Abstract
Melanocortin receptors (MCRs) and their accessory proteins (MRAPs) evolutionarily first appear in the genome of sea lamprey. The most ancient melanocortin system consists of only two melanocortin receptors (slMCa and slMCb) and one MRAP2 (slMRAP2) protein, but the physiological roles have not been fully explored in this primitive species. Here, we synthesize and characterize the pharmacological features of slMRAP2 protein on two slMCRs. Our results show that the slMRAP2 protein lacks the long carboxyl terminus; it directly interacts and decreases the surface expression but enhances the α-MSH-induced agonism of slMCa and slMCb. In comparison with higher organisms such as elephant shark and zebrafish, we also demonstrate the constantly evolving regulatory function of the carboxyl terminus of MRAP2 protein, the unique antiparallel topology of slMRAP2 dimer and the homo- and hetero-dimerization of two slMCRs. This study elucidates the presence and modulation of melanocortin receptor by the accessory protein of the agnathans for the first time, which provides a better insight of the melanocortin system in ancient species of chordates.
Key Words: melanocortin 2 receptor accessory protein 2, melanocortin receptors, sea lamprey
Introduction
The melanocortin receptors (MCRs) belong to α group of rhodopsin G-protein-coupled receptors. Mammalian MCRs consist of five subtypes, generally referred to as MC1R, MC2R, MC3R, MC4R and MC5R. A widely reported function of MC1R is for pigmentation, MC2R for adrenocortical steroidogenesis, MC3R for energy homeostasis, MC4R for appetite control and MC5R for exocrine secretion (1). Four pro-opiomelanocortin (POMC)-derived natural agonists (α-, β-, γ-MSH and ACTH) and two antagonists (Agouti and AgRP) exist for the modulation of MCR signaling (1, 2, 3, 4). The melanocortin receptors have been reported to form homo- and heterodimers and could result in profound functional consequences (5, 6, 7). Moreover, trafficking and signaling of the MCRs are endogenously regulated by melanocortin receptor accessory proteins (MRAPs) containing two single transmembrane proteins, named MRAP (MRAP1) and MRAP2 (8). MRAP1 functions as an antiparallel homodimer and is essential for the plasma membrane translocation and ACTH response of MC2R (9, 10). MRAP could also affect dimerization of melanocortin receptors. In fact, MRAP promotes dimerization of MC2R, but prevents MC5R from forming homodimers (11). MRAP2, found as a homologue of MRAP, could also form antiparallel homodimers and modulate all five melanocortin receptors (12, 13). Human MRAP2 shows some or no effect on suppressing MC4R signaling (12, 13). However, mouse MRAP2 is capable of sensitizing the α-MSH-induced agonism of MC4R, and dysfunction of MRAP2 causes severe obesity syndrome (14). Two MRAP2 homologues exist in the zebrafish genome. MRAP2a stimulates somatic growth during larval development by inhibiting MC4R, while MRAP2b enhances ligand sensitivity of MC4R and regulate appetite and growth when feeding begins (15, 16).
The melanocortin system is present in phylum chordate, while the members (especially the melanocortin receptors) diverge among different species (8, 17, 18). A tremendous amount of work has been done in elucidating the roles of the melanocortin system of Chondrichthyes and Osteichthyes (8). However, little attention has been paid to the most ancient fish species in the class of Cyclostomata. Lamprey (Lampetra japonicum) and hagfish (Myxini glutinosa) are the only known existing jawless vertebrates (19). The ancient ancestors of the melanocortin receptors are cloned from the river lamprey, named MCa and MCb (20). Upon second genomic duplication, MC1R, MC2R and MC5R form from MCa, while MC3R and MC4R form from MCb (21). However, Vastermark et al. suggest that MC2R and MC5R do not evolve from a common ancestor. Except for the ancient melanocortin receptors, an MRAP2-like but not MRAP1-like sequence has been identified in sea lamprey indicating MRAP2 is likely to be the ancestor of the MRAPs system (22). Compared to other species, the amino acid sequence of sea lamprey MRAP2 protein is very short because the carboxyl terminus of slMRAP2 is missing (23). More interestingly, no agonist or antagonist has been identified so far in the sea lamprey genome (22) and whether the ancient slMRAP2 could modulate slMCRs remains unknown.
In this study, we synthesize slMCa, slMCb and slMRAP2, and supplement wild-type slMRAP2 with the carboxyl terminus of elephant shark MRAP2, zebrafish MRAP2a or zebrafish MRAP2b, abbreviated as esMRAP2-C, zMRAP2a-C, zMRAP2b-C, respectively. The pharmacological modulation of the slMRAPs protein on slMCa and slMCb and the dimerization of slMRAP2 and slMCRs are then further examined. Our results demonstrate that the slMRAP2 with a very short carboxyl terminus is fully functional in regulating slMCa or slMCb, and the dimerization event exists for the most ancient melanocortin receptors.
Methods and materials
Sequences blast and alignment
The accession numbers of all sequences in this study are given in Table 1. The National Center for Biotechnology Information (NCBI) tblastn was utilized to search the complete protein sequence of slMRAP2 and slMCa in the sea lamprey genomic database and alignment was performed using the software of DNAMAN.
Table 1.
