Summary
Staphylococcus aureus is a major cause of infectious disease. Liver Kupffer cells (KCs) are responsible for sequestering and destroying S. aureus through the phagolysosomal pathway. Proteins belonging to the COMMD family emerge as key intracellular regulators of protein trafficking, but the role of COMMD10 in macrophage-mediated S. aureus eradication is unknown. Here we report that COMMD10 in macrophages was necessary for its timely elimination, as demonstrated with two different S. aureus subspecies. In vivo, COMMD10-deficient liver KCs exhibited impaired clearance of systemic S. aureus infection. S. aureus-infected COMMD10-deficient macrophages exhibited impaired activation of the transcription factor EB, resulting in reduced lysosomal biogenesis. Moreover, S. aureus-initiated phagolysosomal maturation and function were significantly attenuated in COMMD10-deficient macrophages. Finally, expression of COMMD/CCDC22/CCDC93 complex, linked to phagolysosomal maturation, was reduced by COMMD10 deficiency. Collectively, these results support an important role for COMMD10 in instructing macrophage phagolysosomal biogenesis and maturation during S. aureus infection.
Subject Areas: Biological Sciences, Immunology, Microbiology, Molecular Microbiology
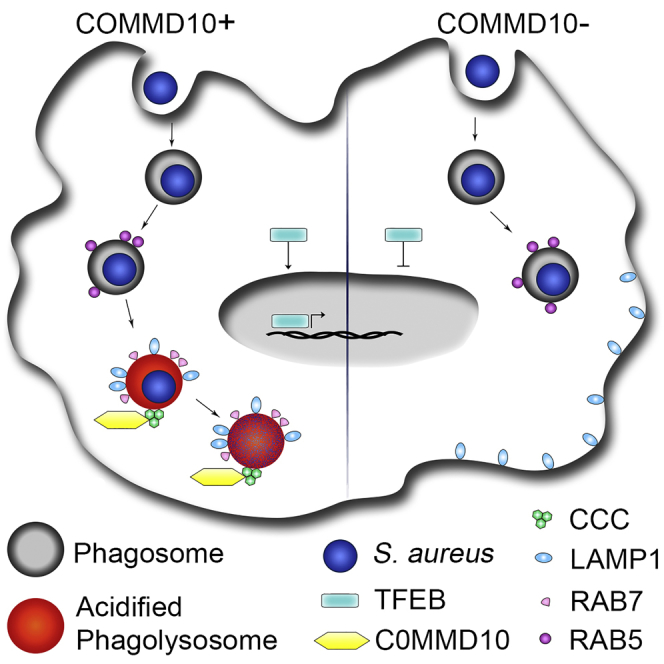
Graphical Abstract
Highlights
-
•
COMMD10 facilitates timely clearance of S. aureus by liver Kupffer cells
-
•
COMMD10 drives transcription program of lysosome biogenesis in infected macrophages
-
•
COMMD10 drives phagolysosomal maturation and acidification in infected macrophages
Biological Sciences; Immunology; Microbiology; Molecular Microbiology
Introduction
Macrophages are immune cells of the myeloid lineage that are strategically positioned in all organs of the body, where they perform tissue-specific functions. A generic macrophage function is to act as immune sentinels in the frontline of tissue defense against infectious invaders, especially at barrier organs, which represent putative entry and colonization sites for pathogens (Varol et al., 2015). Macrophages are armed with a large repertoire of pattern recognition receptors that determine their immunologic and homeostatic potential (Taylor et al., 2005). They are keen in internalizing and destroying invading organisms by trapping them in phagosomes. These structures undergo a sequence of transformations termed phagolysosomal maturation, whereby phagosomal membrane and contents acquire a wide arsenal of microbicidal and lytic features through a strictly choreographed sequence of fusion and fission events with trans-Golgi transport vesicles, endosomes, lysosomes, and autophagosomes (Fairn and Grinstein, 2012, Flannagan et al., 2009).
Staphylococcus aureus is a highly adaptable human pathogen causing significant morbidity and mortality due to both community- and hospital-acquired infections (Lowy, 1998, Magill et al., 2014). The pathogenicity of S. aureus is enhanced by its capacity to cause bacteremia and by the emergence of high-level antimicrobial resistance strains, such as methicillin-resistant S. aureus. Macrophages initiate intracellular microbicidal mechanisms to rapidly kill S. aureus after ingestion. However, when exposed to large inocula of S. aureus, they become progressively exhausted, leading to incomplete phagolysosomal maturation and acidification and to a persisting pool of viable intracellular bacteria (Flannagan et al., 2016, Jubrail et al., 2016). Enabled by a variety of virulence factors and an intricate network of regulators, a few S. aureus survive within phagocytes and disseminate to non-phagocytic cells leading to tissue destruction and persistence of infection (Horn et al., 2018, Pollitt et al., 2018, Strobel et al., 2016). In particular, liver intravascular Kupffer cells (KCs) are primarily responsible for the initial sequestration and killing of circulating S. aureus within minutes. However, a minority of the staphylococci can sometimes overcome KC antimicrobial defenses, survive and proliferate within this intracellular niche, and ultimately escape to colonize other tissues (Pollitt et al., 2018, Surewaard et al., 2016).
Macrophage phagocytic activities must be tightly regulated to allow for rapid escalation in case of S. aureus invasion. Therefore it is essential to define the regulatory pathways that shape the macrophage innate immune responses to S. aureus infection. The COMMD family includes 10 evolutionarily conserved proteins that share a highly conserved and unique motif in their carboxyl terminus termed copper metabolism gene MURR1 domain (COMMD), which functions as an interface for protein-protein interactions. Several functions have been linked to members of the COMMD protein family, which can be broadly grouped into two categories: transcription factor regulation and regulation of intracellular cargo-specific trafficking via interaction with components of the endolysosomal pathway. Previous work in human cell lines has established that all COMMD proteins are negative regulators of the nuclear factor (NF)-κB pathway (Burstein et al., 2005, Maine et al., 2007, Mouhadeb et al., 2018). This was further corroborated in transgenic mouse systems, in which targeting of COMMD1 or COMMD10 deficiencies to myeloid cells resulted in increased NF-κB activation and subsequent exacerbation of lipopolysaccharide (LPS)-induced sepsis and dextran sodium sulfate-induced colitis (Li et al., 2014, Mouhadeb et al., 2018). Additional transcriptional programs that are controlled by COMMD1 include hypoxia-inducible factor (HIF) and E2F1 (Muller et al., 2009, Murata et al., 2017, van de Sluis et al., 2010). Emerging findings also indicate that COMMD proteins play important roles in tuning intracellular signaling and protein trafficking pathways that are highly relevant to macrophage bactericidal activity. In particular, studies in human cells revealed that COMMD proteins are essential components of the COMMD/CCDC22/CCDC93 (CCC) protein complex (Li et al., 2015, Phillips-Krawczak et al., 2015, Starokadomskyy et al., 2013). This complex interacts with the Wiscott-Aldrich and Scar Homolog (WASH) complex (Bartuzi et al., 2016, Phillips-Krawczak et al., 2015), which is intimately linked to bacterial phagocytosis (Buckley et al., 2016) and subsequent lysosomal maturation (King et al., 2013). We have recently shown in human embryonic kidney (HEK) cells that COMMD10 binds the CCDC22 component of the CCC complex (Starokadomskyy et al., 2013). Moreover, COMMD10 has been identified by proteomics to be associated with phagosomes in murine macrophages (Dill et al., 2015). Both these features of COMMD10 prompted us to study its possible involvement in mediating macrophage immune response to S. aureus infection.
Here we report the establishment of transgenic COMMD10 conditional knockout mice allowing the targeting of its deficiency to myeloid cells and specifically macrophages. We show that COMMD10 in macrophages propagates phagolysosome maturation and function to facilitate adequate killing of S. aureus bacteria.
