Abstract
We investigated the microRNA172 (miR172)-mediated regulatory network for the perception of changes in external and endogenous signals to identify a universally applicable floral regulation system in ornamental plants, manipulation of which could be economically beneficial. Transgenic gloxinia plants, in which miR172 was either overexpressed or suppressed, were generated using Agrobacterium-mediated transformation. They were used to study the effect of altering the expression of this miRNA on time of flowering and to identify its mRNA target. Early or late flowering was observed in transgenic plants in which miR172 was overexpressed or suppressed, respectively. A full-length complementary DNA (cDNA) of gloxinia (Sinningia speciosa) APETALA2-like (SsAP2-like) was identified as a target of miR172. The altered expression levels of miR172 caused up-or down-regulation of SsAP2-like during flower development, which affected the time of flowering. Quantitative real-time reverse transcription PCR analysis of different gloxinia tissues revealed that the accumulation of SsAP2-like was negatively correlated with the expression of miR172a, whereas the expression pattern of miR172a was negatively correlated with that of miR156a. Our results suggest that transgenic manipulation of miR172 could be used as a universal strategy for regulating time of flowering in ornamental plants.
Keywords: Flowering time, Transgenic gloxinia, MicroRNA172, APETALA2-like (AP2-like)
1. Introduction
Plants rely on a complex genetic regulatory network for the perception of changes in external and endogenous signals to select the appropriate time of flowering. Within this vast regulatory network, microRNAs (miRNAs) play pivotal roles by controlling the expression of key flowering genes.
All miRNAs are non-coding RNAs of 20–24 nucleotides long that specifically regulate the expression of target genes at the transcriptional or translational level in eukaryotes. Multiple developmental events are regulated by miRNAs in plants (Chen, 2005). Among the numerous miRNA families, only three, namely miR156, miR172, and miR159/miR319, are associated with the regulation of flowering time (Terzi and Simpson, 2008). The overexpression of miR159/miR319 delays flowering: miR159 is mainly involved in gibberellin signaling pathways (Achard et al., 2004; Terzi and Simpson, 2008; Li et al., 2013).
In Arabidopsis, there are eight loci that encode the members of the miR156 family, including miR156a–miR156j (Morea et al., 2016). The expression level of miR156 is highest in plant embryos and plantlets, and it gradually decreases during plant growth (Hong and Jackson, 2015). The main target genes of the miR156 family are the SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-box transcription factors (Ferreira et al., 2014). In Arabidopsis, miR156 regulates the timing of the transition from the juvenile to the adult stage by repressing the transcript levels of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors (Yang et al., 2011). SPL9, a target of miR156, binds directly to the regulatory region of miR172b to promote its expression and induce flowering (Wu et al., 2009). MiR172, which is downstream of miR156, promotes adult epidermal identity. Both miR156 and miR172 are associated with aging in plants.
MiR172 has been predicted to suppress a set of transcription factors, including APETALA2 (AP2) and related proteins, to promote flowering (Zhu and Helliwell, 2011). In Arabidopsis, the overexpression of miR172 caused early flowering and floral defects under long-day conditions. An early-flowering mutant, early activation tagged dominant (eat-D), exhibited a phenotype similar to that of the miR172 overexpression line (Aukerman and Sakai, 2003). The overexpression of TARGET OF EAT1 (TOE1), which is a target gene of miR172, resulted in delayed flowering onset. Mutant plants deficient in TOE1 flowered slightly earlier, but the effect was not significant compared with the wild type (Aukerman and Sakai, 2003). Although flowering occurred significantly earlier in toe1 toe2 double mutants, the extent of early flowering was less significant than in miR172-overexpressing plant lines (Aukerman and Sakai, 2003).
Recent studies have shown that the overexpression of SCHLAFMUTZE (SMZ) and SCHNARCHZAPFEN (SNZ) leads to delayed flowering, whereas smz snz double mutants do not show significant changes in flowering time compared with the wild type (Mathieu et al., 2009). Toe1 toe2 smz snz quadruple mutants flower significantly earlier than toe1 toe2 double mutants, but the extent of early flowering was less significant than in the miR172-overexpressing plant lines (Yamaguchi and Abe, 2012). This indicates that the AP2-like gene family has a certain degree of functional redundancy in the regulation of flowering time (Mathieu et al., 2009). In loss-of-function sextuple mutants in which all AP2-like genes were affected, flowering occurred earlier than in the miR172-overexpressing transgenic plant line, indicating that all AP2-like genes are flowering inhibitors (Mathieu et al., 2009; Yant et al., 2010).
