Abstract
STUDY QUESTION
What is the transcriptional network governed by the androgen receptor (AR) in human epididymis epithelial (HEE) cells from the caput region and if the network is tissue-specific, how is this achieved?
SUMMARY ANSWER
About 200 genes are differentially expressed in the caput HEE cells after AR activation; the AR transcriptional network is tissue-specific and may be mediated in part by distinct AR co-factors including CAAT-enhancer binding protein beta (CEBPB) and runt-related transcription factor 1 (RUNX1).
WHAT IS KNOWN ALREADY
Little is known about the AR transcriptional program genome wide in HEE cells, nor its co-factors in those cells. AR has been best studied in the prostate gland epithelium and prostate cancer cell lines, due to the important role of this factor in prostate cancer. However AR-associated differentially expressed genes (DEGs) and AR co-factors have not yet been compared between human epididymis and prostate epithelial cells.
STUDY DESIGN, SIZE, DURATION
Caput HEE cells from two donors were exposed to the synthetic androgen R1881 at 1 nM for 12–16 h after 72 h of hormone starvation.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Chromatin was prepared from R1881-treated and vehicle control HEE cells. AR-associated chromatin was purified by chromatin immunoprecipitation (ChIP) and AR occupancy genome wide was revealed by deep sequencing (ChIP-seq). Two independent biological replicates were performed. Total RNA was prepared from R1881 and control-treated HEE cells and gene expression profiles were documented by RNA-seq. The interaction of the potential novel AR co-factors CEBPB and RUNX1, identified through in-silico motif analysis of AR ChIP-seq data, was examined by ChIP-qPCR after siRNA-mediated depletion of each co-factor individually or simultaneously.
MAIN RESULTS AND THE ROLE OF CHANCE
The results identify about 200 genes that are differentially expressed (DEGs) in HEE cells after AR activation. Some of these DEGs show occupancy of AR at their promoters or cis-regulatory elements suggesting direct regulation. However, there is little overlap in AR-associated DEGs between HEE and prostate epithelial cells. Inspection of over-represented motifs in AR ChIP-seq peaks identified CEBPB and RUNX1 as potential co-factors, with no evidence for FOXA1, which is an important co-factor in the prostate epithelium. CEBPB and RUNX1 ChIP-seq in HEE cells showed that both these factors often occupied AR-binding sites, though rarely simultaneously. Further analysis at a single AR-regulated locus (FK506-binding protein 5, FKPB5) suggests that CEBPB may be a co-activator. These data suggest a novel AR transcriptional network governs differentiated functions of the human epididymis epithelium.
LARGE SCALE DATA
AR ChIP-seq and RNA-seq data are deposited at GEO: GSE109063.
LIMITATIONS, REASONS FOR CAUTION
There is substantial donor-to-donor variation in primary HEE cells cultures. We applied stringent statistical tests with a false discovery rate (FDR) of 0.1% for ChIP-seq and standard pipelines for RNA-seq so it is possible that we have missed some AR-regulated genes that are important in caput epididymis function.
WIDER IMPLICATIONS OF THE FINDINGS
Our data suggest that a novel AR transcriptional network governs differentiated functions of the human epididymis epithelium. Since this cell layer has a critical role in normal sperm maturation, the results are of broader significance in understanding the mechanisms underlying the maintenance of fertility in men.
STUDY FUNDING/COMPETING INTERESTS
This work was funded by the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Development: R01 HD068901 (PI: Harris). The authors have no competing interests to declare.
Keywords: human epididymis epithelial cells; caput epididymis; androgen receptor, hormonal regulation of gene expression; AR cistrome; AR transcriptional network; CEBPB; RUNX1
Introduction
The epididymis is a tightly coiled tubular structure that forms a major part of the ducts through which sperm pass on their way from the testis to fertilize an oocyte. The luminal environment of the human epididymis is critical for normal sperm maturation (reviewed in Cornwall, 2009). Epithelial cells lining the duct tightly regulate the ionic concentration, pH and protein composition of the luminal fluid. The maintenance of this highly defined and complex environment relies on the coordinated expression of many genes encoding cell structural proteins, ion channels and transporters, and secreted proteins (Zhang et al., 2006; Dube et al., 2007; Thimon et al., 2007; Browne et al., 2016b). The transcriptional networks that integrate gene expression in the human epididymis epithelium are not yet fully defined. Among transcription factors (TFs) that are already identified as important regulators of epididymal luminal environment and sperm maturation in rodents are Reproductive homeobox X-linked 5 (Rhox5) (MacLean et al., 2012), Ladybird-like homeobox gene 2 (Lbx2) (Moisan et al., 2008) and Sonic hedgehog (Shh) (Turner et al., 2006). We recently identified hepatocyte nuclear factor 1 beta (HNF1β) as a regulator of genes involved in human epididymis epithelial (HEE) transport of water, phosphate and bicarbonate (Browne et al., 2016a). The potential importance of HNF1β in HEE function was identified first by mapping open chromatin in caput HEE cells, and then by searching for over-represented TF binding motifs in these regions. Sites of open chromatin are suggestive of active cis-regulatory elements (Bischof et al., 2013; Browne et al., 2014b). The enrichment of the androgen receptor (AR) motif in open chromatin regions that are selective to caput HEE cells also suggests a potential role for AR in the biology of this epithelium (Browne et al., 2014b).
The importance of androgens and AR in the epididymis was established many years ago (Gloyna and Wilson, 1969; Blaquier et al., 1972; Robaire et al., 1977; Cohen et al., 1981) (reviewed in Robaire and Hamzeh, 2011). AR protein is detected in all regions of the epididymis and in most of epididymal cell types (Sar et al., 1990; Zhou et al., 2002; Yamashita, 2004). Transcriptional profiling of the epididymis in orchidectomized mice treated with high doses of dihydrotesterone (DHT) was reported (Chauvin and Griswold, 2004; Snyder et al., 2009). Similarly a list of DHT-upregulated and -downregulated genes were documented after orchidectomy in rats (Hamzeh and Robaire, 2010). A direct role for AR in normal epididymal function was demonstrated in knockout mouse models. A caput epididymal epithelium-specific AR knockout mouse showed abnormal epididymis morphology and impaired epithelial cell function and sperm transit (O’Hara et al., 2011). Similar phenotypes were seen in a mouse with targeted inactivation of AR in the proximal epididymis (Krutskikh et al., 2011). Thus most data on the role of AR in the epididymis derive from rodent models, in which the epididymis structure and function is notably different from human. Here we use primary adult HEE cells (Leir et al., 2015) to identify direct transcriptional targets of AR by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). We focus on the caput region of the epididymis since transcriptome profiling of HEE cells revealed that AR is most abundant in this region (Browne et al., 2016b), which is also most relevant to the process of epididymal sperm maturation (Cornwall, 2009). We show the impact of AR activation by synthetic androgens in HEE cells by RNA-seq and identify direct targets of AR by ChIP-seq. We also identify potential co-factors contributing to epididymis specific AR binding and find that these are different from those seen in the prostate epithelium. Our data reveal an AR-mediated transcriptional network, which is critical for normal epididymis function and sperm maturation.
