ABSTRACT
Ulcerative colitis is a chronic inflammatory disease of the colon that carries a significant disease burden in children. Therefore, new therapeutic approaches are being explored to help children living with this disease. Fecal microbiota transplantation (FMT) has been successful in some children with ulcerative colitis. However, the mechanism of its therapeutic effect in this patient population is not well understood. To characterize changes in gut microbial and metabolomic profiles after FMT, we performed 16S rRNA gene sequencing, shotgun metagenomic sequencing, virome analysis and untargeted metabolomics by gas chromatography-time of flight-mass spectrometry on stool samples collected before and after FMT from four children with ulcerative colitis who responded to this treatment. Alpha diversity of the gut microbiota increased after intervention, with species richness rising from 251 (S.D. 125) to 358 (S.D. 27). In responders, the mean relative abundance of bacteria in the class Clostridia shifted toward donor levels, increasing from 33% (S.D. 11%) to 54% (S.D. 16%). Patient metabolomic and viromic profiles exhibited a similar but less pronounced shift toward donor profiles after FMT. The fecal concentrations of several metabolites were altered after FMT, correlating with clinical improvement. Larger studies using a similar multi-omics approach may suggest novel strategies for the treatment of pediatric ulcerative colitis.
Keywords: microbiome, fecal microbiota transplantation, pediatric ulcerative colitis, inflammatory bowel disease, metagenomics, metabolomics
Gut microbial and metabolomic profiles are altered after fecal microbiota transplantation in pediatric ulcerative colitis patients.
INTRODUCTION
Ulcerative colitis (UC) is characterized by chronic inflammation of the colon. It is an important pediatric disease, since as many as 20%–25% of all cases present during childhood (Turner and Griffiths 2011), and the incidence of the disease is increasing worldwide (Molodecky et al. 2012; Schildkraut et al. 2013; Olen et al. 2017). Immunomodulator and biologic therapies have resulted in stable management for some patients, but many children still develop severe disease, continue to require colectomy, and develop colon cancer (Turner et al. 2011; Levine et al. 2013; Olen et al. 2017). Therefore, there is a great need for more effective and safer therapies.
The enteric microbiota is now accepted as an important factor in the pathogenesis of inflammatory bowel disease (IBD) (Sartor 2008), with data from multiple studies demonstrating a correlation of microbial community composition with the occurrence of IBD (Bakhtiar et al. 2013; Kostic, Xavier and Gevers 2014). The gut microbiota has been suggested as a therapeutic target for IBD, including ulcerative colitis (Shanahan and Quigley 2014). Fecal microbiota transplantation (FMT) has been established as a useful tool for manipulating the gut microbiome and is an effective treatment for recurrent Clostridium difficile infection (van Nood et al. 2013). FMT therapy has been considered for ulcerative colitis patients, with the first reported FMT in the context of UC described in 1989 (Borody, Brandt and Paramsothy 2014). Since then, FMT for UC has been studied in at least three randomized placebo-controlled trials in adults and hasshown efficacy in inducing remission (Moayyedi et al. 2015; Rossen et al. 2015; Paramsothy et al. 2017). Systematic reviews and meta-analyses have shown that FMT is a safe treatment option that is effective in some patients (Colman and Rubin 2014; Shi et al. 2016).
While there have been several controlled studies investigating the efficacy of FMT for adults with UC, relatively few trials have taken place testing FMT in children with UC. These small, uncontrolled studies and case reports (Vandenplas et al. 2015) have had mixed results: one report found no benefit of FMT in a trial with four pediatric UC patients (Suskind et al. 2015), while another study of three patients showed a significant benefit of FMT (Kellermayer et al. 2015). To our knowledge, the trial described by Kunde et al. (2013) is the largest study to date investigating FMT in children with ulcerative colitis. In this open-label, uncontrolled trial, six out of nine patients with mild-to-moderate ulcerative colitis showed clinical improvement four weeks after FMT, with three patients achieving clinical remission. In order to understand the microbial and molecular changes associated with successful FMT in pediatric ulcerative colitis patients, we utilized a multi-omics approach to characterize the gut microbial and metabolomic profiles from seven patients from the Kunde et al. study.
MATERIALS AND METHODS
Patient recruitment and fecal transplant
Patients were recruited as described by Kunde et al. (2013). Briefly, subjects between the ages of 7 and 20 with mild-to-moderate ulcerative colitis were enrolled between April and December 2012 in a single-center pilot study conducted at the outpatient gastroenterology facility at Helen DeVos Children's Hospital in Grand Rapids, Michigan. Stool samples were collected from healthy adult donors (selected by each subject from their family members or close friends) within 6 h prior to FMT. Stool samples were blended in sterile warm saline, filtered and stored in a water bath at 37°C prior to FMT. FMT was delivered as a retention enema for 1 h (60 mL enema every 15 min) daily for five days. Clinical disease activity was tracked for each patient using the pediatric ulcerative colitis activity index (PUCAI) at enrollment, at the time of FMT, and each week for four weeks after FMT. Stool samples were analyzed from seven of the nine patients that completed the Kunde et al. study; samples were not available from two subjects.
Sample collection
Samples were collected as described by Kunde et al. (2013) and stored at –80°C. Briefly, fecal samples were collected from healthy adult donors for FMT (Donor), pediatric subjects with ulcerative colitis before FMT (Baseline), and from the same subjects four weeks after fecal transplant (PostFMT). Each patient underwent FMT with stool from a separate healthy donor, selected from family members or close friends.
DNA extraction
DNA was extracted from fecal samples using the Qiagen DNeasy DNA Extraction Kit (Maryland, USA). Frozen raw stool samples were homogenized in stool stabilization buffer and extracted under clean room conditions.