Gloxinia (Sinningia speciosa) is an herbaceous commercial plant native to south-eastern Brazil, with large, bell-shaped flowers. Because of their long flowering time and colorful flowers, these plants are popular as houseplants. However, the mechanism regulating flowering time is not known. We investigated whether genetic modification of the miR172 gene can provide a universal way to regulate flowering time in ornamental plants by generating transgenic gloxinia plants in which miR172 was overexpressed or suppressed. Candidate target genes of miR172 were identified, and a full-length complementary DNA (cDNA) of AP2-like was cloned to verify whether it is a target of miR172. We investigated whether the altered levels of miR172 modulated the expression of AP2-like genes, and whether the changes would affect the time of flowering under short-day conditions.
2. Materials and methods
2.1. Plant growth conditions
Young leaves cut from cultured plantlets of gloxinia were placed on Murashige and Skoog’s (MS) culture medium (pH 5.8) supplemented with hormones at different concentrations (Table S1). The cultures were placed in a chamber under short-day conditions (8-h light/16-h darkness, photon flux density of 25 μmol/(m2·s)) at 24 °C for differentiation. For regeneration, differentiated plantlets at the same developmental stage were transferred to root-inducing medium (Table S1). After one month, all the plantlets were transferred to a perlite and vermiculite matrix mixture. To assay the roles of miR172a in regulating flowering time, wild-type and transgenic gloxinia plants were grown under short-day conditions (8-h light/16-h darkness, (24±1) °C).
2.2. Plasmid construction
Sequences available in GenBank (http://www.ncbi.nlm.nih.gov/genbank) suggested that the core regions of the miR172 family are highly conserved in plants (Fig. 1). Because no information on the SsmiR172a precursor was available in public databases, we employed the AtmiR172a precursor (Columbia-0, Wassilewskija (Ws) ecotype) to generate mature miR172a which we overexpressed in gloxinia under the regulation of cauliflower mosaic virus 35S (CaMV 35S) promoter. To better understand the roles of miR172a in gloxinia, we used target mimicry (35S:MIM172) to suppress the activity of miR172a under the control of INDUCED BY PHOSPHATE STARVATION1 (IPS1), as described in previous reports (Franco-Zorrilla et al., 2007; Li et al., 2013). On the basis of sequences available in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov), primers were designed for the amplification of miR172a precursor from Arabidopsis (Columbia-0, Ws ecotype) cDNA. The sense (5'-TCCCCCGGGTTCTTCTGGTTTGG AGATGGTTAGG-3') and anti-sense primers (5'-AC GCGTCGACCTTGATAAAGACTGCCAAAAACC TG-3') contained SmaI and SalI sites at their 5'-ends, respectively. The artificial IPS1 fragment was cloned using forward (5'-CTTACGTCGTAGTAGGAGTT CTAAGATTTCTAGAGGGAGATAA-3') and reverse (5'-AATCTTAGAACTCCTACTACGACGTA AGCTTCGGTTCCCCTCG-3') primers designed for its amplification. Polymerase chain reactions (PCRs) were carried out using 2×Taq Master Mix (TaKaRa, Dalian, China) following the protocol provided by the supplier. For each PCR, 5 µL of the template was used in a 50-µL reaction mixture containing 25 µL Taq Master Mix, 200 nmol/L of each primer, and 18 µL of triple-distilled water. PCR was conducted for three biological replications of each sample using the following thermal cycles: initial denaturation at 94 °C for 5 min, followed by 30 cycles, each cycle consisting of 94 °C for 15 s, 50–60 °C for 30 s, and 72 °C for 15 s, and final extension at 72 °C for 10 min.
Fig. 1.