Overview of accession numbers of sequences.
| Accession number | Organism | Description |
|---|---|---|
| ENSPMAT00000000587 | Petromyzon marinus | MRAP2, exons 1 and 2 |
| GL476894.1 | Petromyzon marinus | unplaced genomic scaffold_566 |
| BN001505.1 | Petromyzon marinus | MCa, 5′-end |
| BN001506.1 | Petromyzon marinus | MCa, 3′-end |
| GL477446.1 | Petromyzon marinus | unplaced genomic scaffold_1118 |
| BK007095.1 | Petromyzon marinus | MCb |
| DQ213059.1 | Lampetra fluviatilis | MCa |
| DQ213060.1 | Lampetra fluviatilis | MCb |
| XM_007908433.1 | Callorhinchus milii | MRAP2 |
| XM_001342887.6 | Danio rerio | MRAP2a |
| XM_005168521.3 | Danio rerio | MRAP2b |
| BR000855.1 | Callorhinchus milii | MC1R, partial cds |
| BR000856.1 | Callorhinchus milii | MC2R, partial cds |
| XM_007885593.1 | Callorhinchus milii | MC3R |
| XM_007895520.1/LOC103179728 | Callorhinchus milii | MC4R |
| XM_007895123.1 | Callorhinchus milii | MC5R |
Plasmids and peptides
slMRAP2, slMCa and slMCb were synthesized by Generay Biotechnology (Shanghai, China), sub-cloned into pc-DNA3.1(+) vector. The transmembrane (TM) domain of slMRAP2 was predicted from TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The N-terminal and TM domain of slMRAP2 was then reconstructed with the carboxyl terminus of elephant shark (Callorhinchus milii) MRAP2 (esMRAP2-C), zebrafish (Danio rerio) MRAP2a (zMRAP2a-C) or zebrafish (Danio rerio) MRAP2b (zMRAP2b-C), respectively. Both slMCa and slMCb plasmids carried 3HA tag and all of the MRAP2 plasmids carried V5, HA or 2Flag tag at the N terminus. For live cell imaging assay, slMRPA2, slMCa and slMCb were fused to yellow fluorescent protein (YFP-F1/F2) fragments in the N terminus or C terminus, respectively (slMRAP2-YFP-F1/F2, YFP-F1/F2-slMRAP2, slMCa-YFP-F1/F2, slMCb-YFP-F1/F2). Human α-MSH was purchased from GenScript Corporation Ltd (Nanjing, China) and human AgRP (83–139) were synthesized by Chinese Peptide LLC (Hangzhou, China).
Cell culture and transfection
HEK293T cells were maintained in high-glucose DMEM medium containing 10% FBS and 1% penicillin/streptomycin. CHO cells were maintained in DMEM/F12 medium containing 5% FBS and 1% penicillin/streptomycin. Cells were transfected with indicated plasmids using ViaFect Transfection Reagent (Promega). Total plasmid concentration was kept identical for all transfections by the addition of empty pcDNA3.1 vector.
Co-immunoprecipitation and Western blot
HEK293T cells transiently co-transfected with slMCa or slMCb and four types of MRAP2 were incubated in lysis buffer (0.75% Triton-X, 50 mM Tris–HCl pH 7.9, 150 mM NaCl and proteinase inhibitor cocktail from Roche) for 1 h at 4°C. Lysates were then centrifuged and the supernatants were incubated with anti-HA antibody (Abcam) overnight at 4°C. Immune complexes were collected by adding Protein A/G Agarose beads (Beyotime Biotechnology, Suzhou, China) and rotated at 4°C for 4 h. Beads were washed three times in lysis buffer and resuspended in loading buffer and boiled for 15 min. Initial lysates and immunoprecipitation proteins were separated by SDS-PAGE on 12% polyacrylamide gels and the protein was then transferred onto PVDF membrane and then gently shaken in Tris-buffered saline and Tween (TBST) containing 5% skim milk for 1 h. The membrane was subsequently treated with primary mouse antibody against HA (1:5000, Abcam) at 4°C overnight. Finally, membranes were incubated with the secondary rabbit peroxidase (HRP)-conjugated antibody (Rockland, Limerick, PA, USA) at a dilution of 1:5000 for 2 h at room temperature and washed three times, followed by autoradiography and recording.
cAMP assay
HEK293T cells were plated in 24-well plates and the transient transfections with indicated plasmids were performed the next day. Cell transfection condition was divided into 16 groups: slMCa or slMCb was co-transfected with four types of MRAP2 at the ratio of 1:0, 1:3 and 1:6. Upon 24 h of transfection, cells were treated with different concentrations of α-MSH in DMEM supplemented with 0.1% bovine serum albumin for 4 h at 37°C. For the competition binding experiment, cell transfection condition was divided into 24 groups: slMCa or slMCb was co-transfected with four types of MRAP2 at the ratio of 1:0, 1:3 and 1:6. Cells were then treated with a fixed concentration of α-MSH (2 nM) and different concentrations of AgRP in DMEM supplemented with 0.1% bovine serum albumin for 4 h at 37°C. The cAMP level was measured as described before (24) using the Dual-Glo Luciferase Assay System (Promega). Luminescence was measured with a Spectramax M5 plate reader.
Cell surface ELISA
HEK293T cells were seeded onto poly-l-lysine-coated 12-well plates at 105 cells/well and transfected with indicated plasmid the next day. After 24 h of transfection, cells were washed with D-PBS three times, fixed with 4% polyformaldehyde for 20 min at room temperature, washed, blocked in D-PBS supplemented with 5% milk for 30 min, incubated with 1:5000 anti-HA antibody at 37°C for 2 h, and then washed three times for 5 min with D-PBS, incubated with 1:5000 secondary antibodies at 37°C for 1 h. Then, cells were washed three times for 5 min with D-PBS and incubated with TMB solution (Beyotime Biotechnology, Suzhou, China) for 15–30 min. The reaction was terminated with 2 M sulfuric acid, and then 200 μL were transferred to a 96-well plate, and the absorbance was read at 450 nm on Spectramax M5 plate reader.
