Abstract
An extracellular lipase from Aureobasidium pullulans was obtained and purified with a specific activity of 17.7 U/mg of protein using ultrafiltration and a DEAE-Sepharose Fast Flow column. Characterization of the lipase indicated that it is a novel finding from the species A. pullulans. The molecular weight of the lipase was 39.5 kDa, determined by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme exhibited its optimum activity at 40 °C and pH of 7. It also showed a remarkable stability in some organic solutions (30%, v/v) including n-propanol, isopropanol, dimethyl sulfoxide (DMSO), and hexane. The catalytic activity of the lipase was enhanced by Ca2+ and was slightly inhibited by Mn2+ and Zn2+ at a concentration of 10 mmol/L. The lipase was activated by the anionic surfactant SDS and the non-ionic surfactants Tween 20, Tween 80, and Triton X-100, but it was drastically inhibited by the cationic surfactant cetyl trimethyl ammonium bromide (CTAB). Furthermore, the lipase was able to hydrolyze a wide variety of edible oils, such as peanut oil, corn oil, sunflower seed oil, sesame oil, and olive oil. Our study indicated that the lipase we obtained is a potential biocatalyst for industrial use.
Keywords: Lipase, Aureobasidium pullulans, Purification, Enzymatic characterization
1. Introduction
Lipases (triacylglycerol hydrolases, EC 3.1.1.3) are a class of enzymes that hydrolyze triglycerides at the lipid–water interface and catalyze a series of synthesis reactions, including esterification, transesterification, and interesterification in organic media (Tomke and Rathod, 2015). Lipases are extensively distributed in nature and are found in plants, animals, and microorganisms, but only lipases of microbial origin are commercially significant (Sharma et al., 2001). Microbial lipases are a group of biotechnologically significant enzymes, mainly because of their versatility in application and the ease of mass production (Hasan et al., 2006). Quite a few species of bacteria, yeasts, and molds have been found to produce lipases (Wang et al., 2007; Hasan et al., 2009) and a variety of microbial lipases have been widely used in different industries, such as the food, detergent, and pharmaceutical industries, because of their diversified enzymatic properties and substrate specificities (Hasan et al., 2006).
The use of lipases in various industrial reactions requires a good knowledge of the factors influencing their enzymatic activity, such as temperature, pH values, organic solvents, ions, and reaction media (Aouf et al., 2014). In some cases, the lipase-catalyzed reactions need to be carried out in harsh or even toxic environments. For instance, biodiesels, mostly referred to as methyl esters, are generally synthesized from plant oil and methanol through a transesterification reaction by lipases, where methanol is harmful to lipase activity (Taher and Al-Zuhair, 2017). Microbial lipases that can tolerate harsh environments offer new possibilities for industrial applications, and extensive research has been carried out to find novel lipases with these desired attributes (Salihu and Alam, 2015), mainly encompassing organic solvent-tolerant lipases (Ebrahimpour et al., 2011), thermostable lipases (Ebrahimpour et al., 2011), detergent compatible lipases (Lailaja and Chandrasekaran, 2013), acid-or alkaline-tolerant lipases (Peng et al., 2014), salt-tolerant lipases (Ghasemi et al., 2011), and ionic liquid-compatible lipases (Fauzi and Amin, 2012).
Aureobasidium pullulans is a ubiquitous yeast-like fungus that can be found in different environments and is known as “black yeast” because of its metabolite melanin and its yeast-like colonies (de Hoog, 1993). A. pullulans is particularly famous for its significant role in the production of pullulan, a promising material widely employed in various fields (Singh and Saini, 2008). It also exhibits biotechnological potential in the production of siderophore, single-cell proteins, and various enzymes including amylase, cellulose, lipase, and proteinase, among others (Chi et al., 2009). Regarding its lipase activity, A. pullulans was first found to possess lipolytic activity towards Tween 20, 40, and 80 by Federici (1982). Kudanga et al. (2007) screened 42 A. pullulans isolates from the African tropics for lipase activity and found that 20 isolates showed lipolytic activity. Wang et al. (2007) investigated the diversity of lipase-producing yeasts from marine environments and found that the strain A. pullulans HN2.3 produced extracellular lipase. Later, Liu et al. (2008a, 2008b) characterized the lipase produced by A. pullulans HN2.3 by studying its production, purification, and enzymatic properties, and then isolated and characterized the gene encoding the lipase. The molecular weight of the purified lipase from A. pullulans HN2.3 was 63.5 kDa, and the optimum temperature and pH were 35 °C and 8.5, respectively.
In this work, to purify the extracellular lipase from a lipase-producing strain, ultrafiltration and ion exchange chromatography (DEAE-Sepharose Fast Flow column) were employed. The characterization of the purified lipase under various conditions, such as temperature, pH, organic solvents, ions, surfactants, and edible oils showed that the lipase may have the potential to be used in biodiesels and structural lipids industries.
2. Materials and methods
2.1. Microorganisms
The lipase-producing strains were isolated from stuffed buns steamers, which were reported in our previous work (Li et al., 2015). The microorganisms had been identified as A. pullulans by 26S rRNA gene sequencing. In this study, the previously isolated strain J2, which possessed a relatively high lipase production ability and was selected for current study, was re-identified using internal transcribed spacer (ITS) sequencing for confirmation, and then was renamed as A. pullulans S3. This strain has been deposited in the China Center for Type Culture Collection (CCTCC, Wuhan, China) with the accession number of CCTCC AF 2017014. Its ITS sequence has been deposited in GenBank with an accession number MF948886.
2.2. Lipase production and purification
Pure colonies of strain S3 on potato dextrose agar (PDA) were inoculated into 15 mL medium A solution (per liter: yeast extract 10 g, tryptone 20 g, dextrose 10 g, maltose 10 g, olive oil 10 mL, and Tween 80 0.7 mL) in a 50-mL conical flask and incubated at 30 °C with shaking at 200 r/min for 48 h. Following that, 225 μL of the fermentation liquid was transferred to 15 mL of medium A solution (inoculum size of 1.5%, v/v) in a 50-mL conical flask and incubated at 30 °C with shaking at 200 r/min for 96 h. A lipase solution for the subsequent assays was collected by centrifugation for 5 min at 10 000 r/min at 4 °C.