Alignment of mature miR172a sequences from different plant species
Sequence alignment of mature miR172a from different species: Arabidopsis thaliana (At), Solanum lycopersicum (Sl), Arabidopsis lyrata (Al), Aquilegia coerulea (Aq), Brassica oleracea (Bo), Brassica campestris (Br), Elaeis guineensis (Eg), Glycine max (Gm), Oryza sativa (Os), Populus trichocarpa (Pt), and Vitis vinifera (Vv)
All fragments were cloned into the pCAMBIA13011 plasmid, which contained a CaMV 35S promoter and the terminator of nopaline synthase (Nos), as well as kanamycin-and hygromycin-resistance genes. All recombinant plasmids were transferred into Agrobacterium tumefaciens strain EHA105.
2.3. Screening of transgenic gloxinia plants
Transgenic gloxinia plants were generated by Agrobacterium-mediated transformation as described previously (Zhang et al., 2008; Li et al., 2013). After four cycles of fortnightly selection (Table S1), hygromycin-resistant plantlets were transferred to root-induction medium without hygromycin. PCR was used to screen the transgenic plantlets with gene-specific primers and genomic DNA.
2.4. Genomic DNA isolation
Genomic DNA was isolated from young leaves of transformed plants using a genomic DNA isolation kit (TaKaRa, Dalian, China), following the manufacturer’s protocol, and used for PCR-based screening of the hygromycin-resistance gene.
2.5. Total RNA isolation
Total RNA was extracted from leaves and flower buds using TRIzol reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA were measured using a BioSpectrometer (Eppendorf, Hamburg, Germany). Reverse transcription was carried out with 1 µg RNA following the instructions provided with the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China). Mature miRNAs were detected using stem-loop reverse transcription PCR (RT-PCR) (Varkonyi-Gasic and Hellens, 2010; Li et al., 2013).
2.6. Cloning of full-length gloxinia APETALA2-like (SsAP2-like) cDNA and sequence analysis
Degenerate primers (forward primer: 5'-TAG GTGGATTTGAYACNGCWCATGC-3' and reverse primer: 5'-CCYAAATAAACRTACTKTTTGCCT-3') were designed on the basis of the conserved region in the AP2 sequences of Arabidopsis, tomato, and grape. The PolyATract mRNA Isolation System III (Promega, Madison, WI, USA) was used to purify gloxinia mRNA. On the basis of the partial sequence of the cDNA that was amplified, 5'-rapid amplification of cDNA ends (5'-RACE) and 3'-RACE were performed using the SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) to obtain the full-length cDNA sequences (5'-RACE primer: 5'-CC ACTCCTCTGAATTTAATGGCTGCCC-3'; 3'-RACE primer: 5'-GCCCATTCTAGCTTCCCATCTACCAC-3'). These sequences were aligned using Omega 2.0 (Kramer, 2001), according to the instructions available for the software, to obtain the full-length sequence of SsAP2-like.
2.7. RNA ligase-mediated 5'-RACE
To verify whether SsAP2-like is a direct target of miR172a, modified RNA ligase-mediated (RLM) 5'-RACE was conducted to ascertain the cleavage site of miR172 (Li et al., 2013). A round of 5'-RACE was carried out with a special primer (anti-sense primer: 5'-TCATGAGGGTCTCATGAGAGAGTGGATC-3'). The PCR products were cloned into the T-simple vector and ten independent inserts were sequenced.
3. Results
3.1. Generation of transgenic gloxinia plantlets in which miR172a is overexpressed or suppressed
Distinct bands of predicted size were detected in 24 plantlets of AtmiR172a overexpression lines (35S:miR172a) and in 21 transgenic plantlets overexpressing MIM172 (35S:MIM172), but no bands were detected in wild-type plants (Fig. S1).
