Abstract
Glioblastoma multiforme (GBM) is a lethal brain tumor with a mean survival time of 1 year. One major reason for therapeutic failure is that GBM cells have an extraordinary capacity to invade normal brain tissue beyond the surgical margin, accounting for the lack of treatment efficacy. GBM cells that can infiltrate into the healthy brain possess tumor properties of stemness and invasion, and previous studies demonstrate that Musashi-1 (MSI1), a neural stem cell marker, plays an important role in the maintenance of stem cell status, cellular differentiation, and tumorigenesis in cancers. By analyzing neuronal progenitor cell markers and stemness genes, we predicted that MSI1 might be an important factor in GBM pathogenesis. Because inflammation aids in the proliferation and survival of malignant cells, the inflammatory microenvironment also promotes GBM invasion, and intercellular adhesion molecule-1 (ICAM1), a member of the immunoglobulin superfamily, is involved in inflammation. Our results indicate that the above phenomena are likely due to MSI1 upregulation, which occurred simultaneously with higher expression of ICAM1 in GBM cells. Indeed, MSI1 knockdown effectively suppressed ICAM1 expression and blocked GBM cell motility and invasion, whereas overexpressing ICAM1 reversed these effects. According to RNA immunoprecipitation assays, MSI1-mediated mRNA interactions promote ICAM1 translation. Finally, immunohistochemical analysis showed MSI1 and ICAM-1 to be coexpressed at high levels in GBM tissues. Thus, the MSI1/ICAM1 pathway plays an important role in oncogenic resistance, including increased tumor invasion, and MSI1/ICAM1 may be a target for GBM treatment.
Introduction
One of the most deadly malignant brain tumors in humans is glioblastoma multiforme (GBM), also known as World Health Organization (WHO) Grade IV malignant glioma. GBM appears to be resistant to all current treatments, including surgery, drug intervention and radiotherapy. This malignant tumor can develop from primary GBM or progress from a low-grade glioma. The median survival of GBM is approximately 15 months, and it is highly recurrent even after surgical intervention, chemotherapy, radiation, and immunotherapy [1]. Several mechanisms of GBM have been previously reported. For example, in primary GBM, almost all cells were found to highly express epidermal growth factor receptor (EGFR) and cyclin-dependent kinase inhibitor 2A (CDKN2A) and show loss of heterozygosity (LOH), thereby repressing the level of phosphatase and tensin homolog (PTEN) gene expression. Moreover, aberrations in platelet-derived growth factor receptor (PDGFR) and Tp53 gene expression levels are usually observed in secondary GBM [2]. In addition, some signaling pathways are reported to modulate GBM biological function, such as PI-3 K, NF-κB, vascular endothelial growth factors (VEGF) or STAT3, which are able to mediate GBM cell growth and cell death [3]. Overall, two major factors underlie the poor clinical outcome of GBM: its inflammatory properties and its aggressive invasion into surrounding normal brain tissue.
Musashi-1 (MSI1), a member of the RNA-binding protein family, is highly expressed in the central nervous system. The primary structure and expression pattern of MSI1 have been reported among nematode, fruit fly and ascidian species [4], [5], [6]. In fruit flies, loss of MSI1 gene function causes asymmetrical division of sensory organ precursor cells, indicating that MSI1 plays an important role in brain organ development [7]. In mammals, MSI1 is conserved as an important marker of neural stem cells or progenitor cells. Okano et al. were the first to prove that MSI1 has multiple functions in regulating cell fate decisions, maintenance of the stem cell state, differentiation and tumorigenesis [8]. Moreover, MSI1 has been identified as a functional modulator that mediates Notch signaling by suppressing translation of m-Numb, the intracellular Notch signal inhibitor, thereby maintaining the self-renewal ability of neural stem cells (NSCs) [9]. The MSI1 protein was also reported to be present in the polysomal fraction and to bind directly to a targeting molecule that interacts with both translation initiation factors and mRNAs, repressing the targeting molecule at the translational level [10]. In addition to translational control, MSI1 is important in terms of revealing the full picture of stem cell modulation. Tcf/Lef and the SOX family maintain the cell renewal ability of stem cells, and Tcf/Lef and Sox family binding sequences are present in conserved regions outside of protein-coding regions [11]. As MSI1 is strongly repressed in human colon cancer, it is recognized as a colon cancer stem cell marker in humans [12]. Furthermore, Sureban et al. showed that MSI1 knockdown leads to tumor regression and promotes radiation-induced apoptosis in colon cancer cells, indicating that MSI1 plays important roles in cancer cell proliferation, apoptosis inhibition and tumorigenesis modulation [13]. Additionally, expression of MSI1 correlates with the grade of malignancy and proliferative activity in gliomas [14], and in vitro neurospheres derived from brain tumors express MSI1 [15].
Intercellular adhesion molecule-1 (ICAM1), a single-chain 76–110-kD glycoprotein, is a member of the immunoglobulin supergene family [16]. ICAM1 expression has been found in several cell types, including leucocytes, fibroblasts, endothelial cells, epithelial cells and cultured human fetal brain astrocytes [17]. ICAM1 is involved in several processes, including (1) inflammatory cell trafficking, (2) leukocyte effector activity, (3) adhesion of antigen-presenting cells to T lymphocytes during antigen presentation, (4) microbial pathogenesis and (5) signal transduction pathways through outside-in signaling events. During the development of inflammation, leukocytes will interact with ICAM1 of the endothelium, allowing them to cross the barrier [18]. Yu et al. revealed that secretory phospholipase A(2) plays a substantial role in mediating the inflammatory signals that induce ICAM1 expression in lung cancer cells and that inhibition of the enzyme can significantly decrease ICAM1 expression and subsequent cancer cell invasion [19]. In addition, Lin et al. showed that thalidomide can suppress ICAM1 expression and inhibit ICAM1-mediated cell invasion and metastasis in lung cancer [20]. In human glioma, irradiation reportedly significantly elevated ICAM1 and soluble-ICAM1 levels, promoting migration and invasion phenotypes [21].