The crude lipase solution was filtered using a cellulose membrane filter with a pore diameter of 0.45 μm to remove the cells. The filtered lipase solution was then concentrated by ultrafiltration using a 10-kDa Amicon regenerated cellulose filter (Millipore, Bedford, MA, USA). The concentrated lipase was then lyophilized using a freeze drier (Labconco, Kansas, MO, USA), and 75 mg of the lipase powder was resuspended in 1 mL of buffer A (20 mmol/L Tris-HCl, pH 7.0) and centrifuged at 10 000 r/min for 1 min. The supernatant was loaded into a DEAE-Sepharose Fast Flow column which was pre-equilibrated with buffer A. The lipase was eluted using the buffer A solution with an increasing sodium chloride concentration gradient (0.02, 0.04, 0.10, and 1.00 mol/L). The flow rate was 0.8 mL/min and the collected protein fractions were detected using spectrophotometry. The fraction with the highest lipase activity was concentrated by ultrafiltration, and then freeze-dried for subsequent tests. The purified lipase was detected by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method described by Laemmli (1970) with some modifications. Acrylamide concentrations in the separating and stacking gels were 12% and 5%, respectively. The gel was stained by a Coomassie Blue G250 Stain Solution kit (Tiandz Biotech, Beijing, China) and observed with a gel imaging system (ChampGel 500, Sage Creation, Beijing, China).
2.3. Lipase hydrolysis activity assay
The lipase activity was measured according to the method described by Winkler and Stuckmann (1979) and Yuan et al. (2018) with some modifications. Briefly, 90 mL of buffer B (20 mmol/L Tris-HCl solution containing 0.099 mg gum arabic, pH 8.0) was mixed with 10 mL of isopropanol containing 30 mg p-nitrophenyl palmitate (pNPP; Sigma-Aldrich, CA, USA) to make the substrate solution. The reaction mixture was composed of 600 μL of substrate solution, and 25 μL of an appropriately diluted enzyme was incubated at 40 °C for 15 min along with a control using denatured lipase. The reaction was terminated by adding 500 μL of 95% ethanol. The reaction mixture was centrifuged at 10 000 r/min for 3 min, and the supernatant was used to determine the amount of liberated p-nitrophenol based on the absorbance value at 410 nm. One unit (U) of enzyme activity was defined as the amount of enzyme required for the liberation of 1.0 μmol of p-nitrophenol from pNPP (Sigma-Aldrich, CA, USA) per minute under the assay conditions.
2.4. Protein concentration test
Protein concentration was determined by the Bradford method using bovine serum albumin (BSA) as a standard (Bradford and Williams, 1976).
2.5. Biochemical characterization
2.5.1 Effect of temperature on lipase activity and stability
The purified lipase was resuspended in buffer A and diluted to an appropriate concentration (approximately 0.1 U/mL). The optimum temperature was measured by assessing the hydrolysis activity at various temperatures (10–70 °C). To assess lipase thermostability, the lipase was incubated at different temperatures (10–60 °C) for 1 h. The lipase enzyme incubated at 4 °C served as a control. The lipases were equilibrated to room temperature after a 1-h treatment, and then the residual activity was tested by the pNPP method as described above.
2.5.2 Effect of pH on lipase activity and stability
To investigate the optimum pH, the substrate pNPP, dissolved into isopropanol, was mixed with a series of buffers with different pH values at a ratio of 1:9 (v/v). Buffers used in this study were CH3COONa/ CH3COOH (pH 5.0), NaH2PO4/Na2HPO4 (pH 6.0 and 7.0), Tris-HCl (pH 8.0 and 9.0), and glycine/NaOH (pH 10.0). The reaction was carried out at the optimized temperature. The stability of the lipase in the pH ranged from 4.0 to 10.0 was examined by mixing the lipase with the buffers at a ratio of 1:24 and incubating at 4 °C for 1 h, after which the remaining activity was measured. The lipase suspended in buffer A acted as a control.
2.5.3 Effects of organic solvents, metal ions, detergents, and inhibitors on the stability of the lipase
Several effectors, including 30% organic solvents (methanol, ethanol, n-propanol, isopropanol, acetone, dimethyl sulfoxide (DMSO), hexane, heptane, and chloroform), 10 mmol/L metal ions (Na+, K+, Mg2+, Mn2+, Ca2+, Ba2+, Zn2+, and Fe2+), 0.05% detergents (Tween 20, Tween 80, and Triton X-100), 1 mmol/L SDS, ethylene diamine tetraacetic acid (EDTA), cetyl trimethyl ammonium bromide (CTAB), and dithiothreitol (DTT) were added to the purified lipase and incubated at 4 °C for 1 h. The residual activity was measured by the pNPP method, and the activity of the purified lipase was used as a control.
2.6. Hydrolysis of edible oils
The hydrolysis of edible oils was measured according to the method of Ramani et al. (2010) with some modifications. The oil was emulsified in a 2% polyvinyl alcohol solution at the ratio 1:9 in a disperser homogenizer (IKA, Staufen, Germany). The reaction mixture was placed in a 100-mL conical flask containing 5 mL of oil emulsion, 2 mL of 0.03% Triton X-100, 2 mL of 3 mol/L NaCl, 1 mL of 0.075% (0.75 g/L) CaCl2, and 5 mL of distilled water. The mixture was kept at 40 °C for 5 min, and then 1 mL of lipase was added. All the flasks were kept in a water bath shaker at 40 °C for 10 min. The reaction was immediately terminated by adding 10 mL of 95% ethanol. The liberated fatty acids were titrated against 0.01 mol/L NaOH by adding 2–3 drops of phenolphthalein as an indicator. One unit of hydrolytic activity was defined as the amount of lipase needed to release 1 μmol of free fatty acid per min under the above conditions.