3.2. Influence of altered expression levels of miR172a on the flowering time in gloxinia
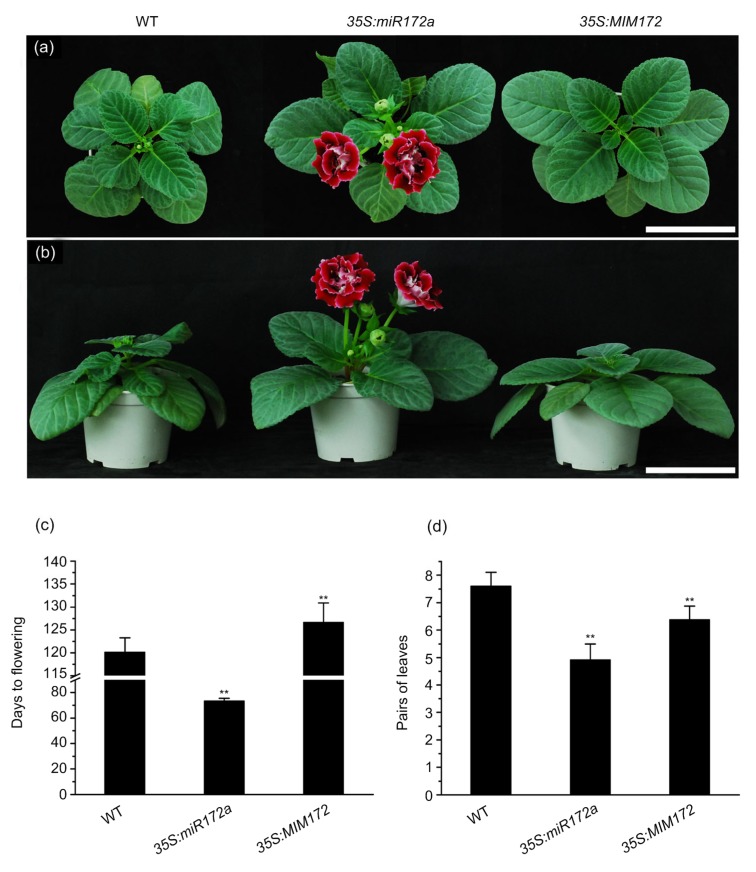
Under identical cultivation conditions, the lines overexpressing miR172a exhibited early flowering, whereas the 35S:MIM172 lines showed a delay in flowering compared with wild-type gloxinia plants (Fig. 2a). The 35S:miR172a transgenic lines flowered at Day 73.20±2.16 after transplantation; i.e., flowering occurred approximately 50 d earlier than in wild-type plants, which flowered at Day 120.20±3.15 after transplantation. In contrast, the 35S:MIM172 lines flowered at Day 126.60±4.28, which was much later than that of the 35S:miR172a lines (Fig. 2c). Phenotypic analysis revealed that the 35S:MIM172 transgenic plants were shorter than wild-type and 35S: miR172 plants. The leaf angles in the 35S:miR172a plants were larger than those in the wild-type and 35S:MIM172 plants (Fig. 2b). 35S:miR172a plants had fewer leaves than wild-type plants (4.91±0.58 and 7.60±0.50, respectively; Fig. 2d). While the overexpression of the AP2 gene family resulted in short plant height and slow growth in Arabidopsis (Todesco et al., 2010), 35S:MIM172 plants had fewer leaves than wild-type plants (6.38±0.50 and 7.60±0.50, respectively) at flowering. These results indicated that alteration of the expression level of miR172a in gloxinia significantly influenced flowering time.
Fig. 2.
Influence of flowering time on transgenic gloxinia plants under short-day conditions
(a, b) Photographs were taken at 127 d after transplantation to a mixed matrix. Top views (a) and side views (b) of wild-type (WT), 35S:miR172a, and 35S:MIM172 plants. Scale bar represents 10 cm. (c) Days to flowering after transplantation. (d) Pairs of leaves at the squaring stage. Data represent the mean±standard deviation (SD) (n>20). Double asterisks indicate a significant difference (ANOVA, P<0.01) between the wild-type and transgenic plants
3.3. Isolation and sequence analysis of SsAP2-like
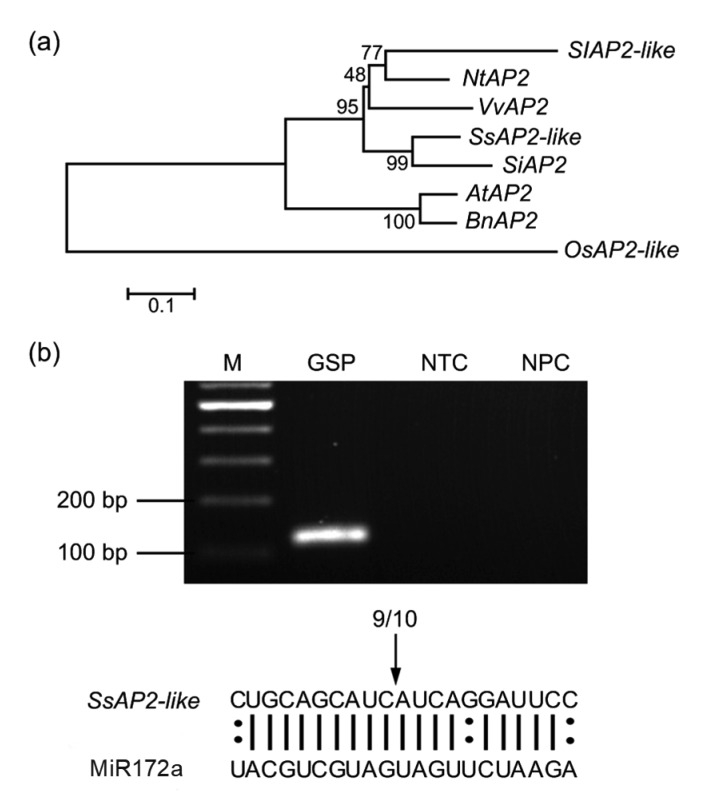
