Abstract
Chitin-glucan (CG) represents a natural carbohydrate source for certain microbial inhabitants of the human gut and may act as a prebiotic for a number of bacterial taxa. However, the bifidogenic activity of this substrate is still unknown. In the current study, we evaluated the ability of chitin-glucan to influence growth of 100 bifidobacterial strains belonging to those species commonly identified within the bifidobacterial communities residing in the infant and adult human gut. Such analyses were coupled with transcriptome experiments directed to explore the transcriptional effects of CG on Bifidobacterium breve 2L, which was shown to elicit the highest growth performance on this natural polysaccharide. In addition, an in vivo trial involving a rat model revealed how the colonization efficiency of this bifidobacterial strain was enhanced when the animals were fed with a diet containing CG. Altogether our analyses indicate that CG is a valuable novel prebiotic compound that may be added to the human diet in order to re-establish/reinforce bifidobacteria colonization in the mammalian gut.
Introduction
Chitin-glucan (CG) is a high-purity bio-polymer composed of two distinct polysaccharides represented by chitin (β-1,4-poly-N-acetyl-D-glucosamine) and β-1,3-D-glucan in a ratio ranging from 20:80 to 40:60 (w/w)1. CG is typically extracted from Aspergillus niger mycelial cell walls. Furthermore, another natural sources of CG are represented by different fungal species and yeasts due to the presence of this bio-polymer in their inner cell walls2,3. Recently, CG has been indicated as food supplements fixing the maximum consumption rate to five gr per day for an average person4.
However, mammalian enzymes are unable to degrade CG and therefore following ingestion this glycopolymer will arrive in its intact form in the large intestine where it may influence growth and/or metabolic activity of different members of the gut microbiota. In this context, there is growing scientific evidence of possible prebiotic effects elicited by CG towards various microorganisms of the mammalian gut5,6. Recently, an in vitro study performed in a dynamic gut simulator (SHIME) illustrated the effect of CG on microbiota composition and activity, leading to a decrease of the Firmicutes/Bacteroidetes ratio and an increase in the relative abundance of Roseburia spp1, which could suggest a potential role exploited by CG in shaping the gut microbiota through a prebiotic effect as previously displayed for other complex carbohydrates7.
The term ‘prebiotic’ includes compounds, such as non-digestible (i.e. cannot be metabolized or degraded by the host) carbohydrates, that are selectively metabolized by beneficial gut bacteria. Prebiotic treatment is a dietary strategy by which the gastrointestinal microbiota can be modified, both in composition and/or activity, for the purpose of conferring health benefits to the host8. In fact, it has been demonstrated that prebiotics can reduce symptoms associated with inflammatory bowel disease, the duration of infectious and antibiotic-associated diarrhea, and the risk of cardiovascular diseases. Furthermore, prebiotics may also promote satiety and weight loss, preventing obesity and enhancing protective effects against the onset of colon cancer9.
Bifidobacteria are very prevalent and abundant human gut microbiota members, especially during the first months following birth10, though their numbers decrease following weaning and in elderly. Bifidobacterial abundance in the human microbiota is markedly reduced following gastrointestinal diseases11, suggesting that this taxon plays a positive role in the promotion of host health12. Prebiotics, such as dietary fibers, have been used to counteract the reduction of bifidobacterial abundance in the human gut13.
So far, a very limited scientific evidence is available regarding the possible prebiotic effect of CG towards members of the Bifidobacterium genus6. In the current report, we evaluated possible bifidogenic features of CG towards 100 different bifidobacterial strains using an in vitro approach. Such analyses revealed the presence of a highly CG-responsive bifidobacterial strain, Bifidobacterium breve 2L, whose transcriptome when cultivated on CG was investigated in detail by RNAseq experiments. Furthermore, in vivo trials in a rat model fed with CG and B. breve 2L clearly demonstrated that CG increases the abundance of B. breve 2L and several other members of the mammalian gut microbiota.
Results and Discussion
Evaluation of prebiotic capability of chitin-glucan toward bifidobacteria
One hundred bifidobacterial strains previously isolated from the human environment (reported in Table 1), were evaluated for their ability to grow on CG, as unique carbon source. These strains were isolated from infant feces (Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium pseudocatenulatum) as well as from gastrointestinal tract of adult (Bifidobacterium adolescentis, Bifidobacterium angulatum, Bifidobacterium catenulatum, Bifidobacterium dentium, Bifidobacterium longum, Bifidobacterium pseudolongum subsp. globosum). In addition, the bifidobacterial collection used in this project included strains belonging to Bifidobacterium animalis subsp. lactis, isolated from various commercial products sold as probiotics. Despite the fact that CG consists of two different polysaccharides, i.e. chitin (β-1,4-poly-N-acetyl-D-glucosamine) and β-1,3-D-glucan, we focused our study on the biopolymer CG due to its commercial relevance as an extract of Aspergillus niger. Furthermore, we have used the Tyndallization procedure, which operates a series moderate heat treatment steps separated by rapid cooling stages, in order to achieve a sterilization of CG-based medium without causing extensive hydrolysis of CG as might occur when using high-level temperature treatments such as autoclaving. Growth assays were conducted on a modified MRS growth media, i.e. MRS w/o glu + CG (0.5% (w/v) together with a positive control (complete MRS) as well as a negative control (MRS w/o glu). Experiments were performed in triplicates for each growth media. We observed a minimal growth also on MRS w/o glu (≤103 cells/ml), which is probably linked to the presence of components such as meat and yeast extract providing extra organic carbon sources. However, the final cell count of positive control and MRS w/o glu + CG were calculated by subtracting the cells count obtained on MRS w/o glu. Interestingly, all strains were shown to exhibit growth using MRS w/o glu + CG as the unique carbon source. Remarkably, almost all assayed strains displayed growth levels comparable with those obtained in complete MRS (MRS + glucose), i.e. comprised between 107–109 cells/mL (Table 1). Notably, this may be caused by partial degradation of CG in simpler sugars, e.g. glucose monomers from glucan, due to the heat treatment included in the modified Tyndallization approach employed to sterilize the MRS w/o glu + CG medium. Only in case of strains belonging to the B. adolescentis and B. longum species, the observed growth levels were significantly different (p-value < 0.05) (Fig. 1). In detail, B. adolescentis strains exhibited a final cell count of 7.59 × 107 cells/mL, and 4.19 × 108 cells/mL, in MRS w/o glu + CG and in complete MRS, respectively. Regarding B. longum, these strains were able to reach an average final cell count of 4.40 × 108 cells/mL in MRS w/o glu + CG, which is a lower cell yield compared to the final cell count in complete MRS (1.05 × 109 cells/mL). This finding indicates that these bifidobacterial species have a reduced ability to utilize CG as compared to the other assessed bifidobacterial strains. In contrast, B. breve and B. bifidum strains were shown to reach the highest cell numbers on this substrate (cell numbers ranging from 108 to 109 cells/mL), and in some cases surpassing growth yields obtained in complete MRS (Fig. 1). These two bacterial species are typically isolated from infants, and consequently expected to be metabolically adapted to degrade host-specific glycans such as mucin and host glycan constituents like N-acetylglucosamine14,15. Notably, CG is partially composed of polymerized β-1,4-poly-N-acetyl-D-glucosamine, which may explain the reason for the more robust metabolic efficiency of these species toward CG (compared to the other examined bifidobacterial species).
Table 1.
