Abstract
Oxidative stress and a disturbed cellular protein homeostasis (proteostasis) belong to the most important hallmarks of aging and of neurodegenerative disorders. The proteasomal and autophagic-lysosomal degradation pathways are key measures to maintain proteostasis. Here, we report that hippocampal cells selected for full adaptation and resistance to oxidative stress induced by hydrogen peroxide (oxidative stress-resistant cells, OxSR cells) showed a massive increase in the expression of components of the cellular autophagic-lysosomal network and a significantly higher overall autophagic activity. A comparative expression analysis revealed that distinct key regulators of autophagy are upregulated in OxSR cells. The observed adaptive autophagic response was found to be independent of the upstream autophagy regulator mTOR but is accompanied by a significant upregulation of further downstream components of the canonical autophagy network such as Beclin1, WIPI1 and the transmembrane ATG9 proteins. Interestingly, the expression of the HSP70 co-chaperone BAG3, mediator of BAG3-mediated selective macroautophagy and highly relevant for the clearance of aggregated proteins in cells, was found to be increased in OxSR cells that were consequently able to effectively overcome proteotoxic stress. Overexpression of BAG3 in oxidative stress-sensitive HT22 wildtype cells partly established the vesicular phenotype and the enhanced autophagic flux seen in OxSR cells suggesting that BAG3 takes over an important part in the adaptation process. A full proteome analysis demonstrated additional changes in the expression of mitochondrial proteins, metabolic enzymes and different pathway regulators in OxSR cells as consequence of the adaptation to oxidative stress in addition to autophagy-related proteins. Taken together, this analysis revealed a wide variety of pathways and players that act as adaptive response to chronic redox stress in neuronal cells.
Keywords: Autophagy, Protein homeostasis, Adaptation, Oxidative stress, BAG3
Abbreviations: ATG, Autophagy related; BAG1, Bcl-2-associated athanogene 1; BAG3, Bcl-2-associated athanogene 3; BECN1, Beclin1; BNIP3, Bcl-2 interacting protein 3; BafA1, Bafilomycin A1; CHX, Cycloheximide; CTSD, Cathepsin D; CV, Canavanine; DLP1, Dynamin-like protein 1; Hsp70, Heat shock protein 70; LAMP1, Lysosomal‐associated membrane protein 1; LAMP2, Lysosomal‐associated membrane protein 2; LC3, Light chain 3 protein; LETM, Leucine zipper and EF-hand containing transmembrane protein; LFQ, Label-free quantification; mTOR, Mammalian target of rapamycin; MTT, (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide); PIK3C3, Class III PI3‐kinase; OPA1, Optic atrophy 1; PolyUB, Polyubiquitin; PURO, Puromycin; RAB18, Member RAS oncogene; RAPA, Rapamycin; siRNA, Small interfering RNA; TUB, Tubulin; TFEB, Transcription factor EB; TEM, Transmission electron microscopy; WIPI1, WD repeat domain phosphoinositide-interacting protein 1
Graphical abstract
Highlights
-
•
OxSR cells have significantly higher autophagic activity.
-
•
Several positive modulators of autophagy network are highly upregulated in OxSR cells e.g. BECN1, PI3KC3, WIPI1 and RAB18.
-
•
BAG3 is actively involved in maintenance of protein homeostasis and autophagic activity in OxSR cells.
1. Introduction
Oxidative stress, the accumulation of reactive oxygen species which chemically modify and damage proteins, lipids, and DNA, disturbs cellular homeostasis and has been linked to aging and the pathogenesis of diseases such as cancer and neurodegeneration [1]. Neuronal disorders like Alzheimer's Disease (AD), Parkinson's Disease and Amyotrophic Lateral Sclerosis (ALS) show increased levels of oxidation end products in affected brain regions [2,3]. Strikingly, in AD nerve cell death is not equally distributed throughout the brain but it is rather occurring in the pyramidal neurons of the entorhinal and the inferior temporal cortex, the hippocampus and the amygdala while other brain regions such as the cerebellum show resistance [2,4,5]. Some aspects characterizing the basis for this selective vulnerability of neurons in distinct brain regions were addressed before and pinned down to the ability to actually resist oxidative stress [6]. Also, in other neurodegeneration pathologies selective vulnerability (and resistance) is observed [7]. A highly upregulated antioxidant defense is also linked to the progression of certain tumor types [8,9]. Though oxidative stress resistance appears to be disadvantageous in the context of tumor therapy, it could be beneficial in the attempt to support long-term survival of cells that are particularly sensitive to oxidative stress, such as post-mitotic neuronal cells. Therefore, it would be a huge step forward to uncover the molecular basis of an adaptation to oxidative stress and redox stress-associated pathological mechanisms.
To overcome challenges of the cellular homeostasis network during elevated cellular stress, autophagy plays a crucial role. In this process, double-membraned vesicles, called autophagosomes, are formed which sequester large protein complexes and organelles and finally deliver them to lysosomes for degradation [10]. Functionally, autophagy is an intracellular recycling process that guarantees energy supply by generating amino acid building blocks during low nutritional condition [11]. Additionally, autophagy works as an essential stress-adaptive response and rescue mechanism by selective degradation of damaged organelles, protein aggregates and intracellular pathogens [12,13]. Recently, autophagy was found to be a survival response against oxidative stress in bone marrow–derived mesenchymal stromal cells [14] and, moreover, the activation of autophagy can protect against oxidative stress-induced neuronal damage in adult rats [15].
In the context of autophagy and oxidative stress, previously it has been shown that the HSP70 co-chaperone Bcl2-associated athanogene 3 (BAG3) is highly upregulated in many aggressive tumor types and a link between oxidative stress, enhanced autophagy and BAG3 expression is described for several tumor cells [[16], [17], [18], [19]]. BAG3 was identified as a key player in the BAG3-mediated selective macroautophagy [20] and established as an important partner of the cellular proteostasis network under oxidative and proteotoxic stress as well as in aging conditions [[21], [22], [23], [24]].
The concept of oxidative stress adaptation has been successfully applied by different groups employing clonal neuronal cells lines, such as rat pheochromocytoma PC12 and mouse clonal hippocampal HT22 cells [[25], [26], [27], [28], [29]]. Previous studies mainly focusing on the redox stress-resistance phenotype and its reversal in PC12 and HT22 cells revealed key roles for the transcription factor NF-κB, sphingolipids and increased levels of antioxidant enzymes to provide the oxidative stress resistance phenotype [[26], [27], [28]]. In our current study, we now systematically analyzed molecular and functional changes in HT22 cells stably adapted to redox stress as induced by hydrogen peroxide (here called OxSR cells) with a particular focus on the autophagy network. We observed an increased autophagic-lysosomal and a decreased proteasomal activity in OxSR cells and analyzed in detail the expression patterns of key autophagy regulators. In addition, we found that the expression of BAG3 and BAG3-mediated selective macroautophagy is upregulated suggesting BAG3 thus may play a particular role in oxidative stress adapted-cells. Finally, a whole proteome comparison between wildtype and OxSR cells revealed a wide range of alterations of key proteins involved in different cellular pathways in addition to the autophagy regulators demonstrating the massive impact of chronic redox stress on the protein expression pattern during oxidative stress adaptation.
2. Material & methods
2.1. Cell culture
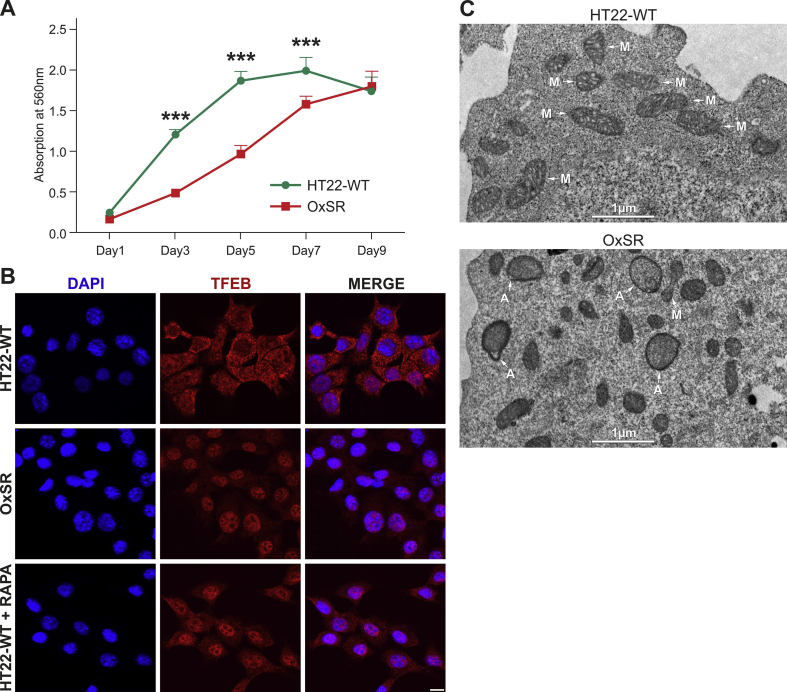
Wildtype HT22 cell line (HT22-WT), a cloned mouse hippocampal neuronal cell line which is very susceptible to oxidative stress [28,30], was used as control cell line. HT22 cells resistant to hydrogen peroxide-induced oxidative stress, here called OxSR cells, were established by clonal selection. The details of the selection procedure have been described elsewhere [31]. Both cell lines were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS), 1 mM sodium pyruvate and 1x penicillin/streptomycin (Invitrogen, Karlsruhe, Germany). To maintain the resistant phenotype, 450 μM of H2O2 f.c. (Sigma, Deisenhofen, Germany) was added twice a week to the OxSR cells. Prior to performing experiments, OxSR cells were cultured for three days without H2O2 and medium was exchanged daily to remove residual toxins. Although oxidative stress-resistant mouse hippocampal HT22 cells have been employed before, for the present study we initially reconfirmed the previously observed characteristics of the cell clones used here. So, the cell proliferation rates of the different cell clones were estimated by MTT assay. Consistent with previous findings [31] the growth rate of the OxSR cells was found to be lower than that of the HT22-WT cells (Suppl. Fig. S1A) confirming that increased vitality and oxidative stress resistance of the selected clones was not simply based on a higher proliferation rate.