The complex functions of AP2 prompted us to investigate the possible regulatory roles of miR172 and AP2 in gloxinia during flower development. Sequence analysis showed that the full-length coding sequence of SsAP2-like was 1476 base pairs (bp) (Data S1). Phylogenetic analysis showed that the sequence of SsAP2-like was similar to that of Sesamum indicum (Fig. 3a). Multiple sequence alignment with other AP2 sequences demonstrated that a putative complementary site of miR172a existed in the region of 1383 to 1403 nucleotides, in which there were only three mismatched nucleotides (Fig. 3b).
Fig. 3.
Analysis of SsAP2-like nucleotide sequence and confirmation of miR172a-guided cleavage site in SsAP2-like mRNA
(a) Phylogenetic analysis of SsAP2-like nucleotide sequences from Solanum lycopersicum (Sl), Nicotiana tabacum (Nt), Vitis vinifera (Vv), Sesamum indicum (Si), Arabidopsis thaliana (At), Brassica napus (Bn), and Oryza sativa (Os). Phylogenetic tree was generated by the neighbor-joining method and the p-distance model using MEGA4 software. The numbers represent the bootstrap values. (b) Experimental confirmation of miR172 cleavage site in SsAP2-like mRNA. The cleavage site was determined by RLM 5'-RACE. M: marker; GSP: gene-specific primer; NTC: no-template control; NPC: no primer control. The arrow indicates the cleavage site of SsAP2-like and the number above the sequences indicates the ratio of 5'-RACE clones
A single band of expected size was amplified by nested PCR using cDNA from flower buds as a template (Fig. 3b). The amplified fragment was inserted into the T-simple vector and ten independent clones were sequenced. The results confirmed that SsAP2-like is a target of miR172a in gloxinia, as nine of the ten cloned fragments were cleaved at the position of the 10th nucleotide (Fig. 3b).
3.4. Expression patterns of miR156a, miR172a, and SsAP2-like in gloxinia
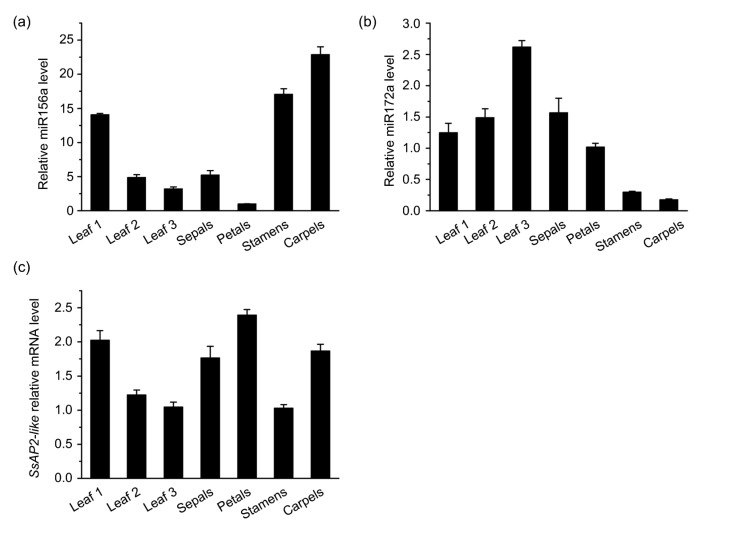
To investigate the expression patterns of miR156 and miR172 in gloxinia, we assessed the accumulation of mature miR156a, miR172a, and SsAP2-like mRNAs in various tissues. The results showed that miR156a was highly expressed in the leaves (leaf 1), stamens, and carpels of the plantlets, as in the case of Arabidopsis. In contrast, the accumulation of miR156a in old leaves (leaves 2 and 3) and sepals was relatively less; in particular, it was nearly absent in the petals (Fig. 4a).
