;43:355–364. doi: 10.1111/j.1872-034X.2012.01090.x. [DOI] [PubMed] [Google Scholar]
- Yousif M., Mudawi H., Bakhiet S., Glebe D., Kramvis A. Molecular characterization of hepatitis B virus in liver disease patients and asymptomatic carriers of the virus in Sudan. BMC Infect. Dis. 2013;13:328. doi: 10.1186/1471-2334-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Yuan Q., Ge S.X., Wang H.Y., Zhang Y.L., Chen Q.R., Zhang J., Chen P.J., Xia N.S. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I”. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009297. [DOI] [PMC free article] [PubMed] [Google Scholar]