In the present study, we found that ICAM1 induced GBM cells to invade healthy brain tissue. MSI1 also appears to regulate ICAM1, and ICAM1 may serve as a surface marker for invasive GBM cells. To our knowledge, this study is the first to demonstrate the crucial role of MSI1/ICAM1 interaction in GBM cell invasion. These results will contribute to our understanding of the therapeutic significance of targeting the MSI1/ICAM1 interaction in future GBM treatment.
Materials and Methods
Cell Line and Culture Conditions
The human glioblastoma cell lines U87MG, 8901, 8401, and 05MG were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), penicillin (10 IU/ml), and streptomycin (10 μg/ml) (Gibco) under standard culture conditions (37 °C, 95% humidified air, and 5% CO2). Culture medium was refreshed every 2 days, and 100 g/mL G418 (Sigma) was only added in the culture selecting plasmid-transfected cells. All cultures were confirmed for no contamination of mycoplasma using MycoAlert PLUS Mycoplasma Detection Kit (Lonza).
RT-PCR
Total RNA isolated using Trizol reagent (Ambion) was subjected to reverse transcription performed using the SuperScript™ III Reverse Transcriptase (Invitrogen, 18080093). RT-PCRs were performed using SYBR Green Master Mix (Thermo Scientific) on the Step One Plus Real-Time PCR instrument (Applied Biosystems). The following primers were used for RT-PCR: MSI1 (NM_002442.3; sense: 5′-GTCTCGAGTCATGCCCTACG-3′; antisense: 5′-ACACGGAATTCGGGGAACTG-3′, 147 bp), ICAM1 (NM_000201.2; sense: 5′- ACGGAG CTCCCAGTCC TAAT -3′; antisense: 5′-CTCCTTCTGGGGAAAGGCAG-3′, 138 bp), and GAPDH (NM_002 046; sense: 5′- GACAGTCAGCCGCATCTTCT-3′; antisense: 5′-GCGCC CAATACGACC AAATC-3′, 104 bp). GAPDH was included as the reference gene for normalization. The ΔΔCt method was used for quantification analysis. The primers were listed in Supplementary Table 1.
RNA Immunoprecipitation (RIP)
RIP was performed in U87MG and 05MG using anti-FLAG M2 magnetic beads (Sigma, M8823). In brief, cells were cross-linked in 0.1% formaldehyde for 10 minutes prior to harvest and lysis. RIP was performed using the Magna RIP RNA-binding protein immune-precipitation kit (Millipore, 17-700) according to the manufacturer’s instructions. RNA in each eluted sample was reverse-transcribed by using SuperScript™ III Reverse Transcriptase (Invitrogen, 18080093) and analyzed by RT-PCR. Each PCR was performed with HiFi-Kapa PCR master mix (Kapa Biosystems, KK2101) and analyzed by electrophoresis. The 1% mRNA of the sample was used as input; meanwhile, the RIP sample by using IgG was used as negative control. Detection was performed using specific primers, including MSI1 (NM_002442.3; sense: 5′-GCCCAAGATGGTGACTCGAA-3′, antisense: 5′-AGGAATGGCTGTAAGCTCG G-3′, 496 bp), ICAM1 (NM_000201.2; sense: 5′-TTGGGCACTGCTGTCTACTG-3′, antisense: 5′-ATGTCCAGACATGACCGCTG-3′, 438 bp), P21 (NM_000389.4; sense: 5′-AGTCAGTTCCTTGTGGAGCC-3′, antisense: 5′-ATCTGTCATGCTGGTCTGCC-3′, 517 bp), and GAPDH (NM_002 046; sense: 5′-CTCATGACCACAGTCCATGC-3′, antisense: 5′-TTCAGCTCTGGGATGACCTT-3′, 548 bp).
Flow Cytometry Analysis
The cells were incubated with the antibodies in phosphate-buffered saline containing 1% bovine serum albumin and 0.1% sodium azide. After incubating with antibody against IL8 (GeneTex, GTX43428), POSTN (BosterBio, M01378), PRDM1 (Merck, 05-1570), SP1 (Merck, FCABS345F), or ICAM1 (GeneTex, GTX20020), the cells were washed with PBS twice and applied into FACS Calibur flow cytometer (BD Biosciences). The corresponding isotype immunoglobulins were used as controls. The data were analyzed with Cell Quest software (BD Biosciences).
mRNA Stability Assays
ICAM1 mRNAs were quantified relative to GAPDH at various times after addition of Actinomycin D (ActD; 6.5 μg/ml; Sigma–Aldrich) to the cell culture medium. After ActD treatment, RNA was extracted from shCtrl or shMSI1 GBM-05MG cells using TRIZOL (Invitrogen), and RNA species were detected by RT-PCR analysis. The primers are the same with the primer used in qRT-PCR.
Western blot assays
Western blot assays were performed by using antibodies, including mouse anti β-Actin (1:1000; Millipore, mab1501), rabbit anti-LIN28 (1:1000; Cell signaling, #3695), rabbit anti-MSI1 (1:1000; Novus Biologicals, NBP1-32812) and mouse anti-ICAM1 (1:1000; OriGene Technologies, AM26247PU-N). The antibodies were listed in Supplementary Table 2.