Immunofluorescence
CHO cells were transfected with indicated plasmid 48 h before the experiment on glass coverslips. Cells were fixed with 4% polyformaldehyde for 20 min at room temperature, washed, blocked in D-PBS supplemented with 5% milk for 30 min. Where noted, nuclei were counterstained with Hoechst 33342 (3 μg/mL) for 10 min at room temperature. A Zeiss confocal microscope (Jena, Germany) with ×60/1.3 NA oil objective and Photometrics CoolSNAP ES camera were employed. Micrographs displayed in a group were exposed and processed identically.
Statistical analysis
All experiments had been repeated three times in separate experiments. Data were shown as the mean ± s.e.m. and analyzed using GraphPad Prism v7.0. cAMP assays were analyzed by two-way ANOVA with Tukey post-test. The Student’s t-test was adopted to analyze the significance between two independent groups. P values <0.05 were considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Characterization of MRAP2 and MCRs in the sea lamprey
Through the NCBI and ensemble database, we found complete slMCb and partial slMCa and slMRAP2 coding sequences (Table 1). The complete slMCa protein sequence was identified with the tblastn tool of the NCBI database. The slMCa and slMCb sequences were aligned to corresponding MCRs of river lamprey and elephant shark. As shown (Fig. 1A and B), slMCa exhibited the highest identity, 94.78% to rlMCa subtypes, while the identity to the esMC1R and esMC2R was lower, 42.03% and 41.74%, respectively. Similarly, slMCb showed the highest identity to rlMCb subtypes (88.74%) and lower identity to esMC3R (55.50%), esMC4R (57.91%) and esMC5R (54.96%). The same approach was utilized to search the complete slMRAP2 sequence, but failed to identify the conserved carboxyl terminus such as those of zebrafish or mammalian MRAP2. Interestingly, we found a stop codon behind the partial sequences near the transmembrane domain of slMRAP2 (Fig. 1C).
Figure 1.
Sequence alignments of sea lamprey MRAP2 and MCRs. (A and B) Sequence alignment of MCRs from sea lamprey(sl), river lamprey(rl) and elephant shark(es). (C) Sequences alignment of MRAP2s from sea lamprey(sl), elephant shark(es) and zebrafish(zf).
The interaction of sea lamprey MRAP2s with MCRs
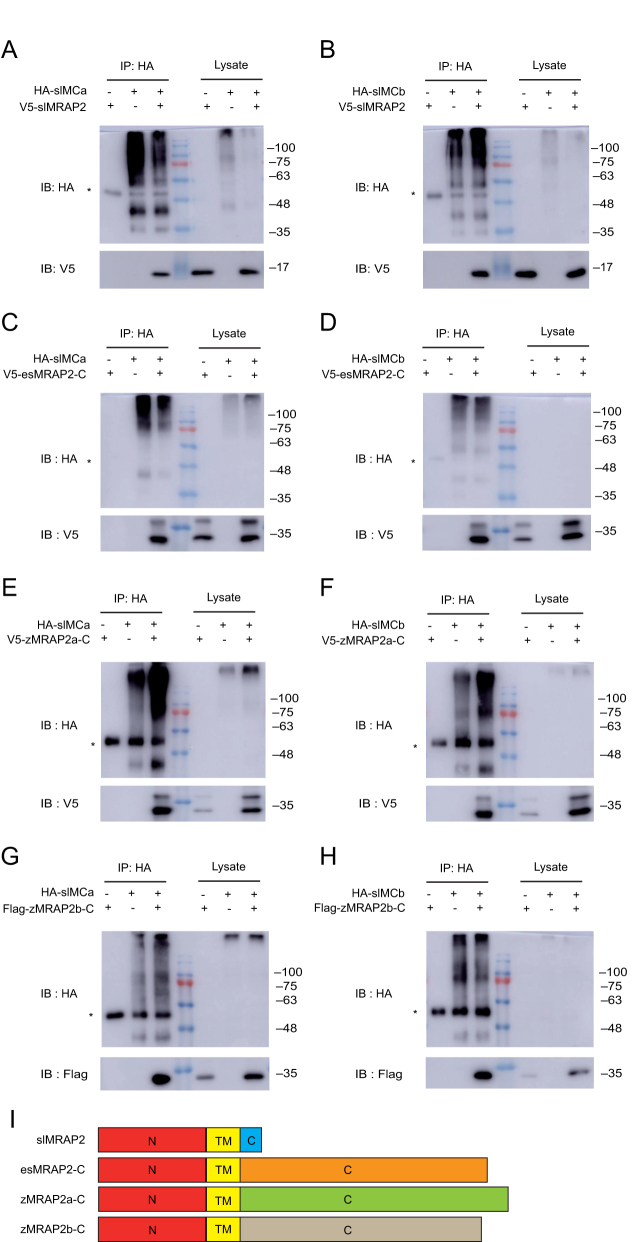
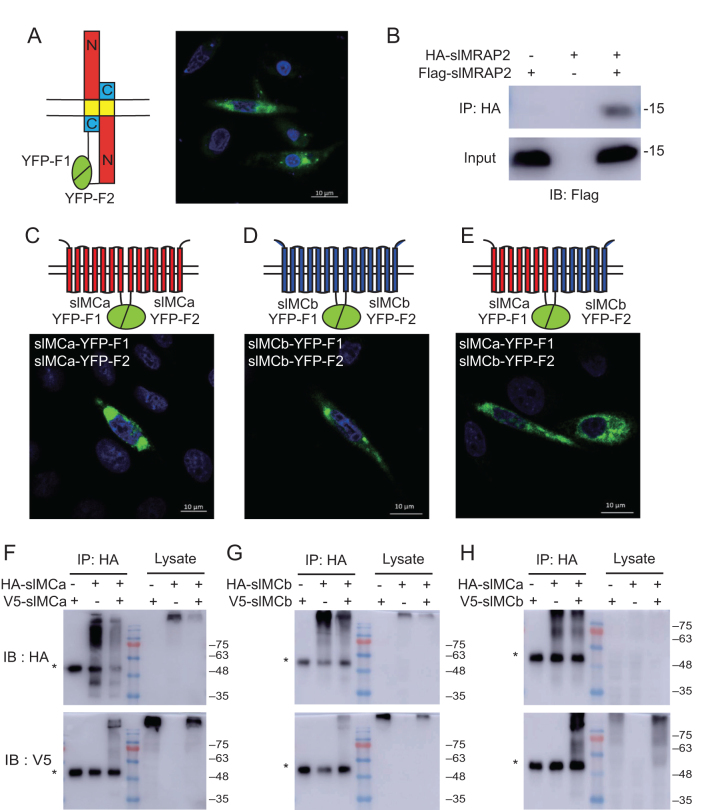