Materials and Methods
Cell culture and immunofluorescence
Human epididymis tissue was obtained with Institutional Review Board approval from patients undergoing inguinal radical orchiectomy for a clinical diagnosis of testicular cancer. None of the epididymides had extension of the testicular cancer. Adult human epididymis epithelial (HEE) cells (caput) were established as described previously (Leir et al., 2015), and grown in CMRL 1066 medium (containing 15% fetal calf serum, 2 mM l-glutamine, 1 μg/ml hydrocortisone, 0.2 U/ml insulin, and 10–10 M cholera toxin (CT)). For experiments on androgen receptor (AR)-mediated pathways, cultures were grown in phenol red-free CMRL 1066 medium containing 15% charcoal stripped FCS without the other supplements described above for at least three days prior to treatment with 1 nM methyltrienolone (R1881; PerkinElmer NLP0050) for 12–16 h. Immunofluorescence detection of AR and TJP1/ZO-1 in HEE monolayers exposed to R1881 or control (ethanol) used standard protocols and the following reagents: Anti-AR (Santa Cruz Biotechnology #sc-816), anti-ZO-1 (Invitrogen #40-2200), Alexa Fluor® 488 AffiniPure Goat Anti-Rabbit IgG (Jackson ImmunoResearch # 111-545-003) and FITC-Conjugated rabbit anti-mouse IgG (Dako #F0313).
Reverse transcription quantitative PCR
Total RNA was extracted from cells with TRIzol (Life Technologies (LT)) following the manufacturers protocols. RT-qPCR was performed by standard protocols. Briefly, cDNA was synthesized by using a TaqMan Reverse Transcription reagents kit (Applied Biosystems) with random hexamers, and qPCR experiments were carried out with SYBR Green master mixes. The sequences of the primer pairs specific for each target gene and the reaction conditions are listed in Supplementary Table SII.
RNA-seq
Total RNA was prepared from four replicates of control (ethanol) or R1881-treated cells. RNA quality was confirmed by Nanodrop measurement of OD 260/280 and 260/230 ratios and Bioanalyzer profiles. RNA-seq libraries were prepared from 2 μg of total RNA using the TruSeq RNA Sample Preparation Kit v2 per the manufacturer’s Low-Throughput protocol (Illumina). The libraries were sequenced on Illumina HiSeq4000 machines. Data were analyzed using TopHat and Cufflinks (Trapnell et al., 2012). All data are deposited at GEO: GSE109063.
ChIP-seq
ChIP-seq experiments for AR, CEBPB and RUNX1 were performed in two biological replicates (donors) after ethanol or R1881 treatment by standard protocols (Fossum et al., 2014). Chromatin was prepared from caput HEE cells as described previously (Browne et al., 2014a). For the immunoprecipitation, 10 μg of AR [Santa Cruz Biotechnology (SCB) sc-815x], CEBPB (sc-150x) or RUNX1 (Abcam, ab23980) antibodies and ~2 × 107 cells were used, and libraries were prepared as described previously (Fossum et al., 2014). Libraries were sequenced on Illumina HiSeq2500 or HiSeq4000 machines. FASTQ files were aligned to the hg19 version of the human genome using Bowtie (Langmead et al., 2009) and ChIP-seq peaks were identified using HOMER (Heinz et al., 2010) with a false discovery rate (FDR) of 0.1%. ChIP-seq data from both replicates were subjected to IDR analysis and only peaks with high confidence (0.1% threshold) were selected for downstream analysis (Fossum et al., 2017). The ChIP-seq datasets are available on GEO (GSE109063). Identification of transcription factor motifs in the data set and peak annotation based on the nearest gene were also performed using HOMER.
ChIP-qPCR
Antibodies were as described for the ChIP-seq experiments above. Chromatin was prepared and ChIP was performed as for ChIP-seq. qPCR was performed by standard protocols (Browne et al., 2016a).
siRNA-mediated depletion of CEBPB and RUNX1
HEE cells at 40-50% confluence were transfected with 40 nM siRNA (Ambion, purchased from ThermoFisher Scientific) as follows: a non-targeting control (Neg Con #1, #4390843), CEBPB (#4392420-s2891), RUNX1, (#4392420-s229352) using Lipofectamine RNAiMax reagent (LT) in OptiMEM. After 24 h, media was replaced with phenol red-free CMRL 1066 medium containing 15% charcoal stripped FCS without the supplements for 24 h prior to treatment with 1 nM R1881 as above for 18 h. Then RNA was extracted using TRIzol® (LT), whole cell lysate was isolated using NET Buffer (Leir et al., 2015), and chromatin was prepared as for ChIP as described above (Browne et al., 2014a). Western blots were probed with antibodies specific for AR (sc-816), CEBPB (sc-7962) and RUNX1 (sc-365644) (from Santa Cruz) and beta-tubulin (T4026, Sigma).
Statistical analysis
GraphPad Prism 6 software was used to analyze statistical differences between experimental groups using students t-test and values of P < 0.05 were considered significant. Data are expressed as mean ± Standard Error of the Mean (SEM).