Immunohistochemistry Staining
GBM clinical samples were stained by using antibodies, including rabbit anti-MSI1 (1:100; Novus Biologicals, NBP1-32812) and mouse anti-ICAM1 (1:100; OriGene Technologies, AM26247PU-N). The antibodies were listed in Supplementary Table 2.
Results
MSI1 Enhances Invasion and Motility in GBM Cells
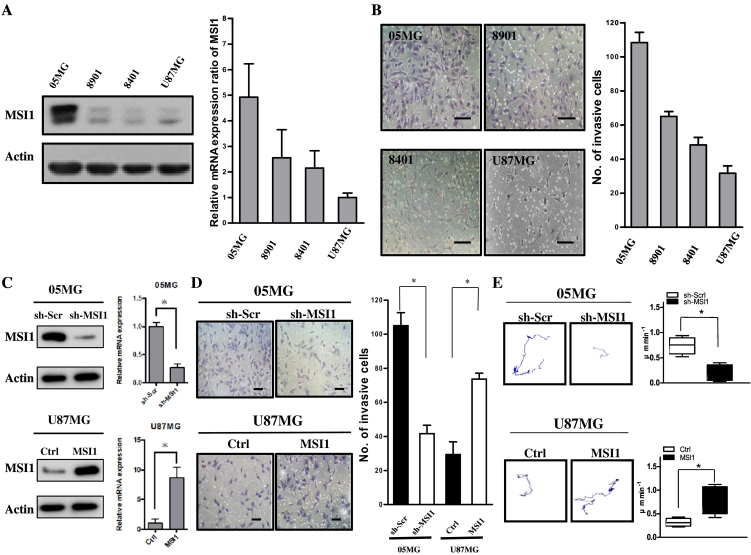
GBM lines were established from tumor tissues of diagnosed WHO grade IV glioblastoma patients and used as a cell model for investigating tumor enrichment [22]. MSI1 silencing attenuated the migration and invasion activities of colon cancer cells, and MSI1 was reported to be associated with differentiation, tumor mass, and invasion of gallbladder adenocarcinoma [23], [24]. However, the role of MSI1 in GBM remains unclear. To determine the relationship between malignancy and MSI1 expression, we compared levels in four GBM cell lines: 05MG, 8901, 8401, and U87MG GBM cells. According to western blot analysis, 05MG cells, with the greatest invasion and migration capacities, exhibited the highest levels of MSI protein among the cell lines (Figure 1A, left panel). Consistent with the immunoblotting results, MSI1 mRNA was 5.01 ± 0.34-fold higher in 05MG cells than in U87MG cells; expression levels of MSI1 mRNA were 2.58 ± 0.41 and 2.24 ± 0.28 in 8901 and 8401 GBM cells, respectively (Figure 1A, right panel). To further compare invasion ability among these cell lines, we performed a transwell assay to examine whether the cells with high MSI1 expression also had higher invasion ability in vitro. Bright-field images showed more 05MG cells escaping from the transwell device compared to the other cell lines (Figure 1B, left). Based on quantification, the numbers for 05MG, 8901, 8401, and U87MG cells were 110 ± 5, 83 ± 4, 47 ± 6, and 30 ± 5, respectively (Figure 1B, right), suggesting that 05MG cells have higher invasion ability. To further identify the effect of MSI1 in regulating tumor invasion, we used short hairpin RNA (sh-MSI1) against MSI1 to knock down MSI1 expression in 05MG cells (Figure 1C, upper). Compared to the control group (sh-Scr), MSI1 knockdown significantly reduced the number of invasive cells (Figure 1D), showing that MSI1 may be essential for GBM invasion. We also overexpressed MSI1 in U87MG cells, with a low in vitro level of MSI1 expression, using lentivirus infection (Figure 1C, lower) and observed that MSI1 overexpression induced a significant increase in invasion cell number (Figure 1D). Next, MSI1-knockdown GBM (sh-MSI1) cells were subjected to a cell tracking assay, which provides a means for comparing motility in a knockdown (sh-Scr vs sh-MSI1 in 05MG) or overexpression (Ctrl vs MSI1 in U87MG) group. The recorded tracks in 2 dimensions (2D) of the knockdown or overexpression group are shown in Figure 1E, and the distance per minute was converted into a migration rate (μm/min). These assays revealed that MSI1 knockdown significantly reduced cell motility but that MSI1 overexpression promoted cell motility. Taken together, the above results demonstrated that MSI1 might manipulate cell invasion and motility in vitro.
Figure 1.
MSI1 promotes invasion and motility in GBM cells. (A) MSI1 expression in different GBM cell lines by western blotting. All data are represented as the mean ± SD, n = 3. (B) GBM-05MG cells had higher invasive abilities than did GBM-8901, GBM-8401 and GBM-U87MG cells. All data are represented as the mean ± SD, n = 3. Scale bars, 100 μm. (C) (Upper) Short hairpin RNA (sh-MSI1) against MSI1 to knock down MSI1 expression in 05MG cells by western blot and qPCR; (Lower) overexpressed MSI1 in U87MG cells by western blot and qPCR. (D) (Upper) Transwell invasion assay using GBM-05MG cells transfected with scrambled shRNA control vector (sh-Scr) versus sh-MSI1; (Lower) transwell invasion assay using GBM-U87MG cells transfected with the vector control (Ctrl) versus ectopic MSI1. All data are represented as the mean ± SD, n = 3, Scale bars, 100 μm. *P < .01 by Student's t-test. (E) (Upper) Representative trajectories and quantification of speed of GBM-05MG cells transfected with sh-Scr vector versus sh-MSI1; (Lower) representative trajectories and quantification of speed of GBM-U87MG cells transfected with the vector Ctrl versus ectopic MSI1. (sh-Scr; n = 10 for each group). *P < .01 by Student's t-test.