In order to examine the influences of the carboxyl terminus of MRAP2 protein on MCR signaling, we constructed four types of MRAP2s: the short carboxyl terminus of slMRAP2 was replaced by the full carboxyl terminus of elephant shark MRAP2 (esMRAP2-C), zebrafish MRAP2a (zMRAP2a-C) or zebrafish MRAP2b (zMRAP2b-C), respectively (Fig. 2I). Co-immunoprecipitation (Co-IP) was applied to monitor the protein interactions of MRAP2s with slMCRs. Results showed that slMRAP2 could interact with either slMCa or slMCb by forming a tight protein complex (Fig. 2A and B). Similarly, esMRAP2-C, zMRAP2a-C and zMRAP2b-C could also interact with slMCa or slMCb (Fig. 2C, D, E, F, G and H). These data suggested that slMRAP2 might be the real functional accessory protein with the potential ability to interact and modulate the signaling of slMCa and slMCb. Interestingly, additional bands were detected in Fig. 2C, D, E and F but not in Fig. 2A and B indicating that the carboxyl terminus should be required for the glycosylation of MRAP2 protein, but not for the interaction with MCRs.
Figure 2.
Interaction of MRAP2 proteins with slMCa or slMCb. Co-IP of slMCa or slMCb with four types of MRAP2 proteins: (A and B) slMRAP2, (C and D) esMRAP2-C, (E and F) zMRAP2a-C and (G and H). *IgG heavy chain. (I) Schematic diagram of four MRAP2 plasmids. N terminus and TM domain of esMRAP2-C, zMRAP2a-C and zMRAP2b-C originate from slMRAP2, and the carboxyl terminus is captured from esMRAP2, zMRAP2a or zMRAP2b.
Pharmacological modulation of MRAP2s on slMCa and slMCb signaling
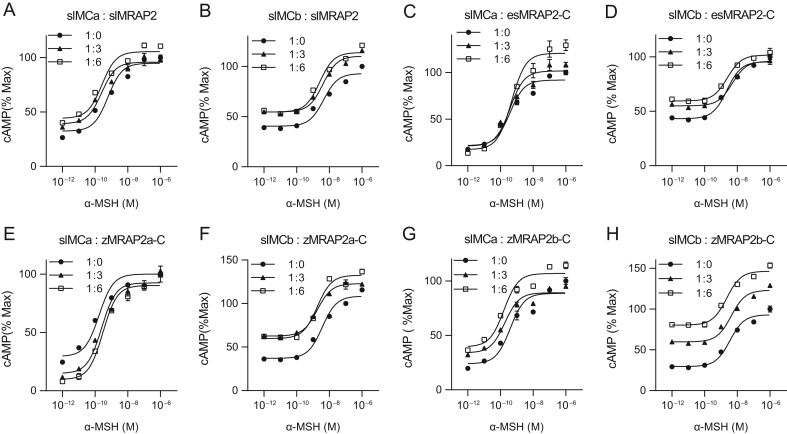
To investigate the impact of four MRAP2s on slMCa and slMCb signaling, the cAMP level was detected with pCre-luciferase reporter assay. Results showed that slMRAP2 could enhance the α-MSH response of slMCa or slMCb (Fig. 3A and B). A similar trend was seen in the presence of esMRAP2-C (Fig. 3C and D) or zMRAP2b-C (Fig. 3G and H). The zMRAP2a-C, however, sensitized the slMCb signaling in a similar way as slMRAP2 (Fig. 3F) but suppressed the response of slMCa (Fig. 3E). Besides, all the four MRAP2s did not significantly change the EC50 of slMCa and slMCb for α-MSH (Table 2).
Figure 3.
Modulation of slMCa and slMCb signaling by MRAP2s. α-MSH stimulated cAMP cascade of slMCa or slMCb is modulated in the presence of (A and B) slMRAP2, (C and D) esMRAP2-C, (E and F) zMRAP2a-C and (G and H) zMRAP2b-C.
