Abstract
We examined the precise localization of dimethylated histone three lysine four (H3K4me2) in mature rat sperm. Within nonintergenic-enriched regions, half of the DNA peaks associated with H3K4me2 retention fell in gene bodies and the other half in promoter regions. The most significant peaks near annotated DNA regions in the composite data included loci known to be associated with RNA metabolism, cell cycle regulation, and spermatogenesis. Regions associated with differential retention of H3K4me2 within gene bodies were significantly enriched for housekeeping gene and cell-cycle functionality. Proximal promoter-associated peaks were enriched for viral reproduction and cell cycle regulation genes, while Promoter1k and Promoter3k peaks were enriched for RNA metabolism functions. Further, homeobox- and kruppel-like factor motifs were among the most significantly enriched de novo and known motifs discovered within gene-associated H3K4me2 peaks. Motif analysis and native chromatin immunoprecipitation followed by sequencing (nChIP-seq) peak calling indicated an instructive role for retained paternal histones in the epigenetic regulation of early embryonic development in the rat.
Keywords: chromatin, histone, sperm
Most investigations of mammalian sperm chromatin and histone retention have used humans and mice [1–6], with few studies using rats, a key model for studying male reproductive biology, physiology, and response to injury. This letter describes the localization of dimethylated histone three lysine four (H3K4me2) in mature Rattus norvegicus sperm using native chromatin immunoprecipitation followed by sequencing (nChIP-seq).
During spermiogenesis, protamines replace over 90% of the canonical histones that are preferentially retained in transcriptional start sites, promoter regions and gene bodies of housekeeping, and developmental regulatory genes [2, 3]. The paternal epigenetic environment is crucial to normal zygotic genome activation with perturbations in sperm histone retention linked to transgenerational epigenetic inheritance [2, 5]. The paternal chromatin landscape reflects epigenetic modifications imparted by environmental conditions, while cross-species difference could reflect evolutionarily distinct developmental timelines and regulation [4].
For this study, epididymal sperm from four adult Fischer 344 rats were processed using a modified nChIP protocol (30U MNase/2 × 106 sperm, 2× digestion time, 5 μg Abcam ab7766 H3K4me2 antibody) [3]. Paired genomic control and immunoprecipitated libraries were sequenced on an Illumina HiSeq2500 (50 bp single end reads, ∼36 million reads per library). DiffReps was used to define enriched regions with default parameters and the built-in R. norvegicus rn4 genome assembly. Genomic regions were designated as gene body (between 3 kb downstream of the transcription start site (TSS) and 1 kb downstream of the transcription end site); proximal promoter (+/– 250 bp of a TSS), promoter1k (+/– 1 kb of a TSS, excluding +/– 250 bp of the TSS), promoter3k (+/– 3 kb of a TSS, excluding +/– 1 kb of the TSS); intergenic; and gene desert. A candidate peak list was refined by q-value and log-fold change to select the top 500 entries in each region. Over-represented biological properties and functional associations within a gene set were assigned statistical significance (p-value) based on QuickGO term finder.
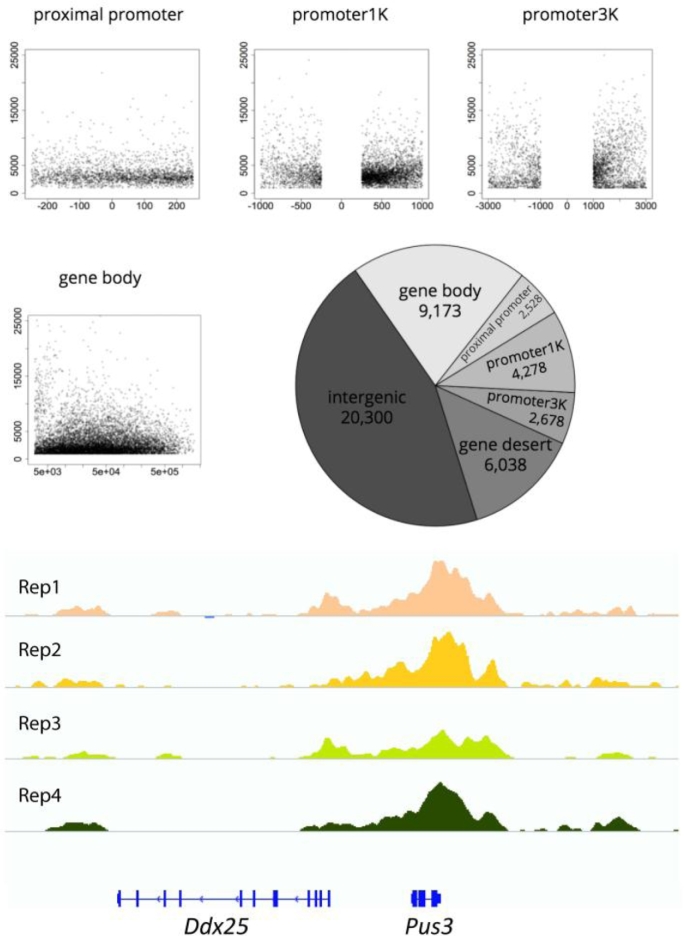
A total of 44,995 significantly enriched peaks were identified. H3K4me2-enriched sites were distributed unevenly around the TSS in promoter-related regions and broadly within gene bodies (Figure 1). Intergenic and gene desert regions contained a little more than half of the H3K4me2 differentially enriched peaks (Figure 1). Of the remaining peaks, about half (9173) were located within a gene body, one quarter (4278) in promoter1k regions, and the remainder in proximal promoter or promoter3k regions (Figure 1). The sample replicates had similar patterns of H3K4me2-enriched peaks, as shown for the chromosomal region containing Ddx25 and Pus3 (Figure 1).