MSI1 Promotes GBM Tumor Invasion via ICAM1 Induction
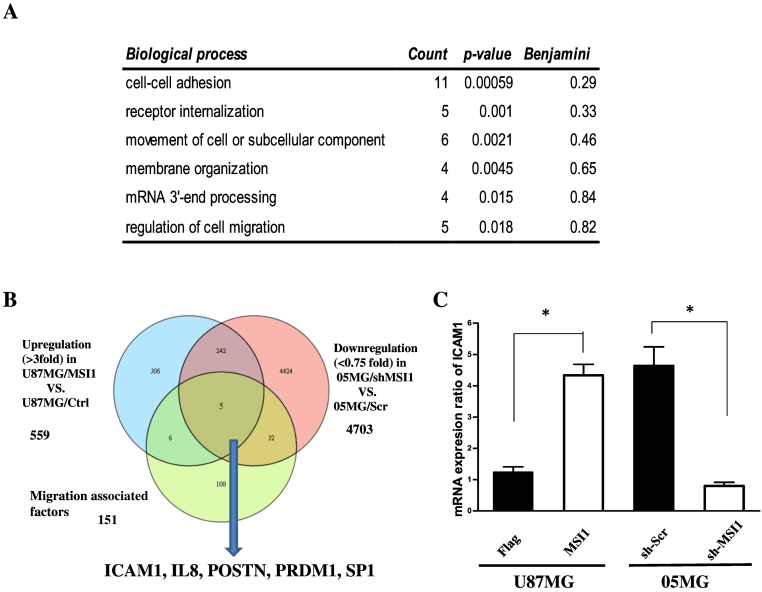
Based on our immunoblotting results (Figure 1A), U87MG cells can be viewed as an MSI1-negative GBM line and 05MG cells as an MSI1-positive GBM line. To investigate the mechanism underlying how MSI1 affects GBM invasion and motility, we aimed to determine which genes are upregulated in MSI1-overexpressing U87MG cells and those oppositely downregulated in MSI1 knockdown 05MG cells using Affymetrix 133 plus a microarray to detect transcriptional changes. We found that 559 genes were upregulated in MSI1-overexpressing GBM-U87MG cells and that 4703 genes were downregulated in MSI1-knockdown GBM-05MG cells. We then performed Gene Ontology (GO) analysis, which revealed that MSI1 is correlated with several biological processes, including cell–cell adhesion, receptor internalization, cell movement or subcellular component, membrane organization, mRNA 3′-end processing, and cell migration regulation (Figure 2A). To identify specific genes downstream of MSI1 and related to migration, we correlated the 559 and 4703 genes with defined migration-associated factors [25] to reveal 247 common genes. Eventually, we obtained five genes from the microarray results: ICAM1, IL8, POSTN, PRDM1, and SP1 (Figure 2B). In addition, among those with altered expression, a number of cancer-associated genes, such ABCC4, BRCA2, and JAK1, were upregulated (Supplemental Table 3), and many tumor suppressors were downregulated, including WT1, NF2, MSH2, MLH1, and MSH6 (Supplemental Table 4). Several studies have revealed that ICAM1 promotes invasion and migration of GBM cells and that ICAM1 silencing prolongs survival of glioblastoma in vivo [21], [26]. Therefore, we employed quantitative polymerase chain reaction (qPCR) to examine the mRNA expression of ICAM1 in both groups (knockdown vs control in 05MG or overexpressed vs control in U87MG). The results show ICAM1 mRNA expression to be upregulated and downregulated in MSI1-overexpressing and -knockdown GBM cells (Figure 2C).
Figure 2.
ICAM1 is more highly expressed in GBM cells expressing greater levels of MSI1-compared to GBM cells expressing lower levels of MSI1. (A) Gene ontology classification. (B) Schema for identifying genes under conditions of >3-fold differences between U87MG/MSI1 and U87MG/Ctrl cells, <0.75-fold differences between 05MG/sh-MSI1 and 05MG/sh-Scr cells, and migration-associated factors. (C) qPCR analysis of ICAM1 in U87MG /Ctrl, U87MG /MSI1, 05MG/sh-Scr, and 05MG /sh-MSI1 cells. *P < .01 by Student's t-test. Data shown are the mean ± SD of three independent experiments.