Table 2.
Statistical analysis of Fig. 3.
| Data analysis | LogEC50 | P value for Vmax comparison | |||||
|---|---|---|---|---|---|---|---|
| 1:0 | 1:3 | 1:6 | 1:0 vs 1:3 | 1:0 vs 1:6 | 1:3 vs 1:6 | ||
| 3A | slMCa: slMRAP2 | −9.22 ± 0.30 | −9.61 ± 0.25 | −9.59 ± 0.27 | 0.5695 | <0.0001 | 0.0001 |
| 3B | slMCb: slMRAP2 | −8.35 ± 0.28 | −8.20 ± 0.39 | −8.60 ± 0.23 | <0.0001 | <0.0001 | 0.0289 |
| 3C | slMCa: esMRAP2-C | −9.42 ± 0.28 | −9.42 ± 0.28 | −9.324 ± 0.27 | 0.0527 | <0.0001 | 0.0003 |
| 3D | slMCb: esMRAP2-C | −8.64 ± 0.18 | −8.13 ± 0.36 | −8.62 ± 0.22 | 0.3386 | 0.0258 | 0.0103 |
| 3E | slMCa: zMRAP2a-C | −9.76 ± 0.34 | −9.60 ± 0.25 | −9.52 ± 0.20 | <0.0001 | <0.0001 | 0.6048 |
| 3F | slMCb: zMRAP2a-C | −8.44 ± 0.20 | −8.63 ± 0.23 | −8.74 ± 0.24 | <0.0001 | <0.0001 | <0.0001 |
| 3G | slMCa: zMRAP2b-C | −9.41 ± 0.35 | −9.49 ± 0.48 | −9.79 ± 0.27 | 0.155 | <0.0001 | <0.0001 |
| 3H | slMCb: zMRAP2b-C | −8.46 ± 0.21 | −8.18 ± 0.39 | −8.65 ± 0.22 | <0.0001 | <0.0001 | <0.0001 |
Two-way ANOVA with Tukey post-test was applied in the statistical analysis.
Effects of MRAP2s on surface expression of slMCa and slMCb
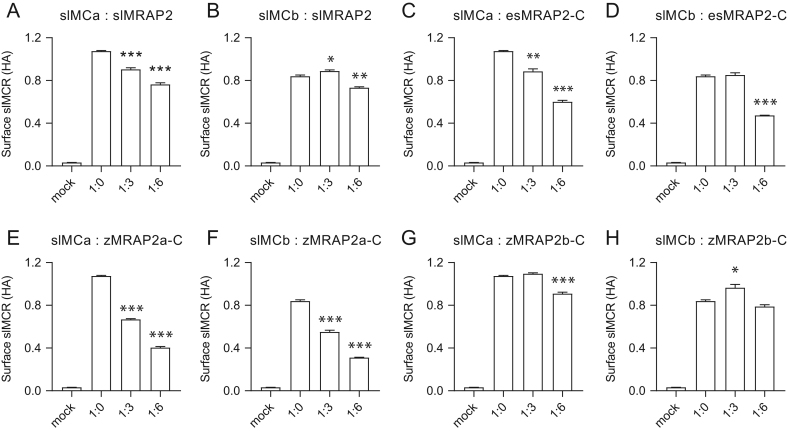
We further explored whether MRAP2 affected MCR signaling by altering the cell surface expression. ELISA assays were employed to monitor the surface level of slMCa and slMCb in the presence of certain ratios of MRAP2 protein. As shown (Fig. 4A, C, E and G), surface expression of slMCa dramatically decreased in the presence of MRAP2s. zMRAP2b-C did not decrease slMCa surface expression at the ratio of 1:3 (Fig. 4G). slMCb surface expression was not suppressed by slMRAP2 as much as slMCa (Fig. 4B). However, the surface level of slMCb significantly decreased when slMRAP2 carboxyl terminus was replaced by esMRAP2 (Fig. 4D) or zMRAP2a carboxyl terminus (Fig. 4F), but not with zMRAP2b-C (Fig. 4H).
Figure 4.
Surface expression regulation of slMCa and slMCb by MRAP2s. Surface expression of slMCa or slMCb is modulated by (A and B) slMRAP2, (C and D) esMRAP2-C, (E and F) zMRAP2a-C and (G and H) zMRAP2b-C. *P < 0.05; **P < 0.01; ***P < 0.001.
Antagonism of slMCb signaling by AgRP
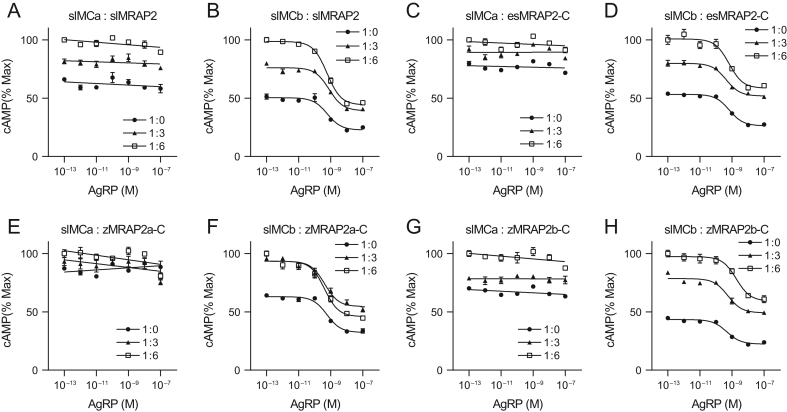
Next, we investigated whether the human AgRP peptide (83–139) could inhibit slMCa and slMCb signaling. Competition binding curves for the slMCa or slMCb regulated by MRAP2s were shown in Fig. 5. Interestingly, the AgRP neither inhibited nor increased the slMCa signaling regardless of the presence of MRAP2s, indicating the lack of antagonism of AgRP on slMCa (Fig. 5A, C, E and G). However, AgRP could clearly inhibit the slMCb signaling (Fig. 5B, D, F and H) with the fact that the IC50 showed no significant difference when co-expressed with four types of MRAP2 proteins (Table 3).
