Abstract
Drug–drug interactions (DDIs) constitute an important concern in drug development and postmarketing pharmacovigilance. They are considered the cause of many adverse drug effects exposing patients to higher risks and increasing public health system costs. Methods to follow-up and discover possible DDIs causing harm to the population are a primary aim of drug safety researchers. Here, we review different methodologies and recent advances using data mining to detect DDIs with impact on patients. We focus on data mining of different pharmacovigilance sources, such as the US Food and Drug Administration Adverse Event Reporting System and electronic health records from medical institutions, as well as on the diverse data mining studies that use narrative text available in the scientific biomedical literature and social media. We pay attention to the strengths but also further explain challenges related to these methods. Data mining has important applications in the analysis of DDIs showing the impact of the interactions as a cause of adverse effects, extracting interactions to create knowledge data sets and gold standards and in the discovery of novel and dangerous DDIs.
Keywords: data mining, drug–drug interactions, adverse drug effects, FAERS, electronic health records
Introduction
Drug–drug interactions (DDIs) are a serious problem in patient safety [1, 2]. Coadministration of two or more drugs at the same time can affect the biological action of the implicated drugs. The interaction can seriously affect efficacy and safety drug profiles. The main types of DDIs include pharmacokinetic and pharmacodynamic interactions [3]. The pharmacokinetic interactions can affect important drug processes that determine bioavailability, such as absorption, distribution, metabolism and excretion [3]. Examples of these interactions are the administration of a medication that increases the motility of the intestine decreasing the absorption of the other drug, competition for the same plasma protein transporter, inhibition of the action of a metabolizing enzyme or even interaction at excretion level affecting the elimination of one of the drugs [4]. On the other hand, pharmacodynamic interactions can occur at the pharmacological receptor level with both drugs interacting with the same protein, at the signaling level affecting different signaling pathways or at the effector levels causing different pharmacological responses [5]. The action can be synergistic or antagonistic or a novel effect can be derived.
DDIs result in many adverse drug effects (ADEs) that can cause severe injuries to the patients, or even be responsible for deaths [6]. It has been reported that DDIs could be responsible for up to 30% of the adverse effects found in the patients [7]. Hospitalizations and emergency department visits because of coadministration of different drugs are estimated around 0.57 and 0.054%, respectively [2]. The problem is so important that in the United States alone, DDIs are estimated to be responsible for up to 195 000 hospital admissions [1]. Between 7 and 22% of the patients in primary or secondary health care are receiving combinations of interacting drugs [2]. This problem is emphasized in the elderly, where this percentage rises to 22–31% [2, 8, 9]. Moreover, the numbers and statistics about DDI impact in health care may underestimate the real public health burden because the statistics are based on known DDIs, and unknown DDIs may be significant. In addition, DDIs that are not serious but have an impact on patients’ quality of life could be underreported [10].
Once a DDI is well described, and depending on the danger associated with the interaction, further actions will be considered that go from a warning notice in the label of the drugs to the withdrawal of some drugs from the pharmaceutical market. For instance, cerivastatin, a drug that lowers cholesterol levels by inhibiting the enzyme HMG-CoA reductase, was withdrawn from the pharmaceutical market worldwide in 2001 because of 52 cases of fatal rhabdomyolysis [11, 12]. The coadministration of cerivastatin with other medications, such as gemfibrozil, was responsible for the death of several patients. Gemfibrozil reduces the metabolization of the statin, increasing its plasma concentration and causing a higher risk of associated adverse effects like myopathy and rhabdomyolysis [13, 14]. Another example of a drug withdrawn from the market is mibefradil, a calcium channel blocker, 1 year after approval by the US Food and Drug Administration (FDA) [15] because of risk of deadly interactions with other drugs [16].
Prevalence of potential DDIs in primary care has been estimated to be high (>60%), although the numbers are lower when clinically significant DDIs are taken into account (3.8–12%) [17, 18]. These reports showed the importance of taking into account by prescribers and healthcare professionals, some well-known DDIs that have a great impact in the generation of ADEs in the patients [19]. Being aware of these dangerous combinations by medical personnel and not prescribing combinations of drugs that cause serious adverse effects is important to improve health systems [20]. A more conscious knowledge of the importance and incidence of DDIs, as well as the most dangerous drug combinations is a crucial feature to avoid their occurrence, reduce their clinical impact and collaborate in the safety of the patients. Systems in clinical decision-making that warn physicians about potential drug combinations are valuable tools to improve health care. However, problems related to alert fatigue, when many potential warnings are given, can undermine the usefulness of the technology [21, 22]. Decision systems should prioritize high-risk DDIs to make the number of alerts manageable by the professionals. Development of a consensus DDI list by experts and endorsement by professional societies, agencies and regulators help to create improved health care decision-making systems [23]. Moreover, avoiding possible DDIs evaluating high-risk factors, such as age, multiple diseases or genetic polymorphisms [24], should be a common practice oriented to enhance patient care. In case multiple and complex therapies are necessary, a better system to specify to the patients which drugs cannot be taken together would be helpful. In this sense, enhancements in the development of tools to facilitate the administration of multiple therapies to patients are welcome to improve clinical decision-making and patient safety.
DDIs are difficult to detect in the different stages of drug development and in postmarketing surveillance [1]. Some DDIs could also be dependent and recognizable at high medication doses [25]. Clinical trials use a limited number of patients and include different criteria for inclusion or exclusion of the participants, with the consequent limitations to deeply examine the effects of polypharmacy. Moreover, human variability can affect the result of DDIs [1], and adverse effects could occur in a certain subgroup of the population not properly represented in a clinical trial. Long-term follow-up provided by data mining of public scientific literature and clinical pharmacovigilance sources could help to overcome some obstacles derived from short-term studies like clinical trials [26]. Improvement in the detection of DDIs is of major interest to regulatory organizations, such as the FDA [15], pharmaceutical companies and a broad group of researchers working in translational medicine and drug safety. Besides a more realistic detection, methods that provide understanding of the mechanisms or principles for the DDIs are also needed. A more detailed description of the different computational and experimental techniques used to discover DDIs is provided in some articles and reviews already published [1, 27–31].
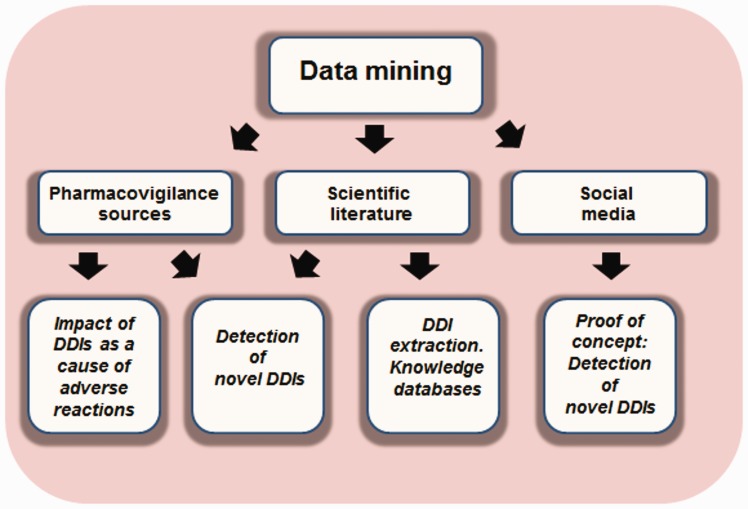
Coadministration of multiple drugs could also be responsible for synergistic effects, such as an increase in the efficacy of the therapy. In fact, this is a common practice in medicine that in some cases offers great results in efficacy, avoiding toxicity or minimizing drug resistance [32, 33]. However, in this review, we focused on the detection of DDIs caused by the coadministration of multiple drug regimens that are responsible for adverse effects in the patients. We reviewed data mining studies that use clinical databases, such as electronic health records (EHRs), the scientific literature and social media tools. We provided a general picture of the usefulness of the different sources and their advantages and limitations in the extraction and detection of DDIs. We showed multiple examples where pharmacovigilance data, scientific literature and social media were used not only in the detection of new DDIs but also to prove the important impact caused in health care and to extract DDI knowledge crucial for the development of reference standards to evaluate detection tools. The different topics discussed in the current article are shown in Figure 1. As a summary, this review contains different sections focusing on data mining of pharmacovigilance sources, scientific literature and social media, along with challenges and limitations of the different DDI sources.
Figure 1.
Flowchart of the different steps described in the review. Applicability of data mining using different sources: applicability showing the importance of DDIs as the cause of ADES, in the detection of novel DDIs and in the development of knowledge databases.
Data mining of pharmacovigilance sources
Impact of DDIs as a cause of ADEs
DDIs are a real problem in clinical practice worldwide. There are different pharmacovigilance studies that reflect the importance of DDIs as the means by which patients develop ADEs. As an example, Montastruc et al. [34] studied DDIs involved in the use of serotonin reuptake inhibitors registered in the spontaneous reports of the French pharmacovigilance system. The authors concluded that around 40% of the reported ADEs using serotoninergic reuptake inhibitors were related to the coadministration of different drugs. Moreover, >50% of the ADEs related to DDIs were considered serious for patients’ health. DDIs were identified using a DDI reference database extracted from different sources, among them is the French National Drug Formulary (Vidal). The authors compared ADEs related to DDIs with ADEs not related to DDIs using χ2 test and Student’s t-test. Importance of DDIs in the French pharmacovigilance database was also confirmed by Fournier et al. [35]. Interactions between antihypertensives and nonsteroidal anti-inflammatory drugs (NSAIDs) led to 24% of the detected ADE reports containing NSAIDs. The most frequent ADE caused by the DDIs was acute renal failure. Moreover, a triple combination therapy receiving two antihypertensive treatments with NSAIDs was associated in a nested case-control study with increased risk of acute kidney injury [36]. In another study, one-third of the interactions detected with cholinesterase inhibitors were estimated to be the cause of adverse effects [37]. Many potential ADEs caused by the concomitant use of drugs were also detected in the spontaneous reporting system (SRS) from Italy [38]. In total, >20% of the patients exposed to potential DDIs were detected to suffer an adverse effect. The study showed the importance of DDIs involving anticoagulant drugs as a risk of serious adverse effects. The adverse effect bleeding was as well evaluated by Schelleman et al. [39] in Medicaid claims data [40] carrying out an observational case-control study. Patients on the anticoagulant warfarin that initiated a concomitant therapy with some antidepressants had a higher risk of hospitalization because of gastrointestinal bleeding. Odds ratios for warfarin users with a combined therapy with the drug citalopram, fluoxetine, paroxetine and amitriptyline were 1.73, 1.63, 1.64 and 1.47, respectively. The impact of DDIs as a cause of adverse effects was additionally reported by Mirosevic Skvrce et al. [41] in the SRS of the Croatian Agency for Medicinal Products and Medical Devices [42]. Almost 40% of the reports including more than one drug showed that potential DDIs could be responsible for ADEs, and 7.8% of the total reports with more than one drug were caused by actual DDIs. The distribution of the different adverse effects caused by the DDIs is shown by Mirosevic Skvrce et al. [41]. Moreover, DDIs have an important impact not only in the generation of adverse effects but also in the efficacy of some of the implicated drugs, with the associated risk for the patients. As an example, there are studies that showed an attenuated effectiveness of antihypertensive drugs when administered with NSAIDs [43]. There are also some excellent reviews available in the literature that showed the impact of the adverse effects caused by DDIs in the health system and clinical care [2, 44].