16S rRNA gene sequencing
The hypervariable V4 region of the bacterial 16S ribosomal RNA (rRNA) gene from stool DNA and negative controls was amplified by PCR using uniquely barcoded primers (515FB: 5′-GTG YCA GCM GCC GCG GTA A-3′; 806RB: 5′-GGA CTA CNV GGG TWT CTA AT-3′) (Apprill et al. 2015), which were modified from the original 515F-806R primer pairs (Caporaso et al. 2011). Triplicate PCR reactions were mixed together for final pooling. Library qualities were assessed on Agilent High Sensitivity DNA Bioanalyzer chips. All samples were pooled and sequenced using custom sequencing primers: Read 1 (5′-TAT GGT AAT TGT GTG YCA GCM GCC GCG GTA A-3′), Read 2 (5′-AGT CAG CCA GCC GGA CTA CNV GGG TWT CTA AT-3′), and Index (5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACG CT-3′). Paired-end sequencing (2 × 150bp) was performed using Illumina MiSeq Reagent Kit v2 flowcell on an Illumina MiSeq System.
16S rRNA gene sequence analysis
The resulting amplicons were prepared for analysis with QIIME version 1.9.0 (Caporaso et al. 2010). Paired end reads were joined and the sequences were analyzed using the default parameters for the open reference operational taxonomic unit (OTU) picking script in QIIME (pick_open_reference_otus.py)—that is, using the uclust OTU picking method (Edgar 2010) and the GreenGenes 13_8 (DeSantis et al. 2006; McDonald et al. 2012) reference sequence database (97% similarity). The resulting OTU abundance table was filtered to remove OTUs comprising less than 0.01% of the total read count, resulting in 3 913 957 total reads, with a mean of 186 379 reads per sample (median of 175 773 reads per sample) and a standard deviation of 73 029. Subsequently, the OTU table was subsampled to a depth equivalent to the minimum read count of all samples (86 345), resulting in a total of 1 813 245 total reads (86 345 per sample).
Shotgun metagenomic sequencing
Barcoded libraries were prepared from stool DNA using the Illumina Nextera XT DNA Library Prep Kit and Illumina Nextera XT Index v2 Kit A and B following the manufacturer's protocols. Library qualities were assessed on Agilent High Sensitivity DNA Bioanalyzer chips. Libraries were pooled and paired-end sequencing (2 × 150bp) was performed using the Illumina NextSeq500 High Output v2 flowcell on an Illumina NextSeq500 System. Reads from all samples including controls were preprocessed and quality filtered using trim_galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Host-derived reads were removed using KneadData (https://bitbucket.org/biobakery/kneaddata/overview).
Shotgun metagenomic sequence analysis
Shotgun metagenomic sequence data were analyzed using the Taxonomic Profiling tool in the Microbial Genomics Module of the CLC Genomics Workbench 10.0.1 (CLC Bio/QIAGEN, Aarhus, Denmark, https://www.qiagenbioinformatics.com/), with the quality limit (Phred score) set to 0.05. Each read was mapped against a reference database downloaded from NCBI on June 13th, 2017, consisting of all 5294 prokaryotic complete single scaffold genomes available in RefSeq at that time. Each mapped read was assigned to a taxon, resulting in a taxonomic abundance table. Additional methods used for shotgun metagenomic sequence analysis can be found in the Supplementary Methods section.
Viromics analysis
To calculate the abundance of known viruses in samples from the responders, we mapped shotgun metagenomic reads against 8403 virus reference genomes (downloaded from NCBI in June 2017) using BWA-MEM (Li 2013) with the default parameters. The relative abundance of a virus in a sample was calculated as the fraction of reads mapped to the virus genome normalized by the genome length. Principal coordinates analysis (PCoA) was performed using Manhattan distances between the virus abundance profiles. Additional viromics analysis methods can be found in the Supplementary Methods section.
Metabolomics
Fecal samples from the four responders were analyzed at the UC Davis West Coast Metabolomics Center using untargeted metabolomics by gas chromatography-time of flight-mass spectrometry (GC-TOF-MS). A standard plasma extraction protocol was used, with 1 mL of 3:3:2 acetonitrile:isopropyl alcohol:water added to 20 µL feces, followed by centrifugation, decanting, desiccation and trimethylsilylation for GC-TOF-MS profiling using Fiehn laboratory standard operating procedures (Kind et al. 2009; Cajka and Fiehn 2016; Kind et al. 2018). Metabolites were identified by comparison to the BinBase database (Lai et al. 2018). Signal intensities were obtained for 494 metabolites including 217 unique structurally identified compounds. All 494 metabolites were included in subsequent analyses.
Quantification of short chain fatty acids
10 mg feces were used for short chain fatty acid analysis. Metabolites were extracted with 700 µL of water, hydrochloric acid and methyl tert-butyl ether (5:1:1). Samples were homogenized using Genogrinder at 1500 rpm for 30 s, shaken for 30 min at room temperature and centrifuged for 2 min at 14 000 rcf. 500 µL supernatant was transferred to a new tube and 0.1 g of anhydrous sodium sulfate was added to the supernatant for dehydration. 25 µL N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA, Sigma-Aldrich) was used for tert-butyldimethylsilylation. Samples were then shaken at 80°C for 30 min and ready for injection. An Agilent 5977A GC-quadrupole mass spectrometer was used for data acquisition in selected ion monitoring (SIM) mode. Raw data were processed using Agilent Mass Hunter Quantitative Analysis software (B.07.00) and data were quantified against authentic standards.
Data analysis
Data analysis was performed in R version 3.4.1 (R Core Team 2017) using RStudio (version 1.0.147). Alpha diversity analysis of microbial abundance data was performed using the ‘vegan’ (version 2.4.4) R package (Oksanen et al. 2017). Taxonomic composition plots were made with the ‘phyloseq’ (version 1.20.0) package (McMurdie and Holmes 2013). Distance matrices for the microbial abundance and metabolomics data were created using the ‘vegdist’ function in vegan. PCoA of distance matrices based on microbial abundances and metabolite intensities was conducted using the ‘cmdscale’ function in the ‘stats’ (version 3.4.1) package (R Core Team 2017). Permutational multivariate analysis of variance (PERMANOVA) was performed on Bray–Curtis dissimilarity matrices using the ‘pairwiseAdonis’ package (Martinez Arbizu 2017), which applies the ‘adonis’ function from vegan to pairwise combinations of sample groups. Bray–Curtis similarity was calculated by subtracting the Bray-Curtis dissimilarity from 1.
