Abstract
Motivation
As part of the NIH Library of Integrated Network-based Cellular Signatures program, hundreds of thousands of transcriptomic signatures were generated with the L1000 technology, profiling the response of human cell lines to over 20 000 small molecule compounds. This effort is a promising approach toward revealing the mechanisms-of-action (MOA) for marketed drugs and other less studied potential therapeutic compounds.
Results
L1000 fireworks display (L1000FWD) is a web application that provides interactive visualization of over 16 000 drug and small-molecule induced gene expression signatures. L1000FWD enables coloring of signatures by different attributes such as cell type, time point, concentration, as well as drug attributes such as MOA and clinical phase. Signature similarity search is implemented to enable the search for mimicking or opposing signatures given as input of up and down gene sets. Each point on the L1000FWD interactive map is linked to a signature landing page, which provides multifaceted knowledge from various sources about the signature and the drug. Notably such information includes most frequent diagnoses, co-prescribed drugs and age distribution of prescriptions as extracted from the Mount Sinai Health System electronic medical records. Overall, L1000FWD serves as a platform for identifying functions for novel small molecules using unsupervised clustering, as well as for exploring drug MOA.
Availability and implementation
L1000FWD is freely accessible at: http://amp.pharm.mssm.edu/L1000FWD.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
As part of the NIH LINCS project, the Broad Institute LINCS Center for Transcriptomics has generated over 1.3 million transcriptomic profiles utilizing the L1000 technology (Subramanian et al., 2017). A large portion of this L1000 data comprises of drug perturbations of human cell lines. The gene expression profiles collected by the L1000 technology can be used to generate hundreds of thousands drug-induced gene expression signatures. Such collection of signatures is a valuable catalog for finding connections between drugs, genes and diseases; and for discovering mechanisms-of-action (MOA) for less understood drugs and small-molecule compounds. The catalog of signatures also provides opportunities for drug repurposing. For instance, by performing network analysis, (Iorio et al., 2010) discovered that densely interconnected drug communities are enriched for drugs with similar MOA. Such drug similarity network was later developed into the web tool Mantra (Carrella et al., 2014). In comparison with L1000FWD, Mantra aggregates signatures across cell line, dose and time points and visualizes a network of only FDA approved drugs. By clustering and visualizing the expression signatures of >2000 pre-clinical investigational compounds together with approved drugs, L1000FWD directly proposes indications and reveals potential MOA for many new potential therapeutics as direct hypotheses that could be further tested. Matching disease signatures with drug signatures has also been systematically evaluated as an effective approach for discovering novel indications for drugs (Cheng et al., 2014). Tools such as DvD (Pacini et al., 2013) and L1000CDS2 (Duan et al., 2016) have been developed to match disease/drug signatures for drug repurposing. In addition to matching signatures based on their similarity, the connectivity mapping approach was shown to be useful for predicting adverse drug reactions using machine learning (Wang et al., 2016). The newly generated LINCS L1000 data are a more comprehensive catalog of expression signatures than the original Connectivity Map (Lamb, 2006). However, it is not trivial to develop methods to enable exploration of this massive dataset. Advances in web-based interactive visualization now enable responsive complex animated displays of millions of data points. In particular, the HTML5 canvas element provides the ability of rendering bitmaps using scripting in the browser. This feature is realized with new JavaScript libraries that implement WebGL (Parisi, 2012), a version of OpenGL for web applications. Here, we present L1000FWD, a web-based data visualization application that interactively visualizes thousands of drug-induced gene expression signatures from the LINCS L1000 dataset using these new technologies.
2 Materials and methods
Preparation of signatures and network construction: Drug/small-molecule perturbation profiles from the quantile normalized LINCS L1000 data (Level 3) were first adjusted for batch effects using a normalization procedure (Iskar et al., 2010). Then, drug/small-molecule perturbation signatures were computed using the Characteristic Direction method (Clark et al., 2014). The quality of the signatures was evaluated using the consistency between drug treatment replicates measured by an empirical permutation test (Niepel et al., 2017). More details about the method can be found in the Supplementary Material. A total of 34 502 significant drug perturbation signatures were used to construct an adjacency matrix of signatures. This adjacency matrix contains the pairwise cosine similarity between signatures in the space of the 978 landmark genes, with negative values set to 0. Edges with cosine similarity larger than the 99.95% percentile (>0.61) were kept, resulting in a weighted undirected graph made of 18 082 nodes (signatures) and 595 177 edges. Connected components with less than 10 nodes were removed from the graph, resulting in a graph made of 16 848 nodes and 594 372 edges. The final graph of signatures covers 68 distinct cell lines, 3237 drugs/compounds, 3 time points and 132 dosages. The graph layout was achieved using the Allegro edge-repulsive strong clustering algorithm implemented in Cytoscape (Shannon, 2003) with 10 000 iterations.
Development of the web application: To build the L1000FWD web application, all the 16 848 signatures, together with their metadata, are stored in a MongoDB database. Python Flask web framework was utilized as the backend to interact with the database through custom object-relational map model (ORMs) pings. To interactively visualize the large network of drug-induced signatures, the HTML5 canvas element was used to layout the network; the JavaScript libraries three.js (Dirksen, 2013) and d3.js (Bostock et al., 2011) were used to handle the zooming, panning, searching, coloring and shaping functionalities and interactivity; the JavaScript library backbone.js (Osmani, 2013) was used to handle event delegations between the visualization and the multiple controllers.