Mining pharmacovigilance sources to discover DDIs
The FDA [15] receives reports of suspected adverse effects from health care professionals, consumers and manufacturers. The FDA facilitates drug surveillance providing access to the FDA’s Adverse Event Reporting System (FAERS) [45]. FAERS is useful for ADEs surveillance as well as the analysis and detection of potential DDIs. Reports in FAERS contain patient, drug and adverse effect information and include many medications with the suspected drug and concomitant drugs (drugs are labeled as primary suspect being the cause of the adverse effect, secondary suspect, etc.).
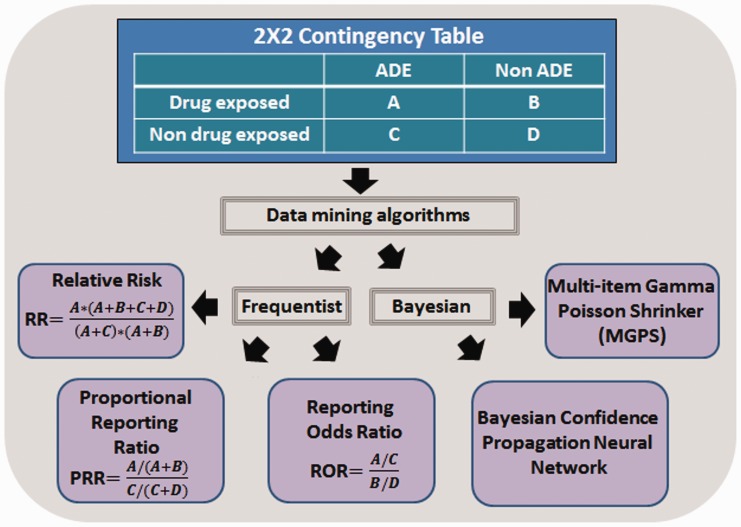
Data mining algorithms (DMAs), such as reporting odds ratio [46] or Multi-item Gamma Poisson Shrinker (MGPS) [47], have been used to generate and rank the different drug-ADE associations or signals found in pharmacovigilance data. Calculations of some algorithms are based on two-dimensional contingency tables (see Figure 2 with a summary of some DMAs). Multivariate modeling, such as logistic regression, has also been used to analyze the effects of drugs [48]. There are in the literature some reviews [49–51] that explain with more detail the different data mining algorithms described in the published studies.
Figure 2.
Common DMAs extracted from 2 × 2 contingency tables.
Besides FAERS, there are other pharmacovigilance data sources, such as the World Health Organization’s (WHO) VigiBase [52], that is used for similar purposes. VigiBase™ is the largest SRS for adverse effects in the world. It contains >12 million reports from >100 member countries compiled since 1968. Moreover, observational data in EHRs can complement adverse event reporting systems in the detection of ADEs and DDIs. In fact, there are several examples describing potential DDIs with reporting sources and completing the study through a validation performed in EHRs [53, 54]. In many cases, the analysis looking for DDIs is based on structured data, although unstructured information that resides in the clinical notes taken by the medical personnel in the EHRs can also be useful. There are some examples that showed that coded data may be insufficient to describe some steps in pharmacovigilance procedures and that the combination with narrative data can yield better results, such as building patient cohorts [55, 56]. The combination of structured and unstructured data can be exploited to detect DDIs and ADEs with higher confidence [57]. In this sense, increasing access to medical records may facilitate the detection and validation of adverse effects caused by the use of concomitant drugs.
Spontaneous reporting systems
There are different studies that showed in early stages the potential of SRS in the identification of DDIs [58–61]. Some studies are designed to analyze an individual adverse effect in small sets of drug combinations, such as ‘delayed withdrawal bleeding’ using itraconazole with oral contraceptives [62], or the increased risk of hypothermia associated with the combination of topiramate and valproic acid [63]. The ability to detect four known DDIs by multiplicative and additive models was also evaluated using FAERS [64]. Nevertheless, there are some studies mining large data sets with the aim of looking for many diverse potential DDIs. As an example of these studies, a DDI signal detection algorithm was published by Tatonetti et al. [53]. The authors divided FAERS into two data sets: reports with only one drug (training) and reports with two drugs (prediction), and constructed eight clinically significant adverse event models. In each model, they described a drug as a profile of the adverse effect frequencies extracted from FAERS and through a logistic regression classifier differentiated between drugs that cause the clinically significant adverse event under study and drugs that do not cause the adverse event. Then, predictions in drug combinations were made for each model. The electronic medical records (EMRs) of the Stanford Hospital, Vanderbilt Hospital and Partners HealthCare were used to provide validation for some drug interaction hypothesis. Their DDI method pointed out that pravastatin and paroxetine could contribute to elevated blood glucose levels, and the possible interaction was assessed in experimental assays [65]. A similar method was applied to identify DDI signals for the prolongation of the QT interval in FAERS and validate the predictions using electrocardiogram laboratory data in EHRs at Columbia University Medical Center [66]. The authors trained a model to detect adverse effect fingerprints with risk of long QT/torsades de pointes for single drugs and applied the predictor to an independent test of combinations of drugs. Their system outperformed an alternative direct evidence model. Novel DDIs related to QT prolongation were provided for further investigation. As an example, ceftriaxone and lansoprazole could prolong the QT interval. Laboratory data measuring the hERG channel block are in agreement with this DDI [67].
Other pharmacovigilance reporting systems were also used in the detection of potential DDIs, such as the WHO database [52]. Norén et al. [68] developed a shrinkage observed-to-expected ratio to identify DDIs in individual case safety reports (ICSRs). The study proposed a disproportionality measure based on an additive risk model. The authors also performed a DDI screening of the whole WHO database showing the feasibility of their approach. A different study of drug hepatic safety [69] after coadministration of multiple medications was also carried out in the WHO VigiBaseTM. Identification of DDIs was carried out through empirical Bayes geometric mean values [47]. Liver event terms were created using the Medical Dictionary for Regulatory Activity [70]. Co-reported therapies were associated with changes in the frequency of hepatic events. All these studies reflect the importance of reporting systems as a source of analysis and follow-up to discover novel DDIs.
Electronic health records
Besides the utilization of EHRs as a source to validate DDI signals extracted from other sources, EHRs were also used as a source of DDI signal generation. There are examples of studies evaluating the effects of a particular DDI, such as the combination between clarithromycin and colchicine in patients with renal insufficiency [71], studies evaluating classes of drugs in a selected cohort of patients, such as potential DDIs between antiretrovirals and hepatitis C virus (HCV) direct acting antivirals in a cohort of HIV/HCV coinfected patients [72] and other studies focused on the inspection of big data and discovery of diverse sets of DDIs. For instance, Iyer et al. [73] used unstructured data to identify DDI signals from >50 million clinical notes available in the EHRs (data sets from Stanford Translational Research Integrated Database Environment and Palo Alto Medical Foundation). The authors used disproportionality ratios to identify DDIs for 1165 drugs and 14 ADEs. In the 2 × 2 contingency table, the exposed group considered patients on both drugs and the nonexposed group considered patients on one of the implicated drugs or no drugs. The association score was AOR025 (lower bound of the 95% CI of the adjusted odds ratio). The results were validated with a gold standard of DDIs along with a complementary study in FAERS extracting DDIs with MGPS algorithm [47]. The study also estimated the rate of adverse effects for the patients on multiple drugs, useful for alert prioritization in DDI surveillance and clinical decision-making. The mentioned study uses an opposite strategy to other publications mining first EHRs and validating the detected DDIs using FAERS.
The clinical data warehouse of the Hôpital Européen Georges-Pompidou (HEGP) was also mined to detect DDIs causing acute kidney injury [74], defined as an increase of ≥ 50% of creatinine basis. The authors tested their algorithm in single drugs (nephrotoxic and non-nephrotoxic), and the system was applied to 45 pairs of non-nephrotoxic drugs finding some combinations interesting to further study. A different approach was described by Pathak et al. [75] that used Semantic Web and Linked Data techniques to study DDIs including cardiovascular and gastroenterology drugs in the EHRs of the Mayo Clinic. They represented patient data as labeled graphs in a Resource Description Framework. Another example of mining adverse effects caused by DDIs using Semantic Web technologies is provided by Jiang et al. [76]. They performed a case study with cardiovascular drugs in FAERS and carried out a signal enrichment using data extracted from medical records. Other examples that demonstrate the usefulness in DDI detection of patient electronic data along with temporal association methodologies are provided in the literature [77].
The development of improved methods to score the DDIs studied in the pharmacovigilance data is important to eliminate possible false-positive cases and prioritize the candidates for further investigation. Banda et al. [78] developed a method to score a set of potential DDIs extracted from EHR (data extracted from a previous study [73]: 5983 DDIs including 345 drugs and 10 ADEs). They used four information sources to score the DDI candidates: public databases, spontaneous reports, literature and non-EHR DDI prediction methods. Their assumption is that DDIs supported by different sources or methods are more probable to be true-positive signals. Our research group also provided alternative systems to prioritize DDIs extracted from clinical data through molecular and biological profile similarity [79].
High-order drug interactions
Most studies focus on the analysis of DDIs caused by the coadministration of two drugs. However, there are some examples of studies where relationships between high-order drug interactions are described. High-order interactions refer to drug interactions because of the administration of more than two drugs to the patients, including multiple drug combinations.