2.2. Pharmacological agents and antibodies
Stock solutions of Bafilomycin A1 (LC Laboratories, B-1080), MG132 (Calbiochem, 474790), Cycloheximide (Sigma, 01810) and Rapamycin (Enzo, BML-A275-0025) were prepared in DMSO (Roth, A994.2). Stock solution of Canavanine (Santa Cruz Biotech, sc-202983A) and Puromycin (Sigma, P8833) was prepared in distilled H2O.
Antibody sources were as follows: for Actin (Sigma, A5060), BAG1 (Abcam, ab7976), BAG3 (Proteintech Group, 10599-1-AP), BECN1 (Cell Signaling, 3495), CTSD (Abcam, ab75852), DLP1 (BD Transduction Laboratories, 611113), LAMP2 (DSHB Biology, ABL-93), LC3B (Nanotools, 0260-100), LC3B (Sigma, L7543), OPA1 (BD Transduction Laboratories, 612607), Phospho mTOR (Abcam, ab109268), Puromycin (Millipore, MABE343), mTOR (Calbiochem, OP97), p62 (Progen, GP62-C), PIK3C3 (Cell Signaling, 4263), Poly-Ubiquitin (Dako, Z0458), RAB18 (Sigma, SAB4200173), Tubulin (Millipore, MAB1637), Tubulin (Sigma, T9026), TFEB (Proteintech Group, 13372-1-AP), Vimentin (SCBT, sc-373717), WIPI1 (Sigma, HPA007493).
2.3. Plasmids, siRNAs and transfection method
Expression plasmid for mouse FLAG tagged BAG3 (pFLAG-BAG3) was constructed by cloning partial mouse BAG3 cDNA containing the whole CDS into pEGFP‐N1 (Clontech). Forward primer sequence used to clone BAG3 plasmid is 5′-AAAGGATCCAGCGCCGCCACCC-3′ and the reverse primer sequence is 5′-GACTCTAGATCACTAGGGAGCCACCAGGTTGC-3'. After vector linearization with BamHI and XbaI, PCR products were inserted using the In‐Fusion reaction according to manufacturer's protocol (Clontech).
Two independent sets of siRNA against BAG3 were purchased from Eurofins MWG Operon with 3′‐dTdT overhangs. Sequence of used siRNA1 is 5′-AAUACCUGAUGAUCG AAGA-3′ and siRNA2 is 5′-AUACCUGAUGAUCGAAGAG-3'. Generally, cells were transfected by mixing 20 μg of each of the siRNA as duplex. In all knockdown experiments, same amounts of siRNA targeting a nonsense sequence (5′-AUUCUCCGAACGUGUCACG-3′) were transfected as control.
Cells were transfected by electroporation using the Amaxa Nucleofector I (program U‐24) and standard electroporation cuvettes (Sigma). Electroporation buffer details are mentioned elsewhere [21].
2.4. Immunoblotting and immunocytochemistry
Western blot analysis and inmmunocytochemistry were carried out as previously described [21]. Blots were developed with the Fusion-SL 3500 WL system (Peqlab, Erlangen, Germany) and densitometry analysis was performed using Aida Image Analyzer v.4.26 software (Raytest, Straubenhardt, Germany). For the labelling of the lysosomes and acidic vacuoles, the fluorescent dye LysoTracker was used. Cells (cultured on 24-well plates with coverslips) were incubated with 100 nM LysoTracker Red DND-99 (Thermo scientific, L7528) in culture medium for 1 h under incubator conditions and then fixed with 4% paraformaldehyde. Cells for immunocytochemistry were analyzed using a confocal laser-scanning microscope (LSM710, Zeiss) and images were processed with Adobe Photoshop CS5 (San Jose, CA, USA).
2.5. Transmission electron microscopy
Sample preparation and transmission electron microscopy (TEM) analysis were carried out as previously described [21] and images were processed with Adobe Photoshop CS5.
2.6. Proteomic analysis
Proteomic analysis was executed in different distinct steps such as protein extraction, sample preparation, 1-dimensional gel electrophoresis (1DE), mass spectrometry (MS) based discovery proteomics analysis, bioinformatics and functional annotation and pathways analyses. General approaches used for the analysis were adapted from previously published papers [[32], [33], [34], [35], [36], [37], [38], [39]]. Details of the individual techniques employed for this paper are described (see Suppl. File S4A).
2.7. Quantitative real time PCR array
RNA extraction, cDNA synthesis and quantitative real‐time PCR (qPCR) for HT22-WT and OxSR cells were performed by the protocol previously described [40]. A commercial ready to use primer mix in 96-well format (Biomol, MATPL-1) containing total 88 different genes involved in autophagy were used for qPCR array. PCR cycles were set according to the manufacturer's protocol. CT values obtained from quantitative real‐time PCR data were applied to REST software for calculating the relative fold change in expression of each gene [41]. Actin was used as a reference gene. A fold change of +1.5 or −0.6 is considered as regulated gene.
2.8. Measurement of proteasome activity
This procedure is adapted from previous publication [42]. Briefly, cells were washed with ice cold PBS and lysed in lysis buffer (50 mM HEPES, pH 7.8, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, and 5 mM DTT)at 4 °C. The homogenates were centrifuged at 10 000 g for 15 min at 4 °C. Supernatants were collected and normalized to protein content. Enzymatic reaction was started by mixing active cell extracts (containing 15 μg protein determined by BCA) with 100 μl of assay buffer. Assay buffer composition is same as lysis buffer only supplemented with ATP (2 mM) and proteasome substrate (100 μM, Suc‐LLVY‐AMC, Sigma). AMC fluorescence released by cleavage of substrate by proteolytic activity was recorded in a black 96‐well plate after 60 min incubation at 37 °C using the Victor 3 V Multilabel counter (Perkin Elmer). Specific proteasomal activity was determined by subtracting unspecific AMC fluorescence obtained in the presence of proteasome inhibitor MG132 (20 μM). Results are expressed as % AMC release.
2.9. Cell proliferation assay
The growth rate of HT22-WT and OxSR cells were determined by the analysis of cellular metabolic activity. The ability of cells to reduce yellow coloured water soluble MTT to a blue violet coloured water insoluble formazan crystals were measured colourimetrically over nine alternate days. The detailed procedure is described elsewhere [31].
2.10. SUnSET (surface sensing of translation) assay
Protein translation rates of HT22-WT and OxSR cells were determined with the SUnSET assay as previously described [43]. Briefly, cells were treated with 5 μg/ml puromycin for 30 min in absence or presence of 60 μg/ml cycloheximide. After cell lysis, protein extracts were analyzed by SDS-PAGE and immunoblotting with an anti-puromycin antibody to detect incorporated puromycin in translation products. The amount of puromycin-labelled proteins is corresponding to the amount of translation products synthesized during puromycin pulse.
2.11. Statistical analysis
Statistical significance of all quantified Western blots, LysoTracker staining and qPCR data was determined by unpaired t‐test using GraphPad Prism 7 (GraphPad Inc., San Diego, USA). Growth curve for cell proliferation assay was analyzed by two-way ANOVA with the same software. Statistical significance was accepted at a level of p < 0.05. The results are expressed as mean ± S.E.M. The proteomics data were statistically analyzed with Perseus software (version 1.6.1.0). The degree of variances between the biological replicates and groups were assessed by Pearson's correlation analysis and Student's two-sided t-test was used for the groups' comparison to identify the significantly differentially abundant proteins. Details are maintained in Suppl. File S4.