Fig. 4.
Analysis of the expression patterns of mature SsmiR156a, SsmiR172a, and SsAP2-like in various tissues by RT-qPCR
(a) Accumulation of mature SsmiR156a in various tissues of gloxinia plants. (b) Accumulation of mature SsmiR172a in different tissues of gloxinia plants. (c) Relative mRNA levels of SsAP2-like in various tissues of gloxinia plants. Leaf 1: juvenile-phase leaf; Leaf 2: adult-phase leaf; Leaf 3: squaring-stage leaf. Mean values were obtained from three independent samples. Error bars represent SD
The expression of miR172a was negatively correlated with that of miR156a. With the growth of the plants, the expression level of miR172a in the leaves increased, with the highest accumulation being observed in the squaring-stage leaves (leaf 3). However, miR172a accumulated relatively less in sepals, petals, and stamens; it was nearly absent in the carpels (Fig. 4b).
SsAP2-like was highly expressed in leaf 1, sepals, petals, and carpels of the plantlet, whereas in the old leaves (leaves 2 and 3) and stamens, the accumulation of SsAP2-like was relatively less (Fig. 4c). The expression pattern of SsAP2-like was in accordance with that of miR156a, but was negatively correlated with the expression of miR172a in gloxinia.
3.5. Effects of overexpression and suppression of miR172a on SsAP2-like expression in transgenic gloxinia plants
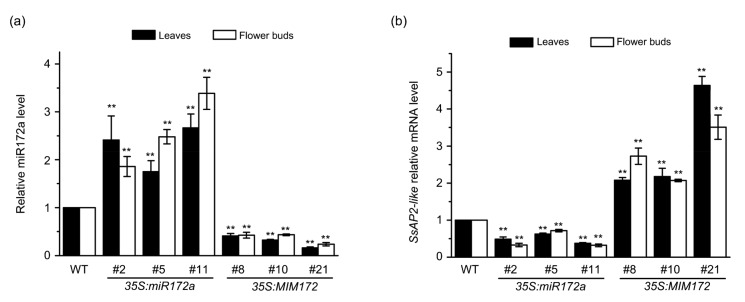
Compared with the wild type, the relative expression levels of mature miR172a in leaves and flower buds were dramatically increased in the 35S:miR172a lines and decreased in the 35S:MIM172 lines (Fig. 5a). These results revealed that we had successfully generated transgenic gloxinia plants with a substantial alteration in the miR172a level in various tissues.
Fig. 5.
Relative expression levels of mature miR172a and SsAP2-like in wild-type (WT) and transgenic gloxinia plants
(a) Transcript levels of mature miR172a in adult-stage leaves and flower buds. (b) Expression levels of SsAP2-like in adult-stage leaves and flower buds. Mean values were obtained from three independent samples. Error bars represent the SD. Double asterisks indicate significant differences (ANOVA, P<0.01) between the transgenic lines and WT plants
The expression levels of SsAP2-like in the 35S:miR172a lines were significantly decreased, whereas the levels of SsAP2-like were substantially increased in the 35S:MIM172 lines; this was consistent with the expression pattern of SsAP2-like, which was negatively regulated by miR172a (Fig. 5b). These results indicate that the accumulation of SsAP2-like mRNA is associated with decreasing expression of miR172a in transgenic gloxinia plants, suggesting that the expression level of SsAP2-like is negatively regulated by miR172 in gloxinia.