Figure 5.
AGRP competitively inhibits slMCb not slMCa signaling. Competition binding inhibition curves for the slMCa or slMCb regulated by (A and B) slMRAP2, (C and D) esMRAP2-C, (E and F) zMRAP2a-C and (G and H) zMRAP2b-C.
Table 3.
Statistical analysis of Fig. 5.
| Data analysis | LogIC50 | P value for Vmax comparison | |||||
|---|---|---|---|---|---|---|---|
| 1:0 | 1:3 | 1:6 | 1:0 vs 1:3 | 1:0 vs 1:6 | 1:3 vs 1:6 | ||
| 5B | slMCb: slMRAP2 | −9.22 ± 0.63 | −9.11 ± 0.42 | 9.23 ± 0.26 | <0.0001 | <0.0001 | 0.0518 |
| 5D | slMCb: esMRAP2-C | −9.17 ± 0.28 | −9.36 ± 0.47 | 9.18 ± 0.65 | <0.0001 | <0.0001 | 0.005 |
| 5F | slMCb: zMRAP2a-C | −9.24 ± 0.45 | −9.38 ± 0.64 | 9.39 ± 0.55 | <0.0001 | <0.0001 | 0.0127 |
| 5H | slMCb: zMRAP2b-C | −9.32 ± 0.42 | −9.26 ± 0.62 | 8.73 ± 0.63 | <0.0001 | <0.0001 | <0.0001 |
Two-way ANOVA with Tukey post-test was applied in the statistical analysis.
Dimerization of sea lamprey MRAP2 and MCRs
To determine whether slMRAP2 could form an antiparallel homodimer, such as those of mouse or human MRAP2 proteins, a complementary YFP luminescent assay was conducted. A schematic representation of configurations in which slMRAP2 molecules fused with YFP fragments on the cell membrane is shown in Fig. 6A, and the interaction of slMRAP2 homodimers were proved by Co-IP as shown in Fig. 6B. In this study, we also checked the dimerization of slMCa and slMCb. Clear YFP fluorescence on the plasma membrane (Fig. 6C, D and E) and Co-IP results (Fig. 6F, G and H) provided strong evidence that slMCa and slMCb could form either homodimer or heterodimer in vitro.
Figure 6.
Dimerization of sea lamprey MRAP2, MCa and MCb. (A) YFP fluorescence emitted by slMRAP2 antiparallel homodimers. Scale bar: 10 μm. (B) Co-IP of slMRAP2 homodimers. (C, D and E) YFP fluorescence emitted by slMCR homo- and heterodimers. Scale bar: 10 μm. (F, G and H) Co-IP of slMCRs homo- and heterodimers. *IgG heavy chain.
Discussion
In this study, we focused on whether the regulatory effect of MRAP2 on MCRs existed in the sea lamprey, even though no ligand had been identified and slMRAP2 and slMCRs were greatly different from that of mammals. We first compared slMCa, slMCb and slMRAP2 protein sequences with orthologs from river lamprey, elephant shark and zebrafish. Alignment suggested that slMCa and slMCb were highly conserved with rlMCa and rlMCb, respectively (identity >88%). For the ancestral slMRAP2, previous studies had revealed a partial protein sequence (22, 25). The partial slMRAP2 was aligned with many other MRAP2 subtypes and reported to be highly conserved at the TM domain (23). However, we found a stop codon located at the third codon behind the TM domain of slMRAP2. We speculated that the carboxyl terminus of slMRAP2 might be naturally shorter than other MRAP2 orthologs. To verify the regulatory function of the short slMRAP2 on slMCRs, we constructed three other MRAP2s with the carboxyl terminus from several other species. Like those of mouse and human MRAP2 (12, 13), slMRAP2 could form a protein complex with both slMCa and slMCb. This result suggested that lack of the carboxyl terminus of slMRAP2 had no effect on its binding to slMCRs. However, the carboxyl terminus of MRAP2 seemed to affect the glycosylation of MRAP2. Co-IP results showed that similar to hMRAP2 (12), two MRAP2 bands were seen for esMRAP2-C and zMRAP2a-C, whereas slMRAP2 and zMRAP2b-C only had one band, indicating the requirement for an unknown key motif within the carboxyl terminus for the glycosylation of MRAP2 protein.