3. Results
3.1. Autophagosomes as well as autophagic-lysosomal activity are increased and mitochondrial fission-fusion balance is altered in OxSR cells
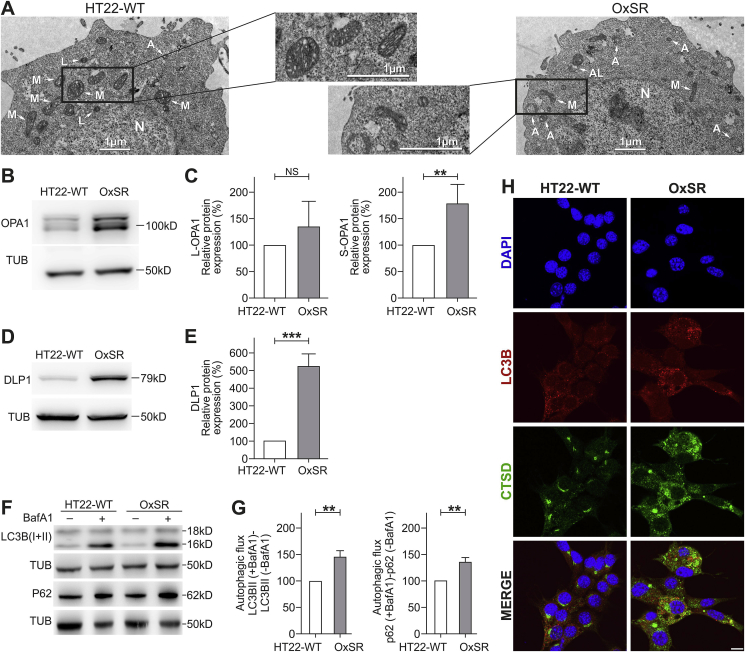
Based on previous findings that HT22 cells resistant to H2O2 (OxSR) show an upregulation of the lysosomal marker LAMP1 compared to oxidative stress-sensitive HT22 wildtype cells (HT22-WT) [28], we analyzed the cellular vesicular ultrastructure in detail. TEM revealed the presence of an intensive vesicular network in OxSR cells and a higher number and size of double-membrane autophagosomes as well as lysosomes in OxSR cells, suggesting an enhanced autophagic-lysosomal activity (Fig. 1A and Suppl. Fig. S1C). In addition, TEM revealed an overall altered mitochondrial morphology in OxSR cells (see Fig. 1A; mitochondria in OxSR cells show an elongated shape) and also an altered mitochondrial dynamics as indicated by the increased expression of DLP1 and OPA1 (S-OPA1 significantly increased) as markers of mitochondrial fission and fusion, respectively (Fig. 1B–E). It is known that endogenous or exogenous stress, such as oxidative stress, disturbs the fission-fusion balance leading to temporary or permanent changes in mitochondrial morphology [44]. The strong impact of chronic oxidative stress on mitochondria in OxSR cells and significant changes in mitochondrial homeostasis and metabolism were detected later in this study by mRNA expression studies and full proteome analysis (see 3.3, 3.7.). Next, we investigated the baseline autophagic flux in OxSR cells in presence of BafA1 which inhibits fusion of autophagosomes and lysosomes. We observed a significantly elevated autophagic flux, the enhanced accumulation of LC3B-II as well as p62 upon BafA1 treatment in OxSR cells compared to HT22-WT cells (Fig. 1F and G). Consistently, immunostainings showed a generally upregulated lysosomal activity in OxSR cells indicated by the lysosomal enzyme marker cathepsin D (Fig. 1H). Nuclear translocation of TFEB is known to have a pivotal role in upregulation of autophagy and lysosomal biogenesis [45]. TFEB was found to be localized mainly in the cytoplasm of HT22-WT cells and almost exclusively translocated to the nucleus in OxSR cells as shown by immunocytochemistry (Suppl. Fig. S1B) pointing towards an ongoing transcription of TFEB target genes in OxSR cells supporting the continuous autophagic activity.
Fig. 1.
OxSR cells show higher autophagic activity. (A) Ultrastructure analysis of HT22-WT and OxSR cells by TEM. OxSR cells clearly showed increased number and size of highly electron dense vacuolar autophagic vesicles at different stages of maturity and changes in overall mitochondrial morphology (representative images are shown; see further images in Suppl. Fig. S1C). Autophagosomes are indicated by A, autolysosomes by AL, nucleus by N and mitochondria by M. (B–E) Protein extracts from untreated samples of HT22-WT and OxSR cells were analyzed by Western blotting for OPA1 (B) and DLP1 (D) expression. Tubulin (TUB) was used as loading control. Bar graphs show changes in L-OPA1 and S-OPA1 (C) as well as in DLP1 protein levels (E) obtained by densitometry analysis and normalization to tubulin. Values represent mean ± S.E.M.; OPA1: n = 5, **p < 0.01; DLP1: n = 3, ***p < 0.001 and NS for no significant statistical difference. Control HT22-WT cells were set to 100%. (F, G) Protein extracts from HT22-WT and OxSR cells have been taken after 6 h of BafA1 (8 μM) treatment and expression of the indicated proteins was detected by Western blotting. The autophagic flux was measured after densitometric analysis. Therefore, tubulin normalized LC3B-II and P62 levels in the absence of the lysosomal inhibitor were subtracted from corresponding levels obtained in the presence of BafA1. Tubulin (TUB) was used as loading control. Values represent mean ± S.E.M., n = 3, **p < 0.01 and control HT22-WT cells were set to 100%. (H) HT22-WT and OxSR cells were immunohistochemically stained against LC3B and Cathepsin D (CTSD). DAPI (blue) was used to stain DNA. Scale bar: 10 μm. Pictures were taken by confocal microscopy. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
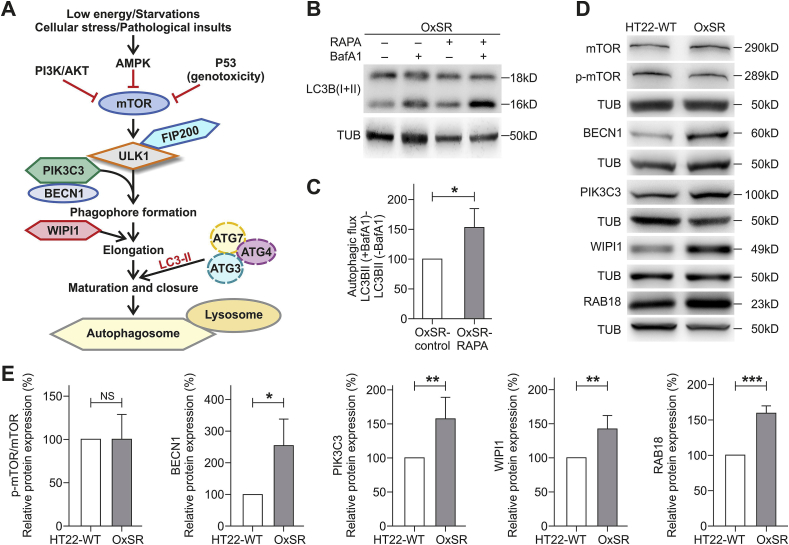
3.2. OxSR cells show enhanced expression of several key autophagy regulators but mTOR expression and phosphorylation status is unchanged
Next, we focused on the expression of certain key regulators of canonical autophagy in OxSR cells (as indicated in the schematic diagram in Fig. 2A). But first, to check the responsiveness of the mTOR-regulated autophagy pathway in OxSR cells, we treated the cells with rapamycin and monitored autophagic flux after co-treatment with BafA1. We found that rapamycin significantly increases autophagic flux in OxSR cells indicating that the mTOR-mediated autophagic pathway is still principally inducible in this cell line (Fig. 2B and C). Interestingly, in the Western blots no changes in the phosphorylation status of the main regulator of canonical autophagy mTOR (ratio of p-mTOR to mTOR as measure of mTOR-dependent autophagy induction) was detectable in the OxSR cells (Fig. 2D and E) indicating that mTOR although inducible is obviously not activated under basal conditions in OxSR cells adapted to redox stress.
Fig. 2.
Adaptation to oxidative stress enhances expression of Beclin1, WIPI1, PIK3C3 and RAB18. (A) Scheme displays major regulators of canonical autophagy. (B, C) OxSR cells were incubated simultaneously with 10 μM Rapamycin (RAPA) and 8 μM of BafA1 for 4 h and expression of the indicated proteins was detected by Western blotting. The autophagic flux was measured after densitometry analysis with LC3B-II levels normalized to tubulin. Values represent mean ± S.E.M., n = 4, *p < 0.05 and autophagic flux of untreated OxSR cells were set as control 100%. (D, E) Protein extracts from untreated HT22-WT and OxSR cells were used to perform Western blot analysis for detection of indicated proteins. The diagrams display the indicated protein levels after normalization to corresponding tubulin. Values represent mean ± S.E.M., n = 3 to 5, *p < 0.05, **p < 0.01, ***p < 0.001 and NS for no significant statistical difference. Control HT22-WT cells were set to 100%.
Another upstream regulator of autophagy is the PIK3C3/BECN1 complex required for the recruitment of PI(3)P during autophagosome generation. We found an almost 2.5-fold higher BECN1 protein level in OxSR cells indicating that the BECN1 complex is highly active in oxidative stress-adapted cells. WIPI1 acts downstream as PI(3)P effectors for proper membrane rearrangement [46,47] and consistent with the observed increased protein level of BECN1, Western blot analysis indeed revealed also upregulated levels of both PIK3C3 and WIPI1 in OxSR cells. Moreover, RAB18 was found to be upregulated in its expression approximately 1.6-fold in OxSR cells (Fig. 2D and E). The small Rab GTPase RAB18 is a known key regulator of ER-Golgi membrane trafficking, involved in cellular lipid metabolism and as recently shown also a positive modulator of autophagy [[48], [49], [50]]. Taken together, in OxSR cells we found a significant upregulation of the expression of distinct key proteins of canonical autophagy regulation comprising BECN1, WIPI1 and PIK3C3 as well as the positive autophagy regulator RAB18.