Results
Genome-wide AR occupancy defined by ChIP-seq
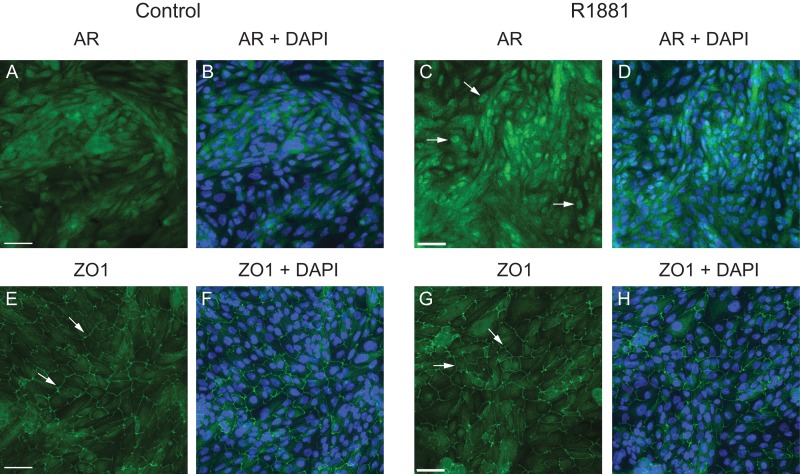
Using an antibody that detects AR specifically, we mapped the genomic locations bound by AR with or without agonist in primary caput HEE cells. The cells were first hormone starved for 72 h before treatment with either 1 nM R1881 (a potent synthetic AR agonist) or vehicle control (ethanol), using a protocol that was optimized in our previous work (Leir et al., 2015). Figure 1 shows relocation of AR into the nucleus after R1881 treatment compared to control (ethanol) and confirms that this protocol has no impact on the structural integrity of the epithelial cell layer, as show by tight junction protein 1 (ZO-1) distribution. After 16 h, cells were harvested for the ChIP-seq experiment. Two independent biological replicates were performed with cells from two individual donors. The sequencing data were aligned by Bowtie 1.0.0 and analyzed with Homer software and IDR (Irreproducible Discovery Rate) analysis was applied to the data in order to identify AR peaks of high confidence.
Figure 1.
Monolayer cultures of primary caput HEE cells treated with R1881 show nuclear relocation of AR and maintenance of epithelial integrity. Immunofluorescence of androgen receptor (AR) (A–D) in primary caput HEE cells treated ethanol (control) or R1881 for 16 h. R1881 treatment induced AR relocation from the cytoplasm to the nucleus (white arrows). 6-Diamino-2-phenylindole (DAPI) was used for nuclear counterstaining. Maintenance of structural integrity of the HEE cell layers after R1881 treatment was shown by ZO-1 staining (E–H; white arrows). Size bar = 50 μM.
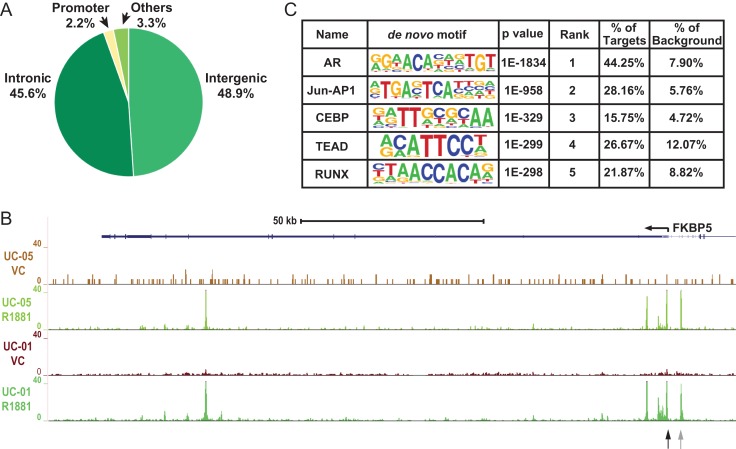
With an IDR threshold of 0.1%, 8748 AR peaks were identified in the R1881 treated cells and only 61 peaks were identified for the vehicle control treated cells. The results indicate that ligand binding of AR is required for its DNA binding activity, and confirmed the specificity of the antibody used for the ChIP experiment. The distribution of AR peaks in the HEE cell genome is shown in Fig. 2A, where 48.9% and 45.6% of peaks are located in intergenic and intronic regions, respectively. The remaining 5.5% of peaks are found in promoters (–1 kb to +100 b with respect to the transcription start sites of genes) or other genomic regions (such as untranslated regions (UTRs) and exons). An example of the ChIP-seq signal enrichment is shown in Fig. 2B at the FKBP5 gene locus, which is a well-characterized AR target in prostate cells (Magee et al., 2006). Specific AR peaks are identified in intron 5 of FKBP5, which is a previously characterized enhancer, as well as at several sites close to the gene promoter (Fig. 1B). Of note, the intron 5 and promoter sites also correspond to regions of open chromatin that we documented in primary HEE cells using DNase-seq (GEO:GSE74709, Yang et al., 2016). These data confirmed specific binding of AR to known targets in the presence of AR ligand. This, combined with the strong correlation in the genome-wide AR binding profile between the two replicates (by Irreproducible Discovery Rate (IDR) analysis), generated confidence in the AR peaks identified by our ChIP-seq experiments.
Figure 2.
Genome-wide AR binding profile in primary caput HEE cells. (A) Pie chart showing the distribution of AR peaks in various genomic regions. (B) UCSC genome browser graphic showing AR ChIP-seq signals in the FKBP5 locus. Data from two donors (UC-01 and UC-05) with treatment of VC (vehicle control) or R1881 are shown. Black and gray arrows denote sites evaluated for AR occupancy in Fig. 5. (C) The top five enriched motifs near AR peaks identified with Homer de-novo motif analysis. The nucleotide frequencies of the genomic sequences aligned at the motif are shown in a sequence logo representation. The percentages of motif incidence in target and background regions are also shown.
De-novo motif analysis of the 8748 AR ChIP-seq peaks was next performed to determine the enrichment of TF binding motifs. The AR binding motif was the most significantly enriched (P = 1e–1834) and ~44% of all peaks contained this motif, providing further support for the robustness of these ChIP-seq datasets (Fig. 2C). Amongst other TF motifs, which are significantly enriched in the ChIP-seq peaks are those of Jun-AP1, CEBP (CAAT-enhancer binding protein), TEAD (TEA domain transcription factor), and RUNX (Runt-related transcription factor), which thus are potential candidates to function as co-factors of AR.