Bifidobacterial strains grown on MRS medium and MRS w/o glucose supplemented with CG and results of in vitro experiments.
| Strainsa | Origin | Average (cells/mL) | SD | Average CG (cells/mL) | CG SD | Average Agar plate (CFU/mL) | Agar plate SD |
|---|---|---|---|---|---|---|---|
| B. adolescentis 1901B | Adult stool sample | 1.03 × 108 | 4.63 × 107 | 3.42 × 107 | 2.67 × 107 | ||
| B. adolescentis 1902B | Adult stool sample | 6.17 × 108 | 2.08 × 108 | 2.92 × 107 | 8.04 × 106 | ||
| B. adolescentis 1903B | Adult stool sample | 2.75 × 108 | 1.52 × 108 | 4.75 × 107 | 2.88 × 107 | ||
| B. adolescentis 1904B | Adult stool sample | 2.50 × 108 | 2.38 × 108 | 3.33 × 108 | 3.64 × 108 | 8.40 × 106 | 2.06 × 106 |
| B. adolescentis 22L | Human milk | 3.67 × 108 | 1.94 × 108 | 2.83 × 107 | 1.18 × 107 | ||
| B. adolescentis 236B | Colonoscopic sample | 2.17 × 108 | 1.59 × 108 | 4.00 × 107 | 4.33 × 106 | ||
| B. adolescentis 382B | Adult stool sample | 5.08 × 108 | 3.13 × 108 | 7.17 × 107 | 9.46 × 106 | ||
| B. adolescentis 388B | Adult stool sample | 1.17 × 108 | 5.20 × 107 | 2.26 × 108 | 1.13 × 108 | 3.75 × 107 | 8.57 × 106 |
| B. adolescentis 42B | Adult stool sample | 2.75 × 108 | 8.66 × 107 | 1.83 × 107 | 1.04 × 107 | ||
| B. adolescentis 487B | Adult stool sample | 4.08 × 108 | 1.89 × 108 | 3.83 × 107 | 1.89 × 107 | ||
| B. adolescentis 532B | Colonoscopic sample | 3.83 × 108 | 1.23 × 108 | 1.02 × 108 | 2.25 × 107 | ||
| B. adolescentis 548B | Colonoscopic sample | 2.67 × 108 | 3.82 × 107 | 6.00 × 107 | 1.73 × 107 | ||
| B. adolescentis 59B | Adult stool sample | 3.08 × 108 | 8.04 × 107 | 2.50 × 107 | 4.33 × 106 | ||
| B. adolescentis 61B | Adult stool sample | 1.58 × 108 | 1.44 × 107 | 2.25 × 107 | 1.98 × 107 | ||
| B. adolescentis 65B | Colonoscopic sample | 5.75 × 108 | 1.95 × 108 | 4.50 × 107 | 1.09 × 107 | ||
| B. adolescentis 679B | Adult stool sample | 1.75 × 108 | 5.00 × 107 | 6.00 × 107 | 6.61 × 106 | ||
| B. adolescentis 703B | Adult stool sample | 9.75 × 108 | 5.91 × 108 | 5.82 × 107 | 4.04 × 107 | ||
| B. adolescentis 70B | Adult stool sample | 4.33 × 108 | 1.76 × 108 | 2.25 × 107 | 1.41 × 107 | ||
| B. adolescentis 711B | Colonoscopic sample | 8.67 × 108 | 5.20 × 107 | 1.75 × 108 | 1.00 × 108 | ||
| B. adolescentis 723B | Colonoscopic sample | 1.57 × 109 | 4.15 × 108 | 8.42 × 107 | 7.88 × 107 | ||
| B. adolescentis 731B | Colonoscopic sample | 7.42 × 108 | 1.38 × 108 | 9.92 × 107 | 5.43 × 107 | ||
| B. adolescentis 734B | Colonoscopic sample | 3.17 × 107 | 1.44 × 107 | 1.13 × 107 | 5.30 × 106 | ||
| B. adolescentis ATCC 15703 | Colonoscopic sample | 7.33 × 107 | 2.50 × 107 | 2.17 × 107 | 5.20 × 106 | ||
| B. adolescentis LMG 10733 | Colonoscopic sample | 1.09 × 109 | 3.17 × 108 | 5.33 × 107 | 3.82 × 106 | ||
| B. adolescentis LMG 10734 | Colonoscopic sample | 1.27 × 108 | 3.16 × 107 | 6.00 × 107 | 0 | ||
| B. adolescentis LMG 18897 | Adult stool sample | 5.83 × 108 | 1.81 × 108 | 2.50 × 107 | 7.07 × 106 | ||
| B. angulatum LMG 11039 | Adult stool sample | 1.25 × 108 | 6.61 × 107 | 3.50 × 108 | 3.68 × 108 | ||
| B. animalis subsp. lactis Bb-12 | Fermented milk product | 7.17 × 108 | 4.25 × 108 | 1.45 × 109 | 2.78 × 108 | 8.30 × 106 | 2.43 × 106 |
| B. animalis subsp. lactis DSM 10140 | Fermented milk | 2.50 × 108 | 4.33 × 107 | 1.08 × 108 | 5.20 × 107 | ||
| B. bifidum 156B | Infant stool sample | 2.58 × 108 | 1.66 × 108 | 6.42 × 108 | 8.78 × 107 | 2.61 × 106 | 2.75 × 105 |
| B. bifidum 324B | Infant stool sample | 1.67 × 108 | 1.04 × 108 | 1.83 × 108 | 5.20 × 107 | 2.58 × 106 | 4.15 × 105 |
| B. bifidum 361B | Adult stool sample | 5.00 × 107 | 2.50 × 107 | 4.00 × 107 | 1.15 × 107 | ||
| B. bifidum 85B | Infant stool sample | 3.25 × 108 | 1.25 × 108 | 3.17 × 108 | 2.89 × 107 | ||
| B. bifidum LMG 11041 | Infant stool sample | 1.83 × 108 | 1.44 × 108 | 4.92 × 107 | 1.44 × 106 | 8.14 × 106 | 9.27 × 105 |
| B. bifidum LMG 11582 | Adult stool sample | 5.42 × 108 | 8.04 × 107 | 5.67 × 108 | 1.42 × 108 | 1.62 × 107 | 4.55 × 106 |
| B. bifidum LMG 11583 | Adult stool sample | 2.08 × 108 | 7.64 × 107 | 1.23 × 109 | 3.44 × 108 | 5.00 × 106 | 5.35 × 105 |
| B. bifidum LMG 13195 | Infant colonoscopic sample | 9.58 × 107 | 2.67 × 107 | 6.42 × 107 | 5.20 × 106 | ||
| B. bifidum LMG 13200 | Infant stool sample | 7.08 × 108 | 3.83 × 108 | 1.04 × 109 | 1.01 × 108 | ||
| B. bifidum PRL2010 | Infant stool sample | 5.42 × 108 | 2.40 × 108 | 1.13 × 109 | 1.95 × 108 | 4.15 × 107 | 4.74 × 106 |
| B. breve 12L | Human milk | 1.30 × 109 | 1.26 × 109 | 1.43 × 109 | 1.32 × 108 | 7.43 × 107 | 6.56 × 106 |
| B. breve 2L | Human milk | 1.23 × 109 | 5.13 × 108 | 1.42 × 109 | 7.52 × 108 | 1.33 × 108 | 2.74 × 107 |
| B. breve 31L | Human milk | 1.17 × 109 | 5.77 × 108 | 3.25 × 108 | 6.61 × 107 | 2.09 × 104 | 1.10 × 104 |
| B. breve 687B | Adult stool sample | 1.55 × 109 | 1.42 × 108 | 1.46 × 109 | 1.68 × 108 | ||
| B. breve 689B | Adult stool sample | 1.73 × 109 | 1.95 × 108 | 2.68 × 108 | 7.39 × 107 | ||
| B. breve 691B | Adult stool sample | 9.08 × 108 | 1.26 × 108 | 1.72 × 109 | 2.88 × 108 | 1.11 × 108 | 5.13 × 106 |
| B. breve LMG 13208 | Infant colonoscopic sample | 2.33 × 108 | 1.44 × 107 | 3.35 × 108 | 9.84 × 107 | 4.34 × 107 | 8.40 × 106 |
| B. catenulatum 1231B | Human gut | 1.00 × 108 | 5.00 × 107 | 6.00 × 107 | 5.00 × 106 | ||
| B. catenulatum 1232B | Human gut | 1.67 × 108 | 8.04 × 107 | 1.09 × 108 | 1.91 × 107 | ||
| B. catenulatum 1233B | Human gut | 2.67 × 108 | 1.42 × 108 | 1.13 × 108 | 3.03 × 107 | ||
| B. catenulatum 1234B | Human gut | 2.08 × 108 | 1.46 × 108 | 8.00 × 107 | 2.70 × 107 | ||