Results
COMMD10 Deficiency Impairs KC-Governed Elimination of S. aureus in the Liver
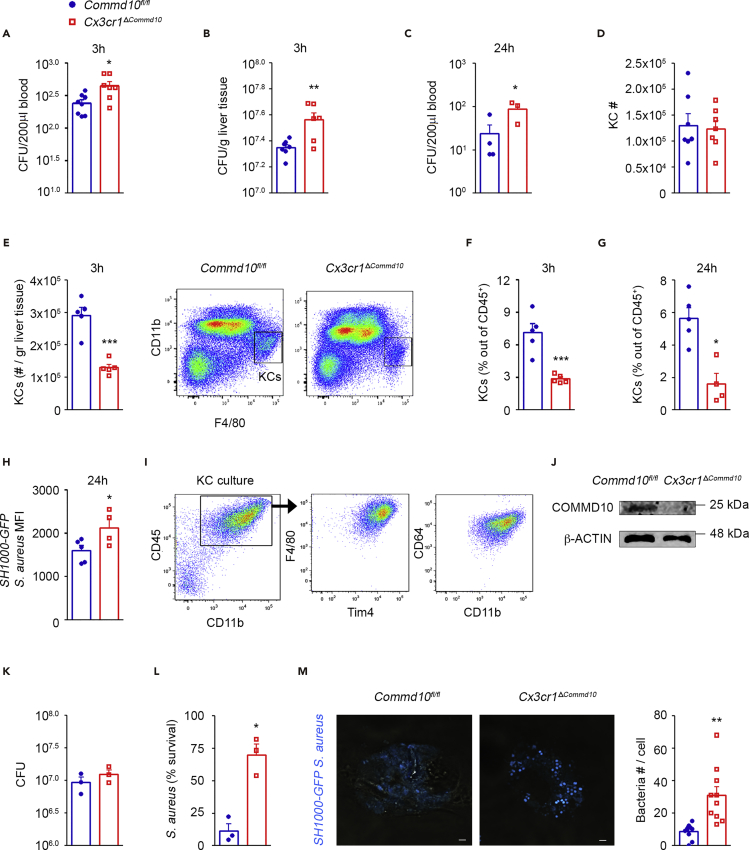
Liver-resident macrophages, namely, KCs, are the key cell population responsible for eliminating S. aureus from the circulation (Surewaard et al., 2016). Hence, we first examined in vivo whether S. aureus handling is influenced by COMMD10 deficiency in KCs. The study of COMMD10 immunoregulation in macrophages has been hampered by embryonic lethality of COMMD10 knockout mice. To overcome this obstacle, we generated transgenic mice that allow specific targeting of COMMD10 deficiency to macrophages by crossing Commd10fl/fl mice (Mouhadeb et al., 2018) with Cx3cr1cre mice (Yona et al., 2013). In these mice, cre-driven recombination (and the ensuing COMMD10 deficiency) is largely restricted to tissue-resident macrophages, and specifically in the liver to KCs (Yona et al., 2013), as well as to recently defined CX3CR1+ liver capsular macrophages (LCMs) (Sierro et al., 2017) and a subset of dendritic cells (David et al., 2016). The resulting Cx3cr1ΔCommd10 mice exhibited normal birth rate and life expectancy (data not shown). Data mining into gene expression databases of sorted steady-state KCs (Zigmond et al., 2014) and LCMs (Sierro et al., 2017) confirmed the expression of Commd10 in both subsets (Figure S1A). Cx3cr1ΔCommd10 mice infected with GFP-tagged SH1000 strain of S. aureus showed a higher burden of bacteria in the blood (Figure 1A) and liver (Figure 1B) 3 h post intravenous inoculation. The increased bacteremia persisted at 24 h post infection (Figure 1C). We next focused on KCs in light of their key role in handling of circulating S. aureus infection. Despite similar abundance of KCs at baseline (Figure 1D), KC numbers were decreased in Cx3cr1ΔCommd10 mice following infection (Figure 1E), and accordingly, the KC fraction out of liver CD45+ leukocytes was lower at 3 and 24 h (Figures 1F and 1G). Looking at the bacterial signal from the remaining KCs, a higher mean fluorescent intensity (MFI) of GFP-tagged S. aureus was observed in infected Cx3cr1ΔCommd10 versus Commd10fl/fl KCs (Figure 1H).
Figure 1.
Impaired Clearance of S. aureus Infection by COMMD10-Deficient KCs
(A–H) Commd10fl/fl (blue, closed circles) or Cx3Cr1ΔCommd10 (red, open squares) mice were intravenously injected with SH1000-GFP S. aureus (5 × 107 CFU per animal). (A and C) Blood and (B) liver specimens were extracted at 3 h (n ≥ 7) and 24 h (n ≥ 3) following S. aureus injection. Colony-forming units (CFU) were determined per 0.2 mL of blood (A and C) or normalized to liver tissue mass (B). (D) Flow cytometry-based assessment of liver-resident KC numbers at steady state (n = 7 from a single experiment). (E) Assessment of liver-resident KC numbers at 3 h (n = 5). Right panel, flow cytometry gating strategy of KCs. (F and G) Assessment of KC fraction out of total CD45+ immune cells at 3 h (F) and at 24 h (G) following S. aureus injection (n = 5). (H) SH1000-GFP S. aureus signal intensity in KCs as depicted by MFI (n = 5).
(I–M) Liver KCs isolated from Commd10fl/fl and Cx3Cr1ΔCommd10 mice were infected with SH1000-GFP or untagged S. aureus at MOI = 5 (n ≥ 3 mice). (I) Flow cytometry images showing isolated KC purity. (J) Immunoblots demonstrating the expression of COMMD10. β-Actin was used as a control (n = 2 biological repeats , each composed of a pool from four mice). (K) CFU of lysed KCs at 30 min following S. aureus injection. (L) Survival of S. aureus in KCs at 2 h following injection assessed by gentamycin protection assay. (M) Confocal microscopic images showing accumulation of SH1000-GFP S. aureus (cyan) at 1 h post infection. Magnification, ×40; scale bar, 2 μM (n ≥ 10 imaged cells per group [n ≥ 3]). Right panel: quantification of bacteria in confocal imaging.
Data in (A), (B), (D–F), and (M) were analyzed by unpaired, two-tailed t test, and data in (C), (G), (H), (K), and (L) were analyzed by non-parametric Mann-Whitney test, comparing each time between Commd10fl/fl and Cx3cr1ΔCommd10 groups. Results are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001
Cx3cr1ΔCommd10 mice infected for 24 h with the untagged Rosenbach strain of S. aureus exhibited increased hepatic damage, as manifested by more abundant and extended subcapsular and parenchymal necrotic lesions (Figures S1B and S1C) and increased serum levels of the liver enzymes alanine and aspartate aminotransferases (Figure S1D). Increased bacteremia (Figure S1E), weight loss (Figure S1F), and serum creatine phosphokinase levels (Figure S1G) further supported aggravated systemic disease in the Cx3cr1ΔCommd10 mice. Similar to infection with SH1000 S. aureus strain, there were significantly reduced numbers and fraction of KCs in the Cx3cr1ΔCommd10 livers, concomitantly with a dramatic increase in the infiltration of Ly6Chi monocytes, but not of neutrophils (Figures S1H and S1I). The prevalence of late apoptotic PI+AnnexinV+ KCs was increased as well in the Cx3cr1ΔCommd10 livers (Figure S1J). Together, these findings suggest increased death of COMMD10-deficient KCs associated with an increased bacterial burden.
To directly test for a role of COMMD10 in KC bacterial handling, we next challenged primary KC isolated from Cx3cr1ΔCommd10 versus Commd10fl/fl mice with S. aureus. KC enrichment was confirmed by combined expression of the lineage-specific markers F4/80, Tim4, and CD64 (FcγR) (Figure 1I), and efficient deficiency of COMMD10 protein was evident in Cx3cr1ΔCommd10 KC (Figure 1J). Cx3cr1ΔCommd10 KCs exhibited similar internalization of S. aureus at 30 min post infection (Figure 1K). However, their clearance of S. aureus was dramatically impaired at 2 h following infection (Figure 1L). Confocal microscopy further validated the increased accumulation of GFP-tagged S. aureus in the COMMD10-deficient KCs (Figure 1M). Together, these findings show that COMMD10 in KC plays an important role in the liver barrier function against invading S. aureus.