3.3. Autophagy qPCR array reveals a high upregulation of ATG9 mRNA in OxSR cells
Employing a qPCR array representing a wide range of genes involved in the direct regulation of various stages of the autophagic process itself as well as candidates that are not part of the core canonical autophagy machinery, we found a highly differential mRNA expression pattern comparing OxSR and WT cells. OxSR cells showed significantly higher mRNA expression of certain ATG proteins, including ATG10, ATG12, ATG5, as well as GABRAPL2 and AKT1S (Table 1). Moreover, consistently with our Western blot findings (see Fig. 2), the qPCR also showed an elevated level of BECN1 and WIPI1 mRNA in OxSR cells. Further, the expression of the lysosomal marker LAMP1 mRNA was upregulated in OxSR cells, confirming earlier observation in H2O2-resistant HT22 cells (Table 1) [28].
Table 1.
Autophagy-associated gene expression profiling of stress-resistant OxSR cells. Total RNA from HT22-WT and OxSR cells was characterized by using the mouse Autophagy Primer Library 1 (MATPL-1) and relative fold change in gene expression profile was analyzed by comparing OxSR cells with HT22-WT cells as control. Numbers in italics indicate an upregulation greater than 1.5-fold, bold numbers indicate a downregulation equal or lower than 0.6-fold. Values represent mean ± S.E.M., n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 and NS for no significant statistical difference.
| OxSR vs HT22-WT |
|||
|---|---|---|---|
| Gene Name | Fold Change | Significance level | |
| ATG1/ULK complex | Ulk2 | 0.4 | * |
| Ambra1 | 2.4 | NS | |
| Other ATG | Atg10 | 2.3 | *** |
| Atg12 | 1.6 | *** | |
| Atg16L2 | 0.6 | * | |
| Atg4A | 3.3 | *** | |
| Atg4B | 1.5 | NS | |
| Atg4C | 2.6 | NS | |
| Atg4D | 2.6 | NS | |
| Atg5 | 2.3 | *** | |
| Atg7 | 2.0 | NS | |
| Atg9A | 1.9 | NS | |
| Atg9B | 36.6 | * | |
| Wipi1 | 1.7 | *** | |
| Gabarapl1 | 1.9 | NS | |
| Gabarapl2 | 1.6 | ** | |
| Map1LC3A | 0.2 | *** | |
| BCL2/PIK3C3 complex | Bcl2L1 | 5.4 | NS |
| Becn1 | 2.1 | ** | |
| PiK3c3 | 1.7 | NS | |
| Autophagy adaptor | Sqstm1 | 3.3 | *** |
| mTOR complex | Raptor | 2.2 | NS |
| Rictor | 1.8 | NS | |
| AKT1s1 | 1.5 | ** | |
| Lysosome associated | Dram | 1.3 | NS |
| Lamp1 | 3.4 | ** | |
| Lamp3 | 2.3 | NS | |
| others | Bire5 | 1.5 | * |
| Bnip3 | 6.1 | *** | |
| Ddit3 | 2.1 | ** | |
| Eif4ebp1 | 2.1 | *** | |
| Eif4g1 | 1.9 | ** | |
| Fkbp15 | 0.5 | NS | |
| Gfi 1 b | 3.2 | NS | |
| Gpsm1 | 1.9 | * | |
| Letm1 | 3.3 | *** | |
| Letm2 | 3.8 | *** | |
| Rasd1 | 2.0 | NS | |
| Sec16A | 2.3 | NS | |
| Sec16B | 1.6 | NS | |
| Sec23A | 2.0 | * | |
| Sec23B | 2.2 | ** | |
| Sh3glb1 | 2.0 | ** | |
| Sh3glb2 | 3.4 | ** | |
| Snx30 | 1.9 | NS | |
| Tpr | 1.6 | * | |
| Tpr53 | 0.5 | * | |
| Ulk3 | 1.8 | NS | |
Most significantly, we also found an over 30-fold upregulation of the mRNA expression of ATG9B in OxSR cells. ATG9 proteins are essential for phagophore maturation (meaning early steps in autophagosome generation) and vesicle elongation and also determines the size of autophagosomes by acting as a lipid carrier delivering lipids to the growing phagophore [[51], [52], [53], [54]]. In addition, also a higher expression of the SEC23a and SEC23b mRNA was observed in OxSR cells (Table 1). SEC protein family members are important for the formation of specialized transport vesicles from ER to autophagosome biogenesis and selection of specific cargo molecules [55,56].
The PCR mRNA array further revealed a significant increase in the mRNA expression of BNIP3 (6-fold) and BNIP3 is known to regulate mitophagy, the autophagic clearance process of damaged mitochondria [57]. LETM1 and 2, which are not directly related to autophagy but required to maintain mitochondrial ion homeostasis and morphology [58,59], were also found to be highly upregulated in OxSR cells in their expression (Table 1), linking oxidative stress adaptation also to altered mitochondrial function; this link is further confirmed by full proteome analysis (see 3.7 below) and these data are consistent with the observed alterations in mitochondrial morphology and dynamics in OxSR cells (Fig. 1A–E).
3.4. OxSR cells can handle more effectively proteotoxicity
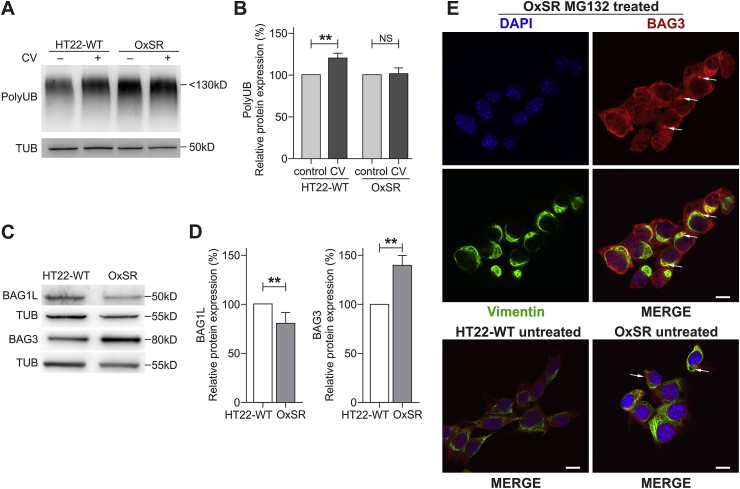
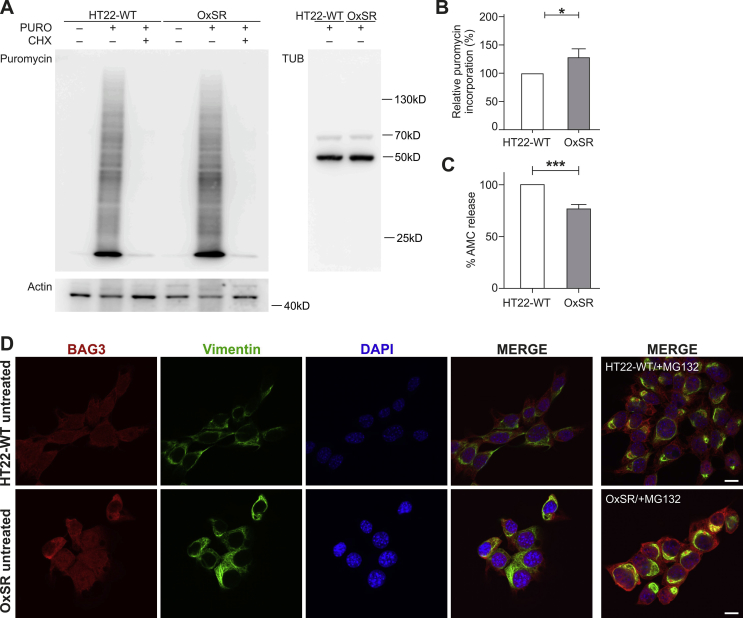
Next, we investigated how effectively OxSR cells can handle disturbances of the proteostasis network. To reproducibly induce protein stress, we employed the compound canavanine (CV), an arginine analogue which disturbs protein homeostasis by interfering with protein translation. Western blot analysis revealed an increased accumulation of polyubiquitinated degradation-prone proteins in HT22-WT but not in OxSR cells after 12 h treatment of the cells with CV (Fig. 3A and B). Interestingly, the lack of accumulation of polyubiquitinated proteins in OxSR cells occurred on the background of an overall even increased protein translation rate in these cells as determined by the SUnSET assay (see Suppl. Figs. S2A and B). The increased ability of the OxSR cells to cope with proteotoxic stress as indicated by the accumulation of polyubiquitinated proteins could be a direct result of an increased autophagic clearance activity.
Fig. 3.