| B. catenulatum LMG 11043 | Colonoscopic sample | 8.48 × 108 | 1.49 × 108 | 2.17 × 108 | 1.18 × 108 | ||
| B. dentium 125B | Adult stool sample | 1.53 × 109 | 4.04 × 108 | 2.25 × 108 | 1.25 × 108 | ||
| B. dentium 181B | Adult stool sample | 3.83 × 108 | 1.53 × 108 | 7.33 × 107 | 2.84 × 107 | ||
| B. dentium 183B | Adult stool sample | 3.00 × 108 | 1.75 × 108 | 6.08 × 107 | 1.66 × 107 | ||
| B. dentium 369B | Adult stool sample | 6.42 × 108 | 2.45 × 108 | 1.08 × 108 | 2.60 × 107 | ||
| B. dentium LMG 11405 | Oral cavity | 1.67 × 108 | 1.26 × 108 | 6.67 × 107 | 1.44 × 107 | 3.19 × 106 | 2.75 × 106 |
| B. gallicum LMG 11596 | Colonoscopic sample | 5.92 × 108 | 2.31 × 108 | 7.67 × 108 | 2.93 × 108 | 5.72 × 107 | 7.25 × 106 |
| B. longum 123B | Colonoscopic sample | 1.50 × 109 | 9.01 × 108 | 4.33 × 108 | 1.38E × 108 | ||
| B. longum 134B | Colonoscopic sample | 1.33 × 108 | 7.64 × 107 | 1.75 × 108 | 7.50 × 107 | 1.85 × 107 | 6.03 × 106 |
| B. longum 159B | Colonoscopic sample | 1.58 × 108 | 5.20 × 107 | 2.58 × 108 | 8.04 × 107 | 1.77 × 106 | 7.33 × 105 |
| B. longum 207B | Colonoscopic sample | 1.67 × 107 | 1.42 × 107 | 6.67 × 107 | 2.27 × 107 | 1.86 × 107 | 3.96 × 106 |
| B. longum 220B | Colonoscopic sample | 3.08 × 108 | 1.66 × 108 | 3.00 × 108 | 2.84 × 108 | ||
| B. longum 224B | Colonoscopic sample | 2.00 × 109 | 4.33 × 108 | 1.57 × 109 | 2.25 × 108 | ||
| B. longum 229B | Colonoscopic sample | 4.92 × 109 | 2.16 × 109 | 9.08 × 108 | 2.01 × 108 | ||
| B. longum 296B | Adult stool sample | 1.42 × 109 | 1.18 × 109 | 6.00 × 108 | 1.75 × 108 | ||
| B. longum 314B | Colonoscopic sample | 2.08 × 108 | 5.20 × 107 | 2.50 × 108 | 0 | 1.78 × 107 | 3.30 × 106 |
| B. longum 319B | Colonoscopic sample | 3.50 × 108 | 3.04 × 108 | 5.25 × 108 | 1.89 × 108 | 3.53 × 107 | 1.74 × 107 |
| B. longum 340B | Adult stool sample | 8.92 × 108 | 1.46 × 108 | 2.17 × 108 | 5.20 × 107 | ||
| B. longum 346B | Adult stool sample | 3.33 × 108 | 2.13 × 108 | 3.25 × 108 | 1.50 × 108 | ||
| B. longum 350B | Adult stool sample | 4.58 × 108 | 1.44 × 107 | 2.42 × 108 | 1.01 × 108 | ||
| B. longum 351B | Adult stool sample | 1.42 × 108 | 7.64 × 107 | 7.17 × 107 | 3.82 × 106 | ||
| B. longum 397B | Adult stool sample | 1.07 × 109 | 7.51 × 107 | 5.25 × 108 | 1.15 × 108 | ||
| B. longum 419B | Adult stool sample | 5.00 × 108 | 1.32 × 108 | 4.25 × 108 | 1.52 × 108 | ||
| B. longum 428B | Adult stool sample | 8.25 × 108 | 3.27 × 108 | 2.75 × 108 | 9.01 × 107 | ||
| B. longum 432B | Adult stool sample | 1.39 × 109 | 2.69 × 108 | 5.08 × 108 | 2.27 × 108 | ||
| B. longum 433B | Adult stool sample | 2.50 × 109 | 5.00 × 108 | 1.15 × 109 | 4.95 × 108 | ||
| B. longum 434B | Adult stool sample | 1.92 × 109 | 1.26 × 109 | 5.50 × 108 | 7.50 × 107 | ||
| B. longum 442B | Adult stool sample | 6.08 × 108 | 5.20 × 107 | 7.33 × 108 | 2.25 × 108 | 7.38 × 105 | 7.61 × 104 |
| B. longum 447B | Adult stool sample | 1.13 × 109 | 4.06 × 108 | 2.83 × 108 | 1.46 × 108 | ||
| B. longum 451B | Adult stool sample | 1.31 × 109 | 3.06 × 108 | 5.08 × 108 | 2.75 × 108 | ||
| B. longum 499B | Colonoscopic sample | 3.00 × 108 | 1.15 × 108 | 5.17 × 107 | 3.82 × 106 | ||
| B. longum 553B | Colonoscopic sample | 2.33 × 109 | 5.20 × 108 | 5.92 × 108 | 2.60 × 108 | ||
| B. longum 606B | Colonoscopic sample | 9.17 × 107 | 9.46 × 107 | 2.50 × 106 | 0 | ||
| B. longum 633B | Adult stool sample | 2.92 × 109 | 1.88 × 109 | 8.75 × 108 | 2.54 × 108 | ||
| B. longum 707B | Adult stool sample | 9.25 × 108 | 7.15 × 108 | 3.75 × 108 | 1.56 × 108 | ||
| B. longum 71B | Adult stool sample | 3.00 × 107 | 8.66 × 106 | 2.42 × 107 | 6.29 × 106 | ||
| B. longum 743B | Adult stool sample | 1.58 × 108 | 1.46 × 108 | 4.92 × 107 | 2.63 × 107 | ||
| B. longum 861B | Adult stool sample | 1.83 × 108 | 5.77 × 107 | 2.42 × 108 | 7.64 × 107 | 8.86 × 105 | 1.54 × 105 |
| B. longum 908B | Colonoscopic sample | 2.07 × 109 | 4.16 × 108 | 8.75 × 108 | 6.61 × 107 | ||
| B. longum subsp. longum LMG 13197 | Colonoscopic sample | 1.42 × 109 | 7.22 × 108 | 5.33 × 108 | 4.47 × 108 | ||
| B. pseudocatenulatum 202B | Colonoscopic sample | 4.45 × 108 | 3.82 × 108 | 3.08 × 108 | 1.28 × 108 | ||
| B. pseudocatenulatum 263B | Adult stool sample | 3.42 × 108 | 2.45 × 108 | 9.67 × 107 | 2.89 × 107 | ||
| B. pseudocatenulatum 289B | Adult stool sample | 2.50 × 108 | 4.33 × 107 | 9.33 × 107 | 6.29 × 106 | ||
| B. pseudocatenulatum 318B | Colonoscopic sample | 3.33 × 108 | 8.04 × 107 | 1.92 × 108 | 1.44 × 107 | ||
| B. pseudocatenulatum LMG 10505 | Infant stool sample | 1.67 × 109 | 1.28 × 109 | 6.67 × 107 | 1.44 × 107 | ||
| B. pseudolongum subsp. globosum 555B | Colonoscopic sample | 1.33 × 108 | 2.89 × 107 | 3.75 × 108 | 2.50 × 107 | 5.52 × 105 | 5.89 × 105 |
| B. pseudolongum subsp. globosum 685B | Adult stool sample | 2.58 × 108 | 1.44 × 108 | 3.17 × 108 | 1.42 × 108 | 4.89 × 107 | 3.86 × 106 |
| B. pseudolongum subsp. globosum 686B | Colonoscopic sample | 3.08 × 108 | 2.04 × 108 | 1.83 × 108 | 5.91 × 107 | ||
| B. pseudolongum subsp. globosum LMG 11596 | Bovine rumen | 5.83 × 107 | 3.82 × 107 | 7.50 × 107 | 3.54 × 107 | 4.05 × 107 | 1.17 × 107 |
| B. stercoris JCM 15918 | Adult stool sample | 7.92 × 107 | 2.13 × 107 | 1.25 × 107 | 2.50 × 106 |
aATCC: American Type Culture Collection, USA; LMG, Belgian Co-ordinated Collection of Microorganisms-Bacterial Collection, Belgium; DSM, German Collection of Microorganism and Cell Cultures, Germany; JCM Japan Collection of Microorganisms, Japan.
bEC: Enzyme Commission.
Values are expressed as the means ± standard errors from three experiments.
Figure 1.