Figure 1.
Size, genomic distribution, and chromosomal profiles of H3K4me2-enriched differential regions. The charts (proximal promoter, promoter1k, promoter3k, and gene body) show the size in kb (y-axis) of H3K4me2-protected sites relative to the TSS (in bp, x-axis). The pie chart shows the distribution of the 44,995 H3K4me2-protected sites by region. The bottom panel shows profiles of the sample replicates of H3K4me2-enriched peaks in the chromosomal region including Ddx25 and Pus3.
As a composite, the top 500 most q-significant sites with the largest log-fold change enrichment relative to input were enriched for RNA metabolic processes, cell cycle regulators, and spermatogenesis-associated genes. Within region types, the top 500 most q-significant sites with highest log-fold change in gene body regions included housekeeping, signal transduction, and cell cycle regulators. Over-enriched biological processes in proximal promoter sites included viral reproduction associated loci and cell cycle regulation. Promoter1k sites were highly enriched for RNA-related genes, with seven of the top ten most p-significant biological processes RNA-related. Similarly, within the top 500 q-significant, largest log-fold change promoter3k sites, seven out of ten of the most-enriched biological processes included RNA metabolism functionality. In all region types, spermatogenesis-associated biological processes appeared within the top 20 enriched networks.
Whole genome motif discovery with HOMER v4.9 and the R. norvegicus rn6 genome assembly yielded a total of 31,665 significant differential regions, including 127 known motifs and 36 de novo motifs. The top four de novo results included JUN family binding sites, a predicted zinc finger protein 354C motif, regulatory factor X5, and zinc finger and BTB domain containing 18. Other significant results of developmental importance included TEAD-binding domains 2 and 4, several homeobox genes, and the nanog-binding motif.
The most significant four known motifs contained elements known to be indispensable for spermatogenesis or recruitment of developmental regulators [7, 8]. These motifs included several kruppel-like factor motifs (KLF1,3,4,5,6,9,10, and 14), SP5 transcription factor, and factors known to regulate cell survival and spermatogenesis (Jun proto-oncogene, AP-1 transcription factor subunit) [7].
In combination with previous studies in mice and men, these new rat data support the hypothesis that patterns of histone retention in sperm are broadly conserved across mammals [2–6]. Brykczynska et al. [9] found that H3K4me2 was enriched near promoter regions of spermatogenesis related genes in human and mouse, and many H3K4me2 peaks in our data were in promoter regions of spermatogenesis associated genes, including Prm2, Spo11, and Spaga16. In addition, in both mice and men [1, 9], histones are retained in sperm near promoters and coding regions of regulatory RNA-related genes (lncRNA, piRNA, and miRNA), presumably providing protection against transposable elements and for regulating RNA processing in the early embryo. We found H3K4me2 peaks near the promoters of regulatory RNA genes, including Piwil1 and Piwil2, indicating a similar role for these histone-enriched genes in rat. Studies in mouse and other metazoan models have shown a high concordance between paternal regulatory histone marks and preimplantation embryonic mRNA expression with patterns analogous to those described here [1, 3, 5].
Our findings provide further support to the emerging concept that canonical histones are retained in sperm near regions of developmental and embryonic importance [2, 5, 10]. In combination, these data strongly implicate patterns of paternal histone retention in the transmission of critical epigenetic information to the early embryo. To fully characterize the extent to which sperm chromatin marks are conserved and/or environmentally malleable, future studies will be needed that alter environmental conditions and then interrogate the resulting biologic response.
Yours sincerely,
Shelby Wilson
Edward Dere
David Klein
Christoph Schorl
Susan Hall
and Kim Boekelheide
Brown University
Department of Pathology and Laboratory Medicine
Providence
RI, USA 02912
Notes
Note: nChIP-seq data deposited on GEO: GSE108182—Localization of dimethylated histone three lysine four in the Rattus norvegicus sperm genome.
Edited by Dr. Sarah Kimmins, PhD, McGill University
Footnotes
Grant Support. This work was supported by the NIEHS Superfund Research Program at Brown University (P42 ES013660). The Genomics Core Facility is supported by the NIGMS P30GM103410, NCRR P30RR031153, P20RR018728 and S10RR02763, NSF EPSCoR 0554548, Lifespan, and the Division of Biology and Medicine, Brown University.
References
- 1. Hammoud SS, Low DHP, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 2014; 15:239–253. [DOI] [PubMed] [Google Scholar]
- 2. Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR.. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hisano M, Erkek S, Dessus-Babus S, Ramos L, Stadler MB, Peters AH. Genome-wide chromatin analysis in mature mouse and human spermatozoa. Nat Protoc 2013; 8:2449–2470. [DOI] [PubMed] [Google Scholar]
- 4. Johnson GD, Jodar M, Pique-Regi R, Krawetz SA. Nuclease footprints in sperm project past and future chromatin regulatory events. Sci Rep 2016; 6: 25864 (pp. 1–17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler, J, Peters, AH et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015; 350:aab2006–aab2006. [DOI] [PubMed] [Google Scholar]
- 6. Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausio J, Taylor J, Corces VG. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Reports 2017; 18:1366–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thepot D, Weitzman JB, Barra J, Segretain D, Stinnakre MG, Babinet C, Yaniv M. Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development 2000; 127:143–153. [DOI] [PubMed] [Google Scholar]
- 8. Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 2008; 10:353–360. [DOI] [PubMed] [Google Scholar]
- 9. Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 2010; 17:679–687. [DOI] [PubMed] [Google Scholar]
- 10. Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, Peters, AH, Gurdon, JB et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res 2016; 26:1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]