2.7. Statistical analysis
The determination of the lipase activity affected by various factors and the hydrolysis of edible oils were performed in triplicate. The results were analyzed using the statistics software SPSS (version 20), and data are given as mean±standard deviation (SD).
3. Results
3.1. Purification of the lipase
The extracellular lipase from the strain S3 was purified to homogeneity using ultrafiltration and ion exchange chromatography (DEAE-Sepharose Fast Flow column). The purification parameters are shown in Table 1. The fraction with the highest activity collected showed a 6.3-fold purity increase compared with the crude lipase, and approximately 15.5% activity was recovered. Following that, the fractions with the highest activity were pooled and condensed by ultrafiltration. Finally, the lipase was purified to a 18.2-fold increase with a 7.2% activity yield.
Table 1.
Summary of the purification schemes of the lipase from A. pullulans
| Purification scheme | Total protein (mg) | Protein yield (%) | Total activity (U) | Activity yield (%) | Specific activity (U/mg protein) | Fold purification |
| Crude lipase | 67.12 | 100.00 | 65.57 | 100.00 | 0.98 | 1.00 |
| Ultrafiltration | 23.22 | 34.59 | 28.25 | 43.08 | 1.22 | 1.25 |
| DEAE-Sepharose Fast Flow | 1.65 | 2.46 | 10.18 | 15.53 | 6.16 | 6.30 |
| Ultrafiltration | 0.27 | 0.40 | 4.75 | 7.24 | 17.74 | 18.16 |
Electrophoretic analysis showed that the purified lipase has a single band with a relative molecular weight of 39.5 kDa on SDS-PAGE gel (Fig. 1), demonstrating that the obtained lipase has a single subunit.
Fig. 1.
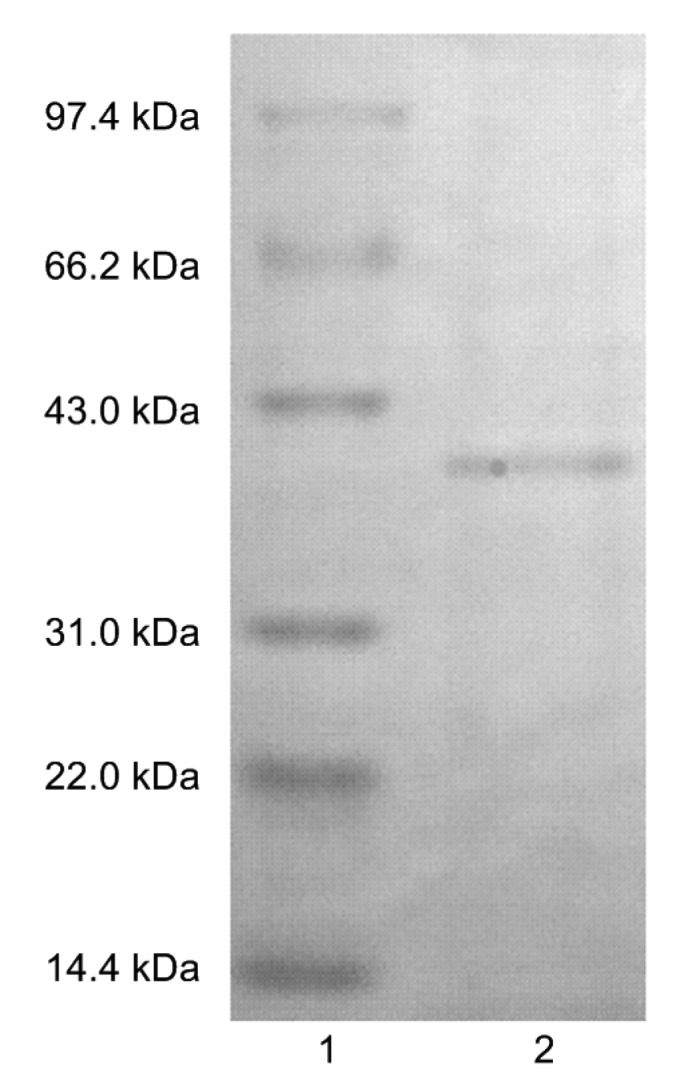
SDS-PAGE analysis of the purified lipase from A. pullulans
Lane 1: marker; Lane 2: purified lipase
3.2. Effects of temperature and pH on lipase properties
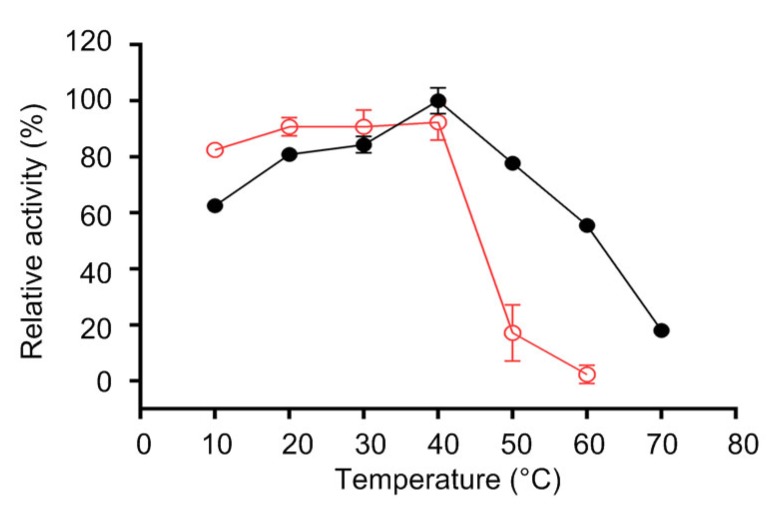
In the range of the temperature test (10–70 °C), maximum hydrolytic activity was observed at 40 °C (Fig. 2). The catalytic activity increased as the temperature increased over the range of 10–40 °C and decreased when the temperature was higher. Notably, the lipase was found to be fairly active in the temperature range of 20–50 °C, although there was a decline after 40 °C. Similarly, its activity remained relatively stable over the range of 10–40 °C, retaining approximately 85% of its initial activity, but deteriorated sharply when the temperature was higher than 40 °C, and most of the activity was lost at 50 °C.