High-order methods can be useful when multiple medications are prescribed to the patients, a common situation in clinical practice. Du et al. [80] mined directional effects of high-order drug interactions causing myopathy in EHRs. They estimated individual risk for each new drug added to the treatment. Moreover, they generated a visualization system of the multiple directional interaction effects. A companion paper was also published where the authors developed a mixture model between constant risk and dose (number of drug combinations)-response risk models [81]. The study identified high-order drug combinations, between two to six, associated with myopathy and detected that the risk increased with the drug order. Previous studies with multiple high-order drug combinations in FAERS were published by Harpaz et al. [82, 83]. The authors applied a method known as association rule mining to generate multiple drug combinations associated with multiple adverse effects. These methods can be computationally expensive because of the big amount of data and possible multidrug combinations necessary to process. To overcome this problem, Xiang et al. [84] generated ADE–multidrug associations using frequent closed itemset mining and filtering. Their method eliminated redundant data and reduced computational cost. All the mentioned analysis showed the potential of data mining pharmacovigilance databases to detect novel and serious DDIs with clinical significance [85].
Table 1 shows a summary of some studies provided in the section ‘Mining pharmacovigilance sources to discover DDIs’ that take into account SRS and EHRs in the detection of novel DDIs. Table 1 is useful to compare the different conclusions and statistics.
Table 1.
Summary of the studies described in the section ‘Mining pharmacovigilance sources to discover DDIs’
| Authors | Year | Pharmacovigilance data | Method | Conclusion |
|---|---|---|---|---|
| Van Puijenbroek EP et al. [58] | 2000 | Reporting system of the Netherlands Pharmacovigilance Foundation Lareb (9822 reports) | Logistic regression analysis, odds ratio | Decreased efficacy of diuretics combined with NSAIDs |
| Ellis RJ et al. [61] | 2000 | FAERS (64 cases) and University of Kansas Medical Center (two cases) | Statistical comparison, Fisher’s exact test | Ciprofloxacin–warfarin coagulopathy |
| Van Puijenbroek EP et al. [62] | 1999 | Reporting system of the Netherlands Pharmacovigilance Foundation Lareb (5503 reports) | Logistic regression analysis, odds ratio | Delayed withdrawal bleeding using itraconazole and oral contraceptives |
| Knudsen JF et al. [63] | 2008 | FAERS | MGPS algorithm: empirical Bayes geometric mean values | Hypothermia using topiramate and valproic acid |
| Thakrar BT et al. [64] | 2007 | FAERS (study of four known DDIs and four non-DDIs) | Multiplicative and additive model, proportional reporting | Potential to recognize four known DDIs |
| Tatonetti NP et al. [65] | 2011 | FAERS (adverse event profiles with 12 627 reports for 37 drugs) | Latent signal detection algorithm, analysis of covariance | Paroxetine and pravastatin increase blood glucose levels |
| Lorberbaum T et al. [66–67] | 2016 | FAERS and EHRs (1.8 million reports, 1.6 million electrocardiograms from 380 000 patients in EHR) | Model to identify an adverse effect fingerprint for risk of torsades de pointes | Ceftriaxone and lansoprazole could prolong QT interval |
| Noren GN et al. [68] | 2008 | ICSRs, WHO database | Shrinkage observed-to-expected ratio, disproportionality measure based on an additive risk model | Development of a method to detect DDIs |
| Suzuki A et al. [69] | 2015 | WHO VigiBaseTM (2275 co-reported drugs with four drugs that cause hepatotoxicity) | Empirical Bayes geometric mean algorithm, logistic regression | Co-reported medications were associated with changes in liver event reporting frequency |
| Hung IFN et al. [71] | 2005 | Care teaching hospital: 116 patients on clarithromycin and colchicine | Fisher’s exact test, Student’s t-test, Mann–Whitney U test, calculation of relative risk and P-values | Clarithromycin increases colchicine toxicity |
| Poizot-Martin I et al. [72] | 2015 | HIV-coinfected or HCV-coinfected patients in French Dat’AIDS cohort (2511 HIV-coinfected or HCV-coinfected patients) | Pearson’s χ2 test, t-test, two-tailed significance testing | Potential DDIs between direct acting antivirals with antiretrovirals |
| Iyer SV et al. [73] | 2014 | EHRs (9 million notes with 1 million patients, over 50 million clinical notes with 1.2 million patients; 1165 drugs and 14 adverse effects) | Adjusted disproportionality ratios (2 × 2 contingency table) | Potential of textual notes from EHRs to detect DDIs |
| Girardeau Y et al. [74] | 2015 | Data warehouse of the HEGP (training with 10 nephrotoxic and 10 non-nephrotoxic drugs and test with 45 drug pairs) | Logistic regression model, unadjusted odds ratio and P-values | Potential DDIs causing kidney injury |
| Pathak J et al. [75] | 2013 | EHRs of the Mayo Clinic (6758 patients, drugs included clopidogrel, warfarin and protein pump inhibitors) | Semantic Web and Linked Data techniques. Patient data as labeled graphs in a Resource Description Framework. | Potential DDIs including cardiovascular and gastroenterology drugs |
| Jiang G et al. [76] | 2015 | FAERS and EMR data (601 DDIs with warfarin, clopidogrel and simvastatin) | Semantic Web technologies | Potential DDIs including cardiovascular drugs |
| Banda JM et al. [78] | 2016 | EHR (5983 drug–drug-event associations with 345 drugs and 10 adverse events) | Adjusted disproportionality ratios (2 × 2 contingency table), priorization of DDIs providing evidence from complementary sources | Priorization of DDIs from EHRs using four sources of information |
| Du L et al. [80] | 2015 | Indiana Patient Care Data (INPC) (case-control data set with 125 275 case events and 6 263 399 control events, 212 drugs) | Odds ratios, frequent itemset mining algorithm | High-order drug interactions causing myopathy |
| Zhang P et al. [81] | 2015 | INPC (top 20 most frequently distributed drugs) | A mixture dose–response model and an empirical Bayes method | High-order drug interactions causing myopathy |
| Harpaz R et al. [82] | 2010 | FAERS (162 744 reports of suspected ADEs, the method identified 1167 DDIs) | Association rule mining, relative reporting ratios | Multiple potential DDIs |
| Xiang Y et al. [84] | 2014 | FAERS (data in the study containing 134 508 records) | Frequent closed itemset mining and filtering | Method to study high-order DDIs with reduced computational cost |
Data mining of the scientific literature
Information about drug interactions in the scientific literature is increasing rapidly. The knowledge hidden in the scientific literature could be analyzed to find connections between articles. For this reason, this resource to extract and discover DDIs has been exploited lately in different studies. Data mining has applications in the construction of knowledge databases extracted from the literature as well as a method to detect novel potential DDIs. Techniques related to information retrieval and natural language processing (NLP) are commonly used to develop these tasks.
Extraction of DDIs
Reviewing the biomedical literature looking for DDIs is time-consuming by the researchers and professionals. For this reason, there is an increasing need to develop novel automatic strategies to extract DDI information from texts. From this point of view, progress on information extraction tools to automatically extract biomedical knowledge from the literature can save time and resources [86, 87]. Moreover, these automatic tools are essential in the construction of more complete drug knowledge databases [86, 87]. Generally, in DDI extraction, methods extract the data as a classification problem providing semantic information to differentiate between DDIs and non-DDIs. The main systems to extract relationships from texts include co-occurrence-based, rule-based and machine learning approaches [88]. Co-ocurrence-based methods are simpler and establish a relationship between two entities based on co-ocurrence. Rule-based methods use linguistic to understand the meaning of a certain relationship. Linguistic rule-based approaches were used to extract DDIs from biomedical texts [89]. On the other hand, machine learning-based methods, such as the shallow linguistic kernel approach [90], can also offer great assistance in DDI extraction from biomedical literature [91]. With the exponential growth of public availability of unstructured biomedical texts, methods based on machine learning have acquired an important role in data extraction tasks. There are mainly two types of machine learning systems: feature-based [92], representing each data instance as a feature vector, and kernel-based methods [91], which exploit structural representations of data instances. Chowdhury et al. [93] applied machine learning approaches including both feature-based and kernel-based methods. Ensemble learning methods based on majority voting schemes are also useful tools with good DDI extraction performance [94]. Moreover, convolutional neural networks exhibited potential in NLP through the extraction of DDIs [95]. In another effort of automatic knowledge extraction, He et al. [96] developed a stacked generalization-based approach. Their method is based on the combination of feature-based, graph and tree kernels. Their approach showed good performance in the DDI Extraction 2011 challenge task [97]. Moreover, other machine learning hybrid systems using feature and kernel-based methods showed good performance in DDI extraction challenges [98]. More examples of applicability of machine learning in the extraction of DDIs from scientific texts are available in the literature [99–102]. The general goal of these studies and challenges is oriented toward the extraction and classification of DDIs and not the extraction of experimental evidence to predict novel interactions. Moreover, another edition of the DDI extraction challenge task was carried out in 2013 [103]. These tasks, and the annotated DDI corpus [104] with >5000 DDIs, have contributed in a relevant way in the extraction and detection of DDIs from the literature [105]. The objective of the new DDI 2013 challenge was the detection and classification of DDIs along with the recognition of pharmacological substances. Fourteen teams participated in the challenge that attracted great interest from the scientific community [103].
Improvements in ontology and well-annotated corpus facilitate the training and development of methodologies for DDI extraction and detection. Besides that, annotated corpora contribute to establish a stable criteria or gold standard of well-known DDIs, an important step to facilitate the assessment of the data mining methods and hence, their performance and reliability. To help in this process, important DDI ontologies, such as DINTO [106] and DIDEO [107], have been of great contribution in the description and categorization of DDIs. Efforts have also been made for the development of models to represent potential DDI knowledge and evidence [108]. Wu et al. [88, 109] constructed a comprehensive pharmacokinetic ontology including in vitro and in vivo experiments related to drug metabolism and transportation. A pharmacokinetic corpus was also constructed as a valuable resource to text mining drug interactions using pharmacokinetic parameters. Following this line of work, Ayvaz et al. [110] combined in a unique data set 14 different sources of potential DDIs including clinical information sources, NLP corpora and bioinformatics and pharmacovigilance information sources. Future work will include the development of new methods to map and integrate the interactions in the different sources and making the final data accessible as Semantic Web Linked Data.