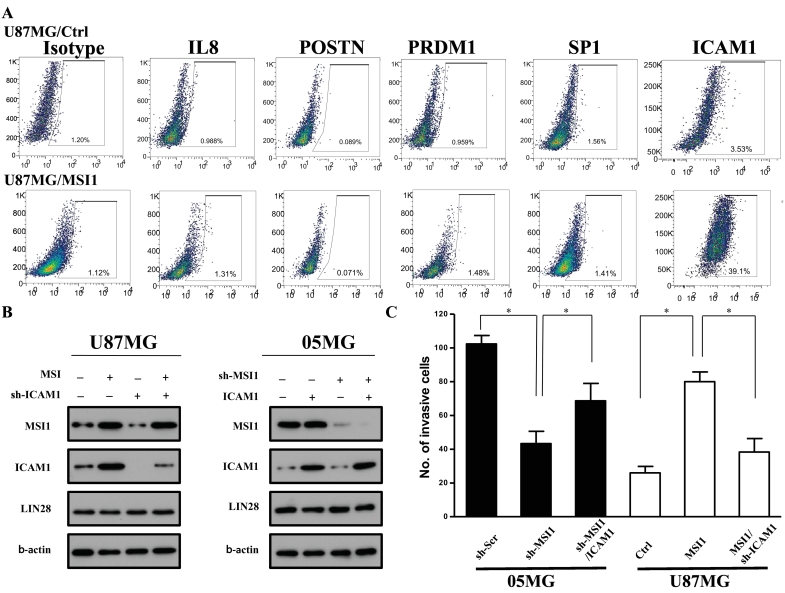
We next examined whether the protein expression of these five genes screened from the microarray analysis was also stimulated by MSI1 overexpression. Flow cytometry analysis revealed that proportions of 1.31 ± 0.98%, 0.071 ± 0.08%, 1.48 ± 0.95%, 1.41 ± 1.46%, and 39.1 ± 3.53% for IL8, POSTN, PRDM1, SP1, and ICAM1, respectively. ICAM1 was significantly upregulated by MSI1 overexpression (Figure 3A), and the population of ICAM1-positive U87MG cells was significantly increased with MSI1 overexpression. Furthermore, we employed western blotting to compare differences in ICAM1 levels between MSI1 knockdown and control cells; to further address the causal effect, we combined ICAM1 knockdown with the presence or absence of MSI1. Immunoblotting revealed that MSI1 overexpression resulted in upregulation of ICAM1 but that ICAM1 overexpression did not cause MSI1 upregulation. Consistently, we found that MSI1 can facilitate ICAM1 expression. ICAM1 knockdown did not significantly suppress the MSI1 level in U87MG cells, and ICAM1 overexpression did not change MSI1 levels in 05MG cells, suggesting that ICAM1 does not influence MSI1 levels (Figure 3B). Moreover, we compared the invasion ability of GBM-U87/MSI1 and GBM-05MG/sh-Scr cells to investigate whether MSI1 affects the invasion ability mediated by ICAM1 signaling. Compared to shMSI1 treatment, ICAM1 overexpression rescued the invasion ability of 05MG cells (Figure 3C). We also observed that shICAM1 significantly suppressed invasion ability in the presence of MSI1 overexpression. The above results indicate that ICAM1 is essential for MSI1-mediating invasion. Because 05MG cells are derived from a patient with GBM who had been treated with irradiation and chemotherapy, we suggest that inability of ICAM1 overexpression to fully rescue suppressed 05MG cell invasion was the result of radio- or drug selection. Significant suppression by shICAM1 was observed in U87MG cells overexpressing MSI1, which also supports our suggestion. However, ICAM1 knockdown suppressed invasion ability in the presence of MSI1, indicating that MSI1 regulates invasion ability via ICAM1.
Figure 3.
ICAM1 is a downstream target of MSI1-modulated cell invasion. (A) ICAM1 is the most highly expressed protein in comparisons of U87/MSI1 with U87/Ctrl GBM cells. (B) Left: Expression of MSI1, ICAM1, and LIN28 in GBM-U87MG/Ctrl, and GBM-U87MG/MSI1. Right: Expression of MSI1, ICAM1, and LIN28 in GBM-05MG/sh-Scr and GBM-05MG/sh-MSI1 cells. ß-Actin was used as a loading control. (C) Left: Transwell invasion assay in GBM-05MG cells transfected with sh-Scr, sh-MSI1 or sh-MSI1/ICAM1. All data are presented as the mean ± SD, n = 10, *P < .01 by Student's t-test. Right: Transwell invasion assay in GBM-U87MG cells transfected with Ctrl, MSI1 and MSI1/sh-ICAM1. All data are presented as the mean ± SD, n = 10, *P < .01 by Student's t-test.
MSI1-Mediated mRNA Interaction Promotes ICAM1 Translation
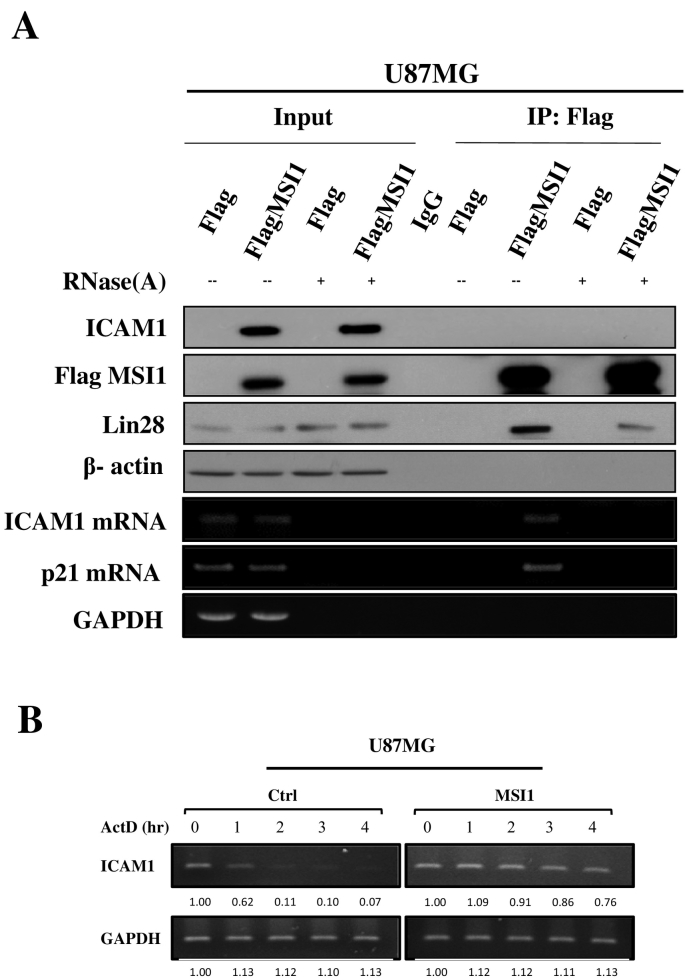
Given that MSI1 is an RNA-binding protein, upregulation of ICAM1 in the presence of MSI1 led us to investigate whether MSI1 modulates ICAM1 expression through an RNA-dependent mechanism. To identify whether MSI1 directly interacts with ICAM1 mRNA in GBM, we conducted RNA immunoprecipitation (RIP) experiments in the U87MG cell line by pulling down MSI1-bound RNAs. Immunoblot analysis revealed that MSI1 overexpression induces ICAM1 upregulation under physiological conditions but does not induce LIN28 upregulation (Figure 4A upper panel). We used the cancer-stemness transcription factor LIN28, which plays a crucial role in GBM malignancy [27], as a positive control for an MSI1-binding protein. Importantly, MSI1 was immunoprecipitated with ICAM1 and p21 mRNA in GBM cells (Figure 4A lower panel). Because p21 mRNA has been reported to be an MSI1 target, this mRNA was utilized a positive control [28], [29]. Furthermore, ICAM1 upregulation was eliminated by RNase A addition, and we observed the complete clearance of ICAM1 mRNA, indicating that the presence of ICAM1 mRNA is critical for MSI1 induction. However, RNase