4. Discussion
The mechanisms of flowering transition in model plants have been extensively investigated and the results have provided a theoretical foundation for research based on the overexpression or inhibition of specific key genes (Zhang et al., 2008). An increasing number of miRNAs have been discovered in plants, and their effects on development and response to environmental change have been studied (Huijser and Schmid, 2011; Teotia and Tang, 2015). However, reports on modifying specific target genes to control flowering time in a species with unknown genomic information are relatively rare and there is a need for more research on the mechanisms of floral transition in economically important ornamental plants. We altered the expression levels of miR172 to affect the accumulation of its target genes, while not manipulating a single gene associated with flowering, and were able to identify an effective pathway for the regulation of flowering time in a species with unknown genomic background.
MiR172 is highly conserved and controls plant growth and development by regulating the expression level of AP2-like mRNA, or by inhibiting the translation of AP2-like genes (Aukerman and Sakai, 2003; Zhu et al., 2009). Multiple reports have revealed that the overexpression of miR172 causes early flowering under long-day conditions (Aukerman and Sakai, 2003; Lee et al., 2014). The role of miR172 on flowering time regulation prompted us to explore its function in gloxinia during flower development under short-day conditions. In most commercial plants, genomic information is still limited, so we employed the AtmiR172 precursor to generate transgenic lines that overexpress miR172a. We used the mimicry target approach to obtain miR172-suppressed transgenic lines in which endogenous miR172 activity was eliminated. We identified a member of the SsAP2-like family in gloxinia that contains a sequence complementary to miR172. AP2 gene family sequences are relatively conserved in numerous species. In plants, they are under the regulation of miR172, and are widespread, being involved in a variety of physiological and biochemical processes, such as plant growth, floral organ development, and signal transduction. Based on sequence alignment and phylogenetic analysis, we found that SsAP2-like and SiAP2 had the closest phylogenetic relationship. The SsAP2-like mRNA expression levels showed a negative relationship with the abundance of miR172 in both vegetative and floral tissues, consistent with previous findings in Arabidopsis and soybean (Lee et al., 2014; Wang et al., 2014). We found that alteration of the expression levels of miR172 significantly influenced flowering time. The overexpression of miR172 accelerated flowering by approximately 50 d, whereas its suppression delayed flowering by approximately six days compared with wild-type gloxinia under short-day conditions. Alteration of the expression levels of miR172 in gloxinia caused drastic changes in the AP2 mRNA levels. These findings reveal that miR172 regulates flowering time via the suppression of the AP2 gene family. In previous studies, alterations in the expression levels of miR172 and AP2 determined floral patterns and fruit defects (Mlotshwa et al., 2006; Yao et al., 2016). However, in the present study, overexpression of miR172 did not cause any change in the flowers.
MiR156 plays a dominant role in the juvenile phase of plant and flower development because it directly promotes the expression of flowering pathway assembly factors and meristem tissue regulators that control flowering time (Xu et al., 2016). MiR156 can control miR172 expression by regulating SBP-box genes, thereby regulating the activity of AP2-like genes in inhibiting flowering. In gloxinia, miR156 and miR172 are expressed alternately. In the juvenile phase, miR156 is expressed at higher levels than miR172. As the plant grows, miR156 expression gradually decreases and miR172 expression gradually increases, which is accompanied by a gradual decrease in AP2 expression. It has been reported that AP2 has different functions in floral morphological transitions and plant development (Yant et al., 2010). Analysis of the expression profiles of miR156, miR172, and AP2 in gloxinia showed that the expression of miR156 was consistent with that of AP2-like genes, but opposite to that of miR172. This is consistent with the expression profiles of miR156, miR172, and AP2 in Arabidopsis, indicating that the mutually influential relationship between miR156 and miR172 is relatively conserved in the evolution of plants.
5. Conclusions
The manipulation of miR172 expression in gloxinia alters flowering time. Our study provides a new genetic approach for regulating time of flowering by altering the expression levels of miR172 in ornamental plants.
List of electronic supplementary materials
Culture medium used in this study
Primers used in the experiments
PCR analysis of Pre-AtmiR172a and MIM172 (35S:MIM172) in transgenic gloxinias
Nucleotide sequence of SsAP2-like
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31171615 and 31401913)
Contributors: Xiao-yan LI performed the experimental research and data analysis, wrote and edited the manuscript. Fu GUO and Sheng-yun MA collected the data. Mu-yuan ZHU, Wei-huai PAN, and Hong-wu BIAN contributed to the study design, data analysis, and editing of the manuscript. All authors read and approved the final manuscript and, therefore, had full access to all the data in the study and take responsibility for the integrity and security of the data.