Fig. 2.
Effects of temperature on activity (●) and thermal stability (○) of the purified lipase
Data are given as mean±SD, n=3. The lipase activity was measured in a buffer of pH 8.0 for 15 min. The maximum activity at 40 °C was defined as 100%
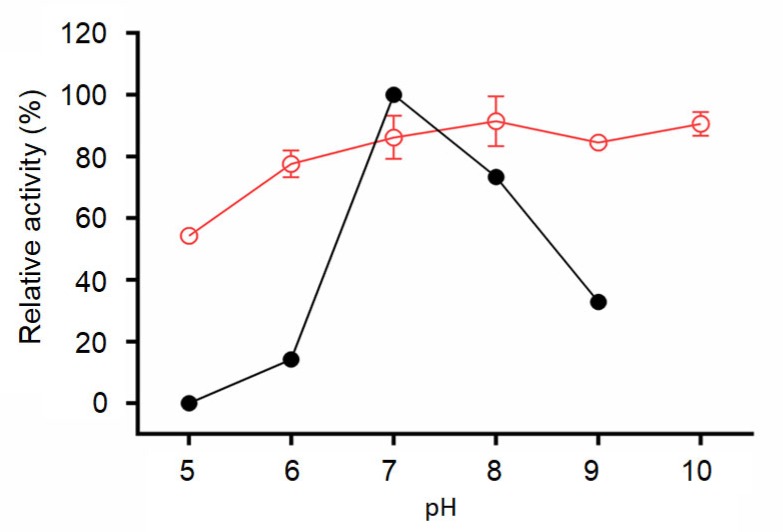
The pH profile is shown in Fig. 3. The lipase exhibited its maximum activity at pH 7.0, and this activity declined rapidly when the pH value diverged from the optimum level. Nevertheless, it has a relatively good tolerance to alkaline conditions and approximately 85% of its original activity was retained in the pH range of 7.0–10.0 after 1 h incubation.
Fig. 3.
Effects of pH on activity (●) and pH stability (○)
Data are given as the mean±SD, n=3. The lipase activity was measured at 40 °C for 15 min. The maximum activity at pH 7.0 was defined as 100%
3.3. Effects of organic solvents on lipase activity
The results of the effects of selected organic solvents (30%, v/v) on the lipase activity are shown in Table 2. Regarding the hydrophilic solvents, the lipase was rather stable after being treated in npropanol, isopropanol, and DMSO with a relative activity of over 100% (the activity of control was defined as 100%). However, the lipase lost part of its activity in the presence of methanol and ethanol. Of all the selected organic solvents in our study, acetone was the most destructive, as it deprived the lipase of 66.1% of its activity after 1 h incubation. For the hydrophobic organic solvents, hexane was found to enhance the lipase activity to 119.1%. Heptane, however, dramatically damaged the activity of the lipase, as well as chloroform.
Table 2.
Stability of the lipase in organic solvents
| Organic solvent | Final concentration (%, v/v) | Relative activity (%) |
| Control | 100.0 | |
| Hydrophilic | ||
| Methanol | 30 | 87.8±5.0 |
| Ethanol | 30 | 55.1±6.6 |
| n-Propanol | 30 | 107.2±4.9 |
| Isopropanol | 30 | 102.4±3.3 |
| Acetone | 30 | 33.9±2.8 |
| DMSO | 30 | 104.6±9.3 |
| Hydrophobic | ||
| Hexane | 30 | 119.1±4.7 |
| Heptane | 30 | 55.7±5.7 |
| Chloroform | 30 | 61.8±8.1 |
Data are given as mean±SD, n=3. The lipase activity was measured at the optimal temperature and pH of 40 °C and 7.0, respectively. The activity of the control (without organic solvents) was defined as 100.0%
3.4. Effects of metal ions, surfactants, and inhibitors on lipase activity
In the metal ions test, 1 mol/L NaCl was found to increase the lipase activity up to 183.2% (Table 3). As seen in Table 3, the lipase activity was not influenced by 10 mmol/L K+, Mg2+, and Ba2+, and its activity was promoted by 10 mmol/L Ca2+ with the relative activity being 116.3%, while it was negatively affected by Mn2+, Zn2+, and Fe2+. The relative activity of lipase treated by the chelating agent EDTA was 106.7%, thus showing little effect.
Table 3.
Effects of metal ions, detergents, and inhibitors on lipase activity
| Parameter | Final concentration | Relative activity (%) |
| Metal ions | ||
| Control | 100.0 | |
| NaCl | 1 mmol/L | 183.2±2.6 |
| KCl | 10 mmol/L | 102.0±0.4 |
| MgCl2 | 10 mmol/L | 100.2±6.5 |
| MnCl2 | 10 mmol/L | 89.7±5.5 |
| CaCl2 | 10 mmol/L | 116.3±4.5 |
| BaCl2 | 10 mmol/L | 107.0±6.0 |
| ZnSO4 | 10 mmol/L | 81.4±4.5 |
| FeSO4 | 10 mmol/L | 96.5±2.2 |
| EDTA | 1 mmol/L | 106.7±5.5 |
|
| ||
| Inhibitors and detergents | ||
| Control | 100.0 | |
| Tween 20 | 0.05% (v/v) | 203.9±2.0 |
| Tween 80 | 0.05% (v/v) | 259.6±6.8 |
| Triton X-100 | 0.05% (v/v) | 112.6±7.7 |
| SDS | 1 mmol/L | 115.9±4.8 |
| CTAB | 1 mmol/L | 22.2±1.9 |
| DTT | 1 mmol/L | 123.1±5.2 |
Data are given as mean±SD, n=3. The lipase activity was measured at the optimal temperature and pH of 40 °C and 7.0, respectively. The activity of the control (without any effectors) was defined as 100%
The effects of detergents and inhibitors on the lipase activity are shown in Table 3. The surfactants Tween 20 and Tween 80 greatly enhanced the activity to 203.9% and 259.6%, respectively. Triton X-100 and SDS enhanced the lipase activity slightly to 112%–116%. In contrast, the CTAB drastically damaged the lipase activity to 22.2%. DTT, as a thiol-reducing agent, enhanced the activity of the lipase to 123.1%.