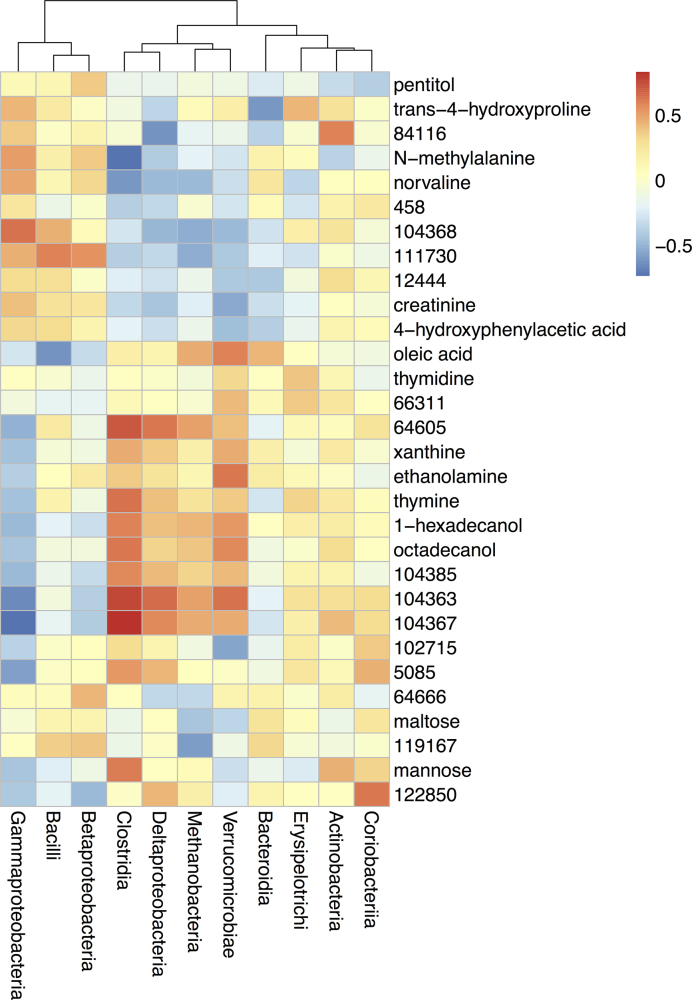
Metabolite intensities were normalized to relative intensities for each sample using the ‘sweep’ function in R. The microbe-metabolite correlation heatmap (Fig. 6) was created by calculating Spearman correlation coefficients for each pairwise combination of microbial taxa abundances and metabolite intensities using the ‘corr.test’ function in the ‘psych’ (version 1.7.8) R package (Revelle 2017).
Figure 6.
Microbe-metabolite correlation heatmap suggesting associations between bacterial classes and the 30 metabolites shown in Fig. 5. Spearman correlation coefficients were calculated for pairwise combinations of microbial abundances at the class level (from 16S rRNA gene sequencing data) and metabolite intensities.
Figures
All plots were created in R (version 3.4.1) using RStudio (version 1.0.147). The PCoA plots were created using R ‘base’ graphics. Heatmaps were created using the ‘pheatmap’ (version 1.0.8) R package (Kolde 2015). Boxplots and barplots were created using the ‘ggplot2’ (version 2.2.1) package (Wickham 2009) along with ‘cowplot’ version 0.9.2 (Wilke 2017). The boxplots display the default statistics for ggplot2 boxplots, where the horizontal line in each box represents the median. The ‘colourpicker’ (version 1.0) R package (Attali 2017) was used to select colors in the stacked barplots in Fig. 2.
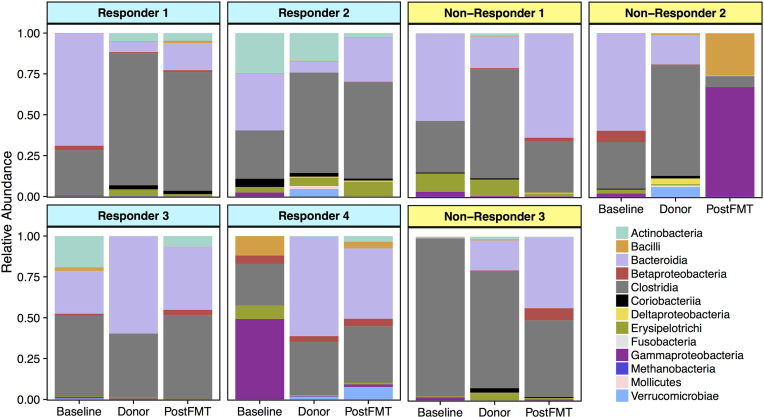
Figure 2.
Taxonomic profiles at the class level based on 16S rRNA gene sequencing. In subjects who showed clinical improvement after FMT (responders), composition of the gut microbiota became more similar to that of their donors four weeks after FMT (PostFMT) compared to baseline, except in Responder 3. In particular, the abundance of bacteria in the class Clostridia (including the genera Blautia, Dorea, and Faecalibacterium) increased toward donor levels after FMT. Microbiome composition also shifted somewhat toward donor profiles in Non-Responder 1 and Non-Responder 3 but not in Non-Responder 2. Bray–Curtis similarity to donor profiles was higher after FMT in responders compared to non-responders.
RESULTS
Microbiome and metabolomic analyses were conducted using samples from seven children with ulcerative colitis. Fecal samples were collected before FMT (Baseline) and four weeks after FMT (PostFMT) as described by Kunde et al. Samples were also obtained from their respective healthy donors (Donor). Subjects were classified as responders (n = 4) or non-responders (n = 3) based on their pediatric ulcerative colitis activity index (PUCAI) score 4 weeks after FMT (in this cohort the responders had a PUCAI ≤ 15).