COMMD10 Is Essential for Timely Clearance of S. aureus by Macrophages
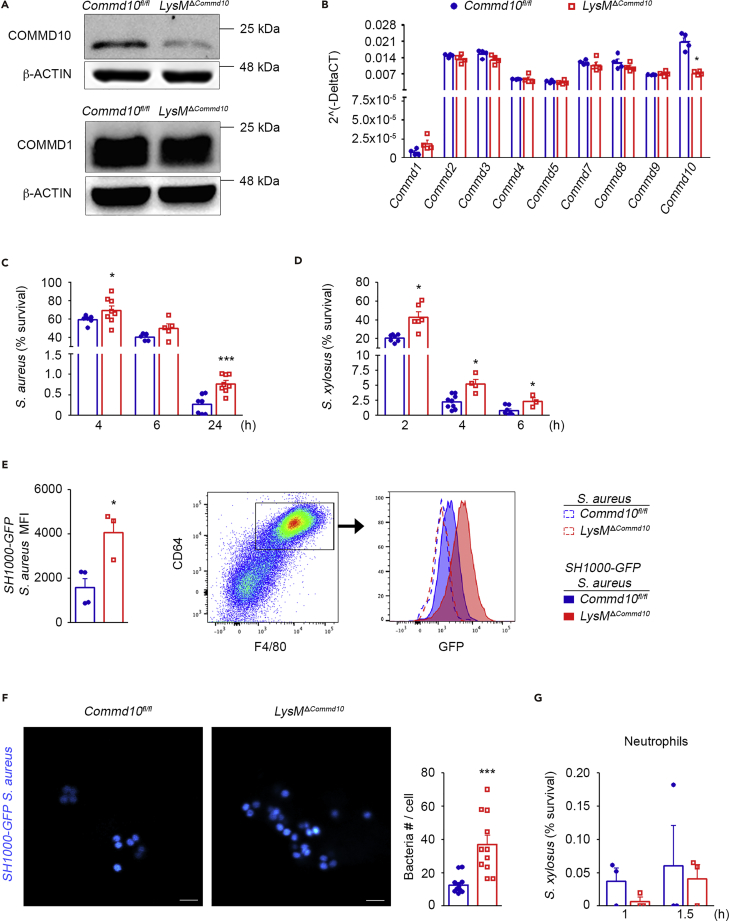
Given the scarcity of cells achieved in KC cultures, we next chose bone marrow (BM)-derived macrophages (BMDM) as a prototype population of primary macrophages for our studies. BMDM were generated from LysMΔCommd10 mice in which COMMD10 deficiency has been targeted to myeloid cells by crossing Lyz2cre mice with Commd10fl/fl mice. Efficient deficiency of COMMD10 protein, but not of COMMD1, was evident in LysMΔCommd10 BMDM infected with S. aureus (Figure 2A). At the transcription level, the LysMΔCommd10 BMDM exhibited specific reduction in Commd10 expression, whereas the expression of all other Commd genes was not significantly altered (Figure 2B). BMDM from Commd10fl/fl or LysMΔCommd10 mice were infected with S. aureus or Staphylococcus xylosus for 30 min, and their intracellular levels were then evaluated at distinct time points using the gentamycin protection assay followed by seeding and colony-forming unit (CFU) counting. Similarly to COMMD10-deficient KCs (Figures 1L and 1M), LysMΔCommd10 BMDM exhibited significantly delayed clearance of S. aureus over time as manifested by increased CFU at 4, 6, and 24 h following infection (Figure 2C). Comparable results were obtained following S. xylosus infection showing impaired clearance at 2, 4, and 6 h following infection (Figure 2D). These results were further corroborated by flow cytometry revealing increased MFI at 2 h following infection with GFP-tagged S. aureus (Figure 2E). Confocal microscopy further demonstrated increased accumulation of GFP-tagged S. aureus bacteria in phagosomal compartments of COMMD10-deficient macrophages at 4 h following infection (Figure 2F). In BM-derived neutrophils, COMMD10 deficiency had no clear effect on the handling of S. xylosus (Figure 2G), suggesting that COMMD10 does not play a role in neutrophil-mediated bactericidal activity against staphylococci. Collectively, these findings highlight that COMMD10 is required for the timely elimination of intracellular S. aureus infection in macrophages.
Figure 2.
Impaired Clearance of S. aureus in COMMD10-Deficient Macrophages
BMDM from Commd10fl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with S. aureus or S. xylosus at MOI = 5.
(A) Immunoblots showing the expression of COMMD10 and COMMD1 proteins. β-Actin was used as a control (n = 2).
(B) qRT-PCR gene expression of various COMMD members (n = 3).
(C and D) Survival over time in BMDM of (C) S. aureus (n ≥ 5) and (D) S. xylosus (n ≥ 3), as assessed by gentamycin protection assay.
(E) Left panel: average mean fluorescent intensity (MFI, calculated by subtracting background MFI of BMDM infected with wild-type S. aureus from MFI of BMDM infected with SH1000-GFP S. aureus) at 2 h post infection (n ≥ 3). Right panel: flow cytometry image showing gating strategy of BMDM (defined as CD64+F4/80+) and a histogram showing overlay of GFP fluorescence in Commd10fl/fl (blue) or LysMΔCommd10 (red) BMDM at 2 h following infection with non-labeled S. aureus (dashed lines) or SH1000-GFP S. aureus (filled lines).
(F) Confocal microscopic images showing internalized GFP-labeled S. aureus (cyan) at 4 h post infection. Magnification, ×40; scale bar, 2 μM (n ≥ 10 imaged cells per group [n ≥ 3]). Right: graph showing bacterial quantification.
(G) Survival of S. xylosus over time in BM-derived neutrophils as assessed by gentamycin protection assay (n = 4).
Data in (B), (E), and (G) were analyzed by non-parametric Mann-Whitney test, and data in (C–D) and (F) were analyzed by unpaired, two-tailed t test comparing each time between Commd10fl/fl and LysMΔCommd10. Results are presented as mean ± SEM with significance: *p < 0.05 and ***p < 0.001.
COMMD10 Does Not Play a Role in Macrophage-Mediated Internalization of S. aureus
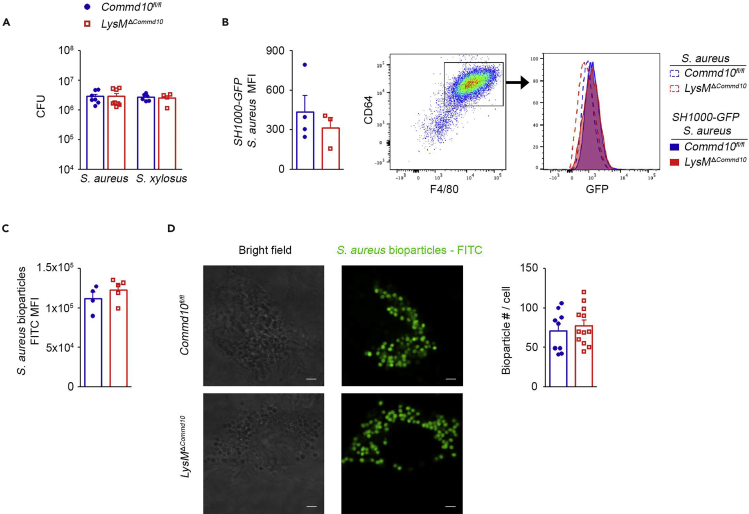
Phagocytosis is a hallmark of anti-bacterial host defense. The attenuated clearance of S. aureus in COMMD10-deficient macrophages in vivo (Figure 1) and in vitro (Figure 2) may be the outcome of its impaired internalization. To study the involvement of COMMD10 in macrophage-governed phagocytosis of staphylococci, BMDM from Commd10fl/fl or LysMΔCommd10 mice were infected with S. aureus or S. xylosus for 30 min, and their intracellular levels were immediately evaluated using the gentamycin protection assay followed by seeding and CFU counting. COMMD10-deficient and COMMD10-proficient BMDM exhibited similar intracellular bacterial counts of both S. aureus and S. xylosus (Figure 3A). In addition, flow cytometry analysis revealed that COMMD10 deficiency had no effect on the MFI of internalized GFP-tagged S. aureus (Figure 3B) or engulfed fluorescently labeled S. aureus-coated bioparticles (Figure 3C). Similar internalization of S. aureus-coated bioparticles was also corroborated by confocal microscopy analysis (Figure 3D). Hence, these results suggest that COMMD10 does not play a critical role in mediating S. aureus internalization.
Figure 3.
COMMD10 Deficiency in Macrophages Does not Affect S. aureus Internalization
BMDM from Commd10fl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with S. aureus or S. xylosus at MOI = 5 for 30 min.
(A) Baseline internalization and survival at 30 min post infection of S. aureus (n = 8) and S. xylosus (n = 6), as assessed by gentamycin protection assay.
(B) Average mean fluorescent intensity (MFI, calculated by subtracting background MFI of BMDM infected with WT S. aureus from MFI of BMDM infected with SH1000-GFP S. aureus) at 30 min post infection (n ≥ 3). Right panel: flow cytometry image showing gating strategy for BMDM (defined as CD64+F4/80+) and a histogram showing overlay of GFP fluorescence in Commd10fl/fl (blue) or LysMΔCommd10 (red) BMDM at 30 min following infection with non-labeled S. aureus (dashed lines) or SH1000-GFP S. aureus (filled lines).
(C) Average MFI of fluorescein-coated S. aureus bioparticles after 3-h incubation (n = 5).
(D) Confocal microscopy images showing internalized fluorescein-coated S. aureus bioparticles (green) at 3 h post incubation. Magnification, ×40; scale bar, 2 μM (n ≥ 10 cells per group). Right: graph showing bacterial quantification.
Data were analyzed by non-parametric Mann-Whitney test, comparing each time between Commd10fl/fl and LysMΔCommd10, and are presented as mean ± SEM.