OxSR cells overcome proteotoxic stress more effectively than HT22-WT cells and show an altered BAG1/BAG3 expression. (A, B) Protein extracts from untreated and canavanine- (CV-) treated samples (at 4 mM) of HT22-WT and OxSR cells were analyzed by Western blot for indicated proteins. Bar graphs represent changes in polyubiquitinated protein levels between untreated control and treated samples obtained by densitometric analysis after normalized to tubulin. Values represent mean ± S.E.M., n = 3, **p < 0.01 and NS for no significant statistical difference. Polyubiquitinated proteins in untreated cells were set as control 100%. (C, D) Protein extracts from untreated HT22-WT and OxSR cells were subjected to Western blot analysis for BAG1 and BAG3 expression; tubulin was used as loading control. The expression of BAG1L and BAG3 was quantified by densitometry analysis and normalization to tubulin. Values represent mean ± S.E.M., n = 3 to 4, **p < 0.01 and control HT22 cells were set to 100%. (E) Untreated HT22-WT and OxSR as well as OxSR cells were treated with MG132 and immunocytochemically analyzed for BAG3-positive aggresomes surrounded with cage like vimentin structure by confocal microscope; arrows indicate aggresomes (see also Suppl. Fig. S2D). DAPI (blue) was used to stain DNA. Scale bars: 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. BAG1/BAG3 expression switch in OxSR cells
In OxSR cells chronically adapted to oxidative stress we observed a switch from BAG1 to BAG3 expression as previously already seen in human primary fibroblasts and neurons under acute redox stress and during aging [21,60]. BAG3 expression is increased in OxSR cells compared to HT22-WT cells, while the expression of BAG1L in OxSR cells was decreased as detected by Western blotting (Fig. 3C and D); BAG1L is the same isoform of BAG1 which was detected to be decreased in aged cells or cells after acute oxidative stress treatment [21]. Taken together, BAG3 and BAG1 (isoform BAG1L) expression are reciprocally regulated in OxSR cells. Next, the proteasomal chymotrypsin‐like activity in OxSR and HT22-WT cells was analyzed and found to be decreased in OxSR cells (Suppl. Fig. S2C). These results suggest that OxSR cells depend more on the autophagic system (driven by BAG3 [21]) rather than on the proteasomal system (driven by BAG1) for protein degradation. To further confirm the recruitment of BAG3 into the proteostasis network of OxSR cells proteasomal activity was inhibited by MG132, an efficient stimulus of protein aggresome formation [21]. The observed colocalization of BAG3 with vimentin-positive perinuclear aggresomes after pharmacological proteasome inhibition in the OxSR cells (Fig. 3E and Suppl. Fig. S2D) suggests an enhanced activity of BAG3-mediated aggresome targeting and autophagy.
3.6. BAG3 overexpression in HT22-WT cells leads to increased autophagic flux and lysosomal rearrangement, BAG3 depletion in OxSR cells results in reduced autophagic flux
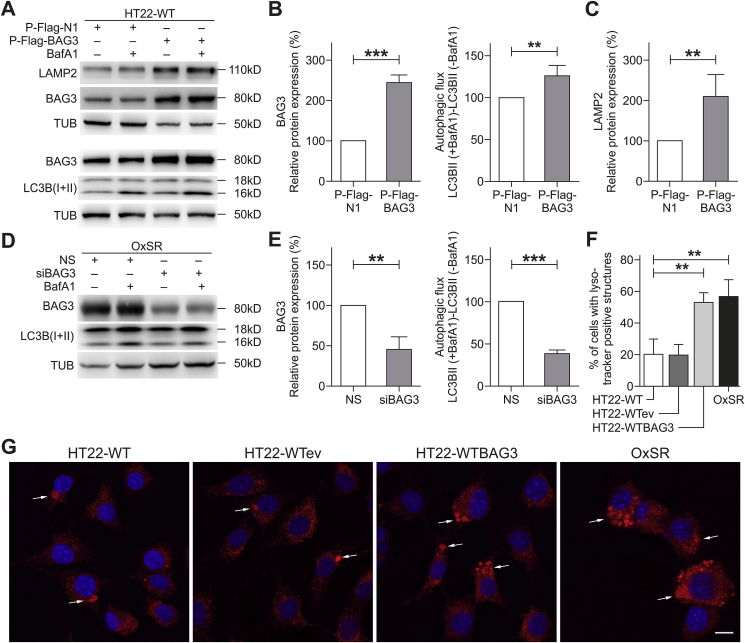
To clarify a direct participation of BAG3-mediated selective macroautophagy in the enhanced overall autophagic activity and the lysosomal phenotype of OxSR cells, we overexpressed BAG3 in HT22-WT cells. HT22-WT cells following transfection with a BAG3 expression construct showed a significantly increased autophagic flux. Intriguingly, along with overexpression of BAG3 also the protein levels of the lysosomal membrane protein LAMP2 were found to be strongly enhanced (Fig. 4A–C). After knockdown of BAG3 in OxSR cells, autophagic flux was again analyzed by determining the accumulation of LC3B-II after treatment with BafA1 by Western blot. Upon knockdown of BAG3 via siRNA a significant reduction of the autophagic flux was found in OxSR cells (Fig. 4D and E), whereas the downregulation of BAG3 by siRNA showed no change in autophagic flux of HT22-WT cells (data not shown). This suggests that BAG3 expression can have a direct effect on lysosomes. Finally, lysosomal compartments were analyzed by LysoTracker staining in HT22-WT (as controls), HT22-WT cells transfected with empty vector (HT22-WTev), HT22-WT cells overexpressing BAG3 (HT22-WTBAG3) and OxSR cells. Perinuclear lysosomes were found in HT22-WT and HT22-WTeV cells (controls), however, BAG3-overexpressing HT22-WTBAG3 cells showed cellular distribution of LysoTracker-positive staining similar to that in OxSR cells (Fig. 4G). Quantification showed that also the total number of cells showing distinguishable LysoTracker-positive vacuolar structure is significantly elevated in HT22-WTBAG3 compared to HT22-WT and HT22-WTeV and is comparable to OxSR cells (Fig. 4F).
Fig. 4.
BAG3 is required for autophagic activity in OxSR cells and BAG3 overexpression promotes autophagy in HT22-WT cells. (A–C) HT22-WT cells transfected for 48 h with empty vector (P-Flag-N1) or BAG3 overexpression vector (P-Flag-BAG3) and treated with vehicle or with 8 μM BafA1 for 6 h before the cell lysis. Protein extracts were analyzed via Western blotting for indicated proteins. Quantification of LAMP2, BAG3 expression and autophagic flux was prepared by densitometric analysis after normalization of protein levels to tubulin. Values represent mean ± S.E.M., n = 4, **p < 0.01 and ***p < 0.001. Expression in cells transfected with empty vector were set to 100%. (D, E) OxSR cells were transfected with nonsense siRNA (NS) and BAG3 siRNA (siBAG3, +/− BafA1) for 48 h. Protein extracts from vehicle or BafA1-treated cells were subjected to Western blot analysis. Quantification of BAG3 expression and autophagic flux was prepared after densitometric analysis and normalization to tubulin. Values represent mean ± S.E.M., n = 3, **p < 0.01 and ***p < 0.001. Expression in cells transfected with nonsense siRNA (NS) were set to 100%. (F) Quantitative bar graph shows number of cells with LysoTracker-positive vacuolar staining in untreated HT22-WT, HT22 transfected with empty vector (HT22-WTeV), HT22 overexpressing BAG3 (HT22-WTBAG3) and OxSR cells. Values represent mean ± S.E.M., n = 4 with 3–8 images of different microscopic fields was counted in each day for every experimental condition, **p < 0.01. (G) Representative confocal images of LysoTracker red staining (LT) in untreated HT22-WT, HT22-WTeV, HT22-WTBAG3 and OxSR cells are shown indicating the changes in lysosomal phenotype. Arrows indicate cells containing LysoTracker-stained structures. DAPI (blue) was used to stain DNA. Scale bars: 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Taken together, these results strongly support the role of BAG3 and BAG3-mediated selective macroautophagy in the adaptation of OxSR cells to chronic redox stress.
3.7. Total proteome analysis underlines differential expression of proteins involved in autophagy, mitochondrial function and metabolism
Finally, to analyze changes in the global proteome in OxSR cells (as compared to HT22-WT) we performed a bottom-up mass spectrometry-based discovery proteomics analysis. Label-free quantification (LFQ) analysis of the triplicates of both cell lines employing a false discovery rate (FDR) of 1% identified a total of 1121 proteins (the complete dataset is included in Suppl. File S4B). Subsequently, we identified the proteins that exhibited significant variations between OxSR and HT22-WT cells. For this, the experimental reproducibility of the LFQ between the replicates was examined employing Pearson's correlation coefficients, which demonstrated excellent biological reproducibility with an average R value of 0.97 ± 0.01 and R of 0.93 ± 0.01 between the two cell lines. Data are represented in the scatter plots of the intensity distribution in one replicate relative to another (Suppl. File S4C).
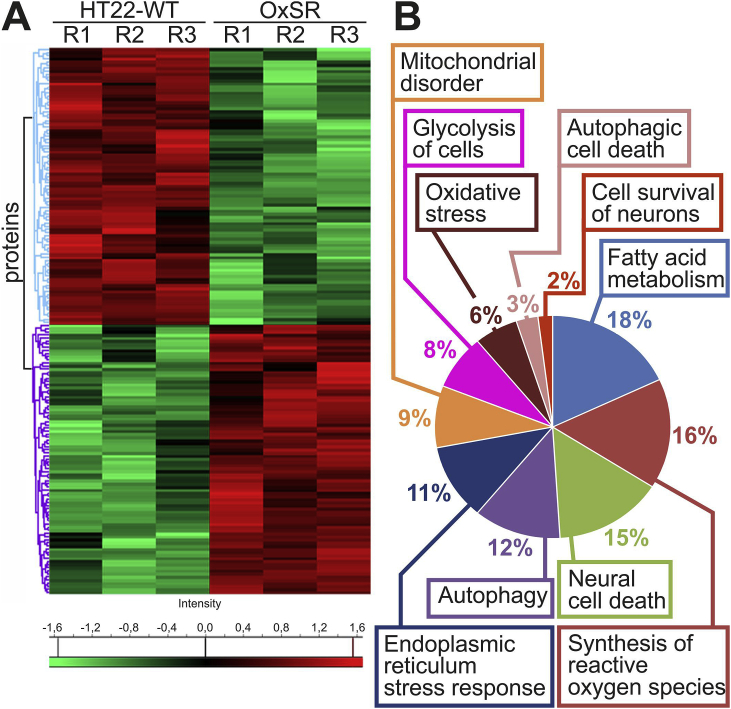
Among the identified proteins, 176 were differentially expressed, with 87 up-regulated and 89 down-regulated proteins (Suppl. File S4D). Further clustering of these differentially expressed proteins depicts the distinct patterns of the proteome presented as a heat map with unsupervised hierarchical clustering (Fig. 5A). Closer examination of these protein clusters revealed significant changes in several cellular functions (Fig. 5B and Suppl. File S4E). Endoplasmic reticulum stress response, fatty acid metabolism, synthesis of reactive oxygen species, glycolysis, mitochondrial disorder, oxidative stress, autophagic cell death, neuronal cell death, autophagy and cell survival of neurons represent the top ten most significant functional changes implicated in OxSR cells in comparison to the oxidative stress-sensitive HT22-WT cells.