We demonstrated, for the first time, that the most ancient known MRAP2 interacted with MCRs. Therefore, it was interesting to see whether the slMRAP2 could affect the pharmacological features of melanocortin receptors. As shown, in common with esMRAP2-C and zMRAP2b-C, slMRAP2 could enhance slMCa or slMCb signaling by elevating their constitutive activities and α-MSH-induced agonism. Mouse MRAP2 was reported to enhance cAMP signaling through MC3R and MC4R (14). However, an exception was observed, in that zMRAP2a-C inhibited the slMCa signaling and increased that of slMCb. This difference might be caused by the different regulatory effects of MRAP2a and MRAP2b. In zebrafish, zMRAP2a blunted MC4R signaling, while zMRAP2b decreased the constitutive activity but promoted the stimulating activity of MC4R (15). To elucidate the mechanism by which slMRAP2 regulates slMCa and slMCb, we next performed ELISA assays and ligand competition-binding assays. The general trends of slMCa and slMCb surface expression were attenuated with a high concentration of slMRAP2, which were in line with esMRAP2-C and zMRAP2a-C. However, zMRAP2b-C showed no effect on the surface expression of slMCb. Our results are consistent with previous work by Webb et al., in that the carboxyl terminus was not required for the interaction or surface trafficking of the MC2R (26), although the carboxyl terminus of zebrafish MRAP2a slightly altered the regulatory capacity of slMRAP2. Taken together, slMRAP2 could boost slMCRs activity without increasing the surface expression of slMCRs. Interestingly, we also found that AgRP, a specific antagonist to MC3R and MC4R (1), could not compete with α-MSH on the slMCa signaling, suggesting that slMCa was more similar to MC1R and MC2R, whereas slMCb was more similar to MC3R and MC4R (20, 21, 22).
MRAPs and MCRs have been reported to form homodimers in vitro (10, 27, 28, 29, 30). Our recent work demonstrated that zebrafish MC5Ra and MC5Rb could form homo- and heterodimers, which could be disrupted by MRAP2a or MRAP2b proteins (24). Therefore, we also conducted YFP luminescent assays and identified the formation of antiparallel homodimers of slMRAP2. Sebag et al. reported that amino acids 31–37 of human MRAP1 were required for its dual topology, which are located just before the TM domain (27). This motif in MRAP1 was found to be highly conserved in the MRAP2 of many species including sea lamprey. In accordance with previous reports (6, 7), slMCa and slMCb were observed to form homodimers and heterodimers as predicted and confirmed by YFP luminescent and Co-IP experiments.
In conclusion, here we have provided the first evidence that the ancient slMRAP2 lacking carboxyl terminus was fully functional and could modulate the signaling of two slMCRs in the form of antiparallel dimers. We also identified the homo- and hetero-dimerization of slMCa and slMCb. This study provides a better insight of the emerging melanocortin system in the agnathans, elucidated the presence and functional modulation of the melanocortin receptor by ancient accessory proteins, which could help us to further investigate the evolutionary perspective of the melanocortin system in the future.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
The work was supported by grants from National Key Research and Development Program of China (Grant No. 2017YFA0103902). The National Natural Science Foundation of China (Grant No. 81570760 and 31771283).
References
- 1.Cone RD. Studies on the physiological functions of the melanocortin system. Endocrine Reviews 2006. 27 736–749. ( 10.1210/er.2006-0034) [DOI] [PubMed] [Google Scholar]
- 2.Blanchard SG, Harris CO, Ittoop OR, Nichols JS, Parks DJ, Truesdale AT, Wilkison WO. Agouti antagonism of melanocortin binding and action in the B16F10 murine melanoma cell line. Biochemistry 1995. 34 10406–10411. ( 10.1021/bi00033a012) [DOI] [PubMed] [Google Scholar]
- 3.Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 1997. 278 135–138. ( 10.1126/science.278.5335.135) [DOI] [PubMed] [Google Scholar]
- 4.Dores RM, Lecaude S, Bauer D, Danielson PB. Analyzing the evolution of the opioid/orphanin gene family. Mass Spectrometry Reviews 2002. 21 220–243. ( 10.1002/mas.10029) [DOI] [PubMed] [Google Scholar]
- 5.Mandrika I, Petrovska R, Wikberg J. Melanocortin receptors form constitutive homo- and heterodimers. Biochemical and Biophysical Research Communications 2005. 326 349–354. ( 10.1016/j.bbrc.2004.11.036) [DOI] [PubMed] [Google Scholar]
- 6.Rediger A, Piechowski CL, Habegger K, Gruters A, Krude H, Tschop MH, Kleinau G, Biebermann H. MC4R dimerization in the paraventricular nucleus and GHSR/MC3R heterodimerization in the arcuate nucleus: is there relevance for body weight regulation? Neuroendocrinology 2012. 95 277–288. ( 10.1159/000334903) [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Hamamoto A, Takahashi A, Saito Y. Dimerization of melanocortin receptor 1 (MC1R) and MC5R creates a ligand-dependent signal modulation: potential participation in physiological color change in the flounder. General and Comparative Endocrinology 2016. 230–231 103–109. ( 10.1016/j.ygcen.2016.04.008) [DOI] [PubMed] [Google Scholar]
- 8.Cortes R, Navarro S, Agulleiro MJ, Guillot R, Garcia-Herranz V, Sanchez E, Cerda-Reverter JM. Evolution of the melanocortin system. General and Comparative Endocrinology 2014. 209 3–10. ( 10.1016/j.ygcen.2014.04.005) [DOI] [PubMed] [Google Scholar]
- 9.Metherell LA, Chapple JP, Cooray S, David A, Becker C, Ruschendorf F, Naville D, Begeot M, Khoo B, Nurnberg P, et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nature Genetics 2005. 37 166–170. ( 10.1038/ng1501) [DOI] [PubMed] [Google Scholar]