Lysosomal Biogenesis Is Impaired in S. aureus-Infected COMMD10-Deficient Macrophages
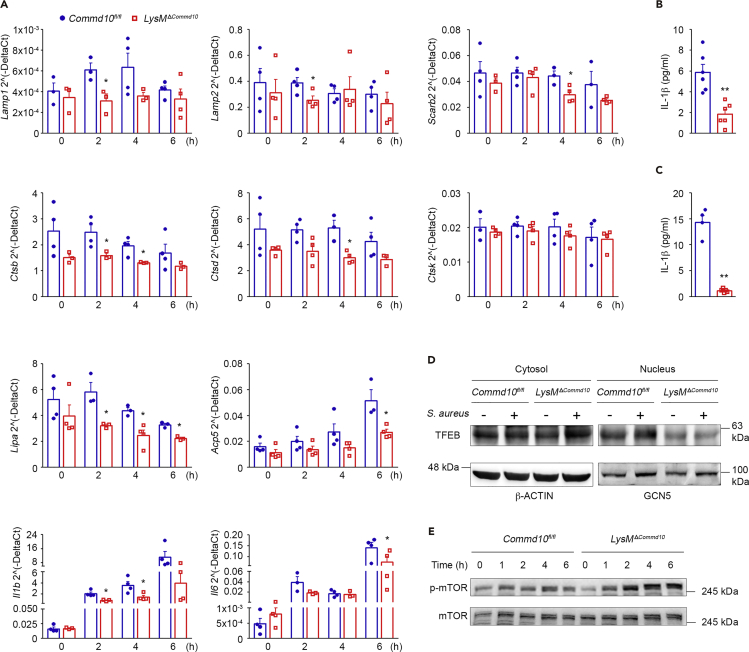
Phagolysosome-mediated cellular clearance processes require the concerted action of hydrolases, the acidification machinery, and membrane proteins. The expression and activity of these components must be coordinated to allow optimal phagolysosomal function during infection and relies on transcriptional regulation of a lysosomal gene network (Settembre et al., 2013). We hypothesized that the attenuated S. aureus clearance in COMMD10-deficient macrophages may be due to impaired lysosomal biogenesis. Indeed, gene expression profiling of BMDM at distinct time points following S. aureus infection uncovered altered expression of structural and functional lysosomal genes, whereas no significant differences were observed at baseline (Figure 4A). Accordingly, late phagosomes and lysosomes are normally enriched in lysosomal-associated membrane proteins (LAMP) 1 and 2 as well as lysosome membrane protein 2 (LIMP2, Scarb2) (Flannagan et al., 2009). COMMD10 deficiency resulted in significantly reduced expression of both Lamp1 and Lamp2 at 2 h and of Scarb2 at 4 h following infection (Figure 4A). With respect to functional lysosomal genes, expression of cathepsins B and D (Ctsb and Ctsd, respectively) was reduced in COMMD10-deficient BMDM at 2 and 4 h following S. aureus infection, whereas cathepsin K (Ctsk) expression remained unchanged (Figure 4A). Gene expression of lysosomal acid lipase A (Lipa) was markedly lower at 2, 4, and 6 h following infection (Figure 4A). Acid phosphatase 5 is a lysosomal di-iron protein important for the clearance of S. aureus infection by mononuclear phagocytes (Bune et al., 2001). Its gene expression was also profoundly reduced at 6 h following infection (Figure 4A). Importantly, BMDM also exhibited reduced gene expression of the inflammatory cytokines interleukin (IL)-1β (Il1b) and IL-6 (Figure 4A). In agreement with this, supernatants from S. aureus- and S. xylosus-infected BMDM contained lower levels of secreted IL-1β (Figures 4B and 4C).
Figure 4.
S. aureus-Infected COMMD10-Deficient Macrophages Exhibit Impaired TFEB-Associated Lysosomal Biogenesis
BMDM from Commd10fl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with S. aureus at MOI = 5 (A and B) or 10 (D and E) or with S. xylosus at MOI = 5 (C). (A) qRT-PCR gene expression over time of lysosomal and inflammatory genes (n = 4). (B–C) Cell-free supernatants were subjected to ELISA analysis of IL-1β following (B) S. aureus and (C) S. xylosus infection (n = 6 for B, n ≥ 4 for C). (D) Immunoblots showing TFEB expression in cytosolic and nuclear fraction lysates of non-infected or S. aureus-infected BMDM. β-Actin and GCN5 antibodies, respectively, were used as controls (n ≥ 5). (E) Immunoblots demonstrating the expression of phospho-mTOR and mTOR in BMDM lysates over a time course post S. aureus infection (n ≥ 3). Data in (A) and (C) were analyzed by non-parametric Mann-Whitney test and data in (B) were analyzed by unpaired, two-tailed t test, comparing each time between Commd10fl/fl and LysMΔCommd10 BMDM. Results are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01.
The transcription factor EB (TFEB) is a master inducer of lysosomal biogenesis (Sardiello et al., 2009). Phagocytosis enhances lysosomal and bactericidal activity in macrophages by activating TFEB translocation from the cytosol to the nucleus to facilitate a boost of the lysosome gene network (Gray et al., 2016). In macrophages infected with S. aureus, TFEB also induces the transcription of pro-inflammatory cytokines (Visvikis et al., 2014). In accordance with the reduced expression of lysosomal genes and of the cytokines IL-1β and IL-6 (Figure 4A), TFEB translocation to the nucleus was impaired in COMMD10-deficient versus COMMD10-proficient macrophages (Figure 4D). Moreover, activation of the master growth regulator mammalian target of rapamycin (mTOR) complex 1 (mTORC1), which inhibits TFEB nuclear translocation (Settembre et al., 2012), was higher in COMMD10-deficient macrophages at 2, 4 and 6 h following S. aureus infection as manifested by increased phosphorylation (Figure 4E). In aggregate, these results indicate a role for COMMD10 in promoting TFEB activation and associated lysosomal biogenesis.
Retarded Lysosomal Maturation in S. aureus-Infected COMMD10-Deficient Macrophages
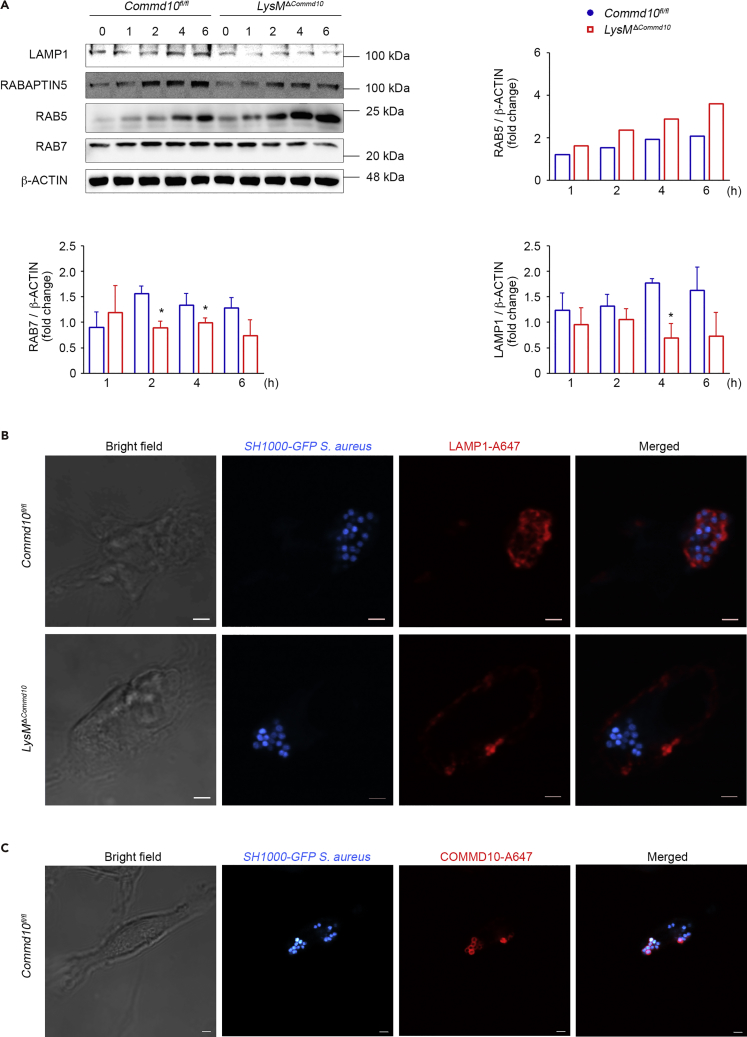
The nascent phagosome acquires the microbicidal and degradative capacities required for pathogen elimination by a process called phagosome maturation (Fairn and Grinstein, 2012, Flannagan et al., 2009). The attenuated clearance of S. aureus in COMMD10-deficient KCs (Figure 1) and macrophages (Figure 2) together with the impaired lysosomal biogenesis (Figure 4) may indicate defective phagolysosomal maturation. The Rab-family GTPases are known to mediate the progression between the early, late, and lysosome fusion stages. Specifically, Rab5 is a characteristic marker of the early phagosome that drives the transition to the late phagosome stage, which is defined by the presence of Rab7 (Harrison et al., 2003, Vieira et al., 2003). The expression of RAB5 was profoundly higher in COMMD10-deficient macrophages at 2 and 4 h following infection, whereas RAB7 protein was significantly reduced (Figure 5A). In addition, there was a decline in the protein expression of Rabaptin-5 (Figure 5A), which is an essential and rate-limiting component for early endosome fusion (Horiuchi et al., 1997, Lippe et al., 2001). The expression of LAMP1, a characteristic marker of late phagosomes and phagolysosomes (Flannagan et al., 2009), was also significantly reduced at 4 h following infection (Figure 5A). Of note, there was no clear difference in the expression of LAMP1, RAB7, and RAB5 at the basal level (Figure S2), suggesting that COMMD10 is not involved with their homeostatic regulation. Confocal microscopic imaging at 1 h following S. aureus infection of macrophages further revealed that the recruitment of LAMP1 to bacteria containing late phagosomes or phagolysosomes depends on COMMD10. Accordingly, whereas in COMMD10-proficient macrophages LAMP1 enveloped S. aureus-containing phagolysosomes, it was mostly mislocalized to the cell surface membrane in the COMMD10-deficient macrophages (Figure 5B). Similarly to LAMP1, staining for COMMD10 localized its expression to the S. aureus-containing phagolysosomal membrane (Figure 5C). Altogether, these results suggest that COMMD10-deficient phagosomes are detained at an early maturation phase.