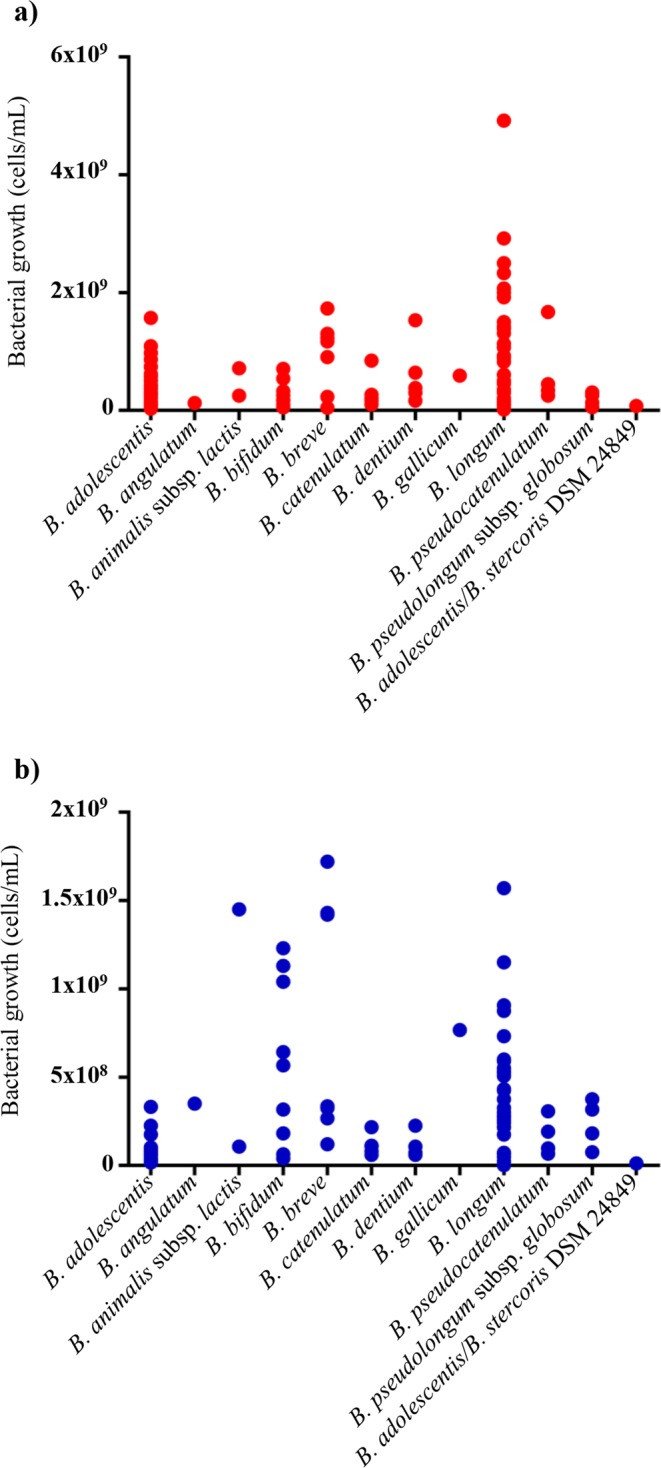
Bifidobacterial growth in MRS and chitin-glucan. The dispersion graphs report bacterial growth for the different bifidobacterial species included in this study, expressed as cells/mL. Panel a shows the growths in complete MRS (in red), while panel b depicts growths in MRS w/o glu + CG (in blue). Each growth reported is the average value of three experiments.
For those bifidobacterial strains showing an identical or higher growth level in MRS w/o glu + CG vs. MRS, growth performances were further evaluated using a different approach, i.e. viable cell count on MRS Agar (Table 1). Moreover, three bifidobacterial strains, i.e., B. bifidum LMG 11041, B. breve 31L and B. dentium LMG 11405, were selected as controls, since these strains showed reduced growth levels in MRS w/o glu + CG when compared to the cell numbers reached when grown on MRS.
Interestingly, all counts obtained through the plating method were lower than those obtained employing the Thoma cell counting chamber. This may not be surprising, since the Thoma chamber utilizes an indirect counting method and is unable to distinguish between living or devitalized cells, while the plating method exclusively evaluates viable cells. Notably, all strains grown using CG as the sole carbon source and then plated on MRS Agar were shown to be able to utilize the substrate tested. Bacterial counts ranged from 2.09 × 104 CFU/mL for B. breve 31L cultures to 1.33 × 108 CFU/mL for B. breve 2L cultures. Thus, B. breve 2L was the strain showing the highest growth performance when CG was provided as the sole carbon source. For this reason, B. breve 2L was chosen as a model bifidobacterial strain to dissect the genetic repertoire responsible for efficient CG metabolism.
Identification of genes induced by CG in the genome of B. breve 2L
In order to identify the genes of B. breve 2L responsible for CG metabolism, we evaluated the transcriptomes of this strain when cultivated on CG by RNAseq analyses. The average transcriptome profile corresponding to the three replicates of B. breve 2L cultivated on MRS w/o glu + CG (0.5%(w/v) as the unique carbon source was compared with the average transcriptome profile of the three replicates of the positive control (B. breve 2L grown on complete MRS). Sequencing reads of B. breve 2L grown on CG as well as on glucose were mapped on the genome sequence of B. breve 2L. Subsequently, evaluation of RPKM (reads per kilobase per million mapped reads) values for each gene revealed high expression (ranging from 1464 to 24685 RPKM) of genes predicted to encode carbohydrate transporters (Table S1, annotation highlighted in red).
Notably, B. breve 2L grown in MRS w/o glu + CG (0.5% (w/v) showed increased transcription, ranging from 8.1 to 106-fold, of various genes, some of which were predicted to be involved in carbohydrate internalization (Table S2, annotation highlighted in red). Furthermore, significant transcriptional up-regulation, ranging from 8.2-fold to 14.6-fold, was observed for three genes encoding enzymes putatively involved in the hydrolysis of glycosidic linkages between hexose sugars (Table S2, annotation highlighted in blue).
A detailed scrutiny of the upregulated B. breve 2L genes upon its cultivation on MRS w/o glu + GC, revealed that the majority of these genes were organized in seven loci, which are predicted to encode carbohydrate transport systems (Table S3, annotation highlighted in red), or are coding for putative glycosyl hydrolases (Table S3, annotation highlighted in blue).
Altogether, these results indicate that the inclusion of CG in the growth medium as the unique carbohydrate source, modulates the expression of genes encoding enzymes toward the degradation and metabolism of CG.
Notably, CG was also observed to increase transcription of tad genes (Table S3, annotation highlighted in green) responsible for the synthesis and assembly of the Type IVb pilus locus, which has been shown in another B. breve strain to mediate the colonization and persistence of bifidobacterial cells in the mammalian gut16–18.
Evaluation of the colonization of B. breve 2L in rats following GC treatment
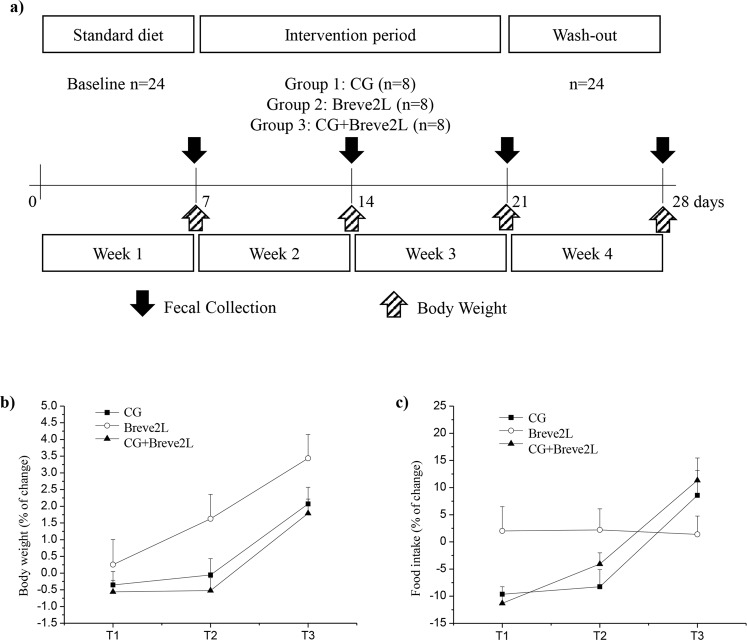
In order to evaluate if CG is able to modulate colonization of B. breve 2L in the mammalian gut, we performed an in vivo trial using Groningen rats (Rattus norvegicus). The trial consisted of three groups of animals, one receiving a daily inoculum of approximately 109 CFU of B. breve 2L, i.e., Breve2L, a second group fed with standard diet supplemented with 10% CG, i.e., CG, and another one treated with the same amount of B. breve 2L cells plus 10% CG, i.e., CG + Breve2L (Fig. 2).
Figure 2.
Design and results of the experimental procedure. Panel a shows the schedule of the experimental procedures. Panel b and c display the body weight and food intake percentage changes relative to the respective ‘T0’ values, during the experiment, respectively. Values are expressed as means ± SEM.