The epididymis specific AR transcriptional regulatory network
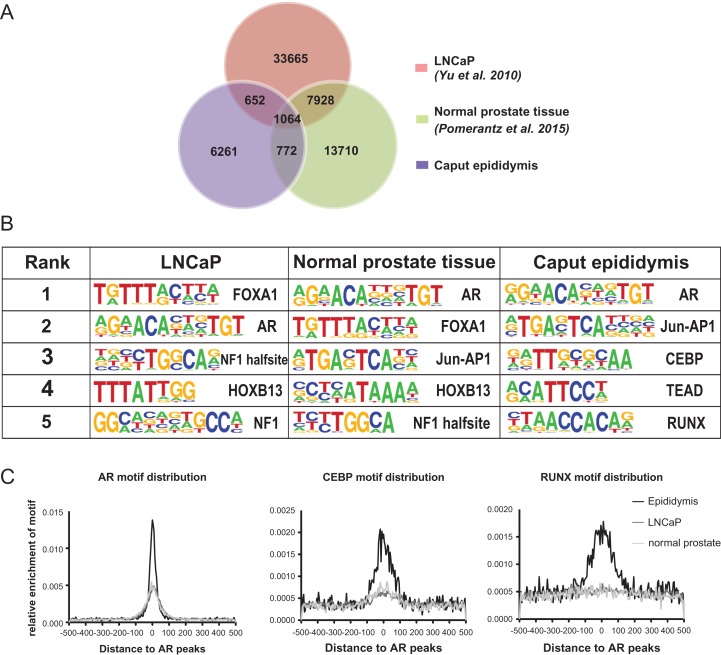
Although AR has an important role in regulating gene expression in multiple tissue types, studies to date on AR in the male reproductive system have largely focused on the prostate gland. Here we characterized the AR cistrome in primary caput HEE cells enabling a comparison between AR functions in the male reproductive ducts and previous data from normal prostate tissue or cells and prostate cancer. To identify epididymis-specific AR-associated pathways, we compared the epididymis AR peaks from our data and publicly available AR cistrome data from LNCaP prostate cancer cells (Yu et al., 2010) or normal prostate tissue (Pomerantz et al., 2015). This comparison revealed a distinct pattern of AR binding in epididymis cells compared to the prostate samples (Fig. 3). Among the 8748 peaks of epididymal AR occupancy, only 19.6% and 21.0% overlap with peaks identified in LNCaP and normal prostate, respectively. In contrast, 38.3% of the normal prostate AR peaks overlap with LNCaP whereas only 7.8% overlap with the epididymis. Details of the overlapping peaks among the three datasets are shown in Fig. 3A.
Figure 3.
Epididymis-specific AR cistrome and co-factors. (A) Venn diagram showing the overlap of AR peaks between LNCaP, normal prostate and caput epididymis cells. (B) The top five enriched de-novo motifs identified near AR peaks in different cell types. (C) Quantification of motif enrichment near AR peaks in different cell types. Enrichment of AR, CEBP and RUNX motifs are shown from left to right.
The motif recognized by AR in epididymis and prostate is very similar, hence it seemed probable that different co-factors interact with AR in the two tissues, resulting in the tissue specific AR binding. To test this hypothesis, de-novo motif analysis was performed for the AR ChIP-seq datasets from epididymis, normal prostate and prostate cancer (LNCaP). The top five enriched motifs from the three datasets are shown in Fig. 3B, where the two prostate datasets show very similar motifs enriched in the sequences surrounding AR peaks. These motifs include FOXA1 (Forkhead box A1), HOXB13 (Homeobox B13), and NF1 (Nuclear Factor 1), which were all reported previously (Yu et al., 2010; Sharma et al., 2013; Jin et al., 2014; Pomerantz et al., 2015). FOXA1 and HOXB13 were also shown to interact with AR and play important roles in prostate cancer progression (Robinson et al., 2014, Pomerantz et al., 2015; Kron et al., 2017). However, in the caput HEE cells FOXA1, HOXB13 and NF1 are not among the enriched motifs, with CEBP, TEAD and RUNX motifs evident instead. The relative enrichment of AR, CEBP, and RUNX motifs around AR peaks was also characterized in the three cell types. The results showed enrichment of the AR motif in all cell types but specific enrichment of both CEBP and RUNX motifs only in the caput HEE cells (Fig. 3C).
Potential cooperation between AR and CEBPB or RUNX1
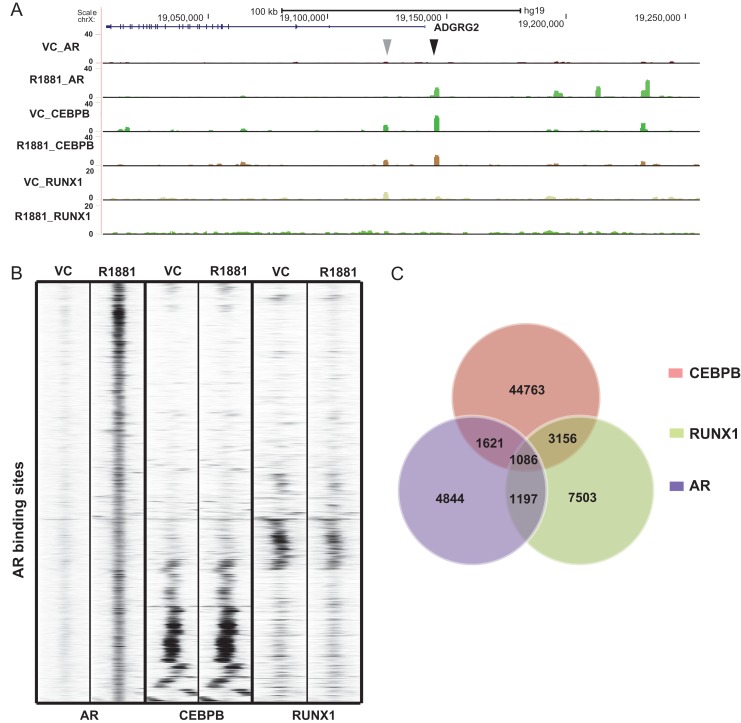
Our RNA-seq data generated previously from cells of the three anatomical regions of the epididymis (caput/head, corpus/body and cauda/tail) (Browne et al., 2016b), were used to predict the specific transcription factors binding to the CEBP and RUNX motifs. CEBP beta and RUNX1 are the most abundant isoforms expressed in the caput HEE cells so these were chosen for further study although other family members are also present (FPKM values are CEBPA: 2.64, CEBPB: 54.14, CEBPG: 21.39 and CEBPE: 0.01; RUNX1 :20.44, RUNX2: 7.75 and RUNX3: 0.008; Browne et al., 2016b). ChIP-seq experiments were performed to map the occupancy of CEBPB and RUNX1 in the caput HEE cell genome with or without R1881 treatment. Experimental procedures and data analysis were carried out as described earlier for the AR ChIP-seq. The ChIP-seq signals of AR, CEBPB, and RUNX1 around the Adhesion G Protein-Coupled receptor G2 (ADGRG2) gene (also known as GPR64), a potential AR target in the epididymis, are shown in Fig. 4A. ADGRG2 encodes an epididymis-selective transmembrane receptor that is pivotal in epididymis function (Patat et al., 2016) and which we know is expressed in Caput HEE cells (Browne et al., 2016b and Supplementary Table SI). R1881-induced occupancy by AR of a site 5’ to the ADGRG2 promoter (Fig. 4A, black arrow) coincides with a peak of constitutive binding of CEBPB, that is not influenced by the AR agonist. Further, a peak of CEBPB and RUNX1 occupancy in the first intron of ADGRG2 lacks AR and is unaffected by R1881 treatment (Fig. 4A, gray arrow). To examine whether AR activation affects binding of these co-factors genome-wide, ChIP-seq signal intensities of AR, CEBPB, and RUNX1 with either control or androgen treatment were compared in a 1 kb region centered around AR peaks (Fig. 4B). As expected, the AR ChIP-seq signal is strongly enriched in the middle of the region with R1881 treatment but not with the vehicle control. In contrast, both CEBPB and RUNX1 showed almost identical binding profiles around AR peaks with or without androgen treatment. These results suggest that the binding of CEBPB and RUNX1 is independent of AR activation and that these co-factors probably function upstream of AR. The analysis shown in Fig. 4B also revealed that CEBPB and RUNX1 share very few common co-localizations with AR peaks, suggesting that there is little cross-talk between the two factors, although both may cooperate with AR individually. Moreover, the co-localization of AR and the co-factors was observed to occur at relatively weak AR binding sites, with almost no overlap between AR and CEBPB or RUNX1 at sites where AR signals are the strongest. Since little difference in CEBPB and RUNX1 binding was observed with or without androgen treatment, we intersected the three datasets all under androgen treated conditions. The numbers of overlapping peaks are shown in Fig. 4C. In agreement with Fig. 4B, a considerable portion of the AR peaks were also bound by CEBPB (30.9%) or RUNX1 (26.1%). Conversely, only a small percentage of CEBPB (5.3%) or RUNX1 (16.9%) peaks were occupied by AR, which likely reflects the more general functions of these factors in comparison to AR.