Alpha diversity of the gut microbiome increases after FMT
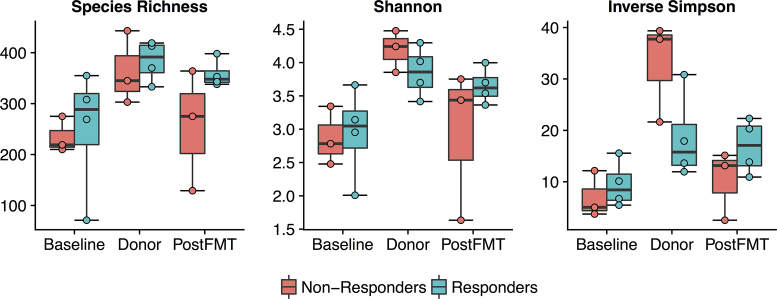
Alpha diversity measures were lower in ulcerative colitis patients at baseline, and increased toward donor levels after FMT (Fig. 1). Mean species richness (total OTUs observed by 16S rRNA gene sequencing) increased 43% in subjects who responded to FMT, from 251 (S.D. 125) before FMT to 358 (S.D. 27) four weeks after treatment (mean species richness in the donor samples was 384 (S.D. 40)). In non-responders, mean species richness increased 9%, from 235 (S.D. 35) to 256 (S.D.119), compared to a mean richness of 364 in their donors (S.D. 72). The Shannon and inverse Simpson indices also increased after FMT. Metagenomic DNA sequencing of samples from the responders showed a similar increase in alpha diversity measures after FMT.
Figure 1.
Microbial alpha diversity as estimated by species richness, the Shannon diversity index, and the Inverse Simpson diversity index based on OTU abundance data from 16S rRNA gene sequencing. All three measures were lower in UC patients before FMT (Baseline), and increased toward donor levels four weeks after FMT (PostFMT). Alpha diversity tended to be higher in responders compared to non-responders both at baseline and after FMT.
FMT alters gut microbiota composition in pediatric ulcerative colitis patients
Taxonomic profiles generated from 16S rRNA gene sequencing differ between baseline patient samples and their respective healthy donors (Fig. 2). Similar profiles were generated using shotgun metagenomic DNA sequence data. In responders, the mean relative abundance of bacteria in the class Clostridia shifted toward donor levels, increasing from 33% (S.D. 11%) to 54% (S.D. 16%) (Fig. S1, Supporting Information). In non-responders, the abundance of Clostridia decreased from 52% (S.D. 38%) to 28% (S.D. 20%) after FMT. The mean relative abundance of the Lachnospiraceae family increased from 11% (S.D. 8%) to 19% (S.D. 6%) after FMT in responders, and fell from 23% (S.D. 17%) to 9% (S.D. 8%) in non-responders (Fig. S1, Supporting Information).
Faecalibacterium prausnitzii increased in abundance after FMT, but the increase was largely driven by a disproportionate increase in one subject (Responder 1) (Fig. S1, Supporting Information). The relative abundance of Bifidobacterium spp. varied considerably in the responder samples at baseline, ranging from 0% to 25% (mean 11% S.D. 13%). After FMT, Bifidobacterium spp. abundance stabilized at a mean abundance of 4% (S.D. 2%) in responders (Fig. S1, Supporting Information). Mean Bifidobacterium spp. abundance was less than 0.2% in samples from non-responders both before and after FMT.
Gut microbial community structure shifts toward donor profiles after FMT in responders
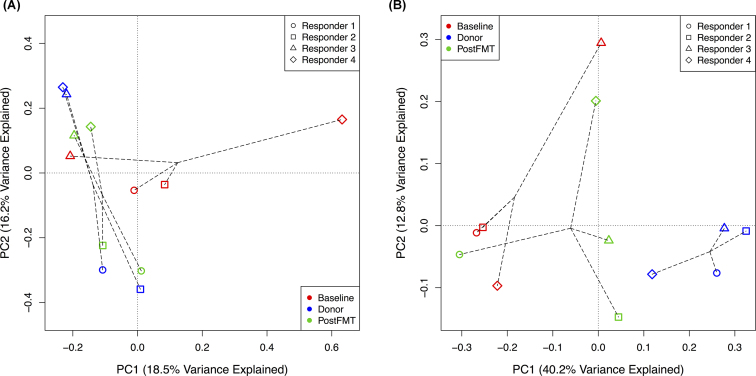
Ordination by PCoA of taxonomic abundances determined by 16S rRNA gene sequencing shows that post-FMT samples tend to cluster closer to the donor samples compared to the baseline samples in responders (Fig. 3A), suggesting that FMT transferred donor microbial communities to the patients. This pattern is recapitulated by PCoA ordinations based on shotgun metagenomic sequencing data, including comparisons of taxonomic abundance profiles using Bray–Curtis dissimilarities (Fig. S2, Supporting Information) and alignment-free metagenomic sequence comparison using the d2S dissimilarity measure (Fig. S3A, Supporting Information). PERMANOVA comparisons of the 16S Bray–Curtis dissimilarity matrix between the Donor, Baseline and PostFMT groups were not statistically significant, potentially due to the small sample size.
Figure 3.
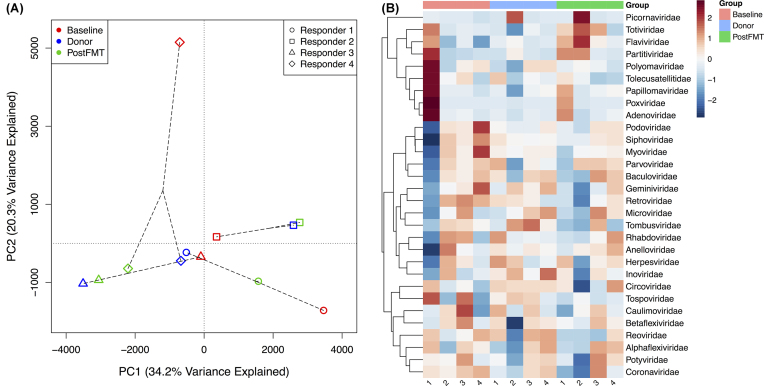
PCoA of (A) microbial taxa abundances from 16S rRNA gene sequence data and (B) metabolite intensities from GC-TOF-MS metabolomic profiling before (Baseline) and four weeks after (PostFMT) fecal transplant in the four responders. The circles, squares, triangles, and diamonds represent Responder 1, Responder 2, Responder 3, and Responder 4, respectively. Red, blue, and green represent Baseline, Donor, and PostFMT samples, respectively.