3.5. Hydrolysis of edible oils by the lipase
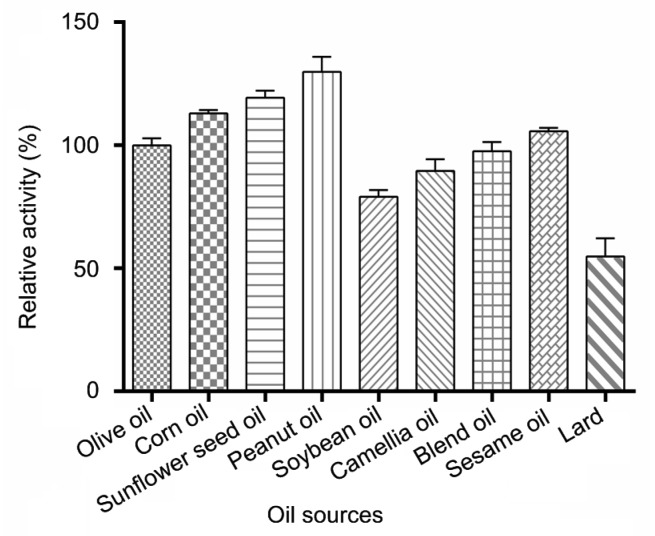
Nine kinds of cooking oils were used to test the hydrolytic activity of the lipase. As shown in Fig. 4, the lipase could hydrolyze all nine edible oils with various degrees of hydrolysis. For a convenient comparison, the hydrolytic activity of the lipase against olive oil was defined as 100.0%. The lipase showed highest hydrolytic activity against peanut oil with a 129.8% relative activity, and higher activity was also shown when against corn oil, sunflower seed oil, and sesame oil than against olive oil. In contrast, the lipase hydrolyzed the animal oil lard more slowly and exhibited the lowest activity of 54.8%, followed by soybean oil at 79.0%.
Fig. 4.
Effects of lipase on the hydrolysis of edible oils
Data are given as mean±SD, n=3. The reactions were kept in a water bath shaker at 40 °C for 10 min
4. Discussion
A variety of techniques have been employed for lipase purification, e.g. precipitation, ultrafiltration, gel filtration, ion exchange chromatography, hydrophobic interaction chromatography, and affinity chromatography (Saxena et al., 2003; Ramani et al., 2010; Shi et al., 2014). Generally, the more techniques or steps used, the less enzyme recovery is obtained; therefore, the selection of purification approaches is practically significant. According to our results, ion exchange chromatography, combined with two ultrafiltration steps, is sufficient for lipase purification, and the enzyme is purified to homogeneity. The purified lipase has a single band with a relative molecular weight of 39.5 kDa on SDS-PAGE gel. The difference in molecular weight with the lipases produced by other A. pullulans strains (Liu et al., 2008a; Leathers et al., 2013) indicated that the lipase reported in our work might be a novel one produced by A. pullulans. In this study, the maximum hydrolytic activity of the lipase was observed at 40 °C, a higher temperature than that of the lipase from strain A. pullulans HN2.3 (Liu et al., 2008a), presumably because of the adaptation of the strain we obtained to the relatively high temperature environment of the steamer. The activity of the lipase remained relatively stable in the range of 10–40 °C. Our results were in accordance with the previous conclusion that lipases from yeasts generally showed maximum activity at a temperature ranging from 30 to 50 °C (Liu et al., 2008a; Li et al., 2013). In addition to temperature, the hydrolytic activity of lipases is greatly influenced by pH values, and lipases are usually stable and active under neutral or alkaline conditions (Sharma et al., 2001; Hasan et al., 2009). In agreement with previous studies, the maximum hydrolytic activity of the lipase was shown at pH 7.0, and this activity declined rapidly when the pH value diverged from the optimum level, which might be due to the destruction of the lipase’s structure or the deterioration of its affinity to the substrate (Hamdy and Abo-Tahon, 2012).
The relative activity of the lipase was over 100% when treated in n-propanol, isopropanol, and DMSO, indicating that the lipase could tolerate these three organic solvents at a relatively high concentration (30%, v/v). A similar result of the tolerance to methanol and ethanol was reported by Tang et al. (2017), but they found that methanol and ethanol at a low concentration (1%) could stimulate the activity of the lipase they studied. The lipase lost most activity when treated in acetone. This may be because acetone changed the secondary conformation of the enzyme by increasing the α-helices and decreasing random coil conformation (Sulong et al., 2006). The activation of lipase by hexane could be explained by the fact that organic solvent molecules could interact with hydrophobic amino acid residues that cover the catalytic site of the enzyme, thereby keeping the enzyme in its open conformation and conducive to catalysis reactions (Rúa et al., 1993). In practice, many reactions catalyzed by lipases occur in the presence of organic solvents. However, most lipases are not stable in organic solvents, especially hydrophilic organic solvents, because of the tendency of organic solvents to strip water molecules from the enzyme surface, thereby leading to the inactivation of the enzyme (Yang et al., 2004). The tolerance of lipase to organic solvents is essential for industrial applications, such as the synthesis of fatty acid esters and biodiesels (Joseph and Ramteke, 2013; Ugur et al., 2014). However, the ability of lipase to resist organic solvents is solvent-dependent. That is to say, one solvent may exert quite a different influence on different lipases, such as stimulating, inhibiting, or having no effect at all (Salihu and Alam, 2015). The lipase in our study is stable, or its stability could be enhanced in n-propanol, isopropanol, DMSO, and hexane; therefore, it has the potential to be employed in industrial fields involving these solvents.