Figure 4.
Genome wide occupancy of AR co-factors. (A) UCSC genome browser graphic showing ChIP-seq data for AR, CEBPB and RUNX1 around the ADGRG2 gene. Black and gray arrowheads denote ChIP-seq peaks discussed in the text. (B) Heatmap showing the ChIP-seq signal intensity of AR, CEBPB and RUNX1 around AR peaks in caput epididymis cells with or without R1881 treatment. (C) Venn diagram showing the numbers of overlapping peaks between AR, CEBPB and RUNX1 in caput HEE cells treated with R1881.
Dependency of AR binding on its co-factors in HEE cells
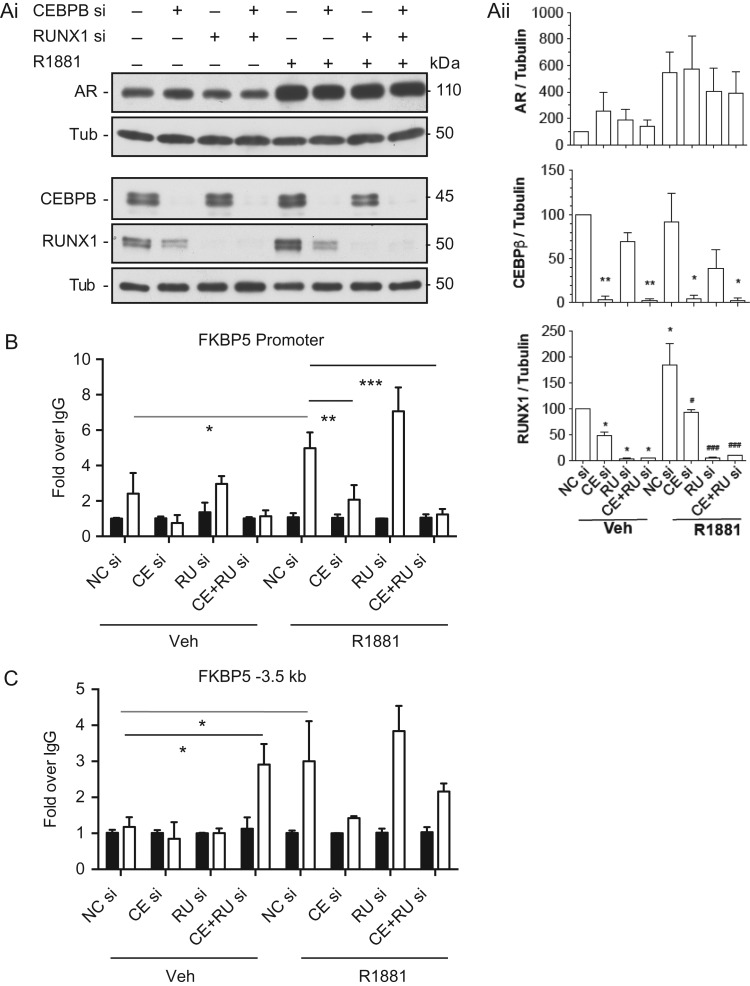
To investigate further the interactions of CEBPB and RUNX1 at R1881-dependent AR binding sites, we focused on a single locus, the well-characterized FKBP5 locus (Fig. 2B). Since the bioinformatics analysis above predicted that CEBPB and RUNX1 likely functioned upstream of AR binding, we next used specific siRNAs to deplete each factor separately and in combination and then performed ChIP with an antibody specific for AR after vehicle or R1881 exposure of the HEE cells. AR occupancy was assayed at the FKBP5 promoter and a site –3.5 kb 5’ to the promoter (Fig. 2B, black and gray arrows, respectively) by ChIP-qPCR (Fig. 5). As expected R1881 increased the abundance of AR (Fig. 5Ai/ii), but had no impact on CEPBB and very little impact RUNX1 protein levels. siRNA-mediated depletion of CEBPB and RUNX1 was effective though of note depletion of CEBPB also somewhat reduced RUNX1 levels, suggesting that the latter might be a transcriptional target of CEBPB. Stimulation of HEE cells with R1881 significantly increased occupancy of AR at both the FKBP5 promoter (Fig. 5B) and the –3.5 kb site (Fig. 5C). ChIP after CEBPB depletion significantly reduced AR occupancy at the promoter though the reduction was not significant at –3.5 kb. This suggests that AR binding at the FKBP5 promoter is dependent on CEBPB potentially as a co-activator. Combined depletion of CEBP and RUNX1 also significantly reduced R1881 activated AR binding at the promoter. In contrast, depletion of RUNX1 alone slightly increased androgen-activated AR occupancy at the promoter and the –3.5 kb site, though in neither case was the change statistically significant. Also of note was the significant increase in basal AR occupancy at the –3.5 kb element upon combined depletion of CEBPB and RUNX1 without R1881 treatment, suggesting that this distal cis-element might be subject to different regulatory mechanisms than the gene promoter.
Figure 5.