Figure 5.
Impaired Lysosomal Maturation in S. aureus-Infected COMMD10-Deficient Macrophages
BMDM from Commdfl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with (A) untagged or (B) SH1000-GFP S. aureus at MOI = 5.
(A) Immunoblots showing the expression of indicated phagolysosome proteins. β-Actin served as control. Densitometry is presented on the right and bottom panels (n ≥ 3).
(B) Confocal microscopic images showing co-localization of SH1000-GFP S. aureus (cyan) and LAMP-1(red) at 1 h post infection. Magnification, ×100; scale bar, 2 μM.
(C) Confocal microscopy images showing COMMD10 staining (red) together with internalized S. aureus (cyan). Magnification, ×40; scale bar, 2 μM (n ≥ 10 cells per group).
Data were analyzed by unpaired, two-tailed t test, comparing each time between Commd10fl/fl and LysMΔCommd10 BMDM. Results are presented as mean ± SEM with significance: *p < 0.05.
COMMD10-Deficient Macrophages Exhibit Impaired Phagolysosomal Acidification in Response to S. aureus Infection
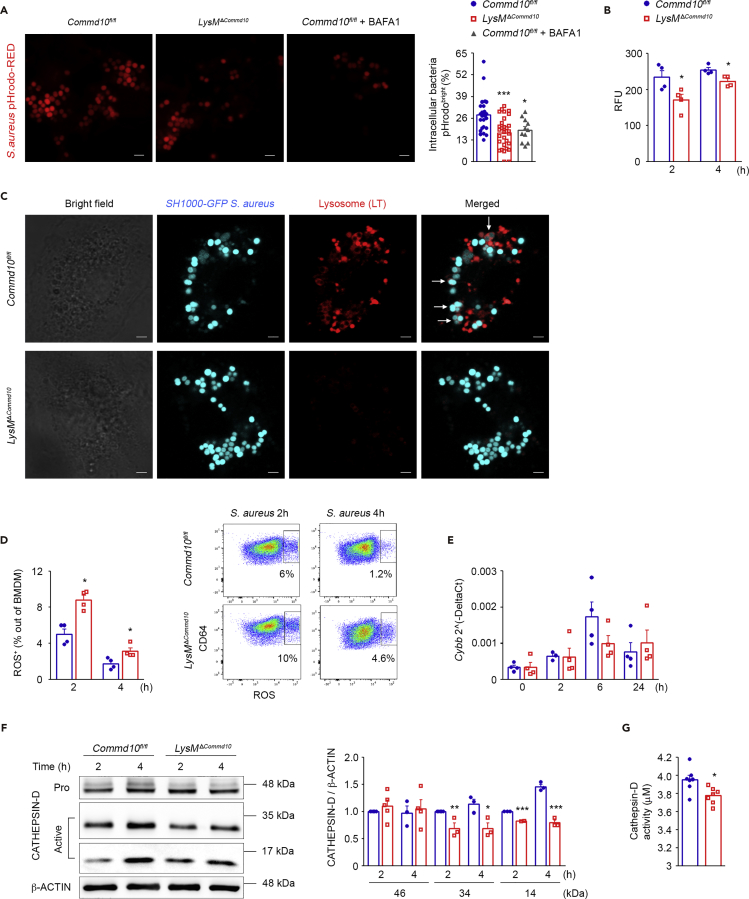
The attenuated phagolysosomal maturation in S. aureus-infected COMMD10-deficient macrophages (Figure 5) may lead to impaired acquisition of bactericidal features. Concomitant with phagosome maturation is the progressive acidification of the phagosome lumen, which aids the killing and digestion of intracellular S. aureus in macrophages (Jubrail et al., 2016). Confocal microscopic analysis of macrophages at 1 h following infection with S. aureus conjugated with the pH-sensitive pHrodo indicator revealed a significantly reduced fraction of pHrodobright phagolysosomes out of total phagolysosomes in the COMMD10-deficient versus COMMD10-proficient macrophages (Figure 6A). Comparable results were achieved with GFP-tagged S. aureus (data not shown). The fraction of pHrodobright S. aureus-containing phagolysosomes was similar to that of Commd10fl/fl macrophages pretreated with the V-ATPase inhibitor bafilomycin A (Figure 6A). This was confirmed by the reduced pHrodo signal in the COMMD10-deficient macrophages at 2 and 4 h following infection (Figure 6B). Labeling with LysoTracker further consolidated the reduction in acidic organelles in the COMMD10-deficient macrophages at 1 h following S. aureus infection (Figure 6C). Activation of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) and the resulting reactive oxygen species (ROS) production neutralizes the acidification of the phagolysosome (Hampton et al., 1998, Savina et al., 2006, Segal et al., 1981). Therefore the impaired phagosome acidification in S. aureus-infected COMMD10-deficient macrophages may be the result of increased NOX activity. Indeed, assessment of ROS production revealed increased fraction of ROS+ macrophages at 2 and 4 h following S. aureus infection (Figure 6D). As NOX-2 complex is a key driver of ROS production (Panday et al., 2015), we next assessed the expression of the cytochrome b (Cybb) subunit following S. aureus infection. Despite the increased production of ROS in COMMD10-deficient BMDM, their expression of Cybb was similar to that of Commd10fl/fl macrophages in steady state and at distinct time points following S. aureus infection (Figure 6E). Acidification is a prerequisite for the function of most lysosomal hydrolases, such as cathepsin D (Sturgill-Koszycki et al., 1996). In the absence of COMMD10, S. aureus-infected macrophages displayed reduced levels of activated cathepsin D (Figure 6F) and attenuated cathepsin D activity (Figure 6G). Altogether, these results uncover that COMMD10 deficiency in macrophages impairs phagolysosomal acidification and function in response to S. aureus infection.
Figure 6.
COMMD10-Deficient Macrophages Exhibit Impaired Lysosomal Killing Machinery upon S. aureus Infection
BMDM from Commd10fl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with pHrodo-labeled (red) (A and B) or GFP-labeled (cyan) (C) S. aureus at MOI = 20 or unlabeled (D–F) S. aureus at MOI = 5. (A) Confocal microscopic images showing intracellular bacteria at 1 h post infection, pHrodo-positive bacteria are bright red. BafilomycinA (BAFA1) was used as positive control in Commd10fl/fl BMDM. Magnification, ×63; scale bar, 2 μM (n ≥ 30 cells per group). Right panel, pHrodobrightS. aureus per cell are presented as percentage of total intracellular bacteria per cell. Data are derived from two independent experiments. (B) Relative fluorescence unit (RFU) of pHrodo-labeled S. aureus at indicated time points post infection as assessed by fluorescent plate reader (n = 4). (C) Confocal microscopic images showing co-localization (as marked by white arrows) of intracellular bacteria (cyan) with acidic compartments (stained with LysoTracker, red) at 1 h post infection. Magnification, ×63; scale bar, 2 μM (n ≥ 30 cells per group). (D) Fraction of ROS+ BMDM at 2 and 4 h post infection, as assessed by flow cytometry using DCFH-DA (n = 4). Right panel, representative flow cytometry images showing ROS fluorescence. (E) Cybb expression at baseline and following S. aureus infection as determined by qRT-PCR (n = 4). (F) Left panel: immunoblots showing expression of pro- (46 kDa) and activated (34 and 14 kDa) cathepsin D over time. β-Actin was utilized as control (n = 5 for 34 and 46 kDa, n = 3 for 14 kDa). Densitometry is presented on the right. (G) Cathepsin D-degrading activity (HiLyte Fluor-488 μM) 2 h post infection as assessed by fluorescent plate reader (n = 7). Data in (A) were analyzed by one-way ANOVA with Tukey post test, comparing between Commd10fl/fl and LysMΔCommd10 or Commd10fl/fl + BAFA1. Data in (B), (D), and (E) were analyzed by non-parametric Mann-Whitney test and data in (F–G) were analyzed by unpaired, two-tailed t test, comparing each time between Commd10fl/fl and LysMΔCommd10 BMDM. Results are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001.