Bray–Curtis similarity between subject-donor pairs (based on 16S abundance data) tended to increase after FMT, but decreased slightly in Responder 3 (Table S1, Supporting Information). Mean Bray–Curtis similarity to donor profiles increased after FMT in both the responders (25.6 (S.D. 19.3) before FMT to 44.2 (S.D. 9.9) after FMT) and non-responders (20.7 (S.D. 1.4) before FMT to 22.1 (S.D. 17.9) after FMT), with higher similarity scores after FMT in the responders (Fig. S4, Supporting Information). The non-responder samples do not exhibit the same clustering pattern on the PCoA ordination of microbial abundance data (Fig. S5, Supporting Information).
Virome profiles shift toward those of the donors in responders after FMT
Virus abundance profiles were generated from metagenomic sequencing data from the responders. PCoA ordination based on Manhattan distances between virus abundance profiles shows that patient samples shifted toward donor profiles after FMT, indicating the successful transfer of viruses (Fig. 4A). The relative abundance in each sample of 30 known virus families is shown in Fig. 4B.
Figure 4.
(A) PCoA of known virus abundances based on metagenomic sequence data from the four responders. The PCoA is based on Manhattan distances between the samples. The circles, squares, triangles, and diamonds represent Responder 1, Responder 2, Responder 3, and Responder 4, respectively. Red, blue, and green represent Baseline, Donor, and PostFMT samples, respectively. (B) Heatmap showing the relative abundances of 30 known virus families in samples from the four responders and their FMT donors. The numbers under each column in the heatmap indicate the corresponding subject for that column (Responders 1–4).
Metabolomic profiles are altered in responders after FMT
Untargeted gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) profiling was performed on samples from the responders before and after FMT. The metabolomic profiles of subjects before FMT were distinct from the donor profiles, which cluster together on the PCoA ordination (Fig. 3B). After FMT, the metabolomic profiles of three of the four subjects (Responders 2, 3 and 4) shifted toward the donor profiles, but still grouped together as a distinct cluster on the ordination, distinguishable from both the baseline and donor samples. The metabolomic profile of Responder 1 moved in the opposite direction, becoming less similar to the donor profile after FMT (Fig. 3B). Bray–Curtis similarity measures between subjects and their donors increased for all pairs except Responder 1 (Table S2, Supporting Information). Mean Bray–Curtis similarity to donor metabolomic profiles increased from 43.2 (S.D. 5.8) at baseline to 49.4 (S.D. 10.7) after FMT. PERMANOVA analysis with Bonferroni correction of the Bray-Curtis dissimilarity matrix found no significant difference between the groups.
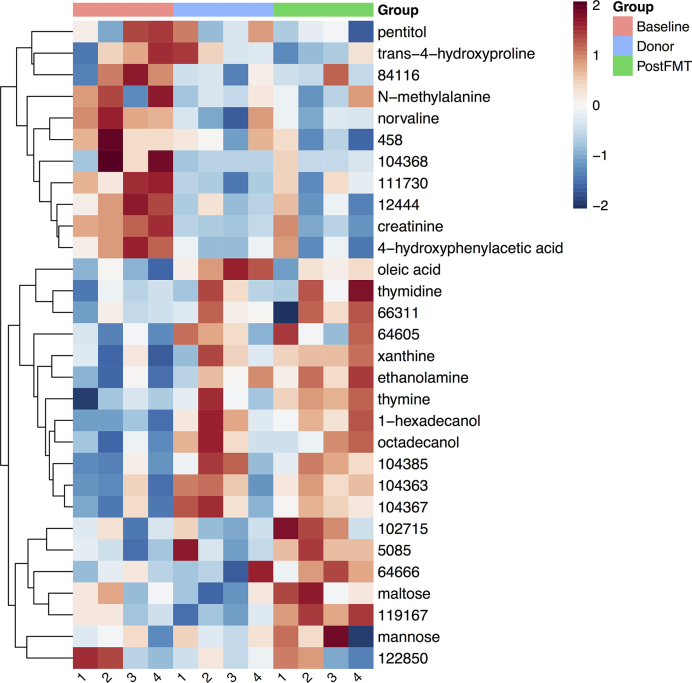
A heatmap of selected metabolites shows that the metabolomic profile of each subject at baseline differs from that of their respective healthy donor, becoming more similar after FMT (Fig. 5). Metabolites with higher intensities at baseline group into a ‘disease-associated’ cluster, and metabolites with higher intensities after FMT group into a ‘healthy’ cluster. Xanthine and oleic acid levels were decreased in baseline samples relative to donors, and increased after FMT (Fig. S6, Supporting Information). Putrescine and 5-aminovaleric acid intensities were higher at baseline compared to four weeks after FMT (Fig. S6, Supporting Information).
Figure 5.
Heatmap of log2-transformed relative metabolite intensities for 30 selected metabolites illustrating changes in the metabolomic profiles of the four responders after FMT. Metabolite intensities are higher in patient samples at baseline in the top (disease-associated) cluster, and higher in donor and/or post-FMT samples in the bottom (healthy) cluster. The numbers under each column in the heatmap indicate the corresponding subject for that column (Responders 1–4).
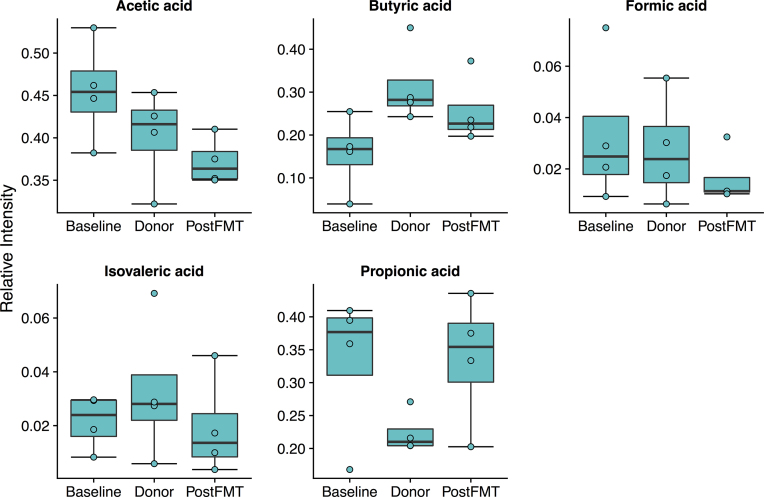
Quantification of short chain fatty acids showed a decrease in acetic acid and an increase in butyric acid levels after FMT in the responders (Fig. 7).