CEBPB and RUNX1 as potential co-factors of AR. Caput HEE cells were transfected with CEBPB and/or RUNX1 siRNA (40 nM) for 48 h prior to treatment with vehicle or R1881 for 18 h. (Ai) Western blots show effective depletion of CEBP and RUNX1 by specific siRNAs and increased abundance of AR, but not CEBPB and a small increase in RUNX1, after R1881 exposure. (Aii) Combined densitometry of western blots by Image J showing the impact of siRNA-mediated depletion of CEBP and RUNX1 or negative control with vehicle or R1881 treatment. Statistics by t-test. *P < 0.05, **P < 0.01, ***P < 0.001 comparing to NC siRNA, Veh or #P < 0.05, ##P < 0.01, ###P < 0.001 comparing to NC siRNA, R1881. (B, C) ChIP-qPCR with an AR antibody to determine occupancy of AR at (B) the FKBP5 promoter or (C) the –3.5 kb element, as shown by arrows in Fig. 1C. Key CE = CEBPB, RU = RUNX1, Veh = vehicle. For both panels the black bars show IgG controls and the open bars show AR occupancy. Enrichment is shown as fold over IgG. Data from donor HEE cultures, statistics by t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
The AR transcriptional network in caput HEE cells
We previously documented the transcriptome of HEE cells from the caput, corpus and cauda regions of the epididymis (Browne et al., 2016b). Here we build on these data to examine the alterations in gene expression effected by androgen-activation of AR in caput HEE cells. Data from four biological replicas were combined to identify 204 significantly differentially expressed genes (DEGs) (Supplementary Table SI).
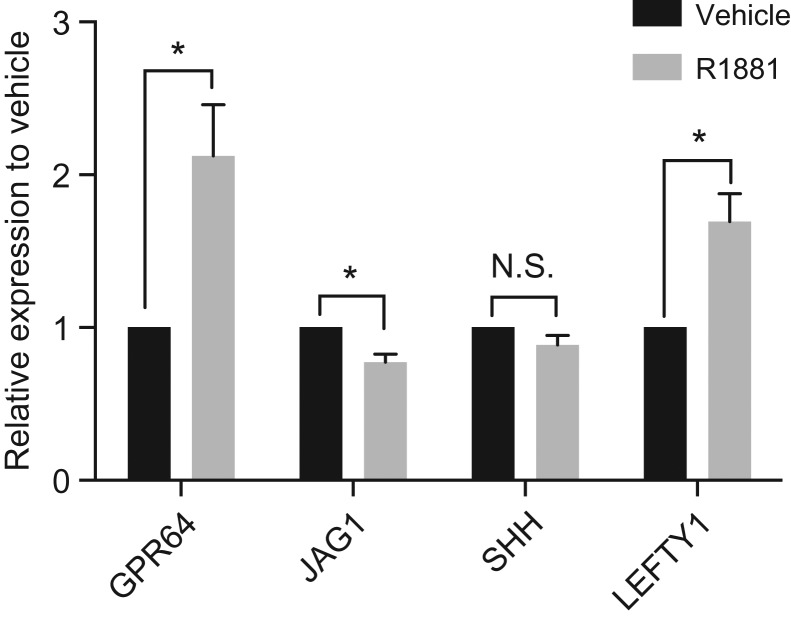
Intersection of genes that were differentially regulated after R1881 treatment with loci with an AR ChIP-seq peak within 20 kb, identified those of particular interest as direct AR targets. These included Jagged 1 (JAG1, a Notch signaling ligand), Sonic Hedgehog (SHH) and Left-right determination factor (LEFTY1), all AR targets in prostate cancer cells (LaTulippe et al., 2002; Bolton et al., 2007). G-protein coupled receptor 64 (GPR64) (Adhesion G Protein-Coupled Receptor G2, ADGRG2) is an epididymis GPR that was not previously known to be AR regulated, though our data above showed R1881- dependent occupancy of AR at the locus. Validation of these AR targets was performed by RT-qPCR (Fig. 6). GPR64 and LEFTY1 are both significantly upregulated by R1881 treatment while JAG1 shows minor repression, consistent with the RNA-seq data. Of note only eight DEGs in HEE cells overlap with loci reported to be androgen-regulated in prostate cancer cells (Supplementary Table SI, marked in red type), reinforcing our earlier observations suggesting that the AR transcriptional program in the epididymis is distinct from that of prostate cells.
Figure 6.
Androgen regulation of genes in HEE cells. HEE cells were exposed to R1881 (1 nM for 16 h), RNA was extracted and RT-qPCR was performed using standard protocols and gene-specific primers. Black bars, vehicle treated; gray bars, R1881-treated. Data are shown as expression level relative to the vehicle control (set to 1) for each gene. n = 4 for each assay. Statistical analysis by two-tailed t-test: *P < 0.05.
Discussion
Relatively little is known about the transcriptional networks controlling the differentiated functions of the human male reproductive ducts. Here we report genome-wide occupancy data for the androgen receptor, one of the key transcription factors in primary human caput epididymis cells. To date, most studies on human AR transcriptional regulation focus on prostate cancer or normal prostate tissue. The functions of AR in the human epididymis are poorly understood in large part due to the lack of experimental materials. Although AR loss-of function models provided important insights into the roles of AR in development and function of the rodent male genital tract, there are profound interspecies differences. The establishment of robust cell cultures from adult human epididymis (Leir et al., 2015) enabled us to interrogate the AR transcriptional network in human cells.
By comparing our AR ChIP-seq data in caput HEE cells to the published data generated in prostate cells, we found that AR distribution varies substantially in the two cell types. The distinct AR binding profiles in epididymis and prostate cells suggests that AR may regulate genes involved in epididymis specific functions, especially those involved in regulating epididymal luminal environment. For example, we note occupancy of AR in an epididymis-selective region of open chromatin 5’ to the promoter of the cystic fibrosis transmembrane conductance regulator gene (CFTR). CFTR encodes an apical membrane chloride ion channel that has a pivotal role in establishing the luminal fluid of the epididymis (Pollard et al., 1991; Leir et al., 2015). Of note, loss of function mutations in CFTR are associated with defects in epididymis and vas deferens that result in the infertility of males with CF. Despite the tissue selective binding profile of AR, it responds to the agonist similarly and recognizes the same motif in both prostate and epididymis cells. To search for mechanisms underlying the cell-specificity of AR binding, we first looked at potential divergence in its co-factors in the two tissues. Several studies previously showed reprograming of AR binding by co-factors during prostate oncogenesis with STAT family members, MYC, E1F and FOXA1 likely having key roles in different tumor types (Sharma et al., 2013; Jin et al., 2014). Here, de-novo motif analysis in AR binding regions identified enriched motifs unique to epididymis cells, including CEBP, TEAD, and RUNX motifs. CEBPB belongs to the CCAAT/enhancer binding proteins (CEBPs) family. CEBPs all have a highly conserved DNA-binding domain, the basic region-leucine zipper motif (bZIP) and are well studied in liver gene regulation and energy metabolism (Roesler, 2001). RUNX1 is one of the RUNX family members which are mainly involved in cellular differentiation and cell cycle progression (Durst and Hiebert, 2004; de Bruijn and Dzierzak, 2017; Deltcheva and Nimmo, 2017) and can function both as repressors or activators. The best-characterized roles of RUNX1 are in hematopoiesis and leukemia (reviewed in Ito et al., 2015). However, the function of these factors in the epididymis is largely unknown.