COMMD10-Deficient Macrophages Display Reduced Stability of the CCC Complex
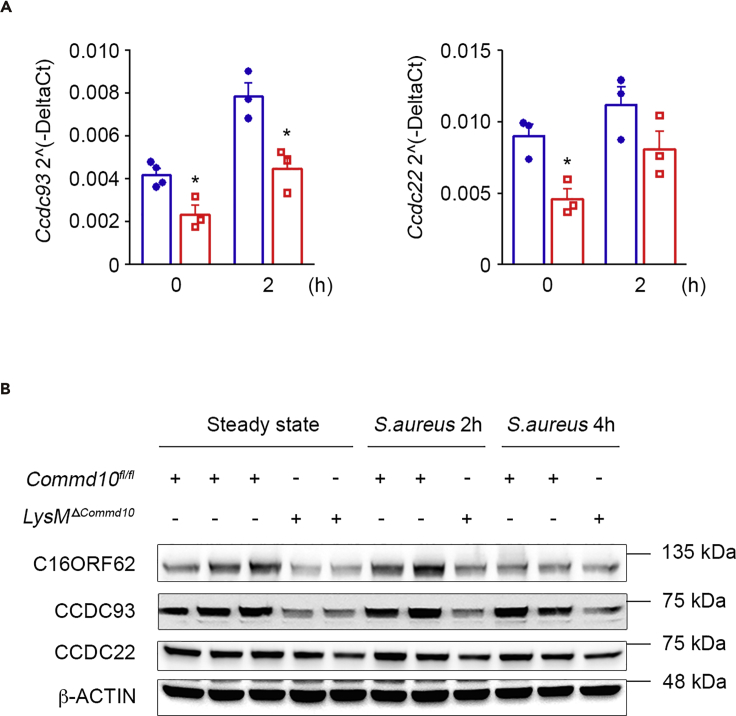
Different COMMD proteins were found, in human cells, to be part of the CCC protein complex, which facilitates intracellular cargo-specific trafficking via interaction with the endolysosomal system (Li et al., 2015, Phillips-Krawczak et al., 2015, Starokadomskyy et al., 2013). In particular, CCC interacts with the WASH complex (Bartuzi et al., 2016, Phillips-Krawczak et al., 2015), which is involved with both bacterial phagocytosis (Buckley et al., 2016) and lysosomal recycling (King et al., 2013). Therefore the impaired phagolysosomal maturation and clearance of S. aureus in COMMD10-deficient macrophages may be related to defects in CCC complex stability. Indeed, analysis of COMMD10-deficient macrophages revealed reduced gene and protein expression of CCDC93 and CCDC22, components of the CCC complex, as well as of the CCC-associated protein C16ORF62, both at steady state and following S. aureus infection (Figures 7A and 7B). Therefore COMMD10-CCC complex interactions may be involved in the trafficking and fusion events required for S. aureus elimination in macrophages.
Figure 7.
COMMD10-Deficient Macrophages Display Reduced Stability of the CCC Complex
BMDM from Commd10fl/fl (blue, closed circles) or LysMΔCommd10 (red, open squares) mice were infected with S. aureus at MOI = 5.
(A) Ccdc93 and Ccdc22 expression at baseline and following S. aureus infection as determined by qRT-PCR (n = 4).
(B) Immunoblots demonstrating expression of CCDC93, CCDC22, and C16ORF62 in BMDM lysates at baseline and post S. aureus infection. β-Actin was utilized as control (n = 2 independent experiments).
Data were analyzed by non-parametric Mann-Whitney test. Results are presented as mean ± SEM with significance: *p < 0.05.
Discussion
We report here by using two different S. aureus subspecies that COMMD10 in macrophages is pivotal for timely handling of the Grampos bacteria S. aureus. We show that COMMD10 is important for the barrier function of KCs against circulating S. aureus. Both COMMD10-deficient primary KCs and BMDM exhibited impaired clearance of S. aureus infection. The failure of KCs to eliminate S. aureus infection in vivo was translated into exacerbated hepatic damage and systemic disease. S. aureus internalization was unaltered by COMMD10 deficiency, whereas phagolysosomal biogenesis and maturation were significantly attenuated. In particular, there was reduced activation of the master lysosomal biogenesis transcription regulator TFEB concomitantly with reduced expression of its associated genes encoding for lysosomal structural and functional proteins. Moreover, there was a delay in the acquisition of mature phagolysosomal markers. COMMD10-deficient macrophages also exhibited reduced phagolysosomal acidification coupled with augmented production of lysosome-neutralizing ROS. Finally, we show reduced representation of the CCC complex genes and proteins in macrophages deficient in COMMD10.
COMMD proteins have been linked to the endolysosomal system. COMMD1, the most studied prototype of this family, functions as part of the CCC complex. Similarly, studies in HEK cells have shown that COMMD10 binds the CCDC22 component of the CCC complex (Starokadomskyy et al., 2013). This complex localizes to endosomes and interacts with the WASH complex (Bartuzi et al., 2016, Phillips-Krawczak et al., 2015). The latter regulates the actin cytoskeleton by modulating Arp2/3-governed actin polymerization in immune cells and thus is important to multiple aspects of immune cell function (Thrasher and Burns, 2010). Specifically, it is intimately linked to bacterial phagocytosis by mononuclear phagocytes, for instance, via its modulation of phagocytic cup formation (Lorenzi et al., 2000, Tsuboi and Meerloo, 2007). Yet, we did not observe an effect for COMMD10 deficiency on S. aureus internalization. WASH is also required for efficient phagolysosomal maturation by governing delivery of lysosomal hydrolases. Moreover, in cells lacking WASH, cathepsin D becomes trapped in a late endosomal compartment, unable to be recycled to nascent phagosomes and autophagosomes (King et al., 2013). We see reduced gene and protein expression of CCC complex members in the COMMD10-deficient BMDM at steady state and following S. aureus infection. These cells also display reduced levels of activated cathepsin D. These findings suggest that COMMD10-CCC-WASH interactions may be required for adequate maturation and recycling of S. aureus containing phagosomes.
TFEB is a master transcription factor that induces transcription of lysosomal biogenesis genes that share a common CLEAR motif such as LAMP1 and 2 and cathepsins (Sardiello et al., 2009). The inhibition of mTOR is required for cytosolic dephosphorylation of TFEB, facilitating its nuclear translocation (Settembre et al., 2012). TFEB activation occurs in response to nutritional shortage leading to autophagy (Settembre et al., 2011), and also in macrophages exposed to LPS or bacterial infection (Gray et al., 2016, Visvikis et al., 2014, Vural et al., 2016). In particular, during S. aureus infection, macrophages with constitutively high TFEB activity have improved survival (Vural et al., 2016). Consistent with the impaired ability of COMMD10-deficient macrophages to clear S. aureus infection, we found that they exhibit repressed TFEB nuclear translocation and suppressed transcription of TFEB-mediated lysosomal genes. Furthermore, TFEB-mediated transcription is not limited to lysosomal biogenesis but is directed at over 400 target gene sites governing, among other processes, endocytosis, autophagy, essential protein degradation, and immune responses (Palmieri et al., 2011). Regarding the latter, in macrophages exposed to S. aureus, TFEB drives the production of inflammatory cytokines essential for adequate host defense such as IL-6 and IL-1β as well as antimicrobial peptides (Visvikis et al., 2014). In line with their repressed TFEB nuclear translocation, COMMD10-deficient macrophages infected with S. aureus had less expression and secretion of inflammatory cytokines, potentially compromising their ability to kill bacteria.
Nascent phagosomes must undergo maturation to acquire microbicidal properties required for elimination of engulfed pathogens (Fairn and Grinstein, 2012, Flannagan et al., 2009). Our results highlight a pivotal role for COMMD10 in timely and proper phagolysosomal maturation following S. aureus infection. Accordingly, COMMD10-deficient macrophages displayed impaired exchange of RAB5 to RAB7 and reduced expression and mislocation of LAMP1 to the cell membrane rather than the phagosomal membrane. Peripheral lysosomes have reduced RAB7 density and are associated with reduced acidification and impaired proteolytic activity when compared with juxtanuclear ones (Johnson et al., 2016). Given the peripheral localization of LAMP1 and reduced RAB7 levels in the COMMD10-deficient BMDM, these results may indicate that their phagolysosomes are less bactericidal.
A hallmark of phagolysosome formation is the marked acidification of the phagosome lumen due to the activity of the vacuolar ATPase (V-ATPase) proton pump (Lukacs et al., 1990). Despite the antimicrobial activity of the macrophage phagolysosome, some S. aureus survive within mature phagolysosomes by virtue of various virulence factors and regulators, where their replication commences before cell death (Flannagan et al., 2016, Flannagan et al., 2018, Pollitt et al., 2018, Surewaard et al., 2016). In fact, it has been recently shown for the S. aureus USA300 strain that its exposure to acidic pH evokes signaling pathways that endow the bacteria with increased resistance to antimicrobial effectors, such as antimicrobial peptides encountered inside macrophage phagolysosomes (Flannagan et al., 2018). Therefore, although the compromised phagolysosomal acidification in COMMD10-deficient macrophages can assist the S. aureus in evading killing, it may in parallel hamper its induction of pH-dependent adaptive survival responses and subsequent replication in these cells. However, replication of S. aureus in macrophages occurs after a significant delay (∼10–12 h) (Flannagan et al., 2016, Surewaard et al., 2016). In contrast, our in vivo data reveal significant reductions in the level of KCs already after 3 h, suggesting that they succumb to the bacteria before their replication commences. Moreover, it remains unclear whether pH-dependent survival mechanisms described for the USA300 S. aureus strain are also a feature of the Rosenbach and SH1000 strains used here. Further studies are required to follow the effect of COMMD10 deficiency on the dynamics of S. aureus survival and replication within KCs.