Regarding its tolerance to ions, 1 mol/L NaCl could increase the lipase activity, which indicated that the lipase could tolerate high concentration of NaCl and ensured that the lipase activity was retained in the elution process of purification. With a concentration of 10 mmol/L, Ca2+ could activate the lipase and many lipases have been found to display enhanced activity in the presence of Ca2+ (Ji et al., 2010; Ramani et al., 2010; Li et al., 2014). A possible explanation for this phenomenon is that Ca2+ binds to the active site of the lipase and changes the conformation of the protein (Rahman et al., 2005). It is worth noting that the effects of metal ions on lipase activity are related to the concentrations of the ions and the sources of the lipases (Hasan et al., 2009). A certain ion may have quite different or even reverse impacts on lipases produced by different microbes. For instance, 1 mmol/L Fe3+ slightly stimulated the activity of the lipase from A. pullulans HN2.3 but greatly inhibited the activity of the lipase of Aspergillus terreus var. africanus. The chelating agent EDTA is a metal chelator and could be used to test whether the lipase is a metalloenzyme or not (Ramírez-Zavala et al., 2004). In this study, EDTA showed little effect on the lipase activity. This demonstrated that the lipase we obtained is not a metalloenzyme.
Lipases hydrolyze triglycerides at the lipid–water interface, and consequently, the presence of surfactants is supposed to affect the activity of lipases (Kanjanavas et al., 2010). In this study, surfactants of all three different categories, namely, cationic, anionic, and non-ionic, exerted a strong influence on the activity of the lipase. The two non-ionic surfactants, Tween 20 and Tween 80, greatly enhanced the activity by almost 2-fold. However, the results from research investigating the influence of these two surfactants on lipases are conflicting, which may be due to the different concentrations the researchers used or the discrepancy of the lipase’s nature (Kanjanavas et al., 2010; Shi et al., 2014). Another non-ionic surfactant, Triton X-100, also enhanced the lipase activity, but to a lesser extent. SDS, an anionic surfactant, also stimulated the activity of the lipase, and SDS’s promoting role for lipase activity has been observed by others (Kanjanavas et al., 2010; Shi et al., 2014). The enhancement of SDS to lipase activity may be explained by: (1) the formation of an SDS/ lipase complex, which led to a conformational alteration of the lipase (Antonov et al., 1988; Martinelle et al., 1995) and was able to bind other lipase molecules which set up their active sites, facilitating the catalytic reactions; (2) at a low concentration, SDS made the lipase destabilized, but not denatured, and the destabilized status might help the lipase better accommodate the substrate molecules (Mogensen et al., 2005). Detergent manufacturing is one of the most important industrial applications of lipases (Aref et al., 2014), and the enhancement of lipase activity indicates that the lipase we obtained could be used with the above surfactants in making various detergents. In contrast, the cationic surfactant CTAB acted as an inhibitor, seriously damaging the lipase activity, and a similar result was reported by Yuan et al. (2016). DTT, as a thiol-reducing agent, can reduce the disulfide bonds in proteins and change protein structures. In this study, DTT enhanced the activity of the lipase, implying the non-existence of disulfide bonds in the lipase active site (Peng et al., 2014). Interestingly, it has been frequently reported that the addition of DTT could increase lipase activity (Peng et al., 2014; Ramakrishnan et al., 2016). This increase may occur because DTT prevents the enzymes from forming intramolecular and intermolecular disulfide bonds and makes the enzyme active sites exposed to more substrates (Peng et al., 2014).
The lipase showed higher hydrolytic activity against peanut oil, corn oil, sunflower seed oil, and sesame oil than against olive oil. As in a previous report on the lipase produced by strain A. pullulans HN2.3 (Liu et al., 2008a), the lipase we obtained also exhibited its highest hydrolytic activity against peanut oil, implying that lipases secreted by A. pullulans may have similar catalytic preferences. Additionally, the reason why peanut oil is preferred is of great interest for further research. In contrast, the lipase hydrolyzed the animal oil lard more slowly and exhibited the lowest activity towards it, followed by soybean oil. Similarly, the lipase produced by the strain A. pullulans HN2.3 also showed a lower activity against lard and soybean oil, but, conversely, the lipase hydrolyzed lard more effectively than soybean in the study by Liu et al. (2008a). Other work has reported that lipases hydrolyzed plant oils more easily than animal oils, presumably because of the difference in the degree of saturation of the oils (Shi et al., 2014). It has also been indicated that most lipases displayed low activity in hydrolyzing marine (fish) oils that are rich in n-3 polyunsaturated fatty acids (Hiol et al., 2000). The variation in lipase activity in relation to different oils may be influenced by their substrate specificity and chain length specificity (Hiol et al., 2000). However, in this study, the ability of the lipase to hydrolyze oils used daily confers on the lipase characteristics allowing it to be potentially applied in the detergent industry and for the digestion of lipids in the fields of food and medicine.
5. Conclusions
An extracellular lipase produced by the species A. pullulans was characterized in terms of its enzymatic properties. Purification methods, including ultrafiltration and DEAE-Sepharose Fast Flow column, were effectively used to obtain the lipase. Electrophoresis analysis showed that it has a molecular weight of 39.5 kDa. Enzymatic characterization of the novel extracellular lipase showed its resistance to some organic solvents, surfactants, and ions. Moreover, it could hydrolyze a series of edible oils. These good characteristics could enable the lipase to be used in some industrial fields, such as detergent production, biodiesel synthetization, and food manufacturing.
Footnotes
Project supported by the Science & Technology Major Project of Zhejiang Province, China (No. 2012C12005-2)
Contributors: Yang LI and Min-jie ZHAO performed the experiments. Yang LI and Tong-jie LIU performed the data analysis and drafted the manuscript. Hui ZHANG and Feng-qin FENG contributed to the study design. All authors read and approved the final manuscript.