Extraction of information from the biomedical literature is an important field that among other tasks contributes to the development of knowledge databases with applications in DDI assessment [97, 103].
Text mining in the detection of novel DDIs
Besides extraction of DDIs from the literature to help in the construction of knowledge databases, applicability of data mining approaches also involves the detection of potential novel DDIs with associated risk to patients. Prediction and detection in a cost-effective manner is a useful feature that can be exploited through the use of these methodologies. Data mining methods applied to the biomedical literature can provide scientific evidences of possible molecular mechanism of action of the potential DDIs. Moreover, as certain degree of disagreement was observed between different DDI resources [111], providing scientific evidences for DDIs can help in the verification of some discrepancies observed in the different DDI compendia. As an example of DDI discovery, Tari et al. [112] proposed an approach that integrates text mining with automated reasoning. Their approach extracts DDIs explicitly mentioned in texts but also detect potential novel DDIs by a reasoning approach based on drug metabolism. Their system takes into account induction or inhibition of the enzymes responsible for the metabolization of the implicated drugs. A manual review of the potential novel DDIs showed that >80% of them were in agreement with supporting evidences. Their approach has implications not only in detection but in the explanation of the mechanism of the DDI. Similar approaches could be extended toward transporter-based information. Moreover, degree of drug metabolization by a particular enzyme constitutes interesting information to be considered in the development of this type of predictors.
A different approach but with applicability in both DDI detection and explanation of the pharmacokinetic molecular mechanism has been proposed by Duke et al. [54]. They predicted an initial set of 13 197 potential DDIs from the literature localizing substrates and inhibitors of cytochrome P450 (CYP450) enzymes. In vitro pharmacology data published in the literature were used. However, in some cases, it is not possible to extrapolate in vitro effects into clinically significant DDIs with serious associated adverse effects. In this sense, the authors narrowed the data with a study carried out in an EMR data source. The initial DDI set was reduced to 3670 drug pairs concomitantly prescribed in the EMR. Among them, text mining identified 196 drug combinations with published clinical DDI testing (123 were confirmed as in vivo DDIs). Special attention in the study was paid to the drug combinations with increased risk of myopathy. The method showed the potential of incorporating data mining of biomedical literature with large clinical sources to predict significant interactions.
Percha et al. [113] exploited the concept that two drugs can interact with the same gene product causing a DDI. The study describes the use of text mining to identify gene–drug relationships available in the literature and create a system to predict DDIs. The authors mined Medline abstracts to extract genes, drugs and the type of relationship, such as metabolize, inhibit, etc., to create a semantic network and infer novel DDIs. They trained a random forest classifier in a set of well-known DDIs using the features extracted from the literature. Mechanistic explanations about the new DDIs were also provided using the semantics of the gene–drug relationships.
As commented in the section related to pharmacovigilance data mining, structure and unstructured data can be combined to generate a more complex model with better performance. As an example, Yan et al. [114] developed different structured models, introducing molecular structure and target information, along with text data models extracting concepts like genes, diseases and Medical Subject Headings (MeSH) terms. In this study, a complex model combining all types of data showed the best results in DDI detection. The authors showed that unstructured text data can improve domain knowledge in the detection of DDIs. To extract or detect DDIs from the literature, it is important to evaluate the ability of the system in the identification of relevant documents related to DDIs. Kolchinsky et al. [115] assessed performance of several classifiers, such as logistic regression, support vector machines (SVMs) or discriminant analysis, among others, to distinguish relevant abstracts from PubMed containing pharmacokinetic DDI evidence. Their approach is also useful to associate causal mechanisms to the potential DDIs. The study showed also the influence of using different data reduction techniques and including dictionaries and named entity recognition tools. The efficacy of data mining to identify PubMed abstracts with DDI pharmacokinetic evidence and extract such evidence was assessed by the same authors in an additional study [116]. Their goal is not only identifying DDIs but also extracting experimental evidence of possible drug interactions.
As mentioned previously, improvements in ontology and annotated corpora facilitate the training of the data mining models and establish a criteria of known DDIs helpful to assess performance and reliability of the methods. A summary of the studies focused on detection of novel DDIs and explained in this section is provided in Table 2 to compare statistics and conclusions.
Table 2.
Summary of the studies focused on detection of novel DDIs described in the section ‘Text mining in the detection of novel DDIs’
| Authors | Year | Text data | Method | Conclusion |
|---|---|---|---|---|
| Tari L et al. [112] | 2010 | Medline abstracts (∼17 million) 265 drugs, extraction of 170 explicit drug interactions and extraction of implicit DDIs: 4154 direct inhibition/induction interactions and 979 interactions based on indirect inhibition or induction of enzymes | Text mining (natural language extraction) and automated reasoning | The approach can identify promising enzyme-based DDIs |
| Duke JD et al. [54] | 2012 | PubMed abstracts and EMR data (Indiana Network for Patient Care: 817 059 patients) Among 1492 drugs, 232 drugs were identified as CYP substrates or inhibitors. 13 197 potential DDIs (3670 drug pairs concomitantly prescribed in the EMR) | Automatic literature mining to predict DDIs based on associated metabolism enzymes. Logistic regression for DDIs causing myopathy, calculation of relative risk and P-values | DDI detection and explanation of the pharmacokinetic molecular mechanism. Potential combinations with increased risk of myopathy |
| Percha B et al. [113] | 2012 | Medline abstracts (17.5 million abstracts and 88 million sentences). In total, 1806 entities were included in a network: 1061 drugs, 532 genes, 172 context terms and 41 relations | Text mining to extract genes, drugs and their relationships (metabolize, inhibit, etc.), and generation of semantic network to infer novel DDIs. Random forest classifier | Identification of DDIs based on gene–drug relationships providing mechanistic explanations |
| Yan S et al. [114] | 2013 | DrugBank DDI data for training. Medline abstracts (from 2006 to 2010, >3.6 million abstracts) | Fingerprints (drug targets and molecular structure). Extraction from Medline: genes, disease names and MeSH concepts. Drug entities: Drug-Entity-Topic model. Drug–drug relations: logistic regression model | Development of structured models, including molecular structure, target information and text data models A complex model combining all types of data showed the best results in DDI detection |
| Kolchinsky A et al. [115–116] | 2013, 2015 | PubMed pharmacokinetics-related abstracts (1213 abstracts) | Six different linear classifiers: Variable Trigonometric Threshold, SVM, logistic regression, binomial Naive Bayes, linear discriminant analysis (LDA) and a ‘diagonal’ version of LDA | Performance of several classifiers to distinguish relevant abstracts from PubMed containing pharmacokinetic DDI evidence |
Data mining of social media resources
Besides pharmacovigilance data and the scientific literature, social media provides different promising resources with large-scale data that can be useful in the identification of ADEs and DDIs. Social media offers the possibility of analyzing big data from a great number of users who post comments about drug outcomes. Moreover, social media sources are useful in biomedical research in the study of interactions between drugs and natural products[117]. Some drug–natural products interactions are poorly explored, and new possibilities are opened because of the use of social media tools. Social media could also be a helpful tool to study some conditions or pathologies with social disapproval that can be underreported using other sources [117].
Different social media platforms offer great potential to monitor public health in the analysis of drugs effects. The potential of social media in pharmacovigilance has been shown through the study of ADEs using sources like Twitter [118–120]. The social platform had already been used to study other health issues, such as influenza and Ebola virus outbreaks [121–123]. Lately, Carbonell et al. [124] studied the mentions of drugs in Twitter to explore the potential of social media in the detection of ADEs and DDIs. The study collected a list of 1242 substances (8368 drug names) and 27 246 DDIs and matched the downloaded mentions in Twitter with the initial drug names. After a filtering process, 99 485 tweets were retained for analysis.The authors downloaded in a period of 3 weeks, 1 456 961 mentions of drugs that matched with 946 drugs and 2406 drug names previously collected. The authors used time series to analyze coevolution of drug mentions and evaluated the correspondency with a set of collected known DDIs. Another example of applicability of Twitter in the identification of DDIs is provided by Hamed et al. [125]. They computed associations between keywords in the same tweet and associations between keywords and hashtags also co-occurring in the same tweet for the construction of association networks. The study developed a new network mining algorithm to detect connections between pairs of drugs. The mentioned studies demonstrate that Twitter is a promising tool to monitor the effects of drugs in the population, including effects caused by DDIs.
Other platforms such as Facebook or Instagram can also offer great opportunities to analyze drug effects. A study was recently published that identifies DDIs and ADEs in Instagram, a platform with >300 million users. Correia et al. [117] analyzed Instagram focusing on drugs that treat depression. They collected a database of posts from Instagram users (complete timelines contain 5 329 720 posts for 6927 users) from 2010 to 2015 to study associations between drugs that treat depression and symptoms. Four different dictionaries, including drugs, pharmacology and adverse effect terminologies, were used to collect the data. The analysis was carried out using co-mentions of drugs in three periods, monthly, weekly and in daily basis. The posts were tagged with the terms in the dictionaries to find matches, and symmetric cooccurrence graphs for the different time windows were computed. Proximity graphs were used to obtain associations between sets of terms and measure the probability that two terms are mentioned in the same period. Their study showed the potential of Instagram and complex network analysis as an important tool to monitor public health care using large-scale quantitative analysis. Instagram users discuss health problems, including diagnosis, drugs prescribed and effects caused by drugs through photos and comments. This is an important and alternative source of valuable information that can be decodified through data analysis to explore new effects caused by drugs.
Nevertheless, besides health care information retrieved from social media platforms, Internet and specific Web sites can be useful to monitor adverse effects. White et al. [126] showed that the search logs provided by Internet users have potential to study ADEs. Moreover, there are health care social media online sites, such as MedHelp [127] or DailyStrength [128], that are useful to study drug outcomes. Consumers and professionals use health care Web sites to provide or seek information. In fact, millions of people use MedHelp every month to find medical answers to their problems. Some of these Web sites could be a good alternative to administrative systems to report adverse effects caused by drugs and DDIs. Different studies showed that health care Web sites provide reliable data to detect ADEs [129, 130] and DDIs [131–133]. A study published in 2013 proposed the detection of DDIs through association mining of online health Web sites [131]. The authors analyzed three DDIs and used measurements such as leverage, lift and interaction ratio. Later, the authors proposed the detection of DDIs by mining a heterogeneous network extracted from health care Web sites [132]. After calculating topological features to the network, a logistic regression model was developed for DDI identification. Combination of weighted heterogeneous network from health care Web sites with supervised learning techniques was also used by the same authors to explore DDIs [133]. Taking advantage of these sources could be important to discover medical knowledge and provide more evidences of health-related problems.