One explanation for the reduced acidification of phagolysosomal compartments in COMMD10-deficient macrophages is their augmented production of ROS, which neutralize the acidification of phagolysosomes (Hampton et al., 1998, Savina et al., 2006, Segal et al., 1981). However, ROS production is an important mechanism in combating S. aureus. Indeed, patients with chronic granulomatous disease having defective ROS production are prone to recurrent life-threatening staphylococcal infections and persistent inflammation (Buvelot et al., 2017). Although increased ROS production would normally be expected to result in more effective bacterial handling, S. aureus may be an exception, as it is known to express anti-oxidant enzymes, such as catalase and superoxide dismutase, that confer resistance to ROS in macrophages (Das and Bishayi, 2009, Das and Bishayi, 2010). It is still unclear how COMMD10 regulates ROS production. Expression of Cybb was unchanged in COMMD10-deficient macrophages. Although expression of other NOX2 complex subunits should be examined, these results suggest the possibility of transcription-independent mechanisms. Alternatively, given the involvement of COMMD proteins in intracellular protein trafficking events, COMMD10 may be important for controlling the assembly and stability of NOX2 multi-domain complex and its translocation from the cytosol to the membrane.
Although our results underscore an important role for COMMD10 in promoting TFEB-governed lysosomal biogenesis at 2–4 h following S. aureus infection, they also indicate profound alterations in phagolysosomal maturation events occurring at earlier time points, as manifested by impaired recruitment of LAMP1 to phagolysosomes, exchange of RAB5 to RAB7, phagolysosomal acidification, and cathepsin D activity. These alterations seem too rapid to be transcription mediated and may be related to the effect of COMMD10 deficiency on protein trafficking events. Indeed, COMMD10 has been associated with phagosomes in macrophages (Dill et al., 2015), and we show here the recruitment of COMMD10 protein to S. aureus containing phagolysosomes already at 1 h following infection. Moreover, macrophages with COMMD10-deficiency exhibit reduced expression of CCC complex genes and proteins. This complex modulates endolysosome architecture and is required for the correct trafficking of diverse transmembrane proteins that traverse this compartment. Hence impaired COMMD10-CCC interactions may be important for the trafficking events driving phagolysosomal maturation, function, and recycling. Given the persistent survival of S. aureus in macrophages (Flannagan et al., 2016, Flannagan et al., 2018, Jubrail et al., 2016, Pollitt et al., 2018, Surewaard et al., 2016), the process of phagolysosomal biogenesis, maturation, and recycling is dynamic and continuous, therefore necessitating both ongoing transcriptional and protein trafficking regulation by COMMD10.
To target COMMD10 deficiency to KCs in vivo we utilized Cx3cr1cre mice (Yona et al., 2013). In these mice, cre-driven recombination (and the ensuing COMMD10 deficiency) is largely restricted in the liver to KCs (Yona et al., 2013), as well as to CX3CR1+ LCMs (Sierro et al., 2017) and a subset of dendritic cells (David et al., 2016). Here we show that LCMs also express COMMD10, and hence their altered activity in the setting of COMMD10 deficiency may contribute to the overall phenotype observed in the Cx3cr1ΔCommd10 livers. In this regard, it has been shown that KCs are mainly responsible for the clearance of bacteria originating from the circulation, whereas LCMs are in charge of handling pathogens traversing the peritoneum (Sierro et al., 2017). Indeed, circulating S. aureus are rapidly killed by KCs (Pollitt et al., 2018, Surewaard et al., 2016). Therefore, although it remains to be examined, we expect that COMMD10 deficiency in LCMs does not directly impair the clearance of blood-borne S. aureus. LCMs express various chemokines that may be involved in the sequential recruitment of immune cells such as neutrophils and Ly6Chi monocytes (Sierro et al., 2017). Our data reveal increased infiltration of Ly6Chi monocytes to S. aureus-infected Cx3cr1ΔCommd10 livers at 24 h, but the respective contributions of KCs and LCMs to their recruitment remain to be determined. Moreover, it is also not clear whether COMMD10 deficiency in LCMs contributes to the overall increase in hepatocyte necrosis observed at the subcapsular zone of Cx3cr1ΔCommd10 livers. We have previously shown that COMMD10 is important for the negative regulation of inflammasome activity in liver-infiltrating Ly6Chi monocytes during endotoxic shock in Lyz2creCommd10fl/fl mice, but not Cx3cr1ΔCommd10 mice (Mouhadeb et al., 2018). This is probably related to the greater activity of cre-recombinase in the ephemeral circulating Ly6Chi monocytes in the Lyz2cre versus Cx3cr1cre mice (Abram et al., 2014). Therefore, we believe that COMMD10 deficiency in Ly6Chi monocytes does not directly contribute to the overall impaired clearance of S. aureus infection in the Cx3cr1ΔCommd10 livers, but may be involved to some extent with the overall augmented hepatic damage observed after 24 h. Macrophages in the liver mediate the initial infecting S. aureus population bottleneck, whereas neutrophils enable the subsequent spread of bacteria to other organs (Pollitt et al., 2018). We have previously shown that circulating neutrophils hardly express COMMD10 (Mouhadeb et al., 2018) and are a priori not a target for cre-recombination in the Cx3cr1cre mice (Abram et al., 2014). Moreover, neutrophil recruitment to the S. aureus-infected liver was not affected in the Cx3cr1ΔCommd10 mice, and LysMΔCommd10 primary neutrophils exhibited unaltered handling of S. xylosus. Therefore these results rule out a contribution of neutrophils to the impaired clearance of S. aureus and collateral damage in the Cx3cr1ΔCommd10 livers.
Altogether, we report a pivotal role for COMMD10 in macrophages in mediating lysosomal biogenesis and maturation in response to S. aureus infection and in upholding KC barrier function against S. aureus bacteremia.
Limitations of the Study
To target COMMD10 deficiency to KCs in vivo we utilized Cx3cr1cre mice (Yona et al., 2013). Besides KCs, cre-driven recombination (and the ensuing COMMD10 deficiency) also targets liver CX3CR1+ LCMs (Sierro et al., 2017) and a subset of dendritic cells (David et al., 2016). This caveat has been extensively elaborated on in the discussion. Although we cannot exclude a contribution of other liver CX3CR1+ cells to in vivo phenotypes observed in this study, we have complemented our in vivo observations by comprehensive in vitro studies directly characterizing the effects of COMMD10 deficiency on S. aureus handling mechanisms in primary cell cultures of KCs and BMDMs.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors acknowledge support from the Israel Science Foundation (grant # 1777/13) and the Israel Ministry of Health (grant # 10052). This research was also supported by a Grant from the GIF, the German-Israeli Foundation for Scientific Research and Development (grant # 2094/09). Work in the Varol laboratory has been supported by the Azrieli Foundation Canada-Israel.
Author Contributions
S.B.S., O.M., C.V., and N.G. conceived the study, designed experiments, and wrote the manuscript. S.B.S., O.M., and K.C. performed the experiments and analyzed the data. N.G and C.V are co-corresponding authors. Both equally supervised the work and are responsible for all data presented and analyzed.
Declaration of Interests
The authors declare no competing interests.
Published: April 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.03.024.
Contributor Information
Chen Varol, Email: chenv@tlvmc.gov.il.
Nathan Gluck, Email: nathang@tlvmc.gov.il.