The putative physiological effect of CG on the Body Weight (BW) of the animals was recorded and compared to the Food Intake (FI) (Fig. 2b,c). Two-way ANOVA for repeated measures provided a significant effect of CG over time for BW changes (p-value < 0.05) and FI/BW (p-value < 0.01). BW increment was significantly higher in the Breve2L group as compared to the CG + Breve2L group at the end of the intervention period (T2) (p-value < 0.05). FI/BW ratio was significantly higher in the Breve2L group compared to either the CG (p-value < 0.05) or CG + Breve2L groups (p-value = 0.01), after the first week of the intervention period (T1). These data suggest that CG stimulates appetite while reducing/limiting body weight increment. In this context, characterization of the modulatory effects of CG toward the mammalian gut microbiota composition is pivotal to understand the physiological and metabolic aspects responsible for the reduction of body weight.
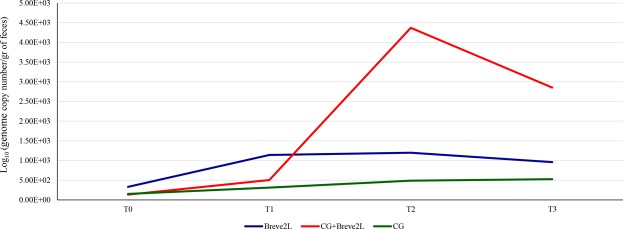
In order to assess the number of B. breve 2L in the fecal samples of the animals enrolled in this study, we applied a qPCR approach (Fig. 3). These analyses highlighted that rats of the CG groups did not show a statistically significant increment of B. breve 2L at the end of the experiment (T3) compared to the other time points. In contrast, a statistically significant increased load of B. breve 2L (p-value < 0.05) was observed at time points T1 (1.14E + 03 CFU/gr) and T2 (1.20E + 03 CFU/gr) compared to T0 (3.30E + 02 CFU/gr) in the Breve2L group, but there were no significant differences with respect to T3 (9.58E + 02 CFU/gr). These data suggest that the daily administration of B. breve 2L allows a transient colonization of the bacterial species as its concentration decreases after the washout week (T3). In this context, when rats were fed with both CG and B. breve 2L, the abundance of the B. breve strain was higher in T2 (4.37E + 03) and T3 (2.85E + 03) as compared to that observed for rats fed with only B. breve 2L, i.e. 1.20E + 03 CFU/gr at T2 and 9.58E + 02 CFU/gr at T3. Thus, supplementation of CG in the standard diet appeared to cause an increase in the abundance of B. breve 2L, thus enhancing gut colonization/persistence of this strain.
Figure 3.
Quantitative PCR evaluations of the load of B. breve 2L in stool samples of rats. The graph reports the average abundance of B. breve 2L observed through qPCR in CG, Breve2L and CG + Breve2L groups at T0, T1, T2 and T3.
Characterization of CG effects on the rat gut microbiota composition
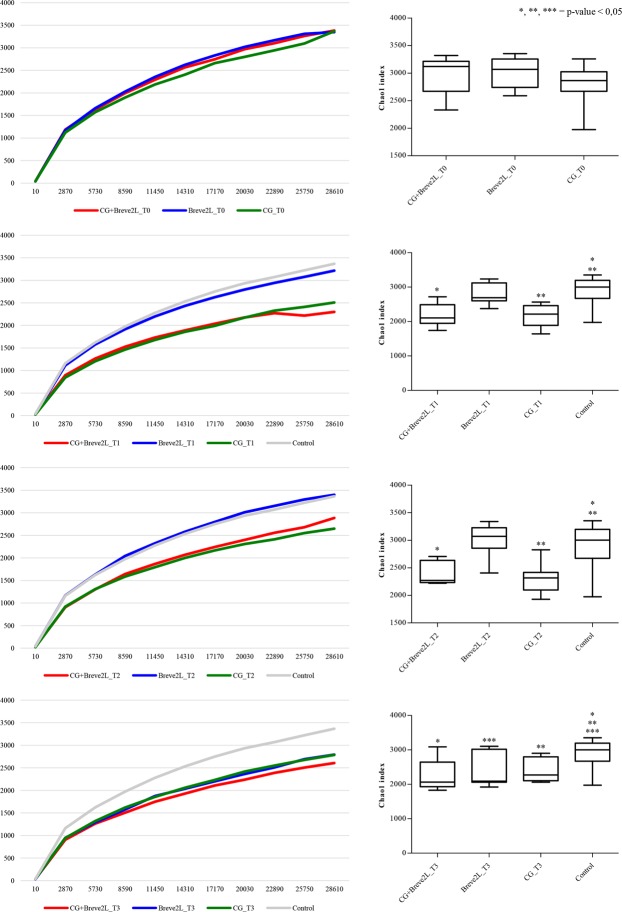
Evaluation of the gut microbiota composition of the animals enrolled in this study was performed by 16 S gene rRNA microbial profiling analyses on fecal samples collected during the trial. Analysis of sequencing data produced a total of 4,581,503 quality-filtered reads with an average of 47,724 reads per sample (Table S4). Evaluation of alpha-diversity, i.e. the biodiversity, of the collected rat fecal samples was performed through analysis of rarefaction curves constructed with 10 sub-sampling of the whole sequenced datasets. Notably, alpha-diversity data revealed, as expected, that at T0 all animal groups possessed similar microbiota diversity (t-test p-values > 0.05) (Fig. 3). T0 samples were used as “Control” non-treated group for analysis of T1, T2 and T3 time points. Intriguingly, analysis of data collected for T1 and T2 revealed that CG supplementation appeared to reduce the microbiota diversity, as observed for CG and CG + Breve2L compared to T0 and Breve2L sample groups (Fig. 4). These observations were confirmed by statistical analysis by means of t-test at 20,000 reads of CG and CG + Breve2L with respect to the same animal at T0, resulting in p-values < 0.05 (Fig. 4). Furthermore, data collected at T3 showed that cessation of CG supplementation does not reverse the biodiversity of the CG and CG + Breve2L groups to pre-treatment levels. These observations indicate that CG modulates the microbiota through selection of specific bacterial taxa, with subsequent reduction of the overall gut microbiota diversity, which persists (at least for the period tested) following termination of CG supplementation. Similar data were previously observed for other prebiotic compounds7.
Figure 4.
Alpha-diversity of CG, Breve2L and CG + Breve2L at T0, T1, T2 and T3 time points. Panels (a–d) report average alpha-diversity obtained using Chao1 index for T0, T1, T2 and T3 time points, respectively.
Beta-diversity analysis did not reveal any statistically significant differences between sample groups at T0 (PERMANOVA p-value > 0.05) and confirmed the modulatory effect exerted by CG at T1 and T2 (Fig. 5). In fact, PERMANOVA statistical analysis between CG and CG + Breve2L groups at T1 and T2 with respect to T0 datasets resulted in all cases in p-values < 0.05 (Fig. 5). Notably, cessation of CG supplementation causes the CG and CG + Breve2L sample groups to cluster together with Breve2L and Control datasets at T3 (Fig. 5). Thus, beta-diversity data confirms the modulatory effect exerted by CG on the animals’ gut microbiota composition.
Figure 5.
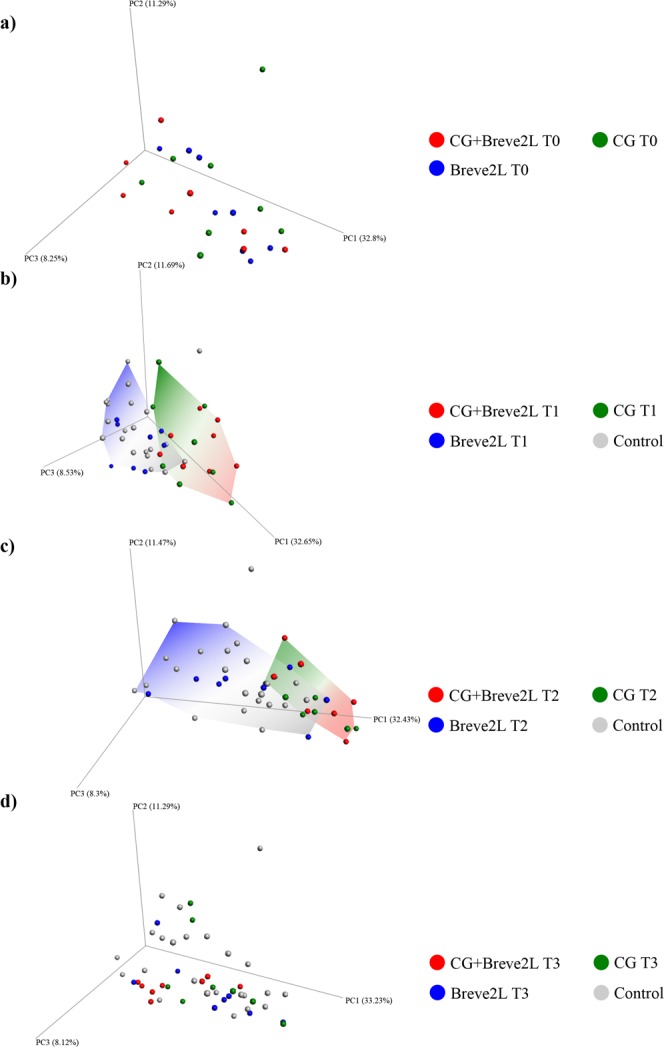
Beta-diversity of CG, Breve2L and CG + Breve2L at T0, T1, T2 and T3 time points. Panels a,b,c and d report three dimensional PCoA obtained using Weighted Unifrac index for T0, T1, T2 and T3 time points, respectively.
