Abstract
O-linked N-acetylglucosamine (O-GlcNAcylation) is an important post-translational modification on serine or threonine of proteins, mainly observed in nucleus or cytoplasm. O-GlcNAcylation regulates many cell processes, including transcription, cell cycle, neural development and nascent polypeptide chains stabilization. However, the facile identification of O-GlcNAc is a major bottleneck in O-GlcNAcylation research. Herein, we report that a lectin, Agrocybe aegerita GlcNAc-specific lectin (AANL), also reported as AAL2, can be used as a powerful probe for O-GlcNAc identification. Glycan array analyses and surface plasmon resonance (SPR) assays show that AANL binds to GlcNAc with a dissociation constant (KD) of 94.6 μM, which is consistent with the result tested through isothiocyanate (ITC) assay reported before (Jiang S, Chen Y, Wang M, Yin Y, Pan Y, Gu B, Yu G, Li Y, Wong BH, Liang Y, et al. 2012. A novel lectin from Agrocybe aegerita shows high binding selectivity for terminal N-acetylglucosamine. Biochem J. 443:369–378.). Confocal imaging shows that AANL co-localizes extensively with NUP62, a heavily O-GlcNAcylated and abundant nuclear pore glycoprotein. Furthermore, O-GlcNAc-modified peptides could be effectively enriched in the late flow-through peak from simple samples by using affinity columns Sepharose 4B-AANL or POROS-AANL. Therefore, using AANL affinity column, we identified 28 high-confidence O-linked HexNAc-modified peptides mapped on 17 proteins involving diverse cellular progresses, including transcription, hydrolysis progress, urea cycle, alcohol metabolism and cell cycle. And most importantly, major proteins and sites were not annotated in the dbOGAP database. These results suggest that the AANL lectin is a new useful tool for enrichment and identification of O-GlcNAcylated proteins and peptides.
Keywords: AANL, HCD/ETD, lectin, O-GlcNAcylation
Introduction
O-Linked β-N-acetylglucosamine (O-GlcNAcylation), which was discovered on lymphocytes in the early 1980s, is an N-acetylglucosamine monomer covalently attached to hydroxyl groups of serine or threonine residues of proteins (Torres and Hart 1984; Holt and Hart 1986). O-GlcNAcylation is ubiquitous, and has been reported in all kinds of organisms, including bacteria (Schirm et al. 2004; Shen et al. 2006), fungi (Woosley et al. 2006) and all metazoans (Hart et al. 2011; Banerjee et al. 2013). Cycling of O-GlcNAc is regulated by two highly conserved and unique enzymes: O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), which respond to stress, hormones or nutrients (Hart, et al. 2007; Hanover et al. 2010; Zeidan and Hart 2010). Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), which is produced via the hexosamine biosynthetic pathway, is the donor substrate for OGT, and OGA regulates O-GlcNAc removal (Dorfman et al. 1955; Ghosh et al. 1960; Marshall et al. 1991; Gao et al. 2001). Knockdown of OGT leads to embryonic lethality in mice (Gao et al. 2001). O-GlcNAcylation has an important role in transcription, protein trafficking and turnover, nutrient sensing, cell cycle, neural development, stress response and nascent polypeptide chains stabilization (Hart et al. 2007, 2011; Zhu et al. 2015). Dysregulation of O-GlcNAcylation contributes to some important diseases, including cardiovascular diseases, cancer, neurodegeneration, diabetes (Marshall et al. 1991; Hart et al. 2007; McClain et al. 2002; Slawson et al. 2010;Paruchuri ; Gong et al. 2012; Dassanayaka and Jones 2014; Yuzwa and Vocadlo 2014).
Although O-GlcNAcylation is ubiquitous and it participates in a multitude of cellular processes, the identification of O-GlcNAcylated proteins and sites is still challenging. Akin to other post-translational modifications (PTMs), O-GlcNAcylation is often substoichiometric, and is commonly found in low-abundance regulatory proteins. Most importantly, O-GlcNAc is uncharged, small and is very labile upon ionization during mass spectrometry (MS) analysis (Hart et al. 2007). Therefore, methods for the enrichment of O-GlcNAcylated proteins or peptides and for the development of mass spectrometric methods are particularly important. For enriching O-GlcNAcylated proteins or peptide, there are some methods, including chemical/enzymatic approaches (Khidekel et al. 2007; Rexach et al. 2008; Wang, Udeshi, O’Malley et al. 2010; Wang, Udeshi, Slawson et al. 2010; Alfaro et al. 2012), O-GlcNAc-specific monoclonal antibodies (Teo et al. 2010; Zhao et al. 2011), β-elimination followed by Michael addition (BEMAD) (Wells et al. 2002; Vosseller et al. 2005; Hahne et al. 2013) and metabolic labeling methods (Zaro et al. 2011; Chuh et al. 2014). Wheat germ agglutinin affinity chromatography is also a useful method for identification of O-GlcNAcylation. Burlingame et al. identified a large number of O-GlcNAc-modified proteins from murine synaptosomes and Arabidopsis (Trinidad et al. 2012; Xu et al. 2017). In 2013, 23 confident O-GlcNAcylated peptides were identified from differentiating MC3T3E1 osteoblasts by using 3 m length LWGA-agarose column (Nagel et al. 2013). CpOGAD298N is a mutant of CpOGA from bacterium Clostridium perfringens, which lose the catalytic activity and retain the binding activity (Mariappa et al. 2015). Psathyrella velutina lectin (PVL) and CpOGAD298N have been as far-western reagents for evaluating O-GlcNAcylation (Cioci et al. 2006; Mariappa et al. 2015; Machon et al. 2017). Furthermore, CpOGAD298N has been used for O-GlcNAcylated proteins in Drosophila embryo (Selvan et al. 2017). All these methods make great progress in O-GlcNAcylation peptides and sites identification and the functional research of O-GlcNAcylated proteins. However, none of these methods would be “the best one” and each of them contributes their different information in identification of O-GlcNAcylation modification. More new methods development is helpful to O-GlcNAcylated peptides identification and function investigation. Here, we reported an Agrocybe aegerita GlcNAc-specific lectin (AANL), could be used as a new tool for enrichment of O-GlcNAcylated peptides or proteins.