Supplemental Information
References
- Abram C.L., Roberge G.L., Hu Y., Lowell C.A. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods. 2014;408:89–100. doi: 10.1016/j.jim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P., Billadeau D.D., Favier R., Rong S., Dekker D., Fedoseienko A., Fieten H., Wijers M., Levels J.H., Huijkman N. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun. 2016;7:10961. doi: 10.1038/ncomms10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C.M., Gopaldass N., Bosmani C., Johnston S.A., Soldati T., Insall R.H., King J.S. WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. Proc. Natl. Acad. Sci. U S A. 2016;113:E5906–E5915. doi: 10.1073/pnas.1524532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bune A.J., Hayman A.R., Evans M.J., Cox T.M. Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disordered macrophage inflammatory responses and reduced clearance of the pathogen, Staphylococcus aureus. Immunology. 2001;102:103–113. doi: 10.1046/j.1365-2567.2001.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E., Hoberg J.E., Wilkinson A.S., Rumble J.M., Csomos R.A., Komarck C.M., Maine G.N., Wilkinson J.C., Mayo M.W., Duckett C.S. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- Buvelot H., Posfay-Barbe K.M., Linder P., Schrenzel J., Krause K.H. Staphylococcus aureus, phagocyte NADPH oxidase and chronic granulomatous disease. FEMS Microbiol. Rev. 2017;41:139–157. doi: 10.1093/femsre/fuw042. [DOI] [PubMed] [Google Scholar]
- Das D., Bishayi B. Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb. Pathog. 2009;47:57–67. doi: 10.1016/j.micpath.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Das D., Bishayi B. Contribution of catalase and superoxide dismutase to the intracellular survival of clinical isolates of Staphylococcus aureus in murine macrophages. Indian J. Microbiol. 2010;50:375–384. doi: 10.1007/s12088-011-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David B.A., Rezende R.M., Antunes M.M., Santos M.M., Freitas Lopes M.A., Diniz A.B., Sousa Pereira R.V., Marchesi S.C., Alvarenga D.M., Nakagaki B.N. Combination of mass cytometry and imaging analysis reveals origin, location, and functional repopulation of liver myeloid cells in mice. Gastroenterology. 2016;151:1176–1191. doi: 10.1053/j.gastro.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Dill B.D., Gierlinski M., Hartlova A., Arandilla A.G., Guo M., Clarke R.G., Trost M. Quantitative proteome analysis of temporally resolved phagosomes following uptake via key phagocytic receptors. Mol. Cell. Proteomics. 2015;14:1334–1349. doi: 10.1074/mcp.M114.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33:397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Cosio G., Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Heit B., Heinrichs D.E. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell. Microbiol. 2016;18:514–535. doi: 10.1111/cmi.12527. [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Kuiack R.C., McGavin M.J., Heinrichs D.E. Staphylococcus aureus uses the GraXRS regulatory system to sense and adapt to the acidified phagolysosome in macrophages. MBio. 2018;9 doi: 10.1128/mBio.01143-18. e01143–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.A., Choy C.H., Dayam R.M., Ospina-Escobar E., Somerville A., Xiao X., Ferguson S.M., Botelho R.J. Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr. Biol. 2016;26:1955–1964. doi: 10.1016/j.cub.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M.B., Kettle A.J., Winterbourn C.C. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92:3007–3017. [PubMed] [Google Scholar]
- Harrison R.E., Bucci C., Vieira O.V., Schroer T.A., Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol. Cell. Biol. 2003;23:6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H., Lippe R., McBride H.M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Horn J., Stelzner K., Rudel T., Fraunholz M. Inside job: Staphylococcus aureus host-pathogen interactions. Int. J. Med. Microbiol. 2018;308:607–624. doi: 10.1016/j.ijmm.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Johnson D.E., Ostrowski P., Jaumouille V., Grinstein S. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 2016;212:677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubrail J., Morris P., Bewley M.A., Stoneham S., Johnston S.A., Foster S.J., Peden A.A., Read R.C., Marriott H.M., Dockrell D.H. Inability to sustain intraphagolysosomal killing of Staphylococcus aureus predisposes to bacterial persistence in macrophages. Cell. Microbiol. 2016;18:80–96. doi: 10.1111/cmi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.S., Gueho A., Hagedorn M., Gopaldass N., Leuba F., Soldati T., Insall R.H. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol. Biol. Cell. 2013;24:2714–2726. doi: 10.1091/mbc.E13-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chan L., Bartuzi P., Melton S.D., Weber A., Ben-Shlomo S., Varol C., Raetz M., Mao X., Starokadomskyy P. Copper metabolism domain-containing 1 represses genes that promote inflammation and protects mice from colitis and colitis-associated cancer. Gastroenterology. 2014;147:184–195.e3. doi: 10.1053/j.gastro.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Koo Y., Mao X., Sifuentes-Dominguez L., Morris L.L., Jia D., Miyata N., Faulkner R.A., van Deursen J.M., Vooijs M. Endosomal sorting of Notch receptors through COMMD9-dependent pathways modulates Notch signaling. J. Cell Biol. 2015;211:605–617. doi: 10.1083/jcb.201505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippe R., Miaczynska M., Rybin V., Runge A., Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi R., Brickell P.M., Katz D.R., Kinnon C., Thrasher A.J. Wiskott-Aldrich syndrome protein is necessary for efficient IgG-mediated phagocytosis. Blood. 2000;95:2943–2946. [PubMed] [Google Scholar]
- Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Lukacs G.L., Rotstein O.D., Grinstein S. Phagosomal acidification is mediated by a vacuolar-type H(+)-ATPase in murine macrophages. J. Biol. Chem. 1990;265:21099–21107. [PubMed] [Google Scholar]
- Magill S.S., Edwards J.R., Fridkin S.K., Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team Survey of health care-associated infections. N. Engl. J. Med. 2014;370:2542–2543. doi: 10.1056/NEJMc1405194. [DOI] [PubMed] [Google Scholar]
- Maine G.N., Mao X., Komarck C.M., Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhadeb O., Ben Shlomo S., Cohen K., Farkash I., Gruber S., Maharshak N., Halpern Z., Burstein E., Gluck N., Varol C. Impaired COMMD10-mediated regulation of Ly6C(hi) monocyte-driven inflammation disrupts Gut barrier function. Front. Immunol. 2018;9:2623. doi: 10.3389/fimmu.2018.02623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., van de Sluis B., Groot A.J., Verbeek D., Vonk W.I., Maine G.N., Burstein E., Wijmenga C., Vooijs M., Reits E. Nuclear-cytosolic transport of COMMD1 regulates NF-kappaB and HIF-1 activity. Traffic. 2009;10:514–527. doi: 10.1111/j.1600-0854.2009.00892.x. [DOI] [PubMed] [Google Scholar]
- Murata K., Fang C., Terao C., Giannopoulou E.G., Lee Y.J., Lee M.J., Mun S.H., Bae S., Qiao Y., Yuan R. Hypoxia-sensitive COMMD1 integrates signaling and cellular metabolism in human macrophages and suppresses osteoclastogenesis. Immunity. 2017;47:66–79.e5. doi: 10.1016/j.immuni.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- Panday A., Sahoo M.K., Osorio D., Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015;12:5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak C.A., Singla A., Starokadomskyy P., Deng Z., Osborne D.G., Li H., Dick C.J., Gomez T.S., Koenecke M., Zhang J.S. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell. 2015;26:91–103. doi: 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt E.J.G., Szkuta P.T., Burns N., Foster S.J. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018;14:e1007112. doi: 10.1371/journal.ppat.1007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Dumenil A.M., Seabra M.C., Raposo G., Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Segal A.W., Geisow M., Garcia R., Harper A., Miller R. The respiratory burst of phagocytic cells is associated with a rise in vacuolar pH. Nature. 1981;290:406–409. doi: 10.1038/290406a0. [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro F., Evrard M., Rizzetto S., Melino M., Mitchell A.J., Florido M., Beattie L., Walters S.B., Tay S.S., Lu B. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity. 2017;47:374–388.e6. doi: 10.1016/j.immuni.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Starokadomskyy P., Gluck N., Li H., Chen B., Wallis M., Maine G.N., Mao X., Zaidi I.W., Hein M.Y., McDonald F.J. CCDC22 deficiency in humans blunts activation of proinflammatory NF-kappaB signaling. J. Clin. Invest. 2013;123:2244–2256. doi: 10.1172/JCI66466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M., Pfortner H., Tuchscherr L., Volker U., Schmidt F., Kramko N., Schnittler H.J., Fraunholz M.J., Loffler B., Peters G. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin. Microbiol. Infect. 2016;22:799–809. doi: 10.1016/j.cmi.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Schaible U.E., Russell D.G. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996;15:6960–6968. [PMC free article] [PubMed] [Google Scholar]
- Surewaard B.G., Deniset J.F., Zemp F.J., Amrein M., Otto M., Conly J., Omri A., Yates R.M., Kubes P. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med. 2016;213:1141–1151. doi: 10.1084/jem.20160334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.R., Martinez-Pomares L., Stacey M., Lin H.H., Brown G.D., Gordon S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- Thrasher A.J., Burns S.O. WASP: a key immunological multitasker. Nat. Rev. Immunol. 2010;10:182–192. doi: 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- Tsuboi S., Meerloo J. Wiskott-Aldrich syndrome protein is a key regulator of the phagocytic cup formation in macrophages. J. Biol. Chem. 2007;282:34194–34203. doi: 10.1074/jbc.M705999200. [DOI] [PubMed] [Google Scholar]
- van de Sluis B., Mao X., Zhai Y., Groot A.J., Vermeulen J.F., van der Wall E., van Diest P.J., Hofker M.H., Wijmenga C., Klomp L.W. COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J. Clin. Invest. 2010;120:2119–2130. doi: 10.1172/JCI40583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Mildner A., Jung S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- Vieira O.V., Bucci C., Harrison R.E., Trimble W.S., Lanzetti L., Gruenberg J., Schreiber A.D., Stahl P.D., Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol. Cell. Biol. 2003;23:2501–2514. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvikis O., Ihuegbu N., Labed S.A., Luhachack L.G., Alves A.F., Wollenberg A.C., Stuart L.M., Stormo G.D., Irazoqui J.E. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural A., Al-Khodor S., Cheung G.Y., Shi C.S., Srinivasan L., McQuiston T.J., Hwang I.Y., Yeh A.J., Blumer J.B., Briken V. Activator of G-protein signaling 3-induced lysosomal biogenesis limits macrophage intracellular bacterial infection. J. Immunol. 2016;196:846–856. doi: 10.4049/jimmunol.1501595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Samia-Grinberg S., Pasmanik-Chor M., Brazowski E., Shibolet O., Halpern Z., Varol C. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J. Immunol. 2014;193:344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.