Social media offers new possibilities to analyze the information provided by million of users. For this reason, Web sites and platforms are currently alternative sources for the study of drugs’ actions, including adverse effects caused by DDIs. In the literature, there are many examples of the potential of social media in the identification of ADEs. However, the examples of detection of DDIs using social media are more limited. More studies are necessary to really prove and understand the potential of social media resources and their role in pharmacovigilance.
Challenges in DDI detection using data mining
Detection of DDIs in developmental drug stages and postmarketing surveillance is challenging. Although FDA is interested in assessing DDIs in development stages and clinical trials [134], it is not feasible to evaluate all possible drug combinations. Many potential DDIs go undetected, and for this reason, improvement in the methods is necessary to provide a better assistance to the patients. Data mining DDIs can provide a useful benchmark to detect novel DDIs. However, information about drugs has to be available in the clinical or scientific literature data, which means that data mining has limited applications in the discovery of new DDIs for experimental drugs still in the discovery and development processes and with no available data in the different sources. Moreover, new DDI safety signals detected by data mining efforts constitute novel hypotheses that need further experimental evaluation or surveillance studies to establish more evidences that corroborate the discovery.
A crucial step in the recognition of DDIs is the normalization of drug names and development of standardized vocabularies [135]. Tools such as RxNorm [136] provided by the National Library of Medicine has become important to support operability between drug vocabularies. RxNorm proposes standard and multipurpose terminology for the representation and identification of drugs. The tool generates normalized names for clinical drugs and also links the names with other medication vocabularies. The idea is to group equivalent terms and codes from different vocabularies into a RxNorm Concept Unique Identifier. Terminologies that code drug properties, such as the National Drug File-Reference Terminology [137], are also integrated in RxNorm. There are related tools useful in drug standardization such as RxTerms, RxNav or MyMedicationList [135]. Our research group has also used NLP systems, such as MedLEE [138], to map drugs to UMLS codes and then RxNorm to obtain generic drug names [57, 139]. Improvements in the normalization of drug names and clinical entities are crucial to correctly identify drugs that can be implicated in DDIs.
Pharmacovigilance data including observational health care databases, such as FAERS, and EHRs are valuable sources to study possible ADEs in the population [1, 140]. However, there are also some challenges in SRS as FAERS that limit their application, such as reporting bias or sampling variance [49, 141, 142]. Methodologies using data as FAERS are based on direct evidence between an adverse effect and the exposure of drugs, and many unexpected events or associations can be unreported [53]. Low reporting numbers could artificially increase the risk estimates for drugs causing false positives. Although some methods try to reduce the false-positive fraction, their capacity to detect adverse effects is at the same time compromised [53]. Besides underreporting, other phenomena include overreporting or reporting duplicity, limited demographic data or failing to provide the number of patients treated with a particular drug [142, 143]. There are also some reports that include a high number of drugs where spurious associations are more likely to be generated. Those facts have influence in the detection of adverse effects caused by drug combinations.
On the other hand, data in EHRs allow real-time surveillance that can transform health care [144]. EHRs are useful to evaluate and improve many processes important for the hospital functionality, to make better decisions and provide more coordinated health care [145]. However, extraction and analysis of adverse effects and DDIs have to be adapted to overcome some limitations. Data from EHRs are complex, and many records provide unstructured narrative [146]. In some cases, the lack of accuracy or even missing data further complicate the analysis. Moreover, confounding factors are an important and challenging problem when using EHRs [147]. Besides associations between drugs as the cause of the adverse effects, indirect associations or confounding associations can be obtained from other events. Signals can be confounded by co-medication, indication and comorbidities. Patients at the hospital often take multiple drugs and have different comorbidities, and this fact complicates the analysis and leads to more confounding. Another limitation is sparse data, and in some cases, a larger number of patients taking both drugs is needed to generate robust signals [148]. Ontology problems and heterogeneity of the information can limit also the analysis and the reproducibility of the results. More precise vocabularies defining concepts, such as drugs, diseases or processes, could help in the analysis and validation. Other limitations in EHRs include restricted public access to medical records to ensure patient privacy [149] and need for external experts to evaluate the importance of the signals in the analysis. Because of these limitations, pharmacovigilance methods are subject to an important number of false-positive cases that reduce their applicability and limit their success [147]. In this sense, initiatives such as OHDSI [150] and the common data model, where drug–condition information is standardized, could facilitate access to large volumes of data and methods, and improve quality, accuracy and reproducibility in data mining of pharmacovigilance resources.
Methods that extract DDIs from the scientific literature showed good performance in different extraction challenges [97, 103]. Co-ocurrence-based methods yield good recall, but the precision is low with many false positives. The precision is improved with rule-based methods, although limitations emerge with complex sentences. Machine learning approaches showed the best performance [88]. However, machine learning needs training with big learning sets, and well-annotated corpora are important for their success. One of the limitations of training models in the detection of DDIs is the absence of true negative examples, i.e. drug pairs that are known to do not interact. In many occasions, the non-DDIs are extracted from drug pairs when the interaction is not described [113, 114]. Moreover, analysis and detection of DDIs in the biomedical texts can be challenging because of the nature of the unstructured data.
Exponential growth of the scientific literature makes difficult the analysis of such amount of data [151]. Computational tools have great applicability to help in the extraction, analysis and classification of the biological information described in all the publications. In fact, during the past years, there have been important advances in biomedical language processing [151]. However, development and improvement of methods to analyze the great amount of information are still an important objective in research- and literature-based discovery. The National Center for Biotechnology Information is constantly improving the PubMed Web service. Moreover, different Web tools that provide literature search service have been developed [152] to help users to make efficient searches and collect relevant data.
Although there are multiple resources of DDIs that can be useful as knowledge databases or gold standards to evaluate methods’ performance, i.e. DrugBank [153], Micromedex [154] and DDI Corpus [97], they showed low consistency and overlapping between them [110]. As it has been discussed previously, improvements in pharmacokinetic ontology and well-annotated corpora along with unified criteria for gold standards could be a crucial step to develop more realistic models to detect novel DDIs. Although we have shown multiple studies demonstrating the applicability of using different sources of data to validate the DDIs, it is also necessary to be cautious when comparing and implementing diverse data sets with different bias and limitations.
Besides pharmacovigilance sources, scientific literature and social media, there are other sources with potential to explore DDIs, such as biomedical images [155]. In the past years, there has been an increasing interest about extracting biomedical information from images. In fact, there are public data sources containing biomedical image data sets extracted from a variety of research projects that can be analyzed from the point of view of data mining. For this reason, there is a need for the development of retrieval systems and platforms that facilitate the extraction of the information from images [155]. There are different technologies with applicability in data mining of biomedical figures: approaches to analyze heterogeneous images or approaches focused on domain-specific images. A more complete description and comparison of the different technologies, development state, accuracies, advantages and limitations have already been discussed in the literature [155]. There are some examples about extraction of text and image based information to study protein-protein interactions [155]. Although no implications in DDIs have been reported, extraction of information through these technologies applied to images could provide important insights for the analysis of DDIs.
Integrative approaches that combine data from the different described resources, including pharmacovigilance data, scientific literature, social media or biomedical images, could be helpful toward the development of more robust models to predict DDIs. Moreover, integration of other type of bioinformatics models, based on protein–protein interaction networks [156] or exploiting similarity between DDIs [157], could represent an excellent strategy to take advantage of all possible resources in the detection of DDIs.
Conclusions
DDIs are the cause of many adverse effects in the patients, and, hence, they have an important impact in health care. Improvements in systems related to clinical decision-making are necessary to decrease the negative impact caused by DDIs in the clinical practice. On the other hand, improvements in the methods used to detect novel DDIs are also needed to discover interactions that are causing harm to the population earlier. Data mining of pharmacovigilance data, such as SRS or EHRs, have shown potential to prove the impact of DDIs as a cause of adverse effects as well as in the detection of novel DDIs. Moreover, data mining can be applied in the scientific literature to extract DDIs and construct powerful knowledge databases useful for the assessment of additional methods. As it was shown by different studies, the biomedical literature can be a source to detect novel DDIs through data mining. There are also examples of the potential of social media as a promising resource for the study of drugs’ actions, including adverse effects caused by DDIs. Although the multiple sources to study DDIs through data mining present some challenges and limitations, they have shown real applicability to study multiple drug combination effects from different perspectives.
Key Points
Methods to discover and follow-up DDIs are a primary aim of drug safety researchers.
The study of DDIs through data mining can be applied in pharmacovigilance sources, the scientific biomedical literature and social media.
Data mining has important applications in the analysis of DDIs: showing the impact in the generation of adverse effects, creating knowledge data sets and gold standards and discovering novel DDIs.
Funding
Discovering and Applying Knowledge in Clinical Databases (grant number R01 LM006910 to G.H.), Pharmacovigilance using Natural Language Processing, Statistics, and the EHR (grant number R01 LM010016 to C.F.). S.V. also thanks `Plan Galego de Investigación, Innovación e Crecemento 2011-2015; Angeles Alvariño program from Xunta de Galicia' and European Social Fund.
Santiago Vilar is a Scientist at the Department of Biomedical Informatics at Columbia University (New York) and the Department of Organic Chemistry at University of Santiago de Compostela (Spain). He is interested in drug design and repurposing, molecular modeling and cheminformatics.
Carol Friedman is a Professor and Graduate Program Director at the Department of Biomedical Informatics at Columbia University. She is interested in natural language processing in the biomedical domain. She developed MedLEE, a natural language extraction system for the clinical field.
George Hripcsak is the Chair of the Department of Biomedical Informatics at Columbia University (New York). He is interested in the storage of clinical information in electronic health records and in the development of next-generation health record systems.