AAL2 is renamed as AANL to avoid confusion with the famous fucose specific lectin Aleuria aurantia lectin (AAL). AANL is a lectin purified from Agrocybe aegerita via affinity chromatography with GlcNAc-coupled Sepharose 6B. AANL was reported to specifically bind to terminal non-reducing GlcNAc residues with higher specificity than WGA and Griffonia simplicifolia lectin-II (GSL-II), which are well-know GlcNAc-binding lectins, and was found to have ability of inducing apoptosis of hepatocellular cells as well as having anti-tumor effect in tumor-bearing mice in vivo (Jiang et al. 2012). It was also used for the identification of an early biomarker in human non-small-cell lung cancer (Jin et al. 2016). AANL monomer has a shape of cylindrical torus, exerting a seven-bladed β-propeller fold. Each blade is in a W-like shape, also named W-motif, all of which are organized around a 7-fold axis of pseudosymmetry and glycan ligands bind to the shallow pockets in the entrance of structure (Ren et al. 2015). Herein, the high affinity for GlcNAc of AANL was further confirmed by surface plasmon resonance (SPR) results. AANL effectively enriched model O-GlcNAc peptides from a peptide mixture, and the lectin also effectively enriched O-GlcNAcylated peptides from Babl/c mouse liver proteins. Enriched peptides were analyzed with combination of higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) MS methods (Alfaro et al. 2012; Zhao et al. 2011). Most of the identified O-GlcNAcylated proteins and sites are novel, suggesting that using AANL could be a novel, alternative strategy for O-GlcNAcylation identification.
Results
AANL has a high affinity for GlcNAc
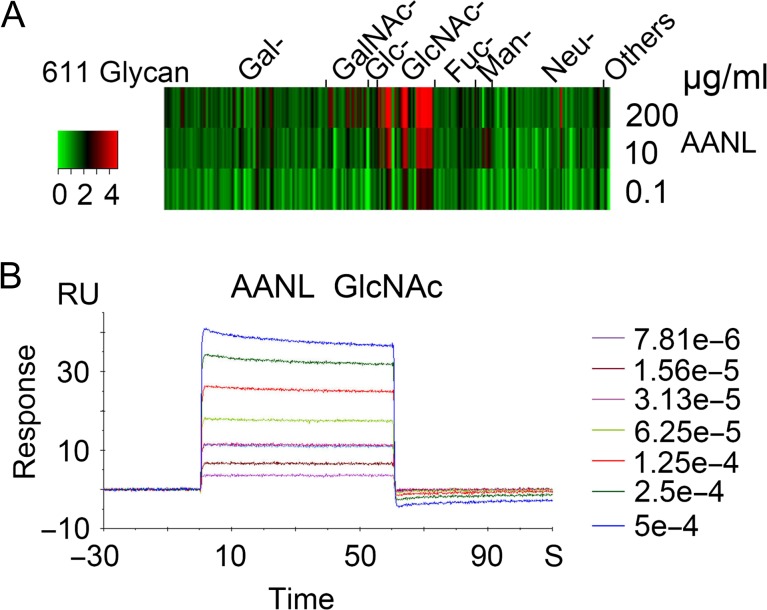
Glycan array analysis was performed at Consortium for Functional Glycomics (CFG) for characterizing the glycan-binding profile of AANL, which was also called AAL2, purified from A. aegerita with GlcNAc-coupled Sepharose 4B column as described before (Jiang et al. 2012). The glycan array data was analyzed through heat map in Figure 1A. The result showed that AANL has a specificity and affinity for glycans with terminal GlcNAc. To further determine the affinity of AANL for GlcNAc, SPR assay was performed. In Figure 1B, the data show that AANL binds to GlcNAc with a dissociation constant (KD) of 94.6 μM, which was consistent with the result tested through ITC assay reported before (Jiang et al. 2012). These results suggest that AANL has high affinity and specificity for GlcNAc and could be a candidate for identification of O-GlcNAcylated proteins.
Fig. 1.
AANL has a high affinity for GlcNAc. (A) The glycan array data of AANL was downloaded from CFG for heap map analysis. (B) SPR analysis of ligands binding to AANL immobilized on the chips. The binding curves measured at different concentration of GlcNAc and the steady-state affinity shown with KD for AANL.
AANL co-localize with O-GlcNAcylated nulear pore glycoprotein NUP62
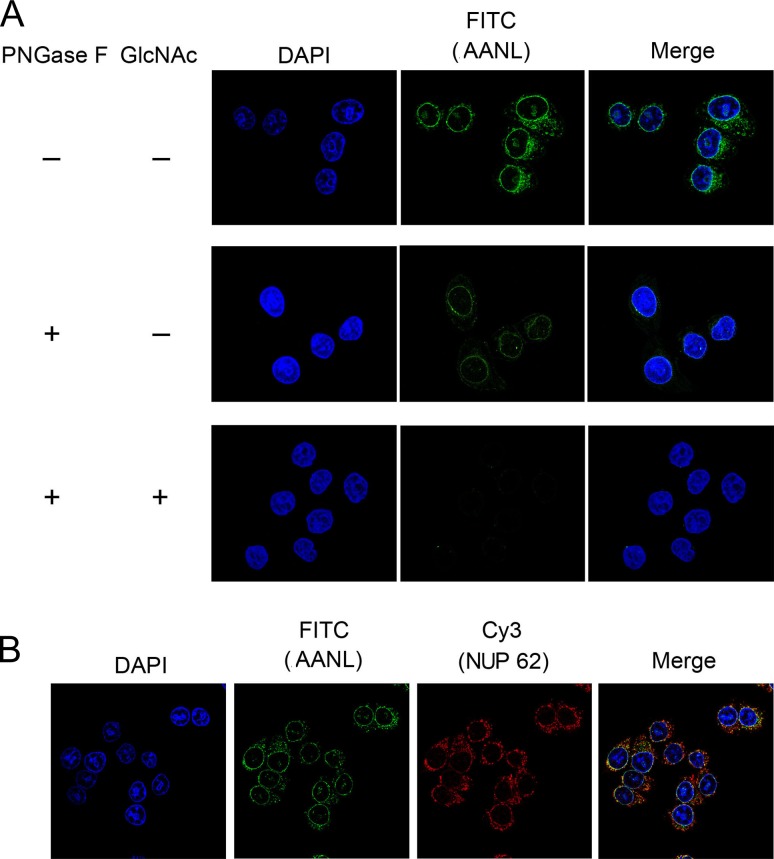
Fixed HeLa cells were incubated with biotinylated AANL, examined by confocal imaging assay analysis. As shown in Figure 2A, AANL recognized proteins around or near to the nuclear membrane. After treating the Hela cells with PNGase F to delete N-linked glycans, the fluorescence intensity weakened and mainly distribute near the nuclear membrane. Furthermore, after treating with GlcNAc, no AANL was observed around the nucleus, which indicated that AANL could be eluted from targeted proteins by GlcNAc. O-GlcNAc is nearly exclusively on nuclear, cytosolic or mitochondrial proteins and highest density of O-GlcNAc is found on nuclear pore and cytoskeletal proteins (Banerjee et al. 2013). Due to the abundance of nuclear pore proteins and their high level of O-GlcNAcylation, the signal from O-GlcNAcylated nucleoporins dominate the image in these types of fluorescent microscopic studies. Therefore, we selected an O-GlcNAcylated nulear pore glycoprotein with no other glycosylation (Uniprot), NUP62, to further verify whether AANL could recognize O-GlcNAcylated proteins. In Figure 2B, AANL was mainly distributed around the nucleus (green), and NUP62 (red) was co-localized with AANL suggesting that AANL could recognize O-GlcNAcylated NUP62. All these data suggest that AANL can bind to glycoproteins around cell nuclear membrane, including GlcNAc terminated N-glycosylated proteins and O-GlcNAcylated proteins.