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1800003) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Xiao-yan LI, Fu GUO, Sheng-yun MA, Mu-yuan ZHU, Wei-huai PAN, and Hong-wu BIAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Achard P, Herr A, Baulcombe DC, et al. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131(14):3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen XM. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579(26):5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira ESG, Silva EM, da Silva Azevedo M. MicroRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014;78(4):604–618. doi: 10.1111/tpj.12493. [DOI] [PubMed] [Google Scholar]
- 5.Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 6.Hong YG, Jackson S. Floral induction and flower formation–the role and potential applications of miRNAs. Plant Biotechnol J. 2015;13(3):282–292. doi: 10.1111/pbi.12340. [DOI] [PubMed] [Google Scholar]
- 7.Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138(19):4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- 8.Kramer JA. Omiga™: a PC-based sequence analysis tool. Mol Biotechnol. 2001;19(1):97–106. doi: 10.1385/MB:19:1:097. [DOI] [PubMed] [Google Scholar]
- 9.Lee YS, Lee DY, Cho LH, et al. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice. 2014;7(1):31. doi: 10.1186/s12284-014-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XY, Bian HW, Song DF, et al. Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann Bot. 2013;111(5):791–799. doi: 10.1093/aob/mct034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu J, Yant LJ, Mürdter F, et al. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7(7):e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mlotshwa S, Yang ZY, Kim YJ, et al. Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana . Plant Mol Biol. 2006;61(4-5):781–793. doi: 10.1007/s11103-006-0049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morea EGO, da Silva EM, e Silva GFF, et al. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol, 16:40. 2016 doi: 10.1186/s12870-016-0716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teotia S, Tang GL. To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant. 2015;8(3):359–377. doi: 10.1016/j.molp.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Terzi LC, Simpson GG. Regulation of flowering time by RNA processing. In: Reddy ASN Golovkin M., editor. Nuclear Pre-mRNA Processing in Plants. Springer, Berlin, Heidelberg; 2008. pp. 201–218. [DOI] [PubMed] [Google Scholar]
- 16.Todesco M, Rubio-Somoza I, Paz-Ares J, et al. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana . PLoS Genet. 2010;6(7):e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varkonyi-Gasic E, Hellens RP. qRT-PCR of small RNAs. In: Kovalchuk I Zemp FJ., editor. Plant Epigenetics: Methods and Protocols. Humana Press, Totowa; 2010. pp. 109–122. [DOI] [PubMed] [Google Scholar]
- 18.Wang YN, Wang LX, Zou YM, et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell. 2014;26(12):4782–4801. doi: 10.1105/tpc.114.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu G, Park MY, Conway SR, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis . Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu ML, Hu TQ, Zhao JF, et al. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana . PLoS Genet. 2016;12(8):e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi A, Abe M. Regulation of reproductive development by non-coding RNA in Arabidopsis: to flower or not to flower. J Plant Res. 2012;125(6):693–704. doi: 10.1007/s10265-012-0513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, Conway SR, Poethig RS. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development. 2011;138(2):245–249. doi: 10.1242/dev.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yant L, Mathieu J, Dinh TT, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22(7):2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao JL, Tomes S, Xu J, et al. How microRNA172 affects fruit growth in different species is dependent on fruit type. Plant Signal Behav. 2016;11(4):e1156833. doi: 10.1080/15592324.2016.1156833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang MZ, Ye D, Wang LL, et al. Overexpression of the cucumber LEAFY homolog CFL and hormone treatments alter flower development in gloxinia (Sinningia speciosa) Plant Mol Biol. 2008;67(4):419–427. doi: 10.1007/s11103-008-9330-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011;62(2):487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
- 27.Zhu QH, Upadhyaya NM, Gubler F, et al. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa) BMC Plant Biol, 9:149. 2009 doi: 10.1186/1471-2229-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Culture medium used in this study
Primers used in the experiments
PCR analysis of Pre-AtmiR172a and MIM172 (35S:MIM172) in transgenic gloxinias
Nucleotide sequence of SsAP2-like