Compliance with ethics guidelines: Yang LI, Tong-jie LIU, Min-jie ZHAO, Hui ZHANG, and Feng-qin FENG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Antonov VK, Dyakov VL, Mishin AA, et al. Catalytic activity and association of pancreatic lipase. Biochimie. 1988;70(9):1235–1244. doi: 10.1016/0300-9084(88)90190-3. [DOI] [PubMed] [Google Scholar]
- 2.Aouf C, Durand E, Lecomte J, et al. The use of lipases as biocatalysts for the epoxidation of fatty acids and phenolic compounds. Green Chem. 2014;16(4):1740–1754. doi: 10.1039/C3GC42143K. [DOI] [Google Scholar]
- 3.Aref HL, Mosbah H, Fekih A, et al. Purification and biochemical characterization of lipase from Tunisian Euphorbia peplus latex. J Am Oil Chem Soc. 2014;91(6):943–951. doi: 10.1007/s11746-014-2444-z. [DOI] [Google Scholar]
- 4.Bradford MM, Williams WL. New, rapid, sensitive method for protein determination. Fed Proc. 1976;35(3):274. [Google Scholar]
- 5.Chi ZM, Wang F, Chi Z, et al. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol. 2009;82(5):793–804. doi: 10.1007/s00253-009-1882-2. [DOI] [PubMed] [Google Scholar]
- 6.de Hoog GS. Evolution of black yeasts: possible adaptation to the human host. Antonie van Leeuwenhoek. 1993;63(2):105–109. doi: 10.1007/BF00872386. [DOI] [PubMed] [Google Scholar]
- 7.Ebrahimpour A, Rahman RNZRA, Basri M, et al. High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresour Technol. 2011;102(13):6972–6981. doi: 10.1016/j.biortech.2011.03.083. [DOI] [PubMed] [Google Scholar]
- 8.Fauzi AHM, Amin NAS. An overview of ionic liquids as solvents in biodiesel synthesis. Renewable Sustainable Energy Rev. 2012;16(8):5770–5786. doi: 10.1016/j.rser.2012.06.022. [DOI] [Google Scholar]
- 9.Federici F. Extracellular enzymatic activities in Aureobasidium pullulans . Mycologia. 1982;74(5):738–743. doi: 10.1080/00275514.1982.12021580. [DOI] [Google Scholar]
- 10.Ghasemi Y, Rasoul-Amini S, Kazemi A, et al. Isolation and characterization of some moderately halophilic bacteria with lipase activity. Microbiology. 2011;80(4):483–487. doi: 10.1134/S0026261711040060. [DOI] [PubMed] [Google Scholar]
- 11.Hamdy HS, Abo-Tahon MA. Extracellular lipase of Aspergillus terreus var. africanus (CBS 130.55): production, purification and characterisation. Ann Microbiol. 2012;62(4):1723–1736. doi: 10.1007/s13213-012-0429-4. [DOI] [Google Scholar]
- 12.Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39(2):235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- 13.Hasan F, Shah AA, Hameed A. Methods for detection and characterization of lipases: a comprehensive review. Biotechnol Adv. 2009;27(6):782–798. doi: 10.1016/j.biotechadv.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Hiol A, Jonzo MD, Rugani N, et al. Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme Microb Technol. 2000;26(5-6):421–430. doi: 10.1016/S0141-0229(99)00173-8. [DOI] [PubMed] [Google Scholar]
- 15.Ji QC, Xiao SJ, He BF, et al. Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J Mol Catal B Enzym. 2010;66(3-4):264–269. doi: 10.1016/j.molcatb.2010.06.001. [DOI] [Google Scholar]
- 16.Joseph B, Ramteke PW. Extracellular solvent stable cold-active lipase from psychrotrophic Bacillus sphaericus MTCC 7526: partial purification and characterization. Ann Microbiol. 2013;63(1):363–370. doi: 10.1007/s13213-012-0483-y. [DOI] [Google Scholar]
- 17.Kanjanavas P, Khuchareontaworn S, Khawsak P, et al. Purification and characterization of organic solvent and detergent tolerant lipase from thermotolerant Bacillus sp. RN2. Int J Mol Sci. 2010;11(10):3783–3792. doi: 10.3390/ijms11103783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudanga T, Mwenje E, Mandivenga F, et al. Esterases and putative lipases from tropical isolates of Aureobasidium pullulans . J Basic Microbiol. 2007;47(2):138–147. doi: 10.1002/jobm.200610207. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lailaja VP, Chandrasekaran M. Detergent compatible alkaline lipase produced by marine Bacillus smithii BTMS 11. World J Microbiol Biotechnol. 2013;29(8):1349–1360. doi: 10.1007/s11274-013-1298-0. [DOI] [PubMed] [Google Scholar]
- 21.Leathers TD, Rich JO, Anderson AM, et al. Lipase production by diverse phylogenetic clades of Aureobasidium pullulans . Biotechnol Lett. 2013;35(10):1701–1706. doi: 10.1007/s10529-013-1268-5. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Yang LR, Xu G, et al. Screening, purification and characterization of a novel cold-active and organic solvent-tolerant lipase from Stenotrophomonas maltophilia CGMCC 4254. Bioresour Technol. 2013;148:114–120. doi: 10.1016/j.biortech.2013.08.101. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Qian P, Wu SG, et al. Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil. Extremophiles. 2014;18(1):171–178. doi: 10.1007/s00792-013-0610-0. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Cai HY, Zhao MJ, et al. Screening and identification of high-yield thermostable lipase producing microorganisms. Biotechnol Bull. 2015;31(1):144–150. (in Chinese) [Google Scholar]
- 25.Liu ZQ, Li XY, Chi ZM, et al. Cloning, characterization and expression of the extracellular lipase gene from Aureobasidium pullulans HN2-3 isolated from sea saltern. Antonie Van Leeuwenhoek. 2008;94(2):245–255. doi: 10.1007/s10482-008-9237-z. [DOI] [PubMed] [Google Scholar]
- 26.Liu ZQ, Chi ZM, Wang L, et al. Production, purification and characterization of an extracellular lipase from Aureobasidium pullulans HN2.3 with potential application for the hydrolysis of edible oils. Biochem Eng J. 2008;40(3):445–451. doi: 10.1016/j.bej.2008.01.014. [DOI] [Google Scholar]