Fig. 2.
AANL co-localized with O-GlcNAcylated nulear pore glycoprotein NUP62. (A) HeLa cells were fixed with methanol/acetone and then treated with PNGase F overnight. Cells were stained with DAPI and further labeled with biotinylated AANL (picture above). After incubating with biotinylated AANL, cells further incubated with 200 mM GlcNAc and then incubated with fluorescein isothiocyanate (FITC)-conjugated streptomycin, which was detected with a fluorescence microscope. (B) HeLa cells were fixed with methanol/acetone, stained with DAPI, and further incubated with NUP62 and biotinylated AANL. Then cells incubated with PE-conjugated secondary antibody and FITC-conjugated streptomycin. The co-localization was viewed with a fluorescence microscope.
AANL affinity column enrichment of model O-GlcNAcylated peptides
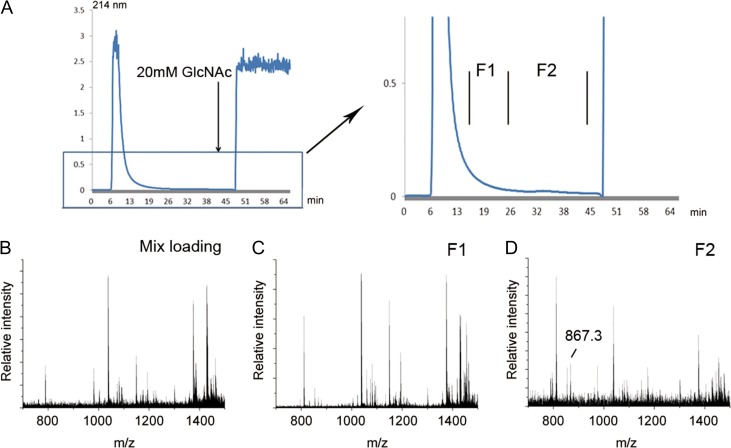
As AANL has selective high affinity for GlcNAc, two affinity columns POROS-AANL column and Sepharose 4B-AANL column were prepared for O-GlcNAcylated peptide enrichment. A peptide mixture, including five model O-GlcNAcylated peptides and tryptic digests of β-casein and κ-casein, were prepared, and loaded on a POROS-AANL column. Three fractions were collected designated as F1–F3 corresponding to two late flow-through fractions and one elution fraction. As shown in Figure 3A, there was an obvious small peak (fraction F1) after the main flow-through peak. The loading sample and collected fractions (F1–F3) were analyzed by MALDI-TOF-MS. As shown in Figure 3B–E, the model O-GlcNAc peptides could not be detected in the unenriched loading sample. However, after chromatographic separation, all five model O-GlcNAc peptides were found in F1. In F2 and F3, there were four and two model peptides found, respectively. These results indicate that the O-GlcNAcylated peptides could be enriched effectively in the late flow-through peak by POROS-AANL column. Another affinity column, Sepharose 4B-AANL column, was also used for O-GlcNAcylated peptides enrichment. Mixed peptides, comprised of tryptic digests of β-casein and κ-casein and five model O-GlcNAcylated peptides, were separated by Sepharose 4B-AANL column and results similar to the POROS-AANL column were obtained (Figure Supplementary data, S1). These data suggest that the O-GlcNAcylated peptides could also be efficiently enriched by Sepharose 4B-AANL column.
Fig. 3.
Enrichment of model O-GlcNAcylated peptides by POROS-AANL column (A) Peptide mixtures containing model O-GlcNAcylated peptides and unmodified peptides migrated through POROS-AANL column and four fractions were obtained. MALDI-TOF analysis of peptide mixture (B), F1 (C), F2 (D), F3 (E) and model O-GlcNAcylated peptides were mainly enriched in the F1 and F2.
Enrichment of O-GlcNAcylated peptides from tryptic digests of the O-GlcNAcylated protein, αA-crystallin
To determine whether AANL can enrich O-GlcNAcylated peptides from native proteins, the model O-GlcNAcylated protein αA-crystallin was digested with trypsin and chromatographed on the POROS-AANL column. The O-GlcNAcylated peptide AIPV(S-O-GlcNAc)R was enriched and readily detected in fraction F2 (Figure 4). All these results suggest that O-GlcNAcylated peptides can be effectively enriched by Sepharose 4B-AANL or POROS-AANL column.
Fig. 4.
Enrichment of O-GlcNAcylated peptides from tryptic digests of model O-GlcNAcylated protein, αA-Crystallin (A) Trypsin digested peptides of αA-crystallin migrated slower through POROS-AANL and two fractions were collected. MALDI-TOF analysis of peptides mixture (B), fraction F1 (C), and fraction F2 (D) and O-GlcNAcylated peptides were enriched in the fraction F2.
Enrichment of O-GlcNAcylated peptides from mouse liver by using AANL affinity columns
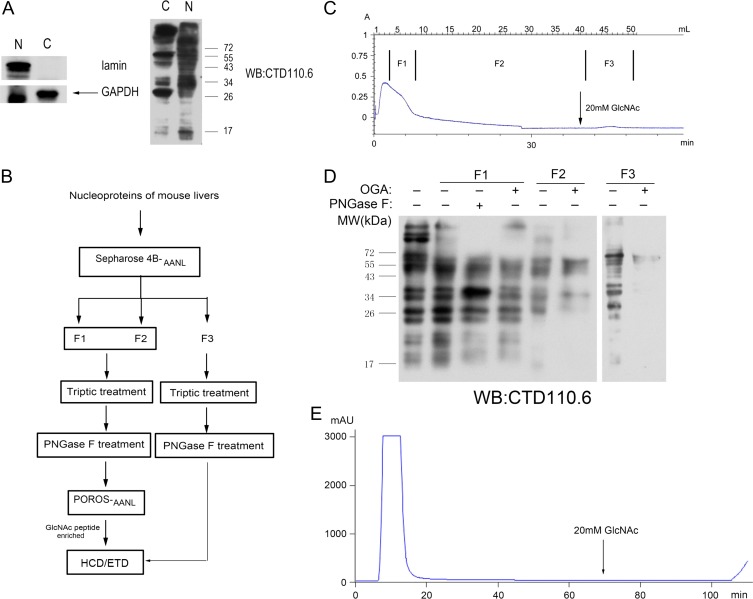
In order to determine if AANL affinity columns could effectively enrich O-GlcNAcylated peptides complex sample obtained from tissues, we used the method to enrich O-GlcNAcylated peptides from mouse liver. O-GlcNAcylation is an important regulatory modification affecting liver physiology (Zhang et al. 2014). As shown in Figure 5A, nuclear proteins and cytoplasmic proteins from liver tissues were separated for O-GlcNAcylation analyses. More O-GlcNAcylated proteins and higher O-GlcNAc modification levels are observed in the nucleus than in cytoplasm. Therefore, nuclear proteins were chosen to test the affinity columns, and two strategies were applied (Figure 5B). The nuclear proteins were separated by Sepharose 4B-AANL affinity column and three fractions (including two late flow-through fractions and one elution fraction) were collected (Figure 5C). After either PNGase F or OGA treatment, O-GlcNAcylated levels of the three fractions were detected by CTD110.6. As shown in Figure 5D, after OGA treatment, the signal of CTD110.6 of F2 and F3 was dramatically reduced, which indicated that O-GlcNAcylated proteins could be enriched by AANL affinity column in the tail of flow-through and elution fractions. Tryptic peptides of F1 and F2 were treated with PNGase F and then migrated slowly though POROS-AANL column for the second enrichment of O-GlcNAcylated peptides and the tail of flow-through were collected for O-GlcNAcylated peptide identification (Figure 5E). Tryptic peptides of F3 were treated with PNGase F to remove N-linked glycans. A total of three enriched fractions from mouse liver tissue were analyzed with LC–MS-MS by using HCD/ETD fragmentation on Orbitrap Fusion mass spectrometer.