3 Results
Within the L1000FWD fireworks display, each node represents a drug-induced gene expression signature. The shape and color of nodes can be changed according to their associated attributes such as cell line, structural scaffold, MOA, time point and batch, through controls placed near the map. Signatures are clustered by their expression similarity. By hovering over a signature, a user can access more information about the signature including the drug name, cell line, concentration, time point and the ID of the signature (‘sig_id’). Each signature is hyperlinked to a landing page that contains information about the drug and the signature. Available information includes common diagnoses and co-prescribed drugs harvested from the Mount Sinai EHR, whereas up and down regulated genes induced by the signature can be piped into Enrichr (Chen et al., 2013) for gene set enrichment analyses. Among the visually identifiable clusters, many clusters contain signatures from the different cell types while sharing common MOA (Fig. 1). For instance, anti-cancer drugs that inhibit cell cycle such as CDK inhibitors and Topoisomerase inhibitors forms a large cluster consisting of 15 cell lines, suggesting that the cell cycle arrest response induced by these small-molecules is universal across cell lines. Interestingly, this cluster also contains small molecules not previously known to be cell cycle inhibitors, for example, BRD-K79222491, a molecular probe compound with unknown function. Hence, many pre-clinical compounds within the clusters that contain diverse cell type, and currently without known, or clear, MOA, could be directly inferred to share similar MOA of the well-studied drugs found in the same cluster. In addition, the L1000FWD map allows users to project their own custom signatures onto the map for identifying where their signature fall in the global expression space. The signature similarity search results are available for sharing and download. In summary, L1000FWD provides an advancement in the interactive visualization of complex high dimensional data, enabling the discovery of MOA for known and novel drugs facilitating pre-clinical drug discovery and drug repositioning opportunities.
Fig. 1.
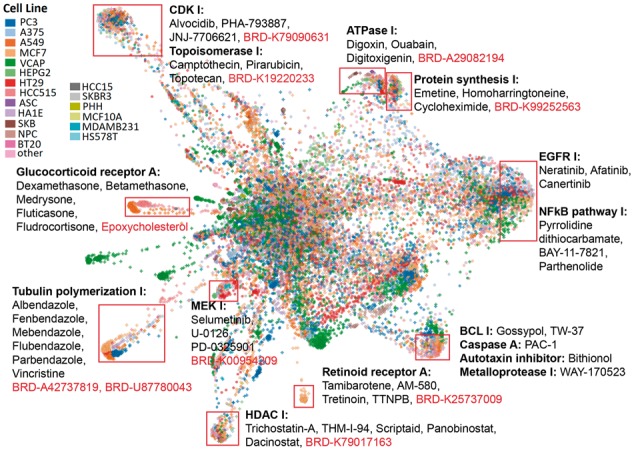
L1000FWD visualization of drug-induced signatures. Signatures are colored by the cell lines. Clusters of drugs that share MOA are highlighted with few example drugs having known MOA (black) as well as examples of small molecules without known MOA (red). I–inhibitors; A–agonists/activators
Supplementary Material
Acknowledgments
Funding
This work was partially supported by the NIH Common Fund grants U54HL127624, U24CA224260 and U54CA189201 to A.M.
Conflict of Interest: none declared.
References
- Bostock M. et al. (2011) Data-driven documents. IEEE Trans.Visualization Computer Graphics, 17, 2301–2309. [DOI] [PubMed] [Google Scholar]
- Carrella D. et al. (2014) Mantra 2.0: an online collaborative resource for drug mode of action and repurposing by network analysis. Bioinformatics, 30, 1787–1788. [DOI] [PubMed] [Google Scholar]
- Chen E.Y. et al. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. et al. (2014) Systematic evaluation of connectivity map for disease indications. Genome Med., 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N. et al. (2014) The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics, 15, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen J. (2013) Learning Three. js: The JavaScript 3D Library for WebGL. Packt Publishing Ltd, Birmingham, UK. [Google Scholar]
- Duan Q. et al. (2016) L1000CDS2: LINCS L1000 characteristic direction signatures search engine. Npj Syst. Biol. Appl., 2, 16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F. et al. (2010) Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci., 107, 14621–14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskar M. et al. (2010) Drug-induced regulation of target expression. PLoS Comput. Biol., 6, e1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. et al. (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science, 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Niepel M. et al. (2017) Common and cell-type specific responses to anti-cancer drugs revealed by high throughput transcript profiling. Nat. Comm., 8, 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. (2013) Developing Backbone. js Applications ‘O’Reilly Media, Inc.’, Sebastopol, CA. [Google Scholar]
- Pacini C. et al. (2013) DvD: an R/Cytoscape pipeline for drug repurposing using public repositories of gene expression data. Bioinformatics, 29, 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi T. (2012) WebGL: Up and Running ‘O’Reilly Media, Inc.’, Sebastopol, CA. [Google Scholar]
- Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. (2017) A next generation connectivity map: L 1000 platform and the first 1, 000, 000 profiles. Cell, 171, 1437–1452.e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2016) Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics, 32, 2338–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