References
- 1. Percha B, Altman RB.. Informatics confronts drug-drug interactions. Trends Pharmacol Sci 2013;34:178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Becker ML, Kallewaard M, Caspers PWJ, et al.Hospitalisations and emergency department visits due to drug-drug interactions: a literature review. Pharmacoepidemiol Drug Saf 2007;16:641–51. [DOI] [PubMed] [Google Scholar]
- 3. Aronson JK. Classifying drug interactions. Br J Clin Pharmacol 2004;58:343–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palleria C, Di Paolo A, Giofrè C, et al.Pharmacokinetic drug-drug interaction and their implication in clinical management. J Res Med Sci 2013;18:601–10. [PMC free article] [PubMed] [Google Scholar]
- 5. Hinder M. Pharmacodynamic drug–drug interactions In: Vogel HG, J Maas, A Gebauer (eds). Drug Discovery and Evaluation: Methods in Clinical Pharmacology. Springer-Verlag Berlin Heidelberg, Germany; 2011, 367–76. [Google Scholar]
- 6. Lazarou J, Pomeranz BH, Corey PN.. Incidence of adverse drug reactions in hospitalized patients—a meta-analysis of prospective studies. JAMA 1998;279:1200–5. [DOI] [PubMed] [Google Scholar]
- 7. Pirmohamed D, Orrne ML.. Drug interactions of clinical importance In: D Davies, RE Ferner, H de Glanville (eds). Davies’s Textbook of Adverse Drug Reactions. London: Chapman & Hall Medical, 1998, 888–912. [Google Scholar]
- 8. Hohl CM, Dankoff J, Colacone A, et al.Polypharmacy, adverse drug-related events, and potential adverse drug interactions in elderly patients presenting to an emergency department. Ann Emerg Med 2001;38:666–71. [DOI] [PubMed] [Google Scholar]
- 9. Smets HLE, De Haes JFF, De Swaef A, et al.Exposure of the elderly to potential nephrotoxic drug combinations in Belgium. Pharmacoepidemiol Drug Saf 2008;17:1014–19. [DOI] [PubMed] [Google Scholar]
- 10. Triplitt C. Drug interactions of medications commonly used in diabetes. Diabetes Spectr 2006;19:202–11. [Google Scholar]
- 11. Staffa JA, Chang J, Green L.. Cerivastatin and reports of fatal rhabdomyolysis. N Engl J Med 2002;346:539–40. [DOI] [PubMed] [Google Scholar]
- 12. Furberg CD, Pitt B.. Withdrawal of cerivastatin from the world market. Curr Control Trials Cardiovasc Med 2001;2:205–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang JS, Neuvonen M, Wen X, et al.Gemfibrozil inhibits CYP2C8-mediated cerivastatin metabolism in human liver microsomes. Drug Metab Dispos 2002;30:1352–6. [DOI] [PubMed] [Google Scholar]
- 14. Backman JT, Kyrklund C, Neuvonen M, et al.Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clin Pharmacol Ther 2002;72:685–91. [DOI] [PubMed] [Google Scholar]
- 15. FDA. U.S. Food and Drug Administration, 2016. http://www.fda.gov/ (30 June 2016, date last accessed).
- 16. Billups SJ, Carter BL.. Mibefradil withdrawn from the market. Ann Pharmacother 1998;32:841–841. [DOI] [PubMed] [Google Scholar]
- 17. Teixeira JJ, Crozatti MT, dos Santos CA, Romano-Lieber NS.. Potential drug-drug interactions in prescriptions to patients over 45 years of age in primary care, Southern Brazil. PLoS One 2012;7:e47062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doubova SV, Reyes-Morales H, Torres-Arreola LDP, et al.Potential drug-drug and drug-disease interactions in prescriptions for ambulatory patients over 50 years of age in family medicine clinics in Mexico City. BMC Health Serv Res 2007;7:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Becker ML, Kallewaard M, Caspers PWJ, et al.Potential determinants of drug-drug interaction associated dispensing in community pharmacies. Drug Saf 2005;28:371–8. [DOI] [PubMed] [Google Scholar]
- 20. Bucher HC, Achermann R, Stohler N, et al.Surveillance of physicians causing potential drug-drug interactions in ambulatory care: a pilot study in Switzerland. PLoS One 2016;11:e0147606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greenberg M, Ridgely MS.. Clinical decision support and malpractice risk. JAMA 2011;306:90–1. [DOI] [PubMed] [Google Scholar]
- 22. Mille F, Schwartz C, Brion F, et al.Analysis of overridden alerts in a drug-drug interaction detection system. Int J Qual Health Care 2008;20:400–5. [DOI] [PubMed] [Google Scholar]
- 23. Tilson H, Hines LE, McEvoy G, et al.Recommendations for selecting drug-drug interactions for clinical decision support. Am J Health Syst Pharm 2016;73:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magro L, Moretti U, Leone R.. Epidemiology and characteristics of adverse drug reactions caused by drug-drug interactions. Expert Opin Drug Saf 2012;11:83–94. [DOI] [PubMed] [Google Scholar]
- 25. Bezabeh S, Mackey AC, Kluetz P, et al.Accumulating evidence for a drug-drug interaction between methotrexate and proton pump inhibitors. Oncologist 2012;17:550–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cerrito P. Application of data mining for examining polypharmacy and adverse effects in cardiology patients. Cardiovasc Toxicol 2001;1:177–9. [DOI] [PubMed] [Google Scholar]
- 27. Hammann F, Drewe J.. Data mining for potential adverse drug-drug interactions. Expert Opin Drug Metab Toxicol 2014;10:665–71. [DOI] [PubMed] [Google Scholar]
- 28. Vilar S, Uriarte E, Santana L, et al.State of the art and development of a drug-drug interaction large scale predictor based on 3D pharmacophoric similarity. Curr Drug Metab 2014;15:490–501. [DOI] [PubMed] [Google Scholar]
- 29. Zhang L, Zhang YC, Zhao P, et al.Predicting drug-drug interactions: an FDA perspective. Aaps J 2009;11:300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fowler S, Zhang H.. In vitro evaluation of reversible and irreversible cytochrome P450 inhibition: current status on methodologies and their utility for predicting drug-drug interactions. Aaps J 2008;10:410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marathe PH, Rodrigues AD.. In vivo animal models for investigating potential CYP3A-and Pgp-mediated drug-drug interactions. Curr Drug Metab 2006;7:687–704. [DOI] [PubMed] [Google Scholar]
- 32. Ejim L, Farha MA, Falconer SB, et al.Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 2011;7:348–50. [DOI] [PubMed] [Google Scholar]
- 33. Sun X, Vilar S, Tatonetti NP.. High-throughput methods for combinatorial drug discovery. Sci Trans Med 2013;5:205rv1.. [DOI] [PubMed] [Google Scholar]
- 34. Montastruc F, Sommet A, Bondon-Guitton E, et al.The importance of drug-drug interactions as a cause of adverse drug reactions: a pharmacovigilance study of serotoninergic reuptake inhibitors in France. Eur J Clin Pharmacol 2012;68:767–75. [DOI] [PubMed] [Google Scholar]
- 35. Fournier J-P, Sommet A, Durrieu G, et al.Drug interactions between antihypertensive drugs and non-steroidal anti-inflammatory agents: a descriptive study using the French pharmacovigilance database. Fundam Clin Pharmacol 2014;28:230–5. [DOI] [PubMed] [Google Scholar]
- 36. Lapi F, Azoulay L, Yin H, et al.Concurrent use of diuretics, angiotensin converting enzyme inhibitors, and angiotensin receptor blockers with non-steroidal anti-inflammatory drugs and risk of acute kidney injury: nested case-control study. BMJ 2013;346:e8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tavassoli N, Sommet A, Lapeyre-Mestre M, et al.Drug interactions with cholinesterase inhibitors—an analysis of the French pharmacovigilance database and a comparison of two national drug formularies (Vidal, British National Formulary). Drug Saf 2007;30:1063–71. [DOI] [PubMed] [Google Scholar]
- 38. Leone R, Magro L, Moretti L, et al.Identifying adverse drug reactions associated with drug-drug interactions data mining of a spontaneous reporting database in Italy. Drug Saf 2010;33:667–75. [DOI] [PubMed] [Google Scholar]
- 39. Schelleman H, Brensinger CM, Bilker WB, et al.Antidepressant-warfarin interaction and associated gastrointestinal bleeding risk in a case-control study. PLoS One 2011;6:e21447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hennessy S, Carson JL, Ray WA, Strom BL.. Pharmacoepidemiology: Medicaid databases In: BL Strom. (ed). Pharmacoepidemiology, 4th edn Chichester: John Wiley and Sons, 2005, 281–94. [Google Scholar]
- 41. Mirosevic Skvrce N, Macolic Sarinic V, Mucalo I, et al.Adverse drug reactions caused by drug-drug interactions reported to Croatian Agency for Medicinal Products and Medical Devices: a retrospective observational study. Croat Med J 2011;52:604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Agency for Medicinal Products and Medical Devices of Croatia (HALMED), 2016. http://www.halmed.hr/ (30 June 2016, date last accessed).
- 43. Ishiguro C, Fujita T, Omori T, et al.Assessing the effects of non-steroidal anti-inflammatory drugs on anti hypertensive drug therapy using post-marketing surveillance database. J Epidemiol 2008;18:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dechanont S, Maphanta S, Butthum B, et al.Hospital admissions/visits associated with drug-drug interactions: a systematic review and meta-analysis. Pharmacoepidemiol Drug Saf 2014;23:489–97. [DOI] [PubMed] [Google Scholar]
- 45. FDA Adverse Event Reporting System (FAERS). FDA U.S. Food and Drug Administration, 2016. http://www.fda.gov/cder/aers/default.htm (30 June 2016, date last accessed).
- 46. Van Puijenbroek EP, Bate A, Leufkens HG, et al.A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol Drug Saf 2002;11:3–10. [DOI] [PubMed] [Google Scholar]
- 47. DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. Am Stat 1999;53:177–90. [Google Scholar]
- 48. Harpaz R, DuMouchel W, LePendu P, et al.Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther 2013;93:539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hauben M, Madigan D, Gerrits CM, et al.The role of data mining in pharmacovigilance. Expert Opin Drug Saf 2005;4:929–48. [DOI] [PubMed] [Google Scholar]
- 50. Harpaz R, DuMouchel W, Shah NH, et al.Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther 2012;91:1010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heba I, Amany A, Ahmed SE, et al.Novel data-mining methodologies for detecting drug-drug interactions: a review of pharmacovigilance literature. Adv Environ Sci Dev Chem 2014:301–14. [Google Scholar]
- 52. Uppsala Monitoring Centre (UMC). WHO Global ICSR database, 2016. http://www.who-umc.org/ (30 June 2016, date last accessed).