Fig. 5.
Enrichment of O-GlcNAcylated peptides from mouse liver nucleoproteins (A) Over O-GlcNAcylation levels in both nuclear fraction and cytoplasm fraction, measured by immunoblotting with CTD110.6. (B) Workflow schematic for enrichment and identification of O-GlcNAcylated proteins. (C) Nuclear proteins migrated through AANL-coupled Sepharose 4B column and three fractions were collected. (D) Three fractions were treated with PNGase F or OGA, and over O-GlcNAcylation levels were detected with CTD110.6. (E) Fractions (F1 and F2) migrated through POROS-AANL column.
Identification of O-linked HexNAc-modified peptides and sites
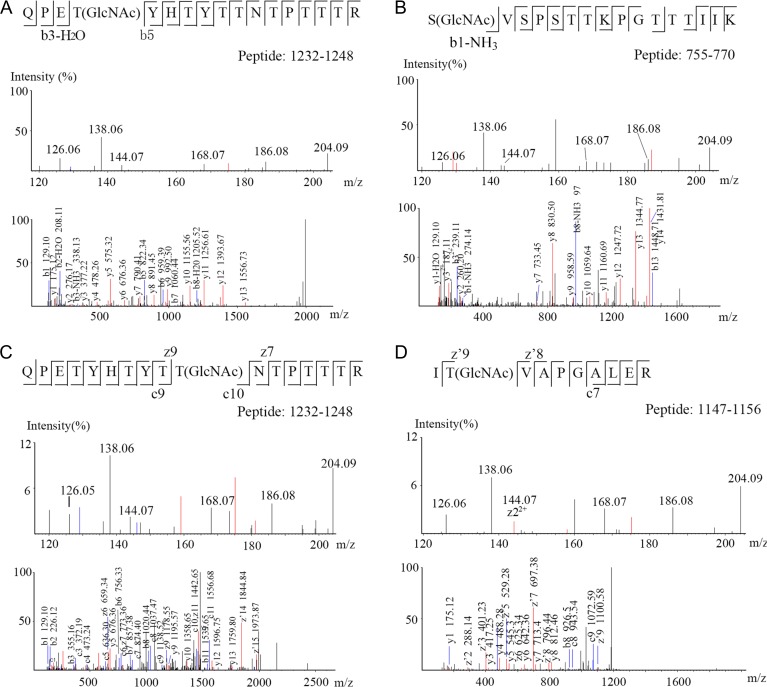
A list of 28 identified O-linked HexNAc-modified peptides in which 23 O-linked HexNAc-modified sites were identified, from liver tissues are presented in Table I, all of them were confirmed by manual inspection of the spectra. About 17 O-linked HexNAc-modified sites and 14 proteins were new (not in dbOGAP). These identified O-linked HexNAc-modified proteins from mouse liver play important roles in diverse cellular progresses, including transcription (e.g., HCF-1), hydrolysis progress (e.g., Mcm2, Ctsd, Cps1, Ctsz and Otud7b), urea cycle (e.g., Cps1), alcohol metabolism (e.g., Aldh2) and cell cycle (e.g., HCF-1 and Mcm2). Six O-GlcNAcylated peptides with six O-GlcNAcylated sites (two new sites) of HCF-1, a highly O-GlcNAcylated protein, were identified, and two sites are new (Figure 6A, B). The spectrum of HCF-1 is presented in the Figure 6. As shown in Figure 6A, the upper figure showed the oxonium ions of GlcNAc, which further established that the peptide “QPETYHTYTTNTPTTTR” is O-GlcNAcylated. The mass difference of 485.19 Da between the b5 and b3-H2O indicates modification of T1235 (Figure 6A) and the mass of 274.14 (b1-NH3) indicating modification of S755 (Figure 6B), which were new O-GlcNAcylated sites. Figure 6C and D shows two O-GlcNAcylated sites of HCF-1, which were previously reported. More than 30 O-GlcNAcylated sites have been reported on HCF-1(Myers et al. 2013), however, the two new sites of six sites were identified in this research suggested these two sites may be the specific sites in liver or are difficult to identify by using other O-GlcNAcylated enrichment methods. Four O-GlcNAcylated peptides and two O-GlcNAcylated sites (one is new) of spectrin β2 were also identified in our study. Cathepsin B, CAP-Gly domain-containing linker protein 2 and La-related protein 4B were identified to possess multiple sites.
Table I.