Fig. 5.
Comparative proteomics analyses of the HT22-WT and OxSR cells. (A) Hierarchical clustering of the 176 differentially expressed proteins depicted as a heat map distinguishes two major clusters of proteins. The first cluster represents significantly down-regulated (green) and the second cluster up-regulated (red) proteins in OxSR cells compared to HT22-WT. R1 to R3 represent the biological replicates. (B) Pie chart displays cellular functions and abundance of proteins which are differentially expressed in OxSR cells compared to HT22-WT cells. The detailed list of proteins in each cellular functional group is provided with Suppl. File S4E. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
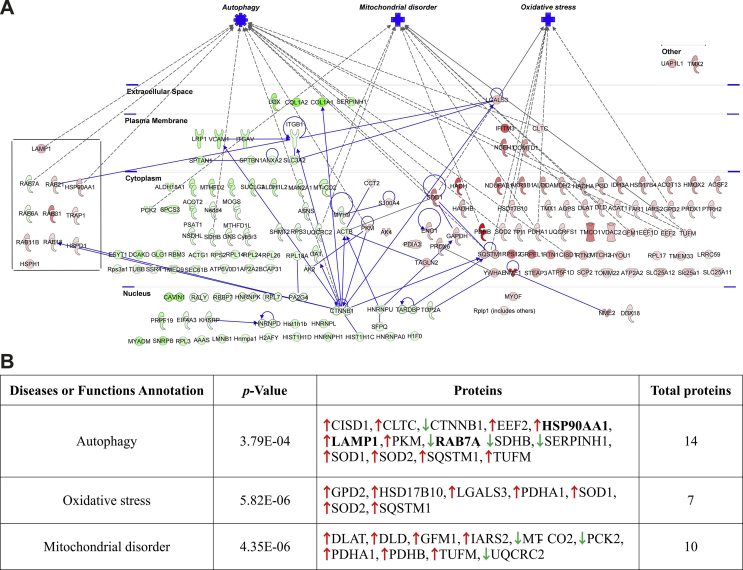
Finally, in an effort to further explore protein-protein interaction pathways (PPIs) of the proteins identified to be differentially expressed, functional pathway enrichment was determined employing the Ingenuity Pathway Analysis (v01-04, IPA; Ingenuity QIAGEN Redwood City, CA) tool. Consistently, a large majority of these proteins were found to be actively involved in the process of autophagy (14 proteins) but also linked to mitochondrial function and disorders (10 proteins) and oxidative stress/redox signaling (7 proteins) (Suppl. Fig. S3 and the complete list to be found in Suppl. File S4F).
4. Discussion
A disturbed protein homeostasis, proteotoxic and oxidative stress are hallmarks of neurodegeneration and aging [[61], [62], [63]]. Autophagy is described as a physiological process involved in antioxidant defense [64] and autophagy deficiency clearly results in neuronal loss and neurodegeneration in mice [65]. On the other hand, induction of autophagy is shown to alleviate oxidative stress in myocardial ischemia/reperfusion injury [66].
The adaptation to oxidative stress is a highly relevant process and some adaptive pathways focusing on antioxidant defense systems and mitochondrial and metabolic adaptations have been described to be constantly activated in neuronal cells resistant to oxidative stress as well as in AD tissue [6,26,29]. Moreover, oxidative stress resistance has been linked to failure of anticancer therapy and here a particular role of upregulated autophagy and BAG3 has been proposed in various tumor types [16,[67], [68], [69]].
Based on the knowledge that dysregulated autophagy is actively involved in neurodegeneration [70,71] combined with previous findings on mouse hippocampal HT22 cells fully adapted to high concentrations of H2O2 (OxSR cells) [28], we describe here in detail the molecular changes associated with redox stress adaptation in neuronal cells. To do so, we engaged different expression and functional studies including morphological parameters obtained by TEM, investigation of autophagic flux and global proteome analysis using mass spectrometry.
We found a significantly increased autophagic activity in the cells adapted to chronic oxidative stress (OxSR cells) (Fig. 1) and on the other hand a reduced proteasomal function (Suppl. Fig. S2) suggesting that autophagy may serve as the preferred protein degradation pathway during oxidative stress challenge. We identified several key proteins involved in autophagy regulation that showed a significantly altered expression (Fig. 2). ROS production i.e. oxidative stress is known to inhibit mTORC1 [66,72], the major regulatory complex for autophagy induction. Interestingly, in OxSR cells the increase in autophagy under chronic oxidative stress is not mediated via mTOR inhibition, whereas the classical BECN1/PIK3C3 complex obviously is involved and highly upregulated in their expression in OxSR cells. Moreover, RAB18 expression is enhanced in OxSR cells (Fig. 2). This finding is consistent with the recent description of RAB18 being a positive autophagy regulator [50] and a role of RAB18 in lipid metabolism, which enables enhanced lipid recruitment for autophagosome formation [73].
In qPCR analysis we observed a massively increased expression of ATG9B in OxSR cells (Table 1). ATG9 is an essential ATG family protein required for phagophore formation and exists in two homologues referred to as ATG9A and ATG9B. Trafficking of ATG9 from the Golgi apparatus to the site of autophagosome formation following autophagy induction (for instance by starvation) is reported to be dependent on PIK3C3 activity and negatively affected by PIK3C3 inhibition [74]. Therefore, it could be argued that upregulated ATG9B may work in concert with PIK3C3 in OxSR cells to promote autophagy. Leow et al. recently suggested that also sub-lethal acute oxidative stress can specifically induce ATG9B expression [75]. Furthermore, it is reported that ATG9 not only just acts as an autophagic membrane protein, but also upregulates autophagy induction via the JNK pathway by interacting with tumor necrosis factor receptor-associated factor 2 (dTRAF2) under oxidative stress condition [76]. In fact, ATG9 could be a key factor for the observed significantly enhanced autophagy activity in OxSR cells, since it can ensure the supply with membrane fragments early in autophagosome formation [77].
During cellular stress, the HSP70 chaperone system is a key player to maintain protein homeostasis together with its co-chaperones BAG1 (for proteasomal degradation) and BAG3 (for autophagic degradation) [24]. OxSR cells demonstrate an upregulated BAG3 accompanied with downregulated BAG1L expression (“BAG1/BAG3-switch”) and cope with a disturbed proteostasis induced by canavanine more effectively. Also, BAG3-positive aggresomes are found upon proteasomal inhibition as previously observed (Fig. 3) [23]. Most interestingly, the overexpression of BAG3 causes an elevated number of LysoTracker-stained structures in HT22-WT cells resembling distribution (and number) of lysosomes seen in OxSR cells (Fig. 4). The observed changes in these LysoTracker-positive structures after BAG3 overexpression may then have also functional consequences on the autophagic flux since LC3B-II flux was increased in HT22-WT cells following overexpression of BAG3. Our results are consistent with recent observations that the lysosomal biogenesis is increased under sub-lethal acute oxidative stress and proposed to be mediated via a selective chaperone-mediated pathway [75]. Furthermore, the BAG3 siRNA knockdown significantly hampers the autophagic activity exclusively in OxSR cells and therefore, identifies BAG3 as one key positive autophagy modulator in OxSR cells (Fig. 4). These data underline a strong role of BAG3 in boosting autophagy in OxSR cells but shows also for the first time that the expression switch from BAG1 to BAG3 and the functional switch from proteasomal pathway to BAG3-mediated macroautophagy can be of permanent nature under stable adaptation to oxidative stress as observed before during acute redox stress and aging [21,60]. It is important to note here that neither knock-down of BAG3 via siRNA nor a general autophagy inhibition with bafilomycin A1 reversed the resistance phenotype (data not shown) strongly arguing that during long-term adaptation to redox stress actually a combination of different pathways may ensure oxidative stress resistance. This is strongly supported by our full proteome analysis as well as by previously identified factors that partially contribute to oxidative stress resistance such as NF-κB, sphingolipids and increased levels of antioxidant enzymes [[26], [27], [28]]. Indeed, employing MS-based global proteome analysis comparing OxSR and HT22-WT cells several proteins were identified to be differentially expressed and significant changes in several key pathways in addition to autophagy were revealed e.g. alteration in the ER stress response, fatty acid metabolism, glycolysis, oxidative stress response and survival of neurons, mitochondrial pathways and disorders. This argues towards the involvement of a wide array of changes involved in redox stress adaptation (Fig. 5) and demonstrates also the plasticity and adaptability of various cellular pathways during chronic redox stress.