- 53. Tatonetti NP, Fernald GH, Altman RB.. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inf Assoc 2012;19:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duke JD, Han X, Wang Z, et al.Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. Plos Comput Biol 2012;8:e1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Birman-Deych E, Waterman AD, Yan Y, et al.Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care 2005;43:480–5. [DOI] [PubMed] [Google Scholar]
- 56. Xu H, Fu Z, Shah A, et al.Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu Symp Proc 2011;1564–72. [PMC free article] [PubMed] [Google Scholar]
- 57. Vilar S, Harpaz R, Santana L, et al.Enhancing adverse drug event detection in electronic health records using molecular structure similarity: application to pancreatitis. PLoS One 2012;7:e41471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Puijenbroek EP, Egberts ACG, Heerdink ER, et al.Detecting drug-drug interactions using a database for spontaneous adverse drug reactions: an example with diuretics and non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol 2000;56:733–8. [DOI] [PubMed] [Google Scholar]
- 59. Almenoff JS, DuMouchel W, Kindman LA, et al.Disproportionality analysis using empirical Bayes data mining: a tool for the evaluation of drug interactions in the post-marketing setting. Pharmacoepidemiol Drug Saf 2003;12:517–21. [DOI] [PubMed] [Google Scholar]
- 60. DuMouchel W, Pregibon D. Empirical Bayes screening for multi-item associations. In: Proceedings of the Seventh ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, 2001. pp. 67–76. ACM New York, NY.
- 61. Ellis RJ, Mayo MS, Bodensteiner DM.. Ciprofloxacin-warfarin coagulopathy: a case series. Am J Hematol 2000;63:28–31. [DOI] [PubMed] [Google Scholar]
- 62. Van Puijenbroek EP, Egberts AC, Meyboom RH, et al.Signalling possible drug-drug interactions in a spontaneous reporting system: delay of withdrawal bleeding during concomitant use of oral contraceptives and itraconazole. Br J Clin Pharmacol 1999;47:689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knudsen JF, Sokol GH, Flowers CM.. Adjunctive topiramate enhances the risk of hypothermia associated with valproic acid therapy. J Clin Pharm Ther 2008;33:513–19. [DOI] [PubMed] [Google Scholar]
- 64. Thakrar BT, Grundschober SB, Doessegger L.. Detecting signals of drug-drug interactions in a spontaneous reports database. Br J Clin Pharmacol 2007;64:489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tatonetti NP, Denny JC, Murphy SN, et al.Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lorberbaum T, Sampson KJ, Woosley RL, et al.An integrative data science pipeline to identify novel drug interactions that prolong the QT interval. Drug Saf 2016;39:433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lorberbaum T, Sampson KJ, Chang JB, et al.Coupling data mining and laboratory experiments to discover drug interactions causing QT prolongation. J Am Coll Cardiol 2016;68:1756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Noren GN, Sundberg R, Bate A, et al.A statistical methodology for drug-drug interaction surveillance. Stat Med 2008;27:3057–70. [DOI] [PubMed] [Google Scholar]
- 69. Suzuki A, Yuen NA, Ilic K, et al.Comedications alter drug-induced liver injury reporting frequency: data mining in the WHO VigiBase (TM). Regul Toxicol Pharmacol 2015;72:481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Medical Dictionary for Regulatory Activities (MedDRA), 2016. http://www.meddra.org/ (15 June 2016, date last accessed)
- 71. Hung IFN, Wu AKL, Cheng VCC, et al.Fatal interaction between clarithromycin and colchicine in patients with renal insufficiency: a retrospective study. Clin Infect Dis 2005;41:291–300. [DOI] [PubMed] [Google Scholar]
- 72. Poizot-Martin I, Naqvi A, Obry-Roguet V, et al.Potential for drug-drug interactions between antiretrovirals and HCV direct acting antivirals in a large cohort of HIV/HCV coinfected patients. PLoS One 2015;10:e0141164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iyer SV, Harpaz R, LePendu P, et al.Mining clinical text for signals of adverse drug-drug interactions. J Am Med Inform Assoc 2014;21:353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Girardeau Y, Trivin C, Durieux P, et al.Detection of drug-drug interactions inducing acute kidney injury by electronic health records mining. Drug Saf 2015;38:799–809. [DOI] [PubMed] [Google Scholar]
- 75. Pathak J, Kiefer RC, Chute CG.. Using linked data for mining drug-drug interactions in electronic health records. Stud Health Technol Inform 2013;192:682–6. [PMC free article] [PubMed] [Google Scholar]
- 76. Jiang G, Liu H, Solbrig HR, et al.Mining severe drug-drug interaction adverse events using Semantic Web technologies: a case study. BioData Min 2015;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ji Y, Ying H, Tran J, et al.A functional temporal association mining approach for screening potential drug-drug interactionsfrom electronic patient databases. Inform Health Soc Care 2016;41:387–404. [DOI] [PubMed] [Google Scholar]
- 78. Banda JM, Callahan A, Winnenburg R, et al.Feasibility of prioritizing drug-drug-event associations found in electronic health records. Drug Saf 2016;39:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vilar S, Lorberbaum T, Hripcsak G, et al.Improving detection of arrhythmia drug-drug interactions in pharmacovigilance data through the implementation of similarity-based modeling. PLoS One 2015;10:e0129974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Du L, Chakraborty A, Chiang CW, et al.Graphic mining of high-order drug interactions and their directional effects on myopathy using electronic medical records. CPT Pharmacometrics Syst Pharmacol 2015;4:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang P, Du L, Wang L, et al.A mixture dose-response model for identifying high-dimensional drug interaction effects on myopathy using electronic medical record databases. CPT Pharmacometrics Syst Pharmacol 2015;4:474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Harpaz R, Chase HS, Friedman C.. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics 2010;11(Suppl 9):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Harpaz R, Haerian K, Chase HS, et al.Statistical mining of potential drug interaction adverse effects in FDA's spontaneous reporting system. AMIA Annu Symp Proc 2010;281–5. [PMC free article] [PubMed] [Google Scholar]
- 84. Xiang Y, Albin A, Ren K, et al.Efficiently mining adverse event reporting system for multiple drug interactions. AMIA Jt Summits Transl Sci Proc 2014;120–5. [PMC free article] [PubMed] [Google Scholar]
- 85. Hampton T. Data mining approach shows promise in detecting unexpected drug interactions. JAMA 2011;306:144. [DOI] [PubMed] [Google Scholar]
- 86. Zweigenbaum P, Demner-Fushman D, Yu H, et al.Frontiers of biomedical text mining: current progress. Brief Bioinform 2007;8:358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Segura-Bedmar I, Crespo M, de Pablo-Sanchez C, et al.Resolving anaphoras for the extraction of drug-drug interactions in pharmacological documents. BMC Bioinformatics 2010;11(Suppl 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wu H-Y, Chiang C-W, Li L.. Text mining for drug-drug interaction. Methods Mol Biol 2014;1159:47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Segura-Bedmar I, Martinez P, de Pablo-Sanchez C.. A linguistic rule-based approach to extract drug-drug interactions from pharmacological documents. BMC Bioinformatics 2011;12(Suppl 2):S1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Giuliano C, Lavelli A, Romano L. Exploiting shallow linguistic information for relation extraction from biomedical literature. In: Proceedings European Chapter of the ACL, Trento, Italy, 2006. pp. 401–8. Association for Computational Linguistics.
- 91. Segura-Bedmar I, Martinez P, de Pablo-Sanchez C.. Using a shallow linguistic kernel for drug-drug interaction extraction. J Biomed Inform 2011;44:789–804. [DOI] [PubMed] [Google Scholar]
- 92. Bui Q-C, Sloot PMA, van Mulligen EM, et al.A novel feature-based approach to extract drug-drug interactions from biomedical text. Bioinformatics 2014;30:3365–71. [DOI] [PubMed] [Google Scholar]
- 93. Chowdhury M, Abacha A, Lavelli A, et al. Two different machine learning techniques for drug-drug interaction extraction. In: Proceedings of DDI Extraction-2011 Challenge Task, Huelva, Spain, 2011. pp. 19-26. The 1st DDI Extraction-2011 challenge task.
- 94. Thomas M, Neves M, Solt I, et al. Relation Extraction for Drug-Drug Interactions using Ensemble Learning. In: Proceedings of DDI Extraction-2011 Challenge Task, Huelva, Spain, 2011. pp. 11–18. The 1st DDI Extraction-2011 challenge task.
- 95. Shengyu L, Buzhou T, Qingcai C, et al.Drug-drug interaction extraction via convolutional neural networks. Comput Math Methods Med 2016;2016:6918381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. He L, Yang Z, Zhao Z, et al.Extracting drug-drug interaction from the biomedical literature using a stacked generalization-based approach. PLoS One 2013;8:e65814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Segura-Bedmar I, Martınez P, Sanchez-Cisneros D. The 1st DDI extraction-2011 challenge task: extraction of drug-drug interactions from biomedical texts. In: Proceedings DDI Extraction-2011 Challenge Task, Huelva, Spain, 2011. pp. 1–9. The 1st DDI Extraction-2011 challenge task.
- 98. Ben Abacha A, Chowdhury MFM, Karanasiou A, et al.Text mining for pharmacovigilance: using machine learning for drug name recognition and drug-drug interaction extraction and classification. J Biomed Inform 2015;58:122–32. [DOI] [PubMed] [Google Scholar]
- 99. Björne J, Airola A, Pahikkala T, et al. Drug-drug interaction extraction from biomedical texts with SVM and RLS classifiers. In: Proceedings DDI Extraction-2011 Challenge Task, Huelva, Spain, 2011. pp. 35-42. The 1st DDI Extraction-2011 challenge task.
- 100. Minard A, Makour L, Ligozat A, et al. Feature selection for drug-drug interaction detection using machine-learning based approaches. In: Proceeding DDI Extraction-2011 Challenge Task, Huelva, Spain, 2011. pp. 43–50. The 1st DDI Extraction-2011 challenge task.
- 101. Zhang Y, Lin H, Yang Z, et al.A single kernel-based approach to extract drug-drug interactions from biomedical literature. PLoS One 2012;7:e48901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kim S, Liu H, Yeganova L, et al.Extracting drug-drug interactions from literature using a rich feature-based linear kernel approach. J Biomed Inform 2015;55:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Segura-Bedmar I, Martıinez P, Herrero-Zazo M. SemEval-2013 Task 9: extraction of drug–drug interactions from biomedical texts. In: Proceedings of the 7th International Workshop on Semantic Evaluation (SemEval 2013), Atlanta, Georgia, USA, 2013. pp. 341–350. Association for Computational Linguistics.