Taxonomic composition of all samples was reconstructed at phylum and genus levels in order to detail the impact of CG on the gut microbiota (Supplementary File 1). A t-test between the relative abundance of each profiled bacterial genus observed in the CG, Breve2L and CG + Breve2L groups was performed and compared between the various time points. Furthermore, statistically significant taxa observed for each comparison were mapped in order to precisely identify those bacterial genera whose relative abundance was increased or decreased upon CG and/or B. breve 2 L supplementation (Fig. 6). Intriguingly, 10 and 12 taxa showed increased (when compared to T0) relative abundance during CG supplementation at T1 and T2, respectively, followed by a decrease in abundance at T3, i.e. after cessation of CG supplementation (Fig. 6). In this context, it is worth mentioning that Prevotella 1, U. m. of Prevotellaceae family and Eubacterium ventriosum group (Lachnospiraceae family) increased their abundance by 100%, 145.4% and 2072.5% in T2 when compared to T0 (Fig. 6) (Supplementary File 1). In contrast, 42 and 38 taxa showed decreased relative abundance at T1 and T2, respectively, followed by an increase in abundance at T3 (Fig. 6) (Supplementary File 1). The observed decreased relative abundance of Bacteroides as well as Ruminococcus (−15.5% and −67.2% in T2 when compared to T0) and the simultaneous increase in relative abundance of Prevotella suggest that CG promotes a shift of the gut microbiota towards a Type 2 enterotype, i.e. Prevotella-driven enterotype19.
Figure 6.
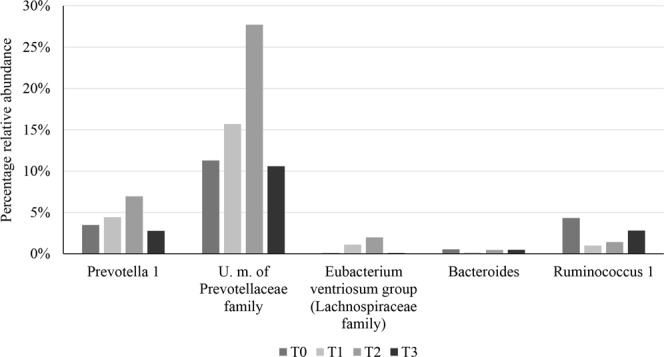
Bacterial genera whose abundance is highly modulated by CG. The bar plot reports the average relative abundance observed in CG samples group of relevant genera modulated by CG supplementation.
Intriguingly, CG was observed to induce increased relative abundance of Akkermansia, a taxon with health-promoting activities20 and reduced relative abundance of Peptoclostridium, a genus encompassing opportunistic pathogens21.
Analysis of the impact of B. breve 2L and CG administration on the animal microbiota could not be accurately performed through 16S rRNA gene microbial profiling due to the low relative abundance of bifidobacteria in the assessed fecal samples. For this reason, we performed a detailed cataloguing of bifidobacterial communities through the use of a previously published bifidobacterial ITS profiling approach22,23, allowing a detailed cataloguing of the bifidobacterial population down to the (sub)species level.
Cataloguing of the rat fecal bifidobacterial communities residing
In order to detail the impact of CG supplementation on the bifidobacterial population colonizing the rat gut, we performed bifidobacterial ITS profiling of DNA extracted from all fecal samples collected in this study. Sequencing produced a total of 316,080 reads, with an average of 3,293 reads per sample (Table S5).
In order to detail the modulatory effects of CG, we performed inspection of (sub)species-level profiles and statistical analysis through t-test of CG, Breve2L as well as CG + Breve2L groups of samples at T1, T2 and T3 compared to T0 (Supplementary File 2). As expected, an increase in the relative abundance of B. breve was observed in the Breve2L and CG + Breve2L groups at T1 and T2, which was followed by a relative decrease in B. breve 2L abundance at T3 (Fig. 7). Moreover, data of samples constituting the CG group revealed that CG supplementation induces a statistically significant increase in relative abundance (p-value < 0.05) of eight bifidobacterial species at T2 (Supplementary File 2). Furthermore, when CG was supplemented in combination with B. breve 2L, this bifidobacterial species (and most likely the B. breve 2L strain) was shown to reach a higher relative abundance as compared to those animals only receiving B. breve 2L (Fig. 7), as also indicated by qPCR results (Fig. 2). These results indicate that CG exerts a species-specific modulation of the bifidobacterial population harbored by the rat gut.
Figure 7.
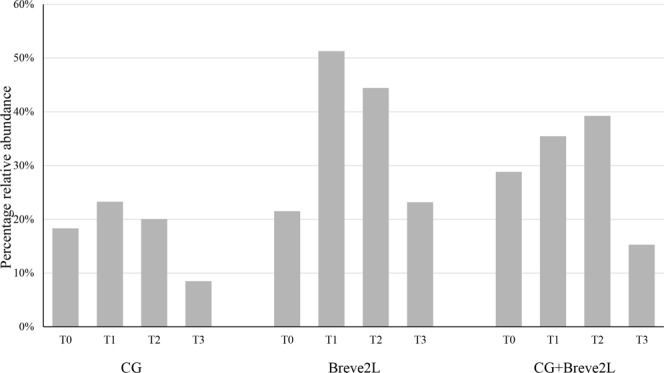
B. breve 2L relative abundance respect to the total bifidobacterial population. The bar plot reports the average relative abundance of B. breve 2L observed in CG, Breve2L and CG + Breve2L at T0, T1, T2 and T3.
Conclusion
Evaluation of the potential prebiotic features of chitin-glucan toward bifidobacteria under in vitro conditions highlighted the ability of 100 bifidobacterial strains to use this substrate as its sole carbon source. The bifidobacterial species that was most effective in utilizing CG for growth was shown to be B. breve. Notably, these results reflect the ability of this typical infant gut colonizer to utilize host-produced glycans, whose molecular structure partially resembles that of CG.
RnaSeq transcriptomics data was performed for the B. breve strain showing the best CG utilization capabilities, i.e. B. breve 2L, pointing out a modulation of the B. breve 2L genes involved in the transport and metabolism of hexose sugars.
The in vivo trials in a rat model clearly supported the notion that CG exploits a clear bifidogenic effect on bifidobacteria and in particular of small number of bifidobacterial species such as B. breve. These findings reinforce the potential of GC in modulating and shaping the bifidobacterial communities especially in ecological conditions where bifidobacteria are depleted (e.g., very often associated with metabolic disorders or gut diseases). In this context, the capability of bifidobacteria to use the CG as carbon source and the subsequent degradation of this bio-polymer in simpler derivatives, i.e. chitooligosaccharides (COS), may support the notion that CG act as a prebiotic. Indeed, it has been largely demonstrated that COS are able to inhibit the growth of pathogenic bacteria24,25. Moreover, CG consumption was shown to reduce body weight increment in rats, thus pointing at CG as an interesting novel prebiotic for prevention and treatment of obesity.
Such in vivo data should be further confirmed by clinical trials performed in human beings consuming CG or CG-based products (e.g., symbiotic products) in those categories of individuals where bifidobacterial abundance is naturally low (e.g., in the elderly) or is depleted as a consequence of metabolic disorder (e.g., constipation) or diseases (auto-immune diseases) as well as antibiotic therapy.
It is also arguable that cross-feeding interactions might be established by the different members of the bifidobacterial communities as well as with the different members of the human gut microbiota for the complete metabolism of CG. This was previously shown in co-participated trophic interactions15,26 where a partner partially metabolizes GC in favor to another microorganism that is genetically incapable to utilize this substrate. Thus, cross-feeding might represent another valuable way exploited by CG to induce a more general prebiotic effect in the human gut.