Identified O-linked HexNAc-modified peptides from mouse liver
| Accession | Protein name | Sequence | Site | New protein/site |
|---|---|---|---|---|
| Q62261 | Spectrin beta chain, non-erythrocytic 1 | AISSAISSDKHDTSASTQSTPASSR | N | |
| SAISSDKHDTSASTQSTPASSgR | S2323 | N/N | ||
| HDTSASTQSTPASSgR | S2323 | N/N | ||
| HDTSASTQSTPASgSR | S2322 | N/Y | ||
| P58871 | 182 kDa tankyrase-1-binding protein | LACSgEAPTDVSK | S260 | Y/Y |
| Q61191 | Host cell factor 1 | QPEtgYHTYTtgNTPTTTR | T1241,T1235 | N/N, Y |
| SgVSPSTTKPGTTTIIK | S755 | N/Y | ||
| ITgVAPGALER | T1148 | N/N | ||
| SPITIITgTK | T800 | N/N | ||
| SPITIITTgK | T801 | N/N | ||
| Q6NXI6 | Regulation of nuclear pre-mRNA domain-containing protein 2 | KEKPVEKPAVSgTGVPTK | S417 | N/N |
| P10605 | Cathepsin B | TIPPCEHHVNGsgRPPCtgGEGDTPR | S194, T199 | Y/Y |
| HVNGSgRPPCTGEGDTPR | S194 | Y/Y | ||
| P18242 | Cathepsin D | YYHGELSYLNVTgR | T263 | Y/Y |
| Q9WUU7 | Cathepsin Z | ECHTIQNYTgLWR | T188 | Y/Y |
| Q02819 | Nucleobindin-1 | AAPPQEDSQATETPDTGLYYHR | Y | |
| P28798 | Granulins | NVECGEGHFCHDNQTgCCK | T528 | Y/Y |
| VECGEGHFCHDNQTgCCK | T528 | Y/Y | ||
| P97310 | DNA replication licensing factor MCM2 | LTASgQR | S108 | Y/Y |
| B2RUR8 | OTU domain-containing protein 7B | STPESGESgDKESVGSSSLGNEGSR | S467 | Y/Y |
| P47738 | Aldehyde dehydrogenase | TgEQGPQVDETQFK | T358 | Y/Y |
| Q9Z0H8 | CAP-Gly domain-comtaining linker protein 2 | ISGTgTgALQEALK | T355, T356 | Y/Y |
| Q9WTQ8 | Mitochondrial import inner membrane translocase subunit Tim23 | STSGLAGFFGAGGAGYSNADLAGVPLTGMNPLSPYLNVDPR | S11, T12 or S13 | Y/Y |
| Q8C196 | Carbamoyl-phosphate synthase [ammonia] | LTSgIDKWFLYK | T872 | Y/Y |
| A2ARZ3 | Fibrous sheath-interacting protein 2 | KPNFTgPFEQVTK | T3270 | Y/Y |
| Q6A0A2 | La-related protein 4B | SLPALTQVPTTgK | T51 | Y/Y |
Within the peptide sequences, small letters indicate the modifications are from different peptides.
Fig. 6.
HCD/ETD MS/MS spectra of O-GlcNAcylated peptides from HCF-1. The O-GlcNAc diagnostic oxonium ions at m/z 204.09, 186.08, 168.07, 144.07, 138.06 and 126.05 (picture above) confidently determined that peptides identified were O-GlcNAc-modified. Annotation of b5 and b3-H2O ions enabled the exact localization of O-GlcNAc on T1235 (A), b1-NH3 ion enabled the exact localization of O-GlcNAc on S755 (B), c- and z-ions enabled the exact localization of O-GlcNAc on T1241 (C) and z6 ion or the only “T” in this peptide enabled the exact localization of O-GlcNAc on T1248 (D).
Discussion
Comparing the affinity for GlcNAc between other GlcNAc-binding proteins and AANL
AANL, also named AAL2, isolated from A. aegerita, was reported could bind to terminal non-reducing GlcNAc residue with high specificity (Jiang et al. 2012). In this study, we evaluated its affinity for GlcNAc monosaccharide through SPR (Figure 1).
rPVL is another recombinant fungal lectin, also has specificity for terminal non-reducing GlcNAc residues. By using SPR assay, the affinity of rPVL for GlcNAc was measured with KD of 134 μM (Audfray et al. 2015) and AANL for GlcNAc with KD of 94.6 μM in this paper, which indicates that AANL has similar affinity for GlcNAc with rPVL. It is reported that Biotin-rPVL and HRP-rPVL is a useful and efficient utilization by lectin blot for selectively detect O-GlcNAcylated proteins (Machon et al. 2017). rPVL may also can be used for GlcNAcylated proteins/peptides enrichment, which is interesting and should be verified.
CpOGAD298N, a catalytically inactive bacterial OGA mutant keeping its GlcNAc-binding activity, was used as a probe for O-GlcNAc protein detect by far-western blotting (Mariappa et al. 2015), which also was used in enriching O-GlcNAc peptides from Drosophila embryo, and 52 high-confidence O-linked HexNAc-modified peptides mapped on 43 proteins were identified (Selvan et al. 2017).
In this paper, we confirmed that AANL co-localizes with O-GlcNAcylated nulear pore glycoprotein NUP62, which has no other form of glycosylation (Figure 2A), and AANL could bind to the proteins around the nucleus and this binding depends on GlcNAc (Figure 2). Furthermore, O-GlcNAcylated peptides were effectively enriched at the tail of the flow-through by using ~25-cm length AANL-coupled affinity column (Figures 3–5). All these results suggested that AANL is a potential new method for enrichment and identification of O-GlcNAcylated proteins or peptides. In the enrichment process, N-glycan with terminal GlcNAc residue is the main disturbing factor. Therefore, PNGase F treatment is necessary and helpful for O-GlcNAcylated peptides enrichment.
Liver plays a key role in glycometabolism including O-GlcNAcylation cycling, which was an important regulatory mechanism underlying normal liver physiology (Zhang et al. 2014). Therefore, we chose mouse liver for O-GlcNAcylation identification although liver does not have a high O-GlcNAcylated level like brain (Vosseller et al. 2006; Chalkley et al. 2009; Trinidad et al. 2012). By using AANL affinity column for enrichment of O-GlcNAcylated proteins or peptides and collecting three fractions, and 28 high-confidence O-linked HexNAc-modified peptides mapped on 17 proteins were identified from mouse liver in our study.
O-linked HexNAc modification of proteins identified from liver nuclear proteins
Recently, HCD/ETD MS-MS, a novel fragmentation mode, combining HCD and ETD, in which oxonium ions from glycan fragments for accurately determining glycopeptides and b-, y-, c- and z-ions could supply confident sequence and modified sites information of peptides, was used for O-GlcNAcylation identification (Zhao et al. 2011; Marino et al. 2015). HCD/ETD MS analyses identified 17 O-linked HexNAc-modified proteins, among which 14 proteins were new (dbOGA; Table I). We also identified 23 O-linked HexNAc-modified sites and 17 were new in single LC–MS-MS analysis, which are manually confirmed and assigned in Table I. The vast majority of O-GlcNAc-modified proteins sites were novel in this study. About 32% peptides agreed with the previously reported sequence predictions, which exhibit a preference for a proline in the −2 or −3 position relative to the modified sites, and 18% peptides agreed with “TTA” motif. In our research, no new obvious consensus sequences of O-GlcNAc modification were identified by motif analysis, about 36% peptides with threonine at +5 position, indicating AANL may be bind to O-GlcNAc with some sequence specificity (Figure Supplementary data, S2). All these results suggested that AANL could be a novel and effective strategy for identifying O-GlcNAcylated peptides, which could make it a useful complement to other methods for O-GlcNAcylated proteins or peptide identification.