In summary, we propose that upregulated autophagy in OxSR cells follows an mTOR-independent pathway. BECN1/PIK3C3 and ATG9B, critical proteins for autophagy induction and downstream execution, respectively, are highly upregulated during adaptation to oxidative stress. Moreover, the HSP70-cochaperone BAG3 is not only increased in its expression, but it is also active to participate in the upregulated selective macroautophagy in OxSR cells and also directly impacts on the lysosomal rearrangement. OxSR cells described here in detail may be a useful model to study these adaptive pathways and may also represent a tool to decipher further factors and interacting pathways able to boost autophagic flux in neuronal cells. Altogether, analyzing the process of adaptation to chronic redox stress reveals potential novel therapeutic targets mediating a robust adaptability to long-term oxidative stress conditions and a disturbed proteostasis as important hallmarks of neurodegenerative disease and aging.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
This study was supported by grants of the CRC1177 of the Deutsche Forschungsgemeinschaft (DFG) and the Peter Beate Heller Foundation to CB and Foundation Fighting Blindness (FFB) (PPA-0717-0719-RAD) to UW. Caroline Manicam is supported by a grant from the DFG (MA 8006/1-1). The authors are grateful to Milena Rossmanith, Katharina Träger for their assistance with sample preparation for mass spectrometry analyses, to Ingrid Koziollek-Drechsler and Heike Nagel for help with the cell culture and Western blotting, to Michael Plenikowski for artwork, and to Andreas Kern, Jana Schepers and Albrecht Clement for critical reading.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101181.
Contributor Information
Debapriya Chakraborty, Email: debapriya222@gmail.com.
Vanessa Felzen, Email: Vanessa.felzen@gmail.com.
Christof Hiebel, Email: christof.hiebel@gmail.com.
Elisabeth Stürner, Email: estuerne@uni-mainz.de.
Natarajan Perumal, Email: nperumal@eye-research.org.
Caroline Manicam, Email: caroline.manicam@unimedizin-mainz.de.
Elisabeth Sehn, Email: sehn@uni-mainz.de.
Franz Grus, Email: fgrus@eye-research.org.
Uwe Wolfrum, Email: wolfrum@uni-mainz.de.
Christian Behl, Email: cbehl@uni-mainz.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Fig. S1.
Results from cell proliferation assay, immunocytochemical analysis of TFEB and TEM ultrastructure analysis. (A) Proliferation/growth rate of HT22-WT and OxSR cells was measured by MTT assay over nine alternate days. Values represent mean ± S.E.M., n=3, *** p<0.001. (B) Immunocytochemical analysis reveals a distinct nuclear translocation of TFEB in OxSR cells compared to cytosolic distribution in HT22-WT under basal condition. HT22-WT cells were treated with 10 μM Rapamycin for 2 h to induce TFEB nuclear translocation as a positive control experiment (HT22-WT+RAPA). Scale bars: 10 μm. Images were acquired by confocal microscopy. (C) Ultrastructure analysis of HT22-WT and OxSR cells by TEM (representative images focusing on autophagosomes are shown; M indicates mirochondria, A autophagosomes).
Supplemental Fig. S2.
Results from SUnSET assay, immunocytochemical analysis and determination of proteasomal activity. (A) HT22-WT and OxSR cells were treated with 5 μg/ml puromycin (PURO) or simultaneously with 60 μg/ml cycloheximide (CHX) for 30 min. Protein extracts were analyzed by Western blotting for puromycin incorporation. Actin and tubulin were used as loading controls. (B) Bar graph shows changes in puromycin incorporation obtained by densitometry analysis and normalization to tubulin. Values represent mean ± S.E.M., n = 3, *p < 0.05. Control HT22-WT cells were set to 100%. (C) Proteasomal activity of HT22-WT and OxSR cells was measured using fluorescence signal by release of AMC from proteasomal substrate Suc-LLVY-AMC by proteasomal enzyme activity. Values represent mean ± S.E.M., n = 5, ***p < 0.001. Control HT22-WT cells were set to 100%. (D) HT22-WT and OxSR cells were immunocytochemically analyzed for BAG3 and vimentin by confocal microscope; additional channels displayed referring to Fig. 3E. DAPI (blue) was used to stain DNA. Scale bars: 10 μm.
Supplemental Fig. S3.
PPI Networks of the significantly differentially expressed proteins in OxSR cells. (A) The protein-protein interaction (PPI) network of the differentially expressed proteins analyzed by IPA analysis demonstrated the involvement of three major protein clusters in autophagy, mitochondrial disorders and oxidative stress-related processes. Red and green colors of the molecules indicate up- and down-regulation of the proteins, respectively, and different intensities correspond to the magnitude of expression change of the individual proteins. The proteins were further distinguished according to their cellular localization and molecular functional classes with different shapes representing enzymes, ion channels, peptidases, transmembrane receptors etc. (B) Additional overview table linking changed expression of proteins to autophagy, oxidative stress, and mitochondrial disorder. The detailed result is also provided in Suppl. File S4E.
References
- 1.Liguori I., Russo G., Curcio F., Bulli G., Aran L., Della-Morte D., Gargiulo G., Testa G., Cacciatore F., Bonaduce D., Abete P. Oxidative stress, aging, and diseases. Clin. Interv. Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hensley K., Hall N., Subramaniam R., Cole P., Harris M., Aksenov M., Aksenova M., Gabbita S.P., Wu J.F., Carney J.M. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J. Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 3.Di Domenico F., Tramutola A., Butterfield D.A. Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 2017;111:253–261. doi: 10.1016/j.freeradbiomed.2016.10.490. [DOI] [PubMed] [Google Scholar]
- 4.Go´ mez-Isla T., Hollister R., West H., Mui S., Growdon J.H., Petersen C.R., Parisi E.J., Hyman B.T. Neuronl loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Am. Neurol. Associat. 1996;41:17–24. doi: 10.1002/ana.410410106. [DOI] [PubMed] [Google Scholar]
- 5.Go´ mez-Isla T., Price J.L., McKeel D.W., Morris J.C., Growdon J.H., Hyman B.T. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J. Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greeve I., Hermans-Borgmeyer I., Brellinger C., Kasper D., Gomez-Isla T., Behl C., Levkau B., Nitsch R.M. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer's disease-associated neurodegeneration and oxidative stress. J. Neurosci. 2000;20:7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nijssen J., Comley L.H., Hedlund E. Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 2017;133:863–885. doi: 10.1007/s00401-017-1708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen E.I., Hewel J., Krueger J.S., Tiraby C., Weber M.R., Kralli A., Becker K., Yates J.R., 3rd, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 9.Hawk M.A., McCallister C., Schafer Z.T. Antioxidant activity during tumor progression: a necessity for the survival of cancer cells? Cancers (Basel) 2016:8. doi: 10.3390/cancers8100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 12.Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 13.Khaminets A., Behl C., Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Song C., Song C., Tong F. Autophagy induction is a survival response against oxidative stress in bone marrow-derived mesenchymal stromal cells. Cytotherapy. 2014;16:1361–1370. doi: 10.1016/j.jcyt.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Singh A.K., Kashyap M.P., Tripathi V.K., Singh S., Garg G., Rizvi S.I. Neuroprotection through rapamycin-induced activation of autophagy and PI3K/Akt1/mTOR/CREB signaling against amyloid-beta-induced oxidative stress, synaptic/neurotransmission dysfunction, and neurodegeneration in adult rats. Mol. Neurobiol. 2017;54:5815–5828. doi: 10.1007/s12035-016-0129-3. [DOI] [PubMed] [Google Scholar]
- 16.Felzen V., Hiebel C., Koziollek-Drechsler I., Reissig S., Wolfrum U., Kogel D., Brandts C., Behl C., Morawe T. Estrogen receptor alpha regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function. Cell Death Dis. 2015;6:e1812. doi: 10.1038/cddis.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosati A., Graziano V., De Laurenzi V., Pascale M., Turco M.C. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falco A., Rosati A., Festa M., Basile A., De Marco M., d'Avenia M., Pascale M., Dal Piaz F., Tavano F., Di Mola F.F., di Sebastiano P., Berloco P.B., Nudo F., Caraglia M., Febbraro A., Barcaroli D., Scarpa A., Pezzilli R., De Laurenzi V., Turco M.C. BAG3 is a novel serum biomarker for pancreatic adenocarcinomas. Am. J. Gastroenterol. 2013;108:1178–1180. doi: 10.1038/ajg.2013.128. [DOI] [PubMed] [Google Scholar]