A-mediated mRNA elimination did not affect upregulation of LIN28, suggesting that increased LIN28 expression is not RNA dependent. This evidence that MSI1 does not directly interact with the ICAMI protein but does interact with its mRNA suggests that MSI1 contributes to a regulatory mechanism via RNA-binding activity and protects or facilitates ICAM1 translation in the presence of mRNA in GBM cells. To investigate the contribution of MSI1 to ICAM1 mRNA stability, mRNA decay was detected by qRT-PCR. Control or MSI1-overexpressing GBM-U87MG cells were treated with actinomycin D (ActD) (6.5 μg/ml) for 0, 1, 2, 3, and 4 h, and ICAM1 mRNA levels were measured at the indicated time point (Figure 4B). This analysis showed that ICAM1 mRNA was very stable at 0 to 4 h under MSI1 overexpression; without overexpression, ICAM1 mRNA was less stable. These results show that MSI1 can stabilize ICAM1 mRNA, which likely contributes to ICAM1 expression.
Figure 4.
MSI1-mediated mRNA interaction promotes ICAM1 translation. Representative immunoblotting analysis of ICAM1, Flag, and LIN28 in (A) U87MG cells. β-Actin was used as the loading control (upper panel). RNA-immunoprecipitation of Flag-control or Flag-MSI1-overexpressing cell lines was performed using an antibody against Flag (IP-Flag). RT-PCR analysis for detecting mRNA expression of ICAM1, p21, and GAPDH (lower panel). To confirm RNA-mediated inhibition of MSI1, RNase A was used to eliminate total RNA in vitro, demonstrating that MSI1 promotes ICAM1 translation in an RNA-dependent manner. Input was 5% of total mRNA, and IgG was used as a negative control. (B) Identification of the effect of MSI1 on stabilizing ICAM1 mRNA in the U87MG cell line. Control or MSI1-overexpressing U87MG cells were treated with the transcriptional inhibitor actinomycin D (Act D, 6.5 μg/ml), and total RNA was harvested at 0, 1, 2 3 and 4 hr. posttreatment. ICAM1 mRNA levels were determined relative to GAPDH by RT-PCR and quantitated using ImageJ software.
MSI1/ ICAM1-Expressing Cells are Enriched in Clinical GBM Samples
Based on the results of the in vitro experiments, we used immunohistochemistry (IHC) staining to evaluate MSI1 and ICAM1 expression in nine GBM patient samples. To emphasize the role of MSI1 and ICAM1 in high-grade GBM, we compared expression among GBM and low-grade glioma (LGG). Compared to LGG, GBM clinical samples showed high MSI1 and ICAM1 expression in two GBM patient samples, as revealed by IHC staining against the MSI1 antigen (Figure 5, A and B). IHC staining of MSI1 and ICAM1 was then applied to further identify the relationship between MSI1 and ICAM1, revealing that MSI1 highly colocalized with ICAM1 in clinical GBM samples. Furthermore, according to the Rembrandt brain database, the survival rate of MSI1high/ICAM1high GBM patients is poor (Figure 5C): among 151 patients, MSI1high (76) and ICAM1high (76) showed significantly lower survival rates. These results indicate that both MSI1 and ICAM1 are critical for patient survival. However, we could not clearly distinguish the survival rates of MSI1-positive (76 patients), ICAM1-positive (76 patients), or double-positive patients (35 patients), suggesting that MSI1 and ICAM1 may share the same mechanism. As the survival rate of double-positive patients was not lower than that of MSI1- or ICAM1-positive patients, MSI1/ICAM1 axis regulation strongly influences the survival rate of double-positive GBM patients. In other words, the survival of double-positive patients appears to be similar to that of each single-positive patient because the major factor affecting the survival rate related to MSI1 is dependent of that related to ICAM1. This indicates that MSI1 affects survival rates through the same pathway as ICAM1. All patients received the full course of concurrent chemoradiotherapy after surgery. Overall, MSI1 possibly mediates translation of ICAM1, whereas MSI1 may serve as a predictive index for ICAM1 expression. In summary, we conclude that MSI1 might mediate ICAM1 translation, which plays an important role in regulating tumor migration and invasion (Figure 5D).
Figure 5.