Various cell processes of O-GlcNAc-modified proteins were observed in this research. Host cell factor 1 (HCF-1) is a transcriptional co-regulator essential for cell cycle progression and cell proliferation (Julien and Herr 2003; Dejosez et al. 2010; Mangone et al. 2010). HCF-1 is a protein highly O-GlcNAcylated and more than 30 modified sites from various cell or tissue types have been reported (Myers et al. 2013). In this study, we have identified six O-GlcNAcylated sites of HCF-1 and two sites are new. Three of these modified sites reside in the basic region, which have important roles in controlling cell proliferation (Mangone et al. 2010). The other three sites were in the proteolytic processing domain with six central 26 amino acid repeats (HCF-1PRO) and cleavage takes place at these repeats resulting in HCF-1N and HCF-1C subunits formation (Wysocka and Herr 2003). The threonine-rich region of HCF-1PRO repeats could bind to OGT, the cleavage of HCF-1 takes place in the same active site of OGT used for glycosylation and UDP-GlcNAc is essential for the cleavage (Capotosti et al. 2011; Daou et al. 2011; Lazarus et al. 2013), so the cleavage, influences cell cycle progression (Goto et al. 1997; Julien and Herr 2003; Tyagi and Herr 2009), is closely linked with glycosylation. HCF-1 could also interact with many transcriptional factors, like E2Fs, which associates with HCF-1 to influence cell cycle progression (Zargar and Tyagi 2012). Most of O-GlcNAc-modified proteins identified here were new. TAB182, a tankyrase-binding protein, regulates the DNA double-strand break repair by regulating interaction of PARP-1/DNA-PKcs (Zou et al. 2015). OTUD7B, identified in our screen, is a deubiquitinase, which could regulate the non-canonical NF-κB pathway through controlling the deubiquitination and stabilization of TRAF3 (Hu et al. 2013). Timm23 was also identified in our study, which is a component of the Tim23 complex that is a mitochondrial inner membrane translocon (Steffen and Koehler 2014). We also identified a RNA-binding protein, LARP4B, which could regulate translation and also as a tumor suppressor of Glioma (Kuspert et al. 2015; Koso et al. 2016). Cps1, a carbamoyl-phosphate synthase, is a rate-limiting enzyme for urea cycle and related with coronary artery disease and weight maintenance (Hartiala et al. 2016; Matone et al. 2016). Aldh2 identified in our study, an aldehyde dehydrogenase, and alcohol dehydrogenase is the principal enzyme in hepatic metabolism of ethanol (Peng and Yin 2009). Aldh2 gene mutation is associated with many cancers, such as hepatocellular carcinoma, gastric cancer and colon cancer. There may be potential value of Aldh2 in the human cancer treatment (Li et al. 2016).
There is an extensive crosstalk between O-GlcNAcylation and phosphorylation and they may influence each other by competing on proteins at the same sites or proximal sites (Butkinaree et al. 2010). Some peptides identified here have a site of O-GlcNAcylation at or near to the phosphorylation site previously reported (Supplementary data, Table SI). The S108 site of DNA replication licensing factor MCM2, the S467 site of OTU domain-containing protein 7B, and the S2322, S2323 sites of spectrin beta chain, non-erythrocytic 1 are also phosphorylated sites annotated in PhosphoSite Plus. There are also some phosphorylated sites proximal to O-GlcNAc sites (within in four amino acides) (Supplementary data, Table SI). These results suggest that there was a potential functional competition/reciprocity between these two modifications.
In summary, O-GlcNcylation is an important PTM and O-GlcNAcylation identification is still a bottleneck for its functional research. In this study, AANL is shown to have a high affinity and specificity for GlcNAc. AANL was used to enrich O-GlcNAcylated proteins and peptides for a single LC–MS-MS analysis, from mouse liver, the tissue not with a large amount of O-GlcNAcylated proteins. About 17 O-linked HexNAc-modified proteins were identified, and most of which were new. The high affinity for GlcNAc, simple and easy protocols are the key strength of AANL, and AANL could be used for identification of O-GlcNAcylated proteins or peptides in a large scale and be an effective and good complement to other methods for O-GlcNAcylated proteins identification.
Materials and Methods
Cell culture and reagents
HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (Carlsbad, CA), and were grown in an incubator at 37°C with 5% CO2. An antibody against to O-GlcNAc (CTD110.6) was purchased from Sigma-Aldrich (St. Louis, MO), and an antibody against to NUP62 was purchased from Santa Cruz (Dallas, Texas). PugNAc, dithiothreitol (DTT), iodoacetamide, β-casein, κ-casein and αA-crystallin were purchased from Sigma-Aldrich (St. Louis, MO). CNBr-activated Sepharose 4B matrix and POROS AL resin were purchased from GE Healthcare (NJ, USA) and Applied biosystems (Massachusetts, USA), respectively. Model O-GlcNAcylated peptides: YSPTgSPS (G-T-CTD1, MW 940.4), YSPTSPSYSPTgSPSYSPTSPSC (G-T-CTD3, MW 2482.04), KTPIAVRFgSTVAGESGC (G-S-Cat, 1662.3), YSPTgSPSYSPTgSPSC (2 G-T-CTD2, 1966.8), 3 G-CTD (YSgPTgSgPSK, 1474.2) were kept in G.W.H.’s Lab.
Glycan array analysis
Glycan array data (611 glycans) of AANL was downloaded from CFG (www.Functionalglycomics.org). R software was used to compare the glycan-binding profile of AANL.
Surface plasmon resonance
Steady-state equilibrium binding of monosaccharide and lectins was monitored at 25°C using a SPR biosensor (Biacore T100). AANL were coupled to a sensor chip (CM5) via amine coupling with 4300 resonance units (RU) in the test flow channels and flow path 1 which was not coupled to any proteins, was used as an activated blank control. GlcNAc was diluted a variety of concentrations, and then were injected at a flow rate of 30 μL/min and implemented with gradient concentrations from low to high. Binding curves were corrected according to the blank control and data analysis results were calculated with Biacore T100 evaluation software.
Sample preparation
AANL was purified from mushroom A. aegerita as previously reported (Jiang et al. 2012) and conjugated with the resin, Sepharose 4B, to be the Sepharose 4B-AANL resin, which were packed to an affinity column. About 10 mg nuclear proteins were loaded onto an affinity column (Sepharose 4B-AANL). TBS was run over the column at 0.5 mL/min and the remaining proteins were eluted with 20 mM GlcNAc and three fractions, respectively, named “F1, F2 and F3” were collected and their volume as reduced using a 3-kD ultrafiltration device. F1–F3 were incubated with 50% acetone/50% ethanol/0.1% acetic acid at −20°C for one day, and then centrifuged at 4000 rpm for 20 min at 4°C. The precipitations were collected and dissolved with denaturing buffer (8 M urea, 0.2 M Tris PH8.0, 4 mM calcium chloride). These samples were incubated for 45 min at 37°C with 10 mM DTT, and then alkylated with 40 mM iodoacetamide in the dark for 30 min. These mixtures were respectively diluted 4-fold with ultrapure water and trypsin (1:50) was added and digested overnight at 37°C. Digested peptides were desalted using C18 Sep Pak cartridge (Waters) and lyophilized.