Figure 7.
Quantification of short chain fatty acids (SCFAs) by GC/MS in the four responders. Levels of butyric acid were lower at baseline and increased toward donor levels after FMT.
Inter-omic correlations suggest associations between microbial taxa and metabolites
Spearman correlation coefficients were calculated for pairwise combinations of microbial abundances at the class level (from 16S rRNA gene sequence data) and metabolite intensities in the four responders. A heatmap including the 30 metabolites shown in Fig. 5 provides a visualization of clusters of correlated microbes and metabolites (Fig. 6). Bacteria in the classes Bacilli and Betaproteobacteria were positively correlated with metabolites from the ‘disease-associated’ cluster such as creatinine and norvaline. Bacteria from the class Clostridia were positively correlated with metabolites from the ‘healthy’ cluster such as xanthine and 1-hexadecanol.
DISCUSSION
Because the Kunde et al. pilot study showed efficacy in some but not all patients, we sought to understand the microbiological and molecular features underlying successful FMT for pediatric ulcerative colitis. Interestingly, the non-responders had higher pediatric ulcerative colitis activity index (PUCAI) scores before FMT (40, 50, 50), while all of the responders had PUCAI scores of 35 or less at baseline (Kunde et al. 2013) (Table S1, Supporting Information), suggesting that the FMT method used in this study may be less effective in patients with more severe disease. Measures of microbial alpha diversity were higher at baseline in the responders compared to non-responders, and increased in both groups after FMT (Fig. 1). Species richness was higher and the Shannon and Inverse Simpson indices were lower in the donors of responders compared to donors of non-responders, suggesting that donor alpha diversity may not predict whether FMT will be successful. However, further investigation of donor factors associated with successful FMT will be important for optimizing this therapy.
16S rRNA gene sequencing revealed that the baseline gut microbial communities of ulcerative colitis patients differed from their respective healthy donors (Fig. 2). However, there was considerable variability across subjects and donors, reflecting the heterogeneity of the gut microbiota in both healthy individuals and ulcerative colitis patients. The decreased levels of Clostridia in the ulcerative colitis subjects at baseline compared to donors is consistent with previous reports (Michail et al. 2012). Colonic inflammation is itself associated with changes in the microbiota, including reduced diversity. Therefore, it is not possible to know whether the changes seen in this study are part of the cause or the result of improvement in colitis. This could be addressed in future studies by performing a similar analysis of the microbiome and metabolome in UC patients before and after receiving treatment with corticosteroids.
The gut microbial profiles of the four patients who responded to therapy shifted toward those of their healthy donors after FMT (Figs 2 and 3A), indicating that the procedure successfully led to engraftment of a microbial community in these patients with features shared by the donor. In the non-responders, microbial community structure also changed after FMT but not to the same degree as in responders (Fig. 3A; Fig. S5, Supporting Information), suggesting that specific alterations in the gut microbiota are important for successful FMT. Interestingly, another study that reported clinical improvement in three pediatric UC patients after FMT found that microbial profiles changed after FMT, but microbial community structure did not shift toward that of the donor (Kellermayer et al. 2015). However, unlike the subjects analyzed here, the patients in that study all received FMT from a singular donor. Interestingly, Bray–Curtis similarity to the donors increased after FMT in two of the three non-responders, despite their lack of response to treatment (Table S1, Supporting Information). This suggests that transfer of donor microbes is not sufficient to induce a clinical response. Response to FMT may depend on both patient and donor factors that have yet to be identified; future studies with larger sample sizes may be able to elucidate these factors and predict the likelihood of clinical response for a given patient–donor pair.
To further understand changes occurring after FMT, we performed shotgun metagenomic sequencing on samples from the four responders. Using an alignment-free sequence comparison approach with shotgun metagenomes, analyses of d2S dissimilarity using different k-mer lengths as well as known and unknown virus abundance analyses recapitulated the pattern seen with 16S amplicon sequencing, where the post-FMT composition became closer to the donor samples compared to baseline (Fig. 4A; Fig. S3, Supporting Information). The consistency of three alignment-free approaches shows the feasibility and power of using either database independent k-mer analysis and predicted or aligned viral community composition to detect patterns from large metagenomic datasets.
Annotation of contigs assembled from shotgun metagenomic sequence data with gene ontology (GO) terms showed an increase in the presence of microbial genes involved in processes including neurotransmitter transport and transcription factor activity after FMT (Fig. S7, Suporting Information). Similar functional metagenomic analysis with a larger dataset may provide insights into the biological functions of the gut microbiota in children with ulcerative colitis.
PCoA ordination of GC-TOF-MS metabolite intensities showed a similar but less striking shift toward donor profiles after FMT (Fig. 3B). Three of the four responders developed metabolomic profiles similar to those of their donors after FMT, while the profile of Responder 1 remained stable, even after successful transfer of microbes by FMT. Interestingly, Responder 1 showed less clinical improvement after FMT, with their PUCAI score decreasing from 30 to 15, compared to the other three responders, whose scores fell to 0 after FMT (remission is a PUCAI score < 10). The relatively decreased clinical response in this patient suggests that sufficient metabolomic alteration may be required for effective FMT. Because this subject's microbiota composition shifted toward that of their donor, the successful transfer of microbes by FMT is likely not the only factor affecting the ability of the therapy to alter the metabolomic profile. Conversely, Responder 3 showed a less profound change in microbiota composition after FMT but exhibited a shift toward the metabolomic profile of the donor. This observation suggests that in addition to modifying the composition of the gut microbiome, FMT may also substantially influence its function and metabolism.