- 104. Herrero-Zazo M, Segura-Bedmar I, Martínez P, et al.The DDI corpus: an annotated corpus with pharmacological substances and drug-drug interactions. J Biomed Inform 2013;46:914–20. [DOI] [PubMed] [Google Scholar]
- 105. Segura-Bedmar I, Martínez P, Herrero-Zazo M.. Lessons learnt from the DDIExtraction-2013 shared task. J Biomed Inform 2014;51:152–64. [DOI] [PubMed] [Google Scholar]
- 106. Herrero-Zazo M, Segura-Bedmar I, Hastings J, et al.DINTO: using OWL ontologies and SWRL rules to infer drug-drug interactions and their mechanisms. J Chem Inf Model 2015;55:1698–707. [DOI] [PubMed] [Google Scholar]
- 107. Brochhausen M, Schneider J, Malone D, et al. Towards a foundational representation of potential drug-drug interaction knowledge. In: The 1st International Drug-Drug Interaction Knowledge Representation Workshop (DIKR 2014). Collocated with the 2014 International Conference on Biomedical Ontology, ICBO, 2014. Houston, Texas. CEUR-WS. [PMC free article] [PubMed]
- 108.A Minimum Information Model for Representing Potential Drug-Drug Interaction Knowledge and Evidence, 2016. http://dbmi-icode-01.dbmi.pitt.edu/dikb-evidence/w3c-ddi/index.html (15 December 2016, date last accessed).
- 109. Wu H-Y, Karnik S, Subhadarshini A, et al.An integrated pharmacokinetics ontology and corpus for text mining. BMC Bioinformatics 2013;14:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ayvaz S, Horn J, Hassanzadeh O, et al.Toward a complete dataset of drug-drug interaction information from publicly available sources. J Biomed Inform 2015;55:206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wong C-M, Ko Y, Chan A.. Clinically significant drug-drug interactions between oral anticancer agents and nonanticancer agents: profiling and comparison of two drug compendia. Ann Pharmacother 2008;42:1737–48. [DOI] [PubMed] [Google Scholar]
- 112. Tari L, Anwar S, Liang S, et al.Discovering drug-drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics 2010;26:i547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Percha B, Garten Y, Altman RB.. Discovery and explanation of drug-drug interactions via text mining. Pac Symp Biocomput 2012;410–21. [PMC free article] [PubMed] [Google Scholar]
- 114. Yan S, Jiang X, Chen Y. Text mining driven drug-drug interaction detection. In: Proceedings IEEE Int Conf Bioinformatics Biomed, 2013. pp. 349–55. IEEE Conference Publications. [DOI] [PMC free article] [PubMed]
- 115. Kolchinsky A, Lourenco A, Li L, et al.Evaluation of linear classifiers on articles containing pharmacokinetic evidence of drug-drug interactions. Pac Symp Biocomput 2013;409–20. [DOI] [PubMed] [Google Scholar]
- 116. Kolchinsky A, Lourenco A, Wu H-Y, et al.Extraction of pharmacokinetic evidence of drug-drug interactions from the literature. PLoS One 2015;10:e0122199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Correia RB, Li L, Rocha LM.. Monitoring potential drug interactions and reactions via network analysis of Instagram user timeliness. Pac Symp Biocomput 2016;21:492–503. [PMC free article] [PubMed] [Google Scholar]
- 118. Sarker A, Gonzalez G.. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J Biomed Inform 2015;53:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ginn R, Pimpalkhute P, Nikfarjam A, et al. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark. In: Fourth Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing. BioTxtM, Reykjavik, Iceland, 2014. LREC 2014 Workshop.
- 120. Jiang K, Zheng Y.. Mining Twitter data for potential drug effects. Advanced Data Mining and Applications 2013;8346:434–43. [Google Scholar]
- 121. Paul MJ, Dredze M. You are what you tweet: analyzing Twitter for public health. In: Proceedings of the Fifth International AAAI Conference on Weblogs and Social Media, Barcelona, Spain, 2011. pp. 265–72. AAAI Press, Menlo Park, CA.
- 122. Chew C, Eysenbach G.. Pandemics in the age of Twitter: content analysis of tweets during the 2009 H1N1 outbreak. PLoS One 2010;5:e14118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chowell G, Nishiura H.. Transmission dynamics and control of Ebola virus disease (EVD): a review. BMC Medicine 2014;12:196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Carbonell P, Mayer MA, Bravo À.. Exploring brand-name drug mentions on Twitter for pharmacovigilance. Stud Health Technol Inform 2015;210:55–9. [PubMed] [Google Scholar]
- 125. Hamed AA, Wu X, Erickson R, et al.Twitter K-H networks in action: advancing biomedical literature for drug search. J Biomed Inform 2015;56:157–68. [DOI] [PubMed] [Google Scholar]
- 126. White RW, Tatonetti NP, Shah NH, et al.Web-scale pharmacovigilance: listening to signals from the crowd. J Am Med Inform Assoc 2013;20:404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. MedHelp. Medical information, forums and communities, 2016. http://www.medhelp.org/(15 December 2016, date last accessed).
- 128. DailyStrength, 2016. https://www.dailystrength.org/ (15 December 2016, date last accessed).
- 129. Yang CC, Jiang L, Yang H, et al. Detecting signals of adverse drug reactions from health consumer contributed content in social media. In: Proceedings of ACM SIGKDD Workshop on Health Informatics, Beijing, China, 2012. ACM Digital Library.
- 130. Yang CC, Yang H, Jiang L, et al. Social media mining for drug safety signal detection. In: Proceedings of ACM CIKM International Workshop on Smart Health and Wellbeing, Maui, Hawaii, 2012. pp. 33--40. ACM, New York, NY.
- 131. Yang H, Yang CC. Harnessing social media for drug-drug interactions detection. In: Proceedings of IEEE International Conference on Healthcare Informatics, 2013. Philadelphia, PA, 2013. IEEE Xplore Digital Library.
- 132. Yang H, Yang CC. Drug-drug interactions detection from online heterogeneous healthcare networks. In: Proceedings of IEEE International Conference on Healthcare Informatics 2014 (ICHI 2014), Verona, Italy, 2014. IEEE Xplore Digital Library.
- 133. Yang H, Yang CC. Mining a weighted heterogeneous network extracted from healthcare-specific social media for identifying interactions between drugs. In: Proceedings of the 6th ICDM Workshop on Biological Data Mining and Its Applications in Healthcare, Atlantic City, NJ, 2015. IEEE International Conference on Data Mining.
- 134. Prueksaritanont T, Chu X, Gibson C, et al.Drug-drug interaction studies: regulatory guidance and an industry perspective. Aaps J 2013;15:629–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nelson SJ, Zeng K, Kilbourne J, et al.Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Unified Medical Language System (UMLS), 2016. https://www.nlm.nih.gov/research/umls/rxnorm/ (15 December 2016, date last accessed).
- 137. Unified Medical Language System (UMLS). 2015AB UMLS national drug file—reference terminology source information, 2016. https://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT/ (15 December 2016, date last accessed).
- 138. Friedman C, Shagina L, Lussier Y, et al.Automated encoding of clinical documents based on natural language processing. J Am Med Inf Assoc 2004;11:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Vilar S, Harpaz R, Chase HS, et al.Facilitating adverse drug event detection in pharmacovigilance databases using molecular structure similarity: application to rhabdomyolysis. J Am Med Inform Assoc 2011;18(Suppl 1):i73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Strandell J, Bate A, Lindquist M, et al.Drug-drug interactions—a preventable patient safety issue? Br J Clin Pharmacol 2008;65:144–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bate A, Evans SJ.. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf 2009;18:427–36. [DOI] [PubMed] [Google Scholar]
- 142. Stephenson WP, Hauben M.. Data mining for signals in spontaneous reporting databases: proceed with caution. Pharmacoepidemiol Drug Saf 2007;16:359–65. [DOI] [PubMed] [Google Scholar]
- 143. Harpaz R, Vilar S, Dumouchel W, et al.Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc 2013;20:413–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Santillana M, Nguyen AT, Louie T, et al.Cloud-based electronic health records for real-time, region-specific influenza surveillance. Sci Rep 2016;6:25732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Birkhead GS, Klompas M, Shah NR.. Uses of electronic health records for public health surveillance to advance public health. Annu Rev Public Health 2015;36:345–59. [DOI] [PubMed] [Google Scholar]
- 146. Johnson SB, Bakken S, Dine D, et al.An electronic health record based on structured narrative. J Am Med Inform Assoc 2008;15:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Li Y, Salmasian H, Vilar S, et al.A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. J Am Med Inform Assoc 2014;21:308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zhao J, Henriksson A, Asker L, et al.Predictive modeling of structured electronic health records for adverse drug event detection. BMC Med Inform Decis Mak 2015;15(Suppl 4):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Menachemi N, Collum TH.. Benefits and drawbacks of electronic health record systems. Risk Manag Healthc Policy 2011;4:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. The Observational Health Data Sciences and Informatics (OHDSI), 2016. http://www.ohdsi.org/ (15 December 2016, date last accessed).
- 151. Hunter L, Cohen KB.. Biomedical language processing: what's beyond PubMed? Mol Cell 2006;21:589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database 2011;2011:baq036.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. DrugBank database, version 4.3, 2016. http://www.drugbank.ca/ (15 June 2016, date last accessed).
- 154. Micromedex ® Healthcare Series. Greenwood Village, CO: Thomson Reuters (Healthcare) Inc, 2016. http://www.micromedexsolutions.com (15 Jun 2016, date last accessed).
- 155. Ahmed Z, Zeeshan S, Dandekar T.. Mining biomedical images towards valuable information retrieval in biomedical and life sciences. Database 2016;2016:baw118. pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Park K, Kim D, Ha S, et al.Predicting pharmacodynamic drug-drug interactions through signaling propagation interference on protein-protein interaction networks. PLoS One 2015;10:e0140816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vilar S, Uriarte E, Santana L, et al.Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat Protoc 2014;9:2147–63. [DOI] [PMC free article] [PubMed] [Google Scholar]