Enrichment of GlcNAcylated peptides
The peptides of F1 and F2 were dissolved with phosphate buffer and 100:1 PNGase F was added and incubated for 3 h, after which peptides were desalted and lyophilized. In addition, PNGase F treated peptides were loaded to another affinity column (POROS-AANL), and the tail of the flow-through were collected and then desalted and lyophilized for MS analysis. For the F3, 100 μg digested peptides mixed with 1 μg PNGase F and incubated for 3 h, after which the samples were transferred to 10-kD ultrafiltration device to remove the PNGase F by centrifugation at 14,000 × g at 18°C for 10 min. The treated peptides of F3 were desalted and lyophilized for MS analysis.
LC–MS-MS analysis
Samples were analyzed with an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an online nano-electrospray ion source. Peptides were separated on a 75 μm × 25 cm Acclaim PepMap C18 analytical column with a linear gradient of 2–40% solvent B for 105 min at a flow rate of 300 nL/min. The Orbitrap Fusion mass spectrometer was operated in the data-dependent mode to switch automatically between MS and MS-MS acquisition, with resolution of 120,000 at m/z 200 in full MS over an m/z range of 350–1500, and AGC setting of 200,000 ions. MS-MS acquisition was performed with resolution of 30,000 at m/z 200, and AGC setting of 50,000 ions. Herein, samples were detected with combination of HCD and ETD. The HCD/ETD were performed when one or more oxonium ions (126.055, 138.055, 203.079 and 204.087) were detected in HCD spectrum. In the method of HCD/ETD, the normalized collision energy of HCD was set to 40%, the scan range was set from 120 to 2000 m/z, and AGC target was set to 100,000.
Data analysis
To obtain comprehensive O-GlcNAcylated peptide identification, the database engines, Peaks Studio 7.5, SEQUEST (Proteome Discoverer ver. 1.4) and MASCOT 2.5 were selected. HCD/ETD data were searched against the SwissProt Mus Musculus with 10 ppm precursor tolerance and 0.05 Da fragment tolerance. The enzyme was set to trypsin and the maximum of missed cleavage sites was set two. The fixed modification was cysteine carbamidomethylation (+57.021 Da), and the variable modifications were HexNAc (T/S) (+203.079372 Da) and methionine oxidation (+15.995 Da). Tandem mass spectra were manually confirmed and were examined for the presence of the neutral loss of HexNAc or oxonium ions generated from HexNAc. The FDR was set to 1%.
Supplementary data
Funding
This work was supported by the Natural Science Foundation of China (NSFC) program [31,370,849, 81,670,531, 31,301,426], the Chinese 111 project [B06018] and the Wuhan University project [No. 204–274,103]. Dr Hart is supported by the USA National Institutes of Health [P01HL107153, R01GM116891, R01DK61671, N01-HV-00,240]
Conflict of interest statement
Dr Hart receives a share of royalty received by the university on sales of the CTD110.6 antibody, which are managed by J.H.U. The other authors confirm that this article content has no conflict of interest.
Supplementary Material
Acknowledgements
We would like to thank Yuanyuan Chen, Zhenwei Yang, Professor Dacheng Wang and Professor Defeng Li (Institute of Biophysics, Chinese Academy of Sciences) for technical help with Biacore experiments.
Abbreviations
- AANL
agrocybe aegerita lectin 2
- O-GlcNAc
O-linked N-acetylglucosamine
- OGT
O-GlcNAc transferase
- OGA
O-GlcNAcase
- UDP-GlcNAc
uridine diphosphate N-acetylglucosamine
- PTM
post-translational modification
- BEMAD
β-elimination followed by Michael addition
- HCD
higher-energy collisional dissociation
- ETD
electron transfer dissociation
- DTT
dithiothreitol
- SPR
surface plasmon resonance
References
- Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG 2nd, Shabanowitz J, Stanley P et al. 2012. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci USA. 109:7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audfray A, Beldjoudi M, Breiman A, Hurbin A, Boos I, Unverzagt C, Bouras M, Lantuejoul S, Coll JL, Varrot A et al. 2015. A recombinant fungal lectin for labeling truncated glycans on human cancer cells. PLoS One. 10:e0128190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Hart GW, Cho JW. 2013. Chemical approaches to study O-GlcNAcylation. Chem Soc Rev. 42:4345–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. 2010. O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 1800:96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W. 2011. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 144:376–388. [DOI] [PubMed] [Google Scholar]
- Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. 2009. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci USA. 106:8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. 2014. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J Am Chem Soc. 136:12283–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci G, Mitchell EP, Chazalet V, Debray H, Oscarson S, Lahmann M, Imberty A, 2006. Beta-propeller crystal structure of Psathyrella velutina lectin: an integrin-like fungal protein interacting with monosaccharides and calcium. J Mol Biol. 357:1575–1591. [DOI] [PubMed] [Google Scholar]
- Daou S, Mashtalir N, Hammond-Martel I, Pak H, Yu H, Sui G, Vogel JL, Kristie TM, Affar EB. 2011. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci USA. 108:2747–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayaka S, Jones SP. 2014. O-GlcNAc and the cardiovascular system. Pharmacol Ther. 142:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. 2010. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman A, Roseman S, Moses FE, Ludowieg J, Mayeda M. 1955. The biosynthesis of hyaluronic acid by group A Streptococcus. II. Origin of the N-acetylglucosamine moiety. J Biol Chem. 212:583–591. [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. 2001. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 276:9838–9845. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Blumenthal HJ, Davidson E, Roseman S. 1960. Glucosamine metabolism. V. Enzymatic synthesis of glucosamine 6-phosphate. J Biol Chem. 235:1265–1273. [PubMed] [Google Scholar]
- Gong CX, Liu F, Iqbal K. 2012. O-GlcNAc cycling modulates neurodegeneration. Proc Natl Acad Sci USA. 109:17319–17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. 1997. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11:726–737. [DOI] [PubMed] [Google Scholar]
- Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DM, Agnew B, Kuster B. 2013. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J Proteome Res. 12:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. 2010. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 1800:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. 2007. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 446:1017–1022. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. 2011. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 80:825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartiala JA, Tang WH, Wang Z, Crow AL, Stewart AF, Roberts R, McPherson R, Erdmann J, Willenborg C, Hazen SL et al. 2016. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat Commun. 7:10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt GD, Hart GW. 1986. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 261:8049–8057. [PubMed] [Google Scholar]
- Hu H, Brittain GC, Chang JH, Puebla-Osorio N, Jin J, Zal A, Xiao Y, Cheng X, Chang M, Fu YX et al. 2013. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature. 494:371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Chen Y, Wang M, Yin Y, Pan Y, Gu B, Yu G, Li Y, Wong BH, Liang Y et al. 2012. A novel lectin from Agrocybe aegerita shows high binding selectivity for terminal N-acetylglucosamine. Biochem J. 443:369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang J, Ye X, Su Y, Yu G, Yang Q, Liu W, Yu W, Cai J, Chen X et al. 2016. Identification of GlcNAcylated alpha-1-antichymotrypsin as an early biomarker in human non-small-cell lung cancer by quantitative proteomic analysis with two lectins. Br J Cancer. 114:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22:2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. 2007. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 3:339–348. [DOI] [PubMed] [Google Scholar]