- 19.Rosati A., Basile A., D'Auria R., d'Avenia M., De Marco M., Falco A., Festa M., Guerriero L., Iorio V., Parente R., Pascale M., Marzullo L., Franco R., Arra C., Barbieri A., Rea D., Menichini G., Hahne M., Bijlsma M., Barcaroli D., Sala G., di Mola F.F., di Sebastiano P., Todoric J., Antonucci L., Corvest V., Jawhari A., Firpo M.A., Tuveson D.A., Capunzo M., Karin M., De Laurenzi V., Turco M.C. BAG3 promotes pancreatic ductal adenocarcinoma growth by activating stromal macrophages. Nat. Commun. 2015;6:8695. doi: 10.1038/ncomms9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behl C. Breaking BAG: the Co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 2016;37:672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Furst D.O., Saftig P., Saint R., Fleischmann B.K., Hoch M., Hohfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Gamerdinger M., Kaya A.M., Wolfrum U., Clement A.M., Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sturner E., Behl C. The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 2017;10:177. doi: 10.3389/fnmol.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behl C., Davis J.B., Lesley R., Schubert D. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 26.Sagara Y., Dargusch R., Klier F.G., Schubert D., Behl C. Increased antioxidant enzyme activity in amyloid beta protein-resistant cells. J. Neurosci. 1996;16:497–505. doi: 10.1523/JNEUROSCI.16-02-00497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lezoualc'h F., Sagara Y., Holsboer F., Behl C. High constitutive NF-kappaB activity mediates resistance to oxidative stress in neuronal cells. J. Neurosci. 1998;18:3224–3232. doi: 10.1523/JNEUROSCI.18-09-03224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clement A.B., Gamerdinger M., Tamboli I.Y., Lutjohann D., Walter J., Greeve I., Gimpl G., Behl C. Adaptation of neuronal cells to chronic oxidative stress is associated with altered cholesterol and sphingolipid homeostasis and lysosomal function. J. Neurochem. 2009;111:669–682. doi: 10.1111/j.1471-4159.2009.06360.x. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer A., Jaeckel M., Lewerenz J., Noack R., Pouya A., Schacht T., Hoffmann C., Winter J., Schweiger S., Schafer M.K., Methner A. Mitochondrial function and energy metabolism in neuronal HT22 cells resistant to oxidative stress. Br. J. Pharmacol. 2014;171:2147–2158. doi: 10.1111/bph.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Maher P., Schubert D. A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron. 1997;19:453–463. doi: 10.1016/s0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 31.Schafer M., Goodenough S., Moosmann B., Behl C. Inhibition of glycogen synthase kinase 3 beta is involved in the resistance to oxidative stress in neuronal HT22 cells. Brain Res. 2004;1005:84–89. doi: 10.1016/j.brainres.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Manicam C., Perumal N., Pfeiffer N., Grus F.H., Gericke A. First insight into the proteome landscape of the porcine short posterior ciliary arteries: key signalling pathways maintaining physiologic functions. Sci. Rep. 2016;6:38298. doi: 10.1038/srep38298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 34.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 35.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Olsen J.V., de Godoy L.M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 40.Hiebel C., Kromm T., Stark M., Behl C. Cannabinoid receptor 1 modulates the autophagic flux independent of mTOR- and BECLIN1-complex. J. Neurochem. 2014;131:484–497. doi: 10.1111/jnc.12839. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valera E., Dargusch R., Maher P.A., Schubert D. Modulation of 5-lipoxygenase in proteotoxicity and Alzheimer's disease. J. Neurosci. 2013;33:10512–10525. doi: 10.1523/JNEUROSCI.5183-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 44.Lee H., Smith S.B., Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J. Biol. Chem. 2017;292:7115–7130. doi: 10.1074/jbc.M116.762567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Napolitano G., Ballabio A. TFEB at a glance. J. Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Moreau K., Narayanan U., Renna M., Siddiqi F.H., Underwood B.R., Winslow A.R., Rubinsztein D.C. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 47.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 48.Liu S., Storrie B. Are Rab proteins the link between Golgi organization and membrane trafficking? Cell. Mol. Life Sci. 2012;69:4093–4106. doi: 10.1007/s00018-012-1021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerondopoulos A., Bastos R.N., Yoshimura S., Anderson R., Carpanini S., Aligianis I., Handley M.T., Barr F.A. Rab18 and a Rab18 GEF complex are required for normal ER structure. J. Cell Biol. 2014;205:707–720. doi: 10.1083/jcb.201403026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldmann A., Bekbulat F., Huesmann H., Ulbrich S., Tatzelt J., Behl C., Kern A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem. Biophys. Res. Commun. 2017;486:738–743. doi: 10.1016/j.bbrc.2017.03.112. [DOI] [PubMed] [Google Scholar]
- 51.Webber J.L., Tooze S.A. New insights into the function of Atg9. FEBS Lett. 2010;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Imai K., Hao F., Fujita N., Tsuji Y., Oe Y., Araki Y., Hamasaki M., Noda T., Yoshimori T. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 2016;129:3781–3791. doi: 10.1242/jcs.196196. [DOI] [PubMed] [Google Scholar]
- 53.Noda T. Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Biochem. Soc. Trans. 2017;45:1323–1331. doi: 10.1042/BST20170128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soreng K., Munson M.J., Lamb C.A., Bjorndal G.T., Pankiv S., Carlsson S.R., Tooze S.A., Simonsen A. SNX18 regulates ATG9A trafficking from recycling endosomes by recruiting Dynamin-2. EMBO Rep. 2018;19 doi: 10.15252/embr.201744837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barlowe C.K., Miller E.A. Secretory protein biogenesis and traffic in the early secretory pathway. Genetics. 2013;193:383–410. doi: 10.1534/genetics.112.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alshudukhi A.A., Zhu J., Huang D., Jama A., Smith J.D., Wang Q.J., Esser K.A., Ren H. Lipin-1 regulates Bnip3-mediated mitophagy in glycolytic muscle. FASEB J. 2018 doi: 10.1096/fj.201800374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Austin S., Tavakoli M., Pfeiffer C., Seifert J., Mattarei A., De Stefani D., Zoratti M., Nowikovsky K. LETM1-Mediated K(+) and Na(+) homeostasis regulates mitochondrial Ca(2+) efflux. Front. Physiol. 2017;8:839. doi: 10.3389/fphys.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamai S., Iida H., Yokota S., Sayano T., Kiguchiya S., Ishihara N., Hayashi J., Mihara K., Oka T. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J. Cell Sci. 2008;121:2588–2600. doi: 10.1242/jcs.026625. [DOI] [PubMed] [Google Scholar]
- 60.Minoia M., Boncoraglio A., Vinet J., Morelli F.F., Brunsting J.F., Poletti A., Krom S., Reits E., Kampinga H.H., Carra S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy. 2014;10:1603–1621. doi: 10.4161/auto.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gan L., Cookson M.R., Petrucelli L., La Spada A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018;21:1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andersen J.K. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004;10(Suppl):S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 64.Giordano S., Darley-Usmar V., Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 66.Zhao D., Yang J., Yang L. Insights for oxidative stress and mTOR signaling in myocardial ischemia/reperfusion injury under diabetes. Oxid. Med. Cell Longev. 2017;2017:6437467. doi: 10.1155/2017/6437467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weng M.S., Chang J.H., Hung W.Y., Yang Y.C., Chien M.H. The interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistance. J. Exp. Clin. Cancer Res. 2018;37:61. doi: 10.1186/s13046-018-0728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velegzhaninov I.O., Ievlev V.A., Pylina Y.I., Shadrin D.M., Vakhrusheva O.M. Programming of cell resistance to genotoxic and oxidative stress. Biomedicines. 2018;6 doi: 10.3390/biomedicines6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Marco M., Basile A., Iorio V., Festa M., Falco A., Ranieri B., Pascale M., Sala G., Remondelli P., Capunzo M., Firpo M.A., Pezzilli R., Marzullo L., Cavallo P., De Laurenzi V., Turco M.C., Rosati A. Role of BAG3 in cancer progression: a therapeutic opportunity. Semin. Cell Dev. Biol. 2018;78:85–92. doi: 10.1016/j.semcdb.2017.08.049. [DOI] [PubMed] [Google Scholar]
- 70.Menzies F.M., Fleming A., Caricasole A., Bento C.F., Andrews S.P., Ashkenazi A., Fullgrabe J., Jackson A., Jimenez Sanchez M., Karabiyik C., Licitra F., Lopez Ramirez A., Pavel M., Puri C., Renna M., Ricketts T., Schlotawa L., Vicinanza M., Won H., Zhu Y., Skidmore J., Rubinsztein D.C. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93:1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 71.Lie P.P.Y., Nixon R.A. Neurobiol Dis; 2018. Lysosome Trafficking and Signaling in Health and Neurodegenerative Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J., Yang X., Zhang J. Bridges between mitochondrial oxidative stress, ER stress and mTOR signaling in pancreatic beta cells. Cell. Signal. 2016;28:1099–1104. doi: 10.1016/j.cellsig.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 73.Pulido M.R., Diaz-Ruiz A., Jimenez-Gomez Y., Garcia-Navarro S., Gracia-Navarro F., Tinahones F., Lopez-Miranda J., Fruhbeck G., Vazquez-Martinez R., Malagon M.M. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi Y., Meyerkord C.L., Hori T., Runkle K., Fox T.E., Kester M., Loughran T.P., Wang H.-G. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2014;7:61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leow S.M., Chua S.X., Venkatachalam G., Shen L., Luo L., Clement M.V. Sub-lethal oxidative stress induces lysosome biogenesis via a lysosomal membrane permeabilization-cathepsin-caspase 3-transcription factor EB-dependent pathway. Oncotarget. 2017;8:16170–16189. doi: 10.18632/oncotarget.14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang H.W., Liao H.M., Peng W.H., Lin H.R., Chen C.H., Chen G.C. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev. Cell. 2013;27:489–503. doi: 10.1016/j.devcel.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 77.Mercer T.J., Gubas A., Tooze S.A. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 2018;293:5386–5395. doi: 10.1074/jbc.R117.810366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.