The metabolomic analysis showed a separation of metabolites into clusters associated with increased disease activity (the baseline samples) and health (the donor and post-FMT samples) (Fig. 5). Metabolites including xanthine, oleic acid, putrescine and 5-aminovaleric acid shifted toward donor levels after intervention (Fig. S6, Supporting Information). 5-aminovaleric acid is a byproduct of the metabolism of cadaverine, which is produced by gut bacteria. 5-aminovaleric acid has been associated with intestinal inflammation in mice (Lin et al. 2010). The increased levels of this metabolite in ulcerative colitis patients may be due to preferential growth of cadaverine-producing bacteria, or intestinal damage that increases the availability of substrates for the production of cadaverine. Xanthine, which is part of the purine salvage pathway, increased after FMT. Interestingly, alterations in metabolites in this pathway are associated with changes in regulatory T cell (T reg cell) function and T cell immunoregulation. Larger future studies would be helpful to address whether FMT-associated changes in T cell regulation contribute to reduced colonic inflammation.
Baseline patient samples showed lower levels of succinic acid and beta-glutamic acid (Fig. S8, Supporting Information), consistent with a study showing that ulcerative colitis patients have lower levels of certain amino acids and TCA cycle-related molecules in colonic lesion tissues (Ooi et al. 2011). Increased fecal amino acids have been associated with pediatric Crohn's disease, and a recent study showed that the transfer of host nitrogen to gut microbes via bacterial urease expression is associated with gut dysbiosis and inflammation in an animal model (Ni et al. 2017). Several amino acids were elevated in baseline samples compared to donors and decreased after FMT, including norvaline, O-acetylserine and trans-4-hydroxyproline (Fig. S8, Supporting Information). However, the levels of most amino acids that were detected varied after FMT. Perhaps unresolved host inflammation supports microbial production of some amino acids, which continues to some extent in patients after FMT.
Depletion of Faecalibacterium prausnitzii in IBD has been reported previously (Miquel et al. 2013; Machiels et al. 2014; Lopez-Siles et al. 2015). Furthermore, animal studies have shown that F. prausnitzii inhibits IL-17 and suppresses Th17 activity (Zhang et al. 2014), suggesting an association between this bacterium and reduced mucosal inflammation. F. prausnitzii was present in samples from all four responders after FMT and increased from baseline levels in 3 out of the 4 (Fig. S1, Supporting Information). F. prausnitzii decreased from baseline after FMT in 2 out of the 3 non-responders. This suggests that FMT is associated with the growth of bacteria that contribute to reduced inflammation. F. prausnitzii and other butyrate-producing bacteria have been shown to cross-feed with bifidobacteria to promote butyrate synthesis in the gut, which is associated with improved gut health (Riviere et al. 2016). Butyrate is a major fuel source for colonic epithelium (Roediger 1980), and its oxidation is impaired in ulcerative colitis patients (Chapman et al. 1994; Den Hond et al. 1998). Topical butyrate is effective as an adjunct therapy for UC (Vernia et al. 2003), and butyrate enemas have been shown to reduce mucosal inflammation in distal UC, indicating a benefit of increased butyrate levels (Scheppach et al. 1992). The anti-inflammatory activity of butyrate in ulcerative colitis has been associated with inhibition of NF-kB activation in lamina propria macrophages, reducing cytokine secretion and inflammation (Luhrs et al. 2002).
While highly variable at baseline (ranging from 0% to 25%), Bifidobacterium spp. levels stabilized at a relative abundance of 4% after FMT in the responders (Fig. S1, Supporting Information). In addition to F. prausnitzii, other butyrate-producing bacteria also increased in abundance after FMT, including Anaerostipes and Roseburia (Fig. S1, Supporting Information). Decreased Roseburia levels have been reported in ulcerative colitis patients previously (Machiels et al. 2014). The concomitant increase of these groups of bacteria in responders suggests that FMT may promote a cross-feeding interaction with Bifidobacterium spp. to produce butyrate and improve gut health. A recent study of FMT in 34 adult UC patients found an association of sustained remission with known butyrate producers and increased butyrate production (Fuentes et al. 2017). Similarly, analysis of short chain fatty acid levels in our samples showed an increase toward donor levels in butyrate after FMT in responders (Fig. 7), suggesting that enrichment of butyrate-producing organisms is one possible mechanism of this therapy in ulcerative colitis.
Using a multi-omic analysis pipeline, we have demonstrated that microbial, viral and metabolomic changes occur in the gut of children with ulcerative colitis after FMT. Larger trials utilizing a similar approach will be required to validate the role of specific microbes and metabolites in mediating the therapeutic effect of FMT, and to identify factors predictive of whether a given patient is likely respond to FMT. Further study of FMT in pediatric ulcerative colitis may allow for the eventual development of targeted therapies involving alteration of specific components of the gut microbiota in lieu of FMT itself.
AVAILABILITY OF DATA AND MATERIAL
The 16S and metagenomic raw sequencing reads are available in the NCBI Sequence Read Archive (SRA) database under SRA accession number SRP135559 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP135559). The metabolomics data and all R scripts used for data analysis are available in the project GitHub repository (https://github.com/djnusbaum/UC-FMT-Omics).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Riddhi Parsana for processing fecal samples, and Meng Li and Dr. Yibu Chen of the Norris Medical Library Bioinformatics Service at the University of Southern California for their assistance in using the CLC Genomics Workbench software for metagenomic DNA sequence analysis.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work was supported by the National Institutes of Health (NIH) [grant # R01 HD081197 awarded to SM and # R01 GM120624 awarded to FS] and by the Higgins Family Foundation. KW was supported by grants from the National Heart, Lung, and Blood Institute (R56HL126754–319 01A1), UC Irvine Biological Sciences and School of Medicine pilot award, and a Gilead Cystic Fibrosis Research Scholars Award (app_00b072). DJN was supported by the 2017 Saban Scholar Summer Research Fellowship from the Saban Research Institute of Children's Hospital Los Angeles.
Conflict of interest. None declared.