(A) IHC staining detected MSI1 in LGG and GBM patient samples. Scale bars, 100 μm. All data are presented as the mean ± SD, n = 10, *P < .01 by Student's t-test. (B) IHC staining detected ICAM1 in LGG and GBM patient samples. Scale bars, 100 μm. All data are presented as the mean ± SD, n = 10, *P < .01 by Student's t-test. (C) MSI1 and ICAM1 protein co-localization in tissues by IHC staining. Scale bars,100 μm. All data are presented as the mean ± SD, n = 10, *P < .01 by Student's t-test. (D) Different Kaplan–Meier plots of GBM patients with high MSI1 expression, high ICAM1expression, and high MSI1/ICAM1 expression. (E) Schematic illustration depicting that MSI1-mediated mRNA interaction promotes ICAM1 translation and tumor invasion.
Discussion
GBM is a high-mortality brain tumor in humans. Surgery, traditional chemotherapy, and radiation therapy have been applied as clinical strategies to treat GBM patients. Recently, “postoperative radiation plus temozolomide (TMZ) followed by adjuvant TMZ chemotherapy has become the standard clinical strategy for treating newly diagnosed GBM and recurrent GBM patients. However, there is a high risk of recurrence and poor treatment outcomes. As GBM cells invading normal brain tissue because of inflammation can escape surgical resection [30], understanding the link between GBM cell invasion and inflammation is critical for developing effective therapies. To our knowledge, the present study is the first to show that MSI1 directly mediates mRNA interaction, promoting ICAM1 translation and tumor invasion.
Tumor invasion consists of several discrete steps, including tumor cell interaction with extracellular matrix (ECM) ligands, hydrolytic destruction of the matrix via the release of proteolytic enzymes, and subsequent migration of the tumor cells through the area of destruction. Of all these steps, recent studies indicate that the ability of tumor cells to digest the ECM by secreting proteolytic enzymes best correlates with tissue invasiveness. Indeed, IHC analysis of the border zone between a glioma and the brain (glial limitans externa) has shown it to contain interstitial collagen, laminin, fibronectin, and type IV collagen. Invasion of most primary human brain tumors is thought to be accomplished at least in part by elevated levels of proteases that breach connective tissue barriers to cause vascular remodeling and destruction of normal brain tissue. For example, Kesanakurti et al. reported that ICAM1 regulates radiation-induced invasion and migration in glioma [21], and blockade of VEGF receptors, such as VCAM1 and ICAM1, by tivozanib exhibited potential antitumor effects on human glioblastoma cells [31]. Yuji et al. revealed that targeting ICAM1 prolongs survival in mice bearing bevacizumab-resistant glioblastoma [26]. Moreover, a previous study showed an essential role for cooperative NF-κB and STAT3 recruitment to ICAM-1 intronic consensus elements in regulating radiation-induced invasion and migration in glioma [21]. Based on these studies and our data, we suggest that elevated ICAM1 expression is responsible for increased invasion by GBM cells.
Based on prior findings that MSI1 is a marker of neural stem cells and progenitor cells [32], our data show that MSI1 is a functional marker of tumor invasion in GBM. In fact, MSI1 is emerging as a regulator of multiple critical biological processes relevant to cancer initiation, progression, and drug resistance [32]. Uren et al. revealed that MSI1 is a central regulator of adhesion pathways in GBM [33], and MSI1 promotes chemoresistant granule formation via PKR/eIF2α signaling in refractory glioblastoma [34]. Our results reveal that the higher MSI1 expression in GBM-05MG cells and the invasive phenotype of these cells correlate with enhanced ICAM1 expression. Nonetheless, the molecular mechanism of MSI1-dependent induction of tumor invasion in GBM has remained unclear. Recent studies have shown that MSI1 directly binds to the promoter region of PYGO2 in esophageal squamous cell carcinoma [35]. Here, we demonstrate that MSI1-mediated mRNA interaction promotes ICAM1 translation and enhances tumor invasion.
In conclusion, our study shows that activating MSI1/ICAM1 signaling promotes migration and invasion abilities. We believe that the MSI1/ICAM1 axis might be a latent therapeutic target for suppressing GBM invasion. Our results provide insight into the development of potential treatments that may be able to overcome GBM tumorigenicity, which is a frequent challenge of current GBM treatment therapies.
Footnotes
Conflicts of Interest: None.
Permissions: obtained. We declare no use of copyrighted material.
Contributors: All authors contributed to 1) the conception and design of the study, acquisition of data, analysis and interpretation of data, 2) drafting the manuscript, and 3) final approval of the version to be submitted.
Acknowledgments: Not applicable.
Founding sources: This work was supported by TSGH-C107-069, TSGH-C108-099 and MOST 107-2314-B-016-002.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.02.006.