- 27.Martinelle M, Holmquist M, Hult K. On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim Biophys Acta. 1995;1258(3):272–276. doi: 10.1016/0005-2760(95)00131-U. [DOI] [PubMed] [Google Scholar]
- 28.Mogensen JE, Sehgal P, Otzen DE. Activation, inhibition, and destabilization of Thermomyces lanuginosus lipase by detergents. Biochemistry. 2005;44(5):1719–1730. doi: 10.1021/bi0479757. [DOI] [PubMed] [Google Scholar]
- 29.Peng Q, Wang X, Shang M, et al. Isolation of a novel alkaline-stable lipase from a metagenomic library and its specific application for milkfat flavor production. Microb Cell Fact, 13:1. 2014 doi: 10.1186/1475-2859-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman RNZRA, Baharum SN, Basri M, et al. High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Anal Biochem. 2005;341(2):267–274. doi: 10.1016/j.ab.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan V, Goveas LC, Suralikerimath N, et al. Extraction and purification of lipase from Enterococcus faecium MTCC5695 by PEG/phosphate aqueous-two phase system (ATPS) and its biochemical characterization. Biocatal Agric Biotechnol. 2016;6:19–27. doi: 10.1016/j.bcab.2016.02.005. [DOI] [Google Scholar]
- 32.Ramani K, Kennedy LJ, Ramakrishnan M, et al. Purification, characterization and application of acidic lipase from Pseudomonas gessardii using beef tallow as a substrate for fats and oil hydrolysis. Process Biochem. 2010;45(10):1683–1691. doi: 10.1016/j.procbio.2010.06.023. [DOI] [Google Scholar]
- 33.Ramírez-Zavala B, Mercado-Flores Y, Hernández-Rodríguez C, et al. Purification and characterization of a lysine aminopeptidase from Kluyveromyces marxianus . FEMS Microbiol Lett. 2004;235(2):369–375. doi: 10.1111/j.1574-6968.2004.tb09612.x. [DOI] [PubMed] [Google Scholar]
- 34.Rúa ML, Díaz-Mauriñob T, Fernández VM, et al. Purification and characterization of two distinct lipases from Candida cylindracea . Biochim Biophys Acta. 1993;1156(2):181–189. doi: 10.1016/0304-4165(93)90134-T. [DOI] [PubMed] [Google Scholar]
- 35.Salihu A, Alam MZ. Solvent tolerant lipases: a review. Process Biochem. 2015;50(1):86–96. doi: 10.1016/j.procbio.2014.10.019. [DOI] [Google Scholar]
- 36.Saxena RK, Sheoran A, Giri B, et al. Purification strategies for microbial lipases. J Microbiol Methods. 2003;52(1):1–18. doi: 10.1016/S0167-7012(02)00161-6. [DOI] [PubMed] [Google Scholar]
- 37.Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization, and applications of lipases. Biotechnol Adv. 2001;19(8):627–662. doi: 10.1016/S0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 38.Shi H, Meng Y, Yang M, et al. Purification and characterization of a hydrolysis-resistant lipase from Aspergillus terreus . Biotechnol Appl Biochem. 2014;61(2):165–174. doi: 10.1002/bab.1142. [DOI] [PubMed] [Google Scholar]
- 39.Singh RS, Saini GK. Pullulan-hyperproducing color variant strain of Aureobasidium pullulans FB-1 newly isolated from phylloplane of Ficus sp. Bioresour Technol. 2008;99(9):3896–3899. doi: 10.1016/j.biortech.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Sulong MR, Rahnian RNZRA, Salleh AB, et al. A novel organic solvent tolerant lipase from Bacillus sphaericus 205y: extracellular expression of a novel OST-lipase gene. Protein Express Purif. 2006;49(2):190–195. doi: 10.1016/j.pep.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Taher H, Al-Zuhair S. The use of alternative solvents in enzymatic biodiesel production: a review. Biofuels Bioprod Biorefin. 2017;11(1):168–194. doi: 10.1002/bbb.1727. [DOI] [Google Scholar]
- 42.Tang LL, Xia YL, Wu XL, et al. Screening and characterization of a novel thermostable lipase with detergent-additive potential from the metagenomic library of a mangrove soil. Gene. 2017;625:64–71. doi: 10.1016/j.gene.2017.04.046. [DOI] [PubMed] [Google Scholar]
- 43.Tomke PD, Rathod VK. Ultrasound assisted lipase catalyzed synthesis of cinnamyl acetate via transesterification reaction in a solvent free medium. Ultrason Sonochem. 2015;27:241–246. doi: 10.1016/j.ultsonch.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 44.Ugur A, Sarac N, Boran R, et al. New lipase for biodiesel production: partial purification and characterization of LipSB 25-4. ISRN Biochem, 2014:289749. 2014 doi: 10.1155/2014/289749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Chi ZM, Wang XH, et al. Diversity of lipase-producing yeasts from marine environments and oil hydrolysis by their crude enzymes. Ann Microbiol. 2007;57(4):495–501. doi: 10.1007/BF03175345. [DOI] [Google Scholar]
- 46.Winkler UK, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens . J Bacteriol. 1979;138(3):663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Dordick JS, Garde S. Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys J. 2004;87(2):812–821. doi: 10.1529/biophysj.104.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan DJ, Lan DM, Xin RP, et al. Screening and characterization of a thermostable lipase from marine Streptomyces sp. strain W007. Biotechnol Appl Biochem. 2016;63(1):41–50. doi: 10.1002/bab.1338. [DOI] [PubMed] [Google Scholar]
- 49.Yuan L, Sadiq FA, Liu TJ, et al. Spoilage potential of psychrotrophic bacteria isolated from raw milk and the thermo-stability of their enzymes. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(8):630–642. doi: 10.1631/jzus.B1700352. [DOI] [PMC free article] [PubMed] [Google Scholar]