REFERENCES
- Apprill A, McNally S, Parsons R et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microb Ecol. 2015;75:129–37. [Google Scholar]
- Attali D. colourpicker: A Colour Picker Tool for Shiny and for Selecting Colours in Plots. R package version 1.0 https://cran.r-project.org/package=colourpicker2017. [Google Scholar]
- Bakhtiar SM, LeBlanc JG, Salvucci E et al. Implications of the human microbiome in inflammatory bowel diseases. FEMS Microbiol Lett. 2013;342:10–7. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Brandt LJ, Paramsothy S. Therapeutic faecal microbiota transplantation: current status and future developments. Curr Opin Gastroenterol. 2014;30:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88:524–45. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Grahn MF, Boyle MA et al. Butyrate oxidation is impaired in the colonic mucosa of sufferers of quiescent ulcerative colitis. Gut. 1994;35:73–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8:1569–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hond E, Hiele M, Evenepoel P et al. In vivo butyrate metabolism and colonic permeability in extensive ulcerative colitis. Gastroenterology. 1998;115:584–90. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Fuentes S, Rossen NG, van der Spek MJ et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017;11:1877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R, Nagy-Szakal D, Harris RA et al. Serial fecal microbiota transplantation alters mucosal gene expression in pediatric ulcerative colitis. Am J Gastroenterol. 2015;110:604–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee DY et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Tsugawa H, Cajka T et al. Identification of small molecules using accurate mass MS/MS search. Mass Spec Rev. 2018;37:513–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.8 https://cran.r-project.org/package=pheatmap2015.
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunde S, Pham A, Bonczyk S et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr. 2013;56:597–601. [DOI] [PubMed] [Google Scholar]
- Lai Z, Tsugawa H, Wohlgemuth G et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods. 2018;15:53–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, de Bie, Turner CI, D et al. Atypical disease phenotypes in pediatric ulcerative colitis: 5-year analyses of the EUROKIDS Registry. Inflamm Bowel Dis. 2013;19:370–7. [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. preprintarXiv:13033997, 2013.
- Lin HM, Barnett MP, Roy NC et al. Metabolomic analysis identifies inflammatory and noninflammatory metabolic effects of genetic modification in a mouse model of Crohn's disease. J Proteome Res. 2010;9:1965–75. [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Abella C et al. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015;81:7582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrs H, Gerke T, Muller JG et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458–66. [DOI] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J et al. A decrease of the butyrate-producing species Roseburia hominisand Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- Martinez Arbizu P. pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. R package version 0.0.12017. [Google Scholar]
- McDonald D, Price MN, Goodrich J et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michail S, Durbin M, Turner D et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Rossi O et al. Faecalibacterium prausnitziiand human intestinal health. Curr Opin Microbiol. 2013;16:255–61. [DOI] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–9.. e106. [DOI] [PubMed] [Google Scholar]
- Molodecky NA, Soon IS, Rabi DM et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.. e42. [DOI] [PubMed] [Google Scholar]
- Ni J, Shen TD, Chen EZ et al. A role for bacterial urease in gut dysbiosis and Crohn's disease. Sci Transl Med. 2017;9:eaah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M et al. vegan: Community Ecology Package. R package version 2.4-4 https://cran.r-project.org/package=vegan2017. [Google Scholar]
- Olen O, Askling J, Sachs MC et al. Childhood onset inflammatory bowel disease and risk of cancer: a Swedish nationwide cohort study 1964–2014. BMJ. 2017;358:j3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi M, Nishiumi S, Yoshie T et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm Res. 2011;60:831–40. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Kamm MA, Kaakoush NO et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–28. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Version 3.4.1. R Foundation for Statistical Computing, Vienna, Austria: https://www.r-project.org/2017. [Google Scholar]
- Revelle W. psych: Procedures for Personality and Psychological Research. R package version 1.7.8. Northwestern University, Evanston, Illinois, USA: https://cran.r-project.org/package=psych2017. [Google Scholar]
- Riviere A, Selak M, Lantin D et al. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen NG, Fuentes S, van der Spek MJ et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015;149:110–8., e114. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. [DOI] [PubMed] [Google Scholar]
- Scheppach W, Sommer H, Kirchner T et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–6. [DOI] [PubMed] [Google Scholar]
- Schildkraut V, Alex G, Cameron DJ et al. Sixty-year study of incidence of childhood ulcerative colitis finds eleven-fold increase beginning in 1990s. Inflamm Bowel Dis. 2013;19:1–6. [DOI] [PubMed] [Google Scholar]
- Shanahan F, Quigley EM. Manipulation of the microbiota for treatment of IBS and IBD-challenges and controversies. Gastroenterology. 2014;146:1554–63. [DOI] [PubMed] [Google Scholar]
- Shi Y, Dong Y, Huang W et al. Fecal microbiota transplantation for ulcerative colitis: a systematic review and meta-analysis. PLoS One. 2016;11:e0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suskind DL, Singh N, Nielson H et al. Fecal microbial transplant via nasogastric tube for active pediatric ulcerative colitis. J Pediatr Gastroenterol Nutr. 2015;60:27–9. [DOI] [PubMed] [Google Scholar]
- Turner D, Griffiths AM. Acute severe ulcerative colitis in children: a systematic review. Inflamm Bowel Dis. 2011;17:440–9. [DOI] [PubMed] [Google Scholar]
- Turner D, Travis SP, Griffiths AM et al. Consensus for managing acute severe ulcerative colitis in children: a systematic review and joint statement from ECCO, ESPGHAN, and the Porto IBD Working Group of ESPGHAN. Am J Gastroenterol. 2011;106:574–88. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–15. [DOI] [PubMed] [Google Scholar]
- Vandenplas Y, Veereman G, van der Werff Ten Bosch J et al. Fecal microbial transplantation in early-onset colitis: caution advised. J Pediatr Gastroenterol Nutr. 2015;61:e12–14. [DOI] [PubMed] [Google Scholar]
- Vernia P, Annese V, Bresci G et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest. 2003;33:244–8. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer Science + Business Media, New York; London: 2009. [Google Scholar]
- Wilke CO. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. R package version 0.9.2 https://cran.r-project.org/package=cowplot2017. [Google Scholar]
- Zhang M, Qiu X, Zhang H et al. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS One. 2014;9:e109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S and metagenomic raw sequencing reads are available in the NCBI Sequence Read Archive (SRA) database under SRA accession number SRP135559 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP135559). The metabolomics data and all R scripts used for data analysis are available in the project GitHub repository (https://github.com/djnusbaum/UC-FMT-Omics).