Methods
Strains and culture conditions
Bifidobacterium strains used in this study are listed in Table 1. Strains were routinely grown anaerobically in De Man, Rogosa, Sharpe (MRS) medium (Scharlau) containing 2% glucose (w/v), which was supplemented with 0.05% L-cysteine-HCl and incubated at 37 °C for 24 h. Anaerobic conditions were achieved by the use of an anaerobic cabinet (Ruskin), in which the atmosphere consisted of 17% CO2, 80% N2, and 2.99% H2.
Ethical statement
All experimental procedures and protocols involving animals were approved by the Veterinarian Animal Care and Use Committee of Parma University (approved protocol 370/2018-PR) and conducted in compliance with the European Community Council Directives dated 22 September 2010 (2010/63/UE).
Chitin-glucan growth assay
CG (KitoZyme, Belgium) was added to MRS without glucose (MRS w/o glu) at a final concentration of 0.5% (w/v), as previously assessed through growth experiments involving various bifidobacteria on different carbon sources14. This growth medium was termed MRS w/o glu + CG and subjected to sterilization employing a modified Tyndallization approach consisting of two thermal cycles at 80 °C for 30 minutes each alternated with cooling in ice.
For all growth tests, cells were recovered from an overnight culture and turbidity was measured at 600 nm, using a biophotometer (Eppendorf). A growth tube containing 6 mL of MRS w/o glu + CG was inoculated with active viable bacterial cells diluted to an OD600nm of ~1.0, obtaining a final inoculum with an OD600nm of ~0.1. Cultures were grown in biologically independent triplicates and the resulting growth data sets were expressed as the means from these replicates. Moreover, positive (MRS) and negative (MRS w/o glu) growth controls were performed. Cultures were incubated under anaerobic conditions at 37 °C for 24 h. Cell growth was monitored using a Thoma cell counting chamber (Herka).
For some bifidobacterial strains, cell growth was also monitored by viable cell count in MRS Agar. For this purpose, strains were inoculated as mentioned above in MRS w/o glu + CG. Then, 1 mL of each grown strain was serially diluted in PBS (Phosphate buffered saline) and plated on MRS agar. Plates were incubated under anaerobic conditions at 37 °C for 48 h. Bacterial growth was assessed by colony counting.
RNA-Seq transcriptomic analysis and identification of genes induced by CG
Bacterial cells were recovered from an overnight culture and turbidity was measured at 600 nm, using a biophotometer (Eppendorf). A growth tube containing 40 mL of complete MRS (reference condition) as well as 40 ml of MRS w/o glu CG was inoculated with viable bacterial cells diluted to an OD600nm of ~1.0, obtaining a final inoculum with an OD600nm of ~0.1. All growth conditions were performed by incubation in anaerobic cabinet at 37 °C. Following inoculation, growth was monitored and when an OD600nm value between 0.6 and 0.8 (exponential phase) cells were centrifuged at 6000 rpm for 5 min. Finally, prior to RNA extraction cells were frozen at −80 °C. Growth assays were carried out in triplicate. Total RNA was isolated from B. breve 2L cultures grown in MRS medium (Scharlau, Italy) as well as MRS w/o glu + GC. The obtained cell pellet was resuspended in 1 ml of QIAZOL (Qiagen, United Kingdom) and placed in a tube containing 0.8 g of glass beads (diameter, 106 μm; Sigma). Cells were lysed by shaking the mix on a Precellys 24 homogenizer (Bertin instruments, France). The mixture was then centrifuged at 13,000 rpm for 15 min, and the RNA-containing upper phase was recovered. RNA was further purified using RNeasy mini kit (Qiagen, UK) as reported in the manufacturer’s instructions.
RNA quality was checked by a Tape station 2200 (Agilent Technologies, USA) analysis. RNA concentration and purity were evaluated by Picodrop microliter spectrophotometer (Picodrop, UK).
For RNA sequencing, 2 μg of total RNA was treated to remove ribosomal RNA by the Ribo-Zero Magnetic Kit (Illumina), followed by purification of the rRNA-depleted sample by ethanol precipitation. RNA was processed according to the manufacturer’s protocol. The efficacy of rRNA depletion was checked by a Tape station 2200 (Agilent Technologies). Then, 500 ng of rRNA-depleted RNAs was fragmented using Bioruptor NGS ultrasonicator (Diagenode, USA) followed by size evaluation using Tape station 2200 (Agilent Technologies). A whole transcriptome library was constructed using the TruSeq Stranded RNA LT Kit (Illumina). Samples were loaded into a Flow cell V2 75 cycles (Illumina) as reported by the technical support guide.
Sequencing reads were mapped with Burrows-Wheeler Aligner (BWA)27 to the genomic sequence of B. breve 2L (NCBI accession number AWUG00000000.1). Reads Per Kilobase per Million mapped reads (RPKM) of each gene were assessed using Artemis software28.
Animal housing
Experiments involved 5-month-old male wild-type Groningen rats (Rattus norvegicus). After weaning, rats were housed in same sex sibling groups in rooms under humidity (50 ± 10%) and temperature-controlled conditions (22 ± 2 °C), a 12-h light-dark cycle (lights on at 7 a.m.), and with food and water available ad libitum.
Experimental design of the in vivo trials
From the initiation of the experiments, rats were housed individually in polymethyl methacrylate (Plexiglas®) cages (39 cm × 23 cm × 15 cm). The first week represented an acclimatization period, during which rats continued to consume a standard chow diet supplemented with an oral administration of 500 µl of sucrose solution (2%) in order to adapt the rats to drink from the syringe. Maintaining their habits, rats could represent the negative control of themselves, acting as the baseline for subsequent microbiota analyses29. For the following two weeks (14 days), rats (n = 24) were randomized to 3 groups: a first group fed with standard diet supplemented with CG [90% standard diet (w/w) + 10% KiOnutrime-CG from KitoZyme, Belgium; CG group], a second group fed with a standard diet and an oral treatment with B. breve 2L (Breve2L group) and a third group fed with standard diet supplemented with CG (same composition of the CG group) and oral treatment with B. breve 2L (CG + Breve2L group) (Table 2). The CG group rats during the intervention period were orally inoculated with 500 µl of sucrose solution (2%) in order to maintain the same condition for the three groups except for the experimental variables. Finally, during the last week of the wash-out period, all animals returned to the standard chow diet.
Table 2.
Fecal samples from rats collected at the different time points.
| Samples | Type of intervention | Time points |
|---|---|---|
| WT1A – WT8A | CG group | T0 |
| WT1B – WT8B | T1 | |
| WT1C – WT8C | T2 | |
| WT1D – WT8D | T3 | |
| WT9A – WT16A | Breve2L group | T0 |
| WT9B – WT16B | T1 | |
| WT9C – WT16C | T2 | |
| WT9D – WT16D | T3 | |
| WT17A – WT24A | CG + Breve2L group | T0 |
| WT17B – WT24B | T1 | |
| WT17C – WT24C | T2 | |
| WT17D – WT24D | T3 |
Standard diet consisted of 54.61% nitrogen-free extract (mainly represented by starch and hemicellulose), 5.54% fibers, 19.42% protein, 11.09% water, 2.58% lipids, and 6.76% ash (non-organic mineral matter) (3.9 kcal/g; 4RF21, Mucedola, Italy). The percentage of supplementation of the substrate CG to standard chow diet was fixed at 10% (w/w) as previously illustrated6.
Food intake (FI) and body weight (BW) were measured daily and weekly, respectively, and the BW changes and FI were calculated as previously described7.
Evaluation of Bifidobacterium breve cell numbers by qPCR
Quantitative PCR (qPCR) was assessed using the species-specific primers BiBre1 (5′-CCGGATGCTCCATCACAC-3′) and BiBre2 (5′-ACAAAGTGCCTTGCTCCCT-3′). qPCR was performed using GoTaq qPCR Master Mix (Promega, USA) on a CFX96 system (BioRad, CA, USA) following previously described protocols30. PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle of 95 °C for 2 min, followed by 40 cycles of 95 °C for 3 s and 56 °C for 30 s. The melting curve was 65 °C to 95 °C with increments of 0.5 °C/s. In each run, negative controls (no DNA) were included. A standard curve was built using the CFX96 software (BioRad).