- Koso H, Yi H, Sheridan P, Miyano S, Ino Y, Todo T, Watanabe S. 2016. Identification of RNA-binding protein LARP4B as a tumor suppressor in glioma. Cancer Res. 76:2254–2264. [DOI] [PubMed] [Google Scholar]
- Kuspert M, Murakawa Y, Schaffler K, Vanselow JT, Wolf E, Juranek S, Schlosser A, Landthaler M, Fischer U. 2015. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA. 21:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, Walker S. 2013. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science. 342:1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhao Z, Sun M, Luo J, Xiao Y. 2016. ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life Sci. 147:59–66. [DOI] [PubMed] [Google Scholar]
- Machon O, Baldini SF, Ribeiro JP, Steenackers A, Varrot A, Lefebvre T, Imberty A. 2017. Recombinant fungal lectin as a new tool to investigate O-GlcNAcylation processes. Glycobiology. 27:123–128. [DOI] [PubMed] [Google Scholar]
- Mangone M, Myers MP, Herr W. 2010. Role of the HCF-1 basic region in sustaining cell proliferation. PLoS One. 5:e9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappa D, Selvan N, Borodkin V, Alonso J, Ferenbach AT, Shepherd C, Navratilova IH, vanAalten DMF. 2015. A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development. Biochem J. 470:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino F, Bern M, Mommen GP, Leney AC, van Gaans-van den Brink JA, Bonvin AM, Becker C, van Els CA, Heck AJ. 2015. Extended O-GlcNAc on HLA Class-I-Bound Peptides. J Am Chem Soc. 137:10922–10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. 1991. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 266:4706–4712. [PubMed] [Google Scholar]
- Matone A, Scott-Boyer MP, Carayol J, Fazelzadeh P, Lefebvre G, Valsesia A, Charon C, Vervoort J, Astrup A, Saris WH et al. 2016. Network Analysis of Metabolite GWAS Hits: Implication of CPS1 and the Urea Cycle in Weight Maintenance. PLoS One. 11:e0150495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC, Hanover JA.. 2002. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA. 99:10695–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Daou S, Affar el B, Burlingame A.. 2013. Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics. 13:982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel AK, Schilling M, Comte-Walters S, Berkaw MN, Ball LE.. 2013. Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation. Mol Cell Proteomics. 12:945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruchuri VD, Zachara NE.. 2011. Defining the heart and cardiovascular O-GlcNAcome: a review of approaches and methods. Circulation. Circ Cardiovasc Genet. 4:710. [DOI] [PubMed] [Google Scholar]
- Peng GS, Yin SJ.. 2009. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Hum Genomics. 3:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Li D, Jiang S, Lan X, Hu Y, Sun H, Wang D.. 2015. Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lectin. PLoS One. 10:e0129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Clark PM, Hsieh-Wilson LC.. 2008. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 4:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Kalmokoff M, Aubry A, Thibault P, Sandoz M, Logan SM.. 2004. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J Bacteriol. 186:6721–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvan N, Williamson R, Mariappa D, Campbell DG, Gourlay R, Ferenbach AT, Aristotelous T, Hopkins-Navratilova I, Trost M, van Aalten DMF.. 2017. A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat Chem Biol. 13:882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Kamp HD, Grundling A, Higgins DE.. 2006. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20:3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Copeland RJ, Hart GW.. 2010. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem Sci. 35:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J, Koehler CM.. 2014. The great escape: Mgr2 of the mitochondrial TIM23 translocon is a gatekeeper tasked with releasing membrane proteins. Mol Cell. 56:613–614. [DOI] [PubMed] [Google Scholar]
- Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ.. 2010. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 6:338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CR, Hart GW.. 1984. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 259:3308–3317. [PubMed] [Google Scholar]
- Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL.. 2012. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 11:215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Herr W.. 2009. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 28:3185–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosseller K, Hansen KC, Chalkley RJ, Trinidad JC, Wells L, Hart GW, Burlingame AL.. 2005. Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics. 5:388–398. [DOI] [PubMed] [Google Scholar]
- Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA et al. 2006. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 5:923–934. [DOI] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, Hart GW.. 2010. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 9:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW.. 2010. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 3:ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW.. 2002. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 1:791–804. [DOI] [PubMed] [Google Scholar]
- Woosley B, Xie M, Wells L, Orlando R, Garrison D, King D, Bergmann C.. 2006. Comprehensive glycan analysis of recombinant Aspergillus niger endo-polygalacturonase C. Anal Biochem. 354:43–53. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Herr W.. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Anal Biochem. 28:294–304. [DOI] [PubMed] [Google Scholar]
- Xu SL, Chalkley RJ, Maynard JC, Wang W, Ni W, Jiang X, Shin K, Cheng L, Savage D, Hühmer AF et al. 2017. Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc Natl Acad Sci USA. 114:e1536–e1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA, Vocadlo DJ.. 2014. O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Circ Cardiovasc Genet. 43:6839–6858. [DOI] [PubMed] [Google Scholar]
- Zargar Z, Tyagi S.. 2012. Role of host cell factor-1 in cell cycle regulation. Transcription. 3:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaro BW, Yang YY, Hang HC, Pratt MR.. 2011. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci USA. 108:8146–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan Q, Hart GW.. 2010. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci. 123:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Yin R, Yang X.. 2014. O-GlcNAc: a bittersweet switch in liver. Front Endocrinol (Lausanne). 5:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Viner R, Teo CF, Boons GJ, Horn D, Wells L.. 2011. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J Proteome Res. 10:4088–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu TW, Cecioni S, Eskandari R, Zandberg WF, Vocadlo DJ.. 2015. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat Chem Biol. 11:319–325. [DOI] [PubMed] [Google Scholar]
- Zou LH, Shang ZF, Tan W, Liu XD, Xu QZ, Song M, Wang Y, Guan H, Zhang SM, Yu L et al. 2015. TNKS1BP1 functions in DNA double-strand break repair though facilitating DNA-PKcs autophosphorylation dependent on PARP-1. Oncotarget. 6:7011–7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.