Appendix A. Supplementary data
Supplementary material
References
- 1.Stupp R, van den Bent MJ, Hegi ME. Optimal role of temozolomide in the treatment of malignant gliomas. Curr Neurol Neurosci Rep. 2005;5:198–206. doi: 10.1007/s11910-005-0047-7. [DOI] [PubMed] [Google Scholar]
- 2.Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selmi T, Martello A, Vignudelli T, Ferrari E, Grande A, Gemelli C, Salomoni P, Ferrari S, Zanocco-Marani T. ZFP36 expression impairs glioblastoma cell lines viability and invasiveness by targeting multiple signal transduction pathways. Cell Cycle. 2012;11 doi: 10.4161/cc.20309. [DOI] [PubMed] [Google Scholar]
- 4.Yoda A, Sawa H, Okano H. MSI-1, a neural RNA-binding protein, is involved in male mating behaviour in Caenorhabditis elegans. Genes Cells. 2000;5:885–895. doi: 10.1046/j.1365-2443.2000.00378.x. [DOI] [PubMed] [Google Scholar]
- 5.Hirota Y, Okabe M, Imai T, Kurusu M, Yamamoto A, Miyao S, Nakamura M, Sawamoto K, Okano H. Musashi and seven in absentia downregulate Tramtrack through distinct mechanisms in Drosophila eye development. Mech Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 6.Kawashima T, Murakami AR, Ogasawara M, Tanaka K, Isoda R, Sasakura Y, Nishikata T, Okano H, Makabe KW. Expression patterns of musashi homologs of the ascidians, Halocynthia roretzi and Ciona intestinalis. Dev Genes Evol. 2000;210:162–165. doi: 10.1007/s004270050024. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 8.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 9.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 10.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 12.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151–2162. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 13.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 14.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 15.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dustin ML, Staunton DE, Springer TA. Supergene families meet in the immune system. Immunol Today. 1988;9:213–215. doi: 10.1016/0167-5699(88)91216-9. [DOI] [PubMed] [Google Scholar]
- 17.Frohman EM, van den Noort S, Gupta S. Astrocytes and intracerebral immune responses. J Clin Immunol. 1989;9:1–9. doi: 10.1007/BF00917121. [DOI] [PubMed] [Google Scholar]
- 18.Frank PG, Lisanti MP. ICAM-1: role in inflammation and in the regulation of vascular permeability. Am J Physiol Heart Circ Physiol. 2008;295:H926–H927. doi: 10.1152/ajpheart.00779.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu JA, Sadaria MR, Meng X, Mitra S, Ao L, Fullerton DA, Weyant MJ. Lung cancer cell invasion and expression of intercellular adhesion molecule-1 (ICAM-1) are attenuated by secretory phospholipase A(2) inhibition. J Thorac Cardiovasc Surg. 2012;143:405–411. doi: 10.1016/j.jtcvs.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Lin YC, Shun CT, Wu MS, Chen CC. A novel anticancer effect of thalidomide: inhibition of intercellular adhesion molecule-1-mediated cell invasion and metastasis through suppression of nuclear factor-kappaB. Clin Cancer Res. 2006;12:7165–7173. doi: 10.1158/1078-0432.CCR-06-1393. [DOI] [PubMed] [Google Scholar]
- 21.Kesanakurti D, Chetty C, Rajasekhar Maddirela D, Gujrati M, Rao JS. Essential role of cooperative NF-kappaB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene. 2013;32:5144–5155. doi: 10.1038/onc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CE, Starr R, Martinez C, Aguilar B, D'Apuzzo M, Todorov I, Shih CC, Badie B, Hudecek M, Riddell SR. Recognition and killing of brain tumor stem-like initiating cells by CD8+ cytolytic T cells. Cancer Res. 2009;69:8886–8893. doi: 10.1158/0008-5472.CAN-09-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Peng X, Yan D, Tang H, Huang F, Yang Y, Peng Z. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 2011;18:2074–2083. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 24.Liu DC, Yang ZL, Jiang S. Identification of musashi-1 and ALDH1 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Cancer Biomark. 2010;8:113–121. doi: 10.3233/DMA-2011-0812. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piao Y, Henry V, Tiao N, Park SY, Martinez-Ledesma J, Dong JW, Balasubramaniyan V, de Groot JF. Targeting intercellular adhesion molecule-1 prolongs survival in mice bearing bevacizumab-resistant glioblastoma. Oncotarget. 2017;8:96970–96983. doi: 10.18632/oncotarget.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu J, Zhao X, Liu Q, Yang J. Knockdown of MSI1 inhibited the cell proliferation of human osteosarcoma cells by targeting p21 and p27. Oncol Lett. 2017;14:5271–5278. doi: 10.3892/ol.2017.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jadhav S, Ajay AK, Trivedi P, Seematti J, Pellegrini K, Craciun F, Vaidya VS. RNA-binding protein musashi homologue 1 regulates kidney fibrosis by translational inhibition of p21 and numb mRNA. J Biol Chem. 2016;291:14085–14094. doi: 10.1074/jbc.M115.713289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klekner A, Hutoczki G, Virga J, Remenyi-Puskar J, Toth J, Scholtz B, Csosz E, Kallo G, Steiner L, Hortobagyi T. Expression pattern of invasion-related molecules in the peritumoral brain. Clin Neurol Neurosurg. 2015;139:138–143. doi: 10.1016/j.clineuro.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Momeny M, Moghaddaskho F, Gortany NK, Yousefi H, Sabourinejad Z, Zarrinrad G, Mirshahvaladi S, Eyvani H, Barghi F, Ahmadinia L. Blockade of vascular endothelial growth factor receptors by tivozanib has potential anti-tumour effects on human glioblastoma cells. Sci Rep. 2017;7:44075. doi: 10.1038/srep44075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudinov AE, Karanicolas J, Golemis EA, Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res. 2017;23:2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uren PJ, Vo DT, de Araujo PR, Potschke R, Burns SC, Bahrami-Samani E, Qiao M, de Sousa Abreu R, Nakaya HI, Correa BR. RNA-binding protein Musashi1 is a central regulator of adhesion pathways in glioblastoma. Mol Cell Biol. 2015;35:2965–2978. doi: 10.1128/MCB.00410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HY, Lin LT, Wang ML, Tsai KL, Huang PI, Yang YP, Lee YY, Chen YW, Lo WL, Lan YT. Musashi-1 promotes chemoresistant granule formation by PKR/eIF2alpha signalling cascade in refractory glioblastoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1850–1861. doi: 10.1016/j.bbadis.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Moghbeli M, Sadrizadeh A, Forghanifard MM, Mozaffari HM, Golmakani E, Abbaszadegan MR. Role of Msi1 and PYGO2 in esophageal squamous cell carcinoma depth of invasion. J Cell Commun Signal. 2016;10:49–53. doi: 10.1007/s12079-015-0314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material