The comparison of ChIP-seq data for AR and the co-factors in HEE cells revealed several interesting findings. Firstly, the binding profiles of both CEBPB and RUNX1 are not affected by androgen treatment (Fig. 4B). In the absence of androgens, very little AR binding is evident whereas the co-factors show equivalent occupancy with vehicle or after R1881 treatment. Thus CEBPB and RUNX1 binding is independent of AR occupancy, indicating that these co-factors may function upstream of AR. Another feature of the ChIP-seq results is that AR peaks often overlap with CEBPB or RUNX1 peaks individually but rarely coincide with both jointly. This suggests that CEBPB and RUNX1 may cooperate with AR independently and may not interact with each other at AR binding sites. The data also showed that localization of AR and CEBPB or RUNX1 occurred preferentially in relatively weak AR binding sites. It is possible that binding of AR at these locations is dependent on the co-factors to stabilize the interactions. Our observation on the loss of AR occupancy at two sites in the FKBP5 locus upon siRNA-mediated depletion of CEBPB would support this hypothesis. However, the potential role of RUNX1 remains to be elucidated.
ADGRG2 (formerly named HE6 or GPR64) belongs to the family of adhesion-GPCRs (G protein-coupled receptors) and is a full orphan receptor with no synthetic or endogenous ligands known (reviewed in Paavola and Hall, 2012). Abundant transcripts of this gene were found in epididymis by differential screening of a human epididymal cDNA library (Osterhoff et al., 1997) and it was later found to have a key role in sperm maturation. Hemizygous mutant male mice with a disrupted ADGRG2 gene (ADGRG2 is X-linked) had severely reduced fertility primarily due to dysregulation of fluid reabsorption, preventing spermatozoa from migrating through the epididymis (Davies et al., 2004). Subsequently several epididymis-specific genes were found to be downregulated in microarray analysis of the same mouse model (Davies et al., 2007). Many of these repressed genes encode secretory and membrane proteins which may contribute to control of the epididymal luminal environment. ADGRG2 functions have also been investigated in breast cancer where it is involved in the adhesion and migration of certain breast cancer cells through non-canonical NFkB pathways (Peeters et al., 2015). Here we show by RNA-seq that the ADGRG2 gene is upregulated in response to R1881 in HEE cells and validated this by RT-qPCR. In androgen-treated cells, four peaks of AR occupancy were present within 100 kb upstream of ADGRG2 gene, including one that is adjacent to the promoter (Fig. 4A), which is also occupied by CEBPB. In combination, our data support a model in which AR modulates the normal function of human caput epididymis cells, including secretion, fluid transport, and cell adhesion through direct transcriptional activation of the ADGRG2 gene. The specific downstream pathways that are regulated by ADGRG2 in HEE cells warrant further investigation.
The current AR ChIP-seq data combined with RNA-seq after androgen stimulation in HEE cells provides evidence for an AR-regulated transcriptional network in the human caput epididymis epithelium. However, additional work is required to identify and confirm other direct targets of AR and to enable the discovery of relevant biological pathways regulated by this factor in normal epididymis function. Further studies are also required to understand the mechanism whereby AR exerts such different functions in epididymis and prostate. Though different co-factors in the epididymis were identified in the current study, our understanding of how they interact and contribute to AR specific binding is incomplete.
Supplementary Material
Acknowledgements
We thank Dr Pieter Faber and his staff at the University of Chicago Genomics Core for RNA-seq library preparation and RNA-seq and ChIP-seq library sequencing. We also thank Shiyi Yin for assistance.
Authors’ roles
R.Y., S.-H.L. and A.H. designed the study; R.Y., J.A.B., S.-H.L., S.E.E. and A.H. acquired, analyzed and interpreted data. R.Y. and A.H. drafted the article. All authors revised and approved the article.
Funding
This work was funded by the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development: R01 HD068901 (PI: Harris).
References
- Bischof JM, Gillen AE, Song L, Gosalia N, London D, Furey TS, Crawford GE, Harris A. A genome-wide analysis of open chromatin in human epididymis epithelial cells reveals candidate regulatory elements for genes coordinating epididymal function. Biol Reprod 2013;89:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaquier JA, Cameo MS, Burgos MH. The role of androgens in the maturation of epididymal spermatozoa in the guinea pig. Endocrinology 1972;90:839–842. [DOI] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 2007;21:2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Harris A, Leir SH. An optimized protocol for isolating primary epithelial cell chromatin for ChIP. PLoS One 2014. a;9:e100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Eggener SE, Leir SH, Harris A. HNF1 regulates critical processes in the human epididymis epithelium. Mol Cell Endocrinol 2016. a;425:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Leir SH, Eggener SE, Harris A. Expression profiles of human epididymis epithelial cells reveal the functional diversity of caput, corpus and cauda regions. Mol Hum Reprod 2016. b;22:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Song L, Crawford GE, Leir SH, Harris A. Open chromatin mapping identifies transcriptional networks regulating human epididymis epithelial function. Mol Hum Reprod 2014. b;20:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin TR, Griswold MD. Androgen-regulated genes in the murine epididymis. Biol Reprod 2004;71:560–569. [DOI] [PubMed] [Google Scholar]
- Cohen J, Ooms MP, Vreeburg JT. Reduction of fertilizing capacity of epididymal spermatozoa by 5 alpha-steroid reductase inhibitors. Experientia 1981;37:1031–1032. [DOI] [PubMed] [Google Scholar]
- Cornwall GA. New insights into epididymal biology and function. Hum Reprod Update 2009;15:213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Baumann C, Kirchhoff C, Ivell R, Nubbemeyer R, Habenicht UF, Theuring F, Gottwald U. Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol 2004;24:8642–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Behnen M, Cappallo-Obermann H, Spiess AN, Theuring F, Kirchhoff C. Novel epididymis-specific mRNAs downregulated by HE6/Gpr64 receptor gene disruption. Mol Reprod Dev 2007;74:539–553. [DOI] [PubMed] [Google Scholar]
- de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 2017;129:2061–2069. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Nimmo R. RUNX transcription factors at the interface of stem cells and cancer. Biochem J 2017;474:1755–1768. [DOI] [PubMed] [Google Scholar]