Fecal bacterial DNA extraction and 16S rRNA/ITS microbial profiling
Fecal samples were subjected to DNA extraction using the QIAmp DNA Stool Mini Kit following the manufacturer’s instructions (Qiagen). Partial 16S rRNA gene sequences were amplified from extracted DNA using primer pair Probio_Uni/Probio_Rev, which targets the V3 region of the 16S rRNA gene sequence31. Partial ITS sequences were amplified from extracted DNA using the primer pair Probio-bif_Uni/Probio-bif_Rev, which targets the spacer region between the 16S rRNA and the 23S rRNA genes within the ribosomal RNA (rRNA) locus22. Illumina adapter overhang nucleotide sequences were added to the partial 16S rRNA gene-specific amplicons and to the generated ITS amplicons of 200 bp, which were further processed using the 16S Metagenomic Sequencing Library Preparation Protocol (Part No. 15044223 Rev. B—Illumina). Amplifications were carried out using a Verity Thermocycler (Applied Biosystems). The integrity of the PCR amplicons was analyzed by electrophoresis on a 2200 TapeStation Instrument (Agilent Technologies, USA). DNA products obtained following PCR-mediated amplification of the 16S rRNA gene sequences were purified by a magnetic purification step involving the Agencourt AMPure XP DNA purification beads (Beckman Coulter Genomics GmbH, Bernried, Germany) in order to remove primer dimers. DNA concentration of the amplified sequence library was determined by a fluorimetric Qubit quantification system (Life Technologies, USA). Amplicons were diluted to a concentration of 4 nM, and 5 µL quantities of each diluted DNA amplicon sample were mixed to prepare the pooled final library. 16S rRNA gene and ITS sequencing were performed using an Illumina MiSeq sequencer with MiSeq Reagent Kit v3 chemicals.
Metagenomics analyses
Following sequencing, the.fastq files were processed using a custom script based on the QIIME software suite32. Paired-end read pairs were assembled to reconstruct the complete Probio_Uni/ Probio_Rev and Probio-bif_Uni/Probio-bif_Rev amplicons. Quality control retained sequences with a length between 140 and 400 bp and mean sequence quality score >20 while sequences with homopolymers >7 bp and mismatched primers were omitted. In order to calculate downstream diversity measures (alpha and beta diversity indices, Unifrac analysis), 16S rRNA Operational Taxonomic Units (OTUs) were defined at ≥99% sequence homology using uclust33 and OTUs with less than 10 sequences were filtered. ITS Operational Taxonomic Units (OTUs) were defined at 100% sequence homology using uclust33. All reads were classified to the lowest possible taxonomic rank using QIIME32 and a reference dataset from the SILVA database34 for 16S rRNA data or an updated version of the bifidobacterial ITS database22 for ITS data. Biodiversity of the samples (alpha-diversity) were calculated with Chao1 and Shannon indexes. Similarities between samples (beta-diversity) were calculated by unweighted uniFrac35. The range of similarities is calculated between the values 0 and 1. PCoA representations of beta-diversity were performed using QIIME32.
Statistical analysis
For in vitro trials, statistical significance between means was analyzed using the Student’s t-test. Values are expressed as the means ± standard errors from three experiments. Statistical calculations were performed using the software program GraphPad Prism 5 (La Jolla, CA, USA).
For in vivo experiments, two-way ANOVA for repeated measures with ‘group’ as between-subject factor (three levels: CG group, Breve2L group and CG + Bbreve2L group) was performed for: (i) BW changes, with ‘time’ as within-subject factor (three levels: T1, T2, T3); (ii) FI-to-BW ratio, with ‘time’ as within-subject factor (four levels: week1, week2, week3 and week4). Follow up analysis was performed using Student’s t-test, with a Bonferroni correction for multiple comparisons.
Furthermore, the difference between the rarefaction curves and relative abundance of taxa, as well as the differential abundance of bacterial genera were statistically analysed by t-test. All statistical analyses were performed through SPSS software (www.ibm.com/software/it/analytics/spss/).
Data Deposition
Raw sequences of 16S rRNA gene profiling and bifidobacterial ITS profiling as well as RNAseq data are accessible through SRA study accession numbers SRP164382 and SRP164394.
Supplementary information
Acknowledgements
This study was supported by KitoZyme, Belgium. Financial support has been provided by a grant from the Walloon Region (ADIPOSTOP Project, convention 7366). This research benefited from the HPC (High Performance Computing) facility of the University of Parma, Italy. We furthermore thank GenProbio srl for financial support of the Laboratory of Probiogenomics.
Author Contributions
G.A., C.M. and S.D. designed and performed the experiments and wrote the manuscript. G.A. and S.D. performed the experiments. C.M., L.M. and F.B. performed the bioinformatics analyses. L.C. and R.S. performed the in vivo experiments. T.R. and S.M. provided the chitin-glucan charbohydrate. M.C.O. and S.M. contributed to the manuscript preparation. F.T. and A.S. participated in the design of the study. D.v.S. participated and supervised the study. M.V. conceived the study, participated in its design and coordination, and contributed to the manuscript preparation. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Giulia Alessandri, Christian Milani and Sabrina Duranti contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42257-z.
References
- 1.Marzorati, M. et al. High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator. Molecular nutrition & food research61, 10.1002/mnfr.201600150 (2017). [DOI] [PubMed]
- 2.Araujo D, Freitas F, Sevrin C, Grandfils C, Reis MA. Co-production of chitin-glucan complex and xylitol by Komagataella pastoris using glucose and xylose mixtures as carbon source. Carbohydr Polym. 2017;166:24–30. doi: 10.1016/j.carbpol.2017.02.088. [DOI] [PubMed] [Google Scholar]
- 3.Meichik NR, Vorob’ev DV. Chitin-glucan complex in cell walls of the Peltigera aphthosa lichen. Prikl Biokhim Mikrobiol. 2012;48:340–345. [PubMed] [Google Scholar]
- 4.Commission, E. Commission Decision of 2 February 2011 authorising the placing on the market of a chitin-glucan from Aspergillus niger as a novel food ingredient under Regulation (EC) No 258/97 of the European Parliament and of the Council. Official Journal of the European Union (2011).
- 5.Berecochea-Lopez A, et al. Fungal chitin-glucan from Aspergillus niger efficiently reduces aortic fatty streak accumulation in the high-fat fed hamster, an animal model of nutritionally induced atherosclerosis. Journal of agricultural and food chemistry. 2009;57:1093–1098. doi: 10.1021/jf803063v. [DOI] [PubMed] [Google Scholar]
- 6.Neyrinck AM, et al. Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J Nutr Biochem. 2012;23:51–59. doi: 10.1016/j.jnutbio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Ferrario C, et al. How to Feed the Mammalian Gut Microbiota: Bacterial and Metabolic Modulation by Dietary Fibers. Frontiers in microbiology. 2017;8:1749. doi: 10.3389/fmicb.2017.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 9.Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013;5:1417–1435. doi: 10.3390/nu5041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turroni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PloS one. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duranti, S. et al. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS microbiology ecology92, 10.1093/femsec/fiw191 (2016). [DOI] [PubMed]
- 12.Ventura M, Turroni F, Lugli GA, van Sinderen D. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. Journal of the science of food and agriculture. 2014;94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 13.Collins, S. & Reid, G. Distant Site Effects of Ingested Prebiotics. Nutrients8, 10.3390/nu8090523 (2016). [DOI] [PMC free article] [PubMed]
- 14.Turroni F, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Applied and environmental microbiology. 2014;80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell Motherway M, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization facto. r. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends in microbiology. 2012;20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Turroni, F. et al. Glycan Utilization and Cross-Feeding Activities by Bifidobacteria. Trends Microbiol, 10.1016/j.tim.2017.10.001 (2017). [DOI] [PubMed]
- 19.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Applied and environmental microbiology. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milani C, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016;6:25945. doi: 10.1038/srep25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milani C, et al. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS microbiology ecology. 2014;90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 23.Milani, C. et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J, 10.1038/ismej.2017.138 (2017). [DOI] [PMC free article] [PubMed]
- 24.Okawa Y, Kobayashi M, Suzuki S, Suzuki M. Comparative study of protective effects of chitin, chitosan, and N-acetyl chitohexaose against Pseudomonas aeruginosa and Listeria monocytogenes infections in mice. Biol Pharm Bull. 2003;26:902–904. doi: 10.1248/bpb.26.902. [DOI] [PubMed] [Google Scholar]
- 25.Simunek J, Koppova I, Filip L, Tishchenko G, Belzecki G. The antimicrobial action of low-molar-mass chitosan, chitosan derivatives and chitooligosaccharides on bifidobacteria. Folia Microbiol (Praha) 2010;55:379–382. doi: 10.1007/s12223-010-0063-0. [DOI] [PubMed] [Google Scholar]
- 26.Egan M, et al. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. Bmc Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Turroni F, et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 2016;10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani C, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milani C, et al. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PloS one. 2013;8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/Nmeth.F.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71:8228–8235. doi: 10.1128/Aem.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.