- 10.Sebag JA, Hinkle PM. Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. PNAS 2007. 104 20244–20249. ( 10.1073/pnas.0708916105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebag JA, Hinkle PM. Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. Journal of Biological Chemistry 2009. 284 22641–22648. ( 10.1074/jbc.M109.022400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan LF, Webb TR, Chung TT, Meimaridou E, Cooray SN, Guasti L, Chapple JP, Egertova M, Elphick MR, Cheetham ME, et al. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. PNAS 2009. 106 6146–6151. ( 10.1073/pnas.0809918106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebag JA, Hinkle PM. Regulation of G protein-coupled receptor signaling: specific dominant-negative effects of melanocortin 2 receptor accessory protein 2. Science Signaling 2010. 3 ra28 ( 10.1126/scisignal.2000593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N, Ramanathan V, Strochlic DE, Ferket P, Linhart K, et al. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science 2013. 341 275–278. ( 10.1126/science.1233000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebag JA, Zhang C, Hinkle PM, Bradshaw AM, Cone RD. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 2013. 341 278–281. ( 10.1126/science.1232995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Elmquist JK, Williams KW. Mrap2: an accessory protein linked to obesity. Cell Metabolism 2013. 18 309–311. ( 10.1016/j.cmet.2013.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan DW, Bryson-Richardson RJ, Pagan KE, Taylor MS, Currie PD, Jackson IJ. The structure and evolution of the melanocortin and MCH receptors in fish and mammals. Genomics 2003. 81 184–191. ( 10.1016/S0888-7543(02)00037-X) [DOI] [PubMed] [Google Scholar]
- 18.Klovins J, Haitina T, Fridmanis D, Kilianova Z, Kapa I, Fredriksson R, Gallo-Payet N, Schioth HB. The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Molecular Biology and Evolution 2004. 21 563–579. ( 10.1093/molbev/msh050) [DOI] [PubMed] [Google Scholar]
- 19.Green SA, Bronner ME. The lamprey: a jawless vertebrate model system for examining origin of the neural crest and other vertebrate traits. Differentiation 2014. 87 44–51. ( 10.1016/j.diff.2014.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haitina T, Klovins J, Takahashi A, Lowgren M, Ringholm A, Enberg J, Kawauchi H, Larson ET, Fredriksson R, Schioth HB. Functional characterization of two melanocortin (MC) receptors in lamprey showing orthology to the MC1 and MC4 receptor subtypes. BMC Evolutionary Biology 2007. 7 101 ( 10.1186/1471-2148-7-101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baron A, Veo K, Angleson J, Dores RM. Modeling the evolution of the MC2R and MC5R genes: studies on the cartilaginous fish, Heterondotus francisci. General and Comparative Endocrinology 2009. 161 13–19. ( 10.1016/j.ygcen.2008.11.026) [DOI] [PubMed] [Google Scholar]
- 22.Vastermark Å, Schioth HB. The early origin of melanocortin receptors, agouti-related peptide, agouti signalling peptide, and melanocortin receptor-accessory proteins, with emphasis on pufferfishes, elephant shark, lampreys, and amphioxus. European Journal of Pharmacology 2011. 660 61–69. ( 10.1016/j.ejphar.2010.10.106) [DOI] [PubMed] [Google Scholar]
- 23.Valsalan R, Krishnan A, Almén MS, Fredriksson R, Schiöth HB. Early vertebrate origin of melanocortin 2 receptor accessory proteins (MRAPs). General and Comparative Endocrinology 2013. 188 123–132. ( 10.1016/j.ygcen.2013.01.004) [DOI] [PubMed] [Google Scholar]
- 24.Zhu M, Wang M, Chen Y, Zhang C. Pharmacological modulation of two melanocortin-5 receptors by MRAP2 proteins in zebrafish. Journal of Molecular Endocrinology 2019. 62 27–36. ( 10.1530/JME-18-0104) [DOI] [PubMed] [Google Scholar]
- 25.Dores RM. Hypothesis and theory: revisiting views on the co-evolution of the melanocortin receptors and the accessory proteins, MRAP1 and MRAP2. Frontiers in Endocrinology 2016. 7 79 ( 10.3389/fendo.2016.00079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb TR, Chan L, Cooray SN, Cheetham ME, Chapple JP, Clark AJL. Distinct melanocortin 2 receptor accessory protein domains are required for melanocortin 2 receptor interaction and promotion of receptor trafficking. Endocrinology 2009. 150 720–726. ( 10.1210/en.2008-0941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebag JA, Hinkle PM. Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. Journal of Biological Chemistry 2009. 284 610–618. ( 10.1074/jbc.M804413200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickolls SA, Maki RA. Dimerization of the melanocortin 4 receptor: a study using bioluminescence resonance energy transfer. Peptides 2006. 27 380–387. ( 10.1016/j.peptides.2004.12.037) [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Laorden BL, Sanchez-Mas J, Martinez-Alonso E, Martinez-Menarguez JA, Garcia-Borron JC, Jimenez-Cervantes C. Dimerization of the human melanocortin 1 receptor: functional consequences and dominant-negative effects. Journal of Investigative Dermatology 2006. 126 172–181. ( 10.1038/sj.jid.5700036) [DOI] [PubMed] [Google Scholar]
- 30.Cooray SN, Almiro Do Vale I, Leung KY, Webb TR, Chapple JP, Egertova M, Cheetham ME, Elphick MR, Clark AJL. The melanocortin 2 receptor accessory protein exists as a homodimer and is essential for the function of the melanocortin 2 receptor in the mouse y1 cell line. Endocrinology 2008. 149 1935–1941. ( 10.1210/en.2007-1463) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a