- Dube E, Chan PT, Hermo L, Cyr DG. Gene expression profiling and its relevance to the blood-epididymal barrier in the human epididymis. Biol Reprod 2007;76:1034–1044. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 2004;23:4220–4224. [DOI] [PubMed] [Google Scholar]
- Fossum SL, Mutolo MJ, Tugores A, Ghosh S, Randell SH, Jones LC, Leir SH, Harris A. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J Biol Chem 2017;292:10938–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossum SL, Mutolo MJ, Yang R, Dang H, O’Neal WK, Knowles MR, Leir SH, Harris A. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res 2014;42:13588–13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyna RE, Wilson JD. A comparative study of the conversion of testosterone to 17-beta-hydroxy-5-alpha-androstan-3-one (Dihydrotestosterone) by prostate and epididymis. J Clin Endocrinol Metab 1969;29:970–977. [DOI] [PubMed] [Google Scholar]
- Hamzeh M, Robaire B. Identification of early response genes and pathway activated by androgens in the initial segment and caput regions of the regressed rat epididymis. Endocrinology 2010;151:4504–4514. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer 2015;15:81–95. [DOI] [PubMed] [Google Scholar]
- Jin HJ, Zhao JC, Wu L, Kim J, Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun 2014;5:3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron KJ, Murison A, Zhou S, Huang V, Yamaguchi TN, Shiah YJ, Fraser M, van der Kwast T, Boutros PC, Bristow RG et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat Genet 2017;49:1336–1345. [DOI] [PubMed] [Google Scholar]
- Krutskikh A, De Gendt K, Sharp V, Verhoeven G, Poutanen M, Huhtaniemi I. Targeted inactivation of the androgen receptor gene in murine proximal epididymis causes epithelial hypotrophy and obstructive azoospermia. Endocrinology 2011;152:689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL, Down T, Rakyan V, Turner D, Flicek P, Li H, Kulesha E et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res 2002;62:4499–4506. [PubMed] [Google Scholar]
- Leir SH, Browne JA, Eggener SE, Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil Steril 2015;103:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA 2nd, Hayashi K, Turner TT, Wilkinson MF. The Rhox5 homeobox gene regulates the region-specific expression of its paralogs in the rodent epididymis. Biol Reprod 2012;86:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 2006;147:590–598. [DOI] [PubMed] [Google Scholar]
- Moisan V, Bomgardner D, Tremblay JJ. Expression of the Ladybird-like homeobox 2 transcription factor in the developing mouse testis and epididymis. BMC Dev Biol 2008;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara L, Welsh M, Saunders PT, Smith LB. Androgen receptor expression in the caput epididymal epithelium is essential for development of the initial segment and epididymal spermatozoa transit. Endocrinology 2011;152:718–729. [DOI] [PubMed] [Google Scholar]
- Osterhoff C, Ivell R, Kirchhoff C. Cloning of a human epididymis-specific mRNA, HE6, encoding a novel member of the seven transmembrane-domain receptor superfamily. DNA Cell Biol 1997;16:379–389. [DOI] [PubMed] [Google Scholar]
- Paavola KJ, Hall RA. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol Pharmacol 2012;82:777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patat O, Pagin A, Siegfried A, Mitchell V, Chassaing N, Faguer S, Monteil L, Gaston V, Bujan L, Courtade-Saidi M et al. Truncating mutations in the adhesion G protein-coupled receptor G2 gene ADGRG2 cause an X-linked congenital bilateral absence of vas deferens. Am J Hum Genet 2016;99:437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters MC, Fokkelman M, Boogaard B, Egerod KL, van de Water B, IJzerman AP, Schwartz TW. The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFkappaB and is involved in cell adhesion and migration. Cell Signal 2015;27:2579–2588. [DOI] [PubMed] [Google Scholar]
- Pollard CE, Harris A, Coleman L, Argent BE. Chloride channels on epithelial cells cultured from human fetal epididymis. J Membr Biol 1991;124:275–284. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaire B, Ewing LL, Zirkin BR, Irby DC. Steroid delta4-5alpha-reductase and 3alpha-hydroxysteroid dehydrogenase in the rat epididymis. Endocrinology 1977;101:1379–1390. [DOI] [PubMed] [Google Scholar]
- Robaire B, Hamzeh M. Androgen action in the epididymis. J Androl 2011;32:592–599. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Hickey TE, Warren AY, Vowler SL, Carroll T, Lamb AD, Papoutsoglou N, Neal DE, Tilley WD, Carroll JS. Elevated levels of FOXA1 facilitate androgen receptor chromatin binding resulting in a CRPC-like phenotype. Oncogene 2014;33:5666–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr 2001;21:141–165. [DOI] [PubMed] [Google Scholar]
- Sar M, Lubahn DB, French FS, Wilson EM. Immunohistochemical localization of the androgen receptor in rat and human tissues. Endocrinology 1990;127:3180–3186. [DOI] [PubMed] [Google Scholar]
- Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY, Mills IG et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 2013;23:35–47. [DOI] [PubMed] [Google Scholar]
- Snyder EM, Small CL, Li Y, Griswold MD. Regulation of gene expression by estrogen and testosterone in the proximal mouse reproductive tract. Biol Reprod 2009;81:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimon V, Koukoui O, Calvo E, Sullivan R. Region-specific gene expression profiling along the human epididymis. Mol Hum Reprod 2007;13:691–704. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TT, Bang HJ, Attipoe SA, Johnston DS, Tomsig JL. Sonic hedgehog pathway inhibition alters epididymal function as assessed by the development of sperm motility. J Androl 2006;27:225–232. [DOI] [PubMed] [Google Scholar]
- Yamashita S. Localization of estrogen and androgen receptors in male reproductive tissues of mice and rats. Anat Rec A Discov Mol Cell Evol Biol 2004;279:768–778. [DOI] [PubMed] [Google Scholar]
- Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, Crawford GE, Kosak ST, Leir SH, Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res 2016;44:3082–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010;17:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Liu Q, Li YM, Hall SH, French FS, Zhang YL. Genome-wide profiling of segmental-regulated transcriptomes in human epididymis using oligo microarray. Mol Cell Endocrinol 2006;250:169–177. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Prins GS, Saunders PT, Katzenellenbogen BS, Hess RA. Localization of androgen and estrogen receptors in adult male mouse reproductive tract. J Androl 2002;23:870–881. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.