Abstract
Different regions of the mammalian nose smell different odors. In the mouse olfactory system, spatially regulated expression of >1000 olfactory receptors (ORs) along the dorsomedial–ventrolateral (DV) axis forms a topological map in the main olfactory epithelium (MOE). However, the locations of most ORs along the DV axis are currently unknown. By sequencing mRNA of 12 isolated MOE pieces, we mapped out the DV locations—as quantified by “zone indices” on a scale of 1–5—of 1033 OR genes with an estimated error of 0.3 zone indices. Our map covered 81% of all intact OR genes and 99.4% of the total OR mRNA abundance. Spatial regulation tended to vary gradually along chromosomes. We further identified putative non-OR genes that may exhibit spatial expression along the DV axis.
Keywords: gene expression, olfactory receptors, olfactory sensory neurons, RNA sequencing, spatial transcriptomics, topological map
Introduction
The olfactory system relies on a topological map of olfactory receptor (OR) expression. The first stage of map formation occurs in the main olfactory epithelium (MOE), where different OR genes are expressed in different, yet partially overlapping, regions along the dorsomedial–ventrolateral (DV) axis. Spatially regulated OR expression was first discovered in rodents (Ressler et al. 1993; Vassar et al. 1993) and later observed in fish (Weth et al. 1996), insects (Vosshall et al. 1999), and primates (Horowitz et al. 2014). This expression pattern, previously known as discrete “zones” (Ressler et al. 1993; Vassar et al. 1993) but later found to be more continuous (Miyamichi et al. 2005), is crucial for downstream projection to the main olfactory bulb (Ressler et al. 1994; Vassar et al. 1994; Mombaerts et al. 1996), and may contribute to the establishment of the “one-neuron-one-receptor” rule (Chess et al. 1994; Malnic et al. 1999; Hanchate et al. 2015; Tan et al. 2015). However, existing spatial information either had a limited resolution (Zhang et al. 2004) or was available only for a small subset of <100 ORs (Miyamichi et al. 2005), and was limited for non-OR genes (Miyawaki et al. 1996; Yoshihara et al. 1997; Norlin et al. 2001; Oka et al. 2003; Tietjen et al. 2003; Gussing and Bohm 2004; Ling et al. 2004; Whitby-Logan et al. 2004; Tietjen et al. 2005; Duggan et al. 2008; Vedin et al. 2009). In this work, we generated a near-complete map of 1033 mouse ORs by mRNA sequencing (Tan et al. 2015; Hanchate et al. 2015; Ibarra-Soria et al. 2014; Saraiva et al. 2015; Ibarra-Soria et al. 2017; Scholz et al. 2016; Kanageswaran et al. 2015; Greer et al. 2016; Plessy et al. 2012; Magklara et al. 2011; Keydar et al. 2013; Shiao et al. 2012; Jiang et al. 2015; Von Der Weid et al. 2015; Fletcher et al. 2017) of isolated MOE pieces, and identified novel non-OR genes that may exhibit spatial expression along the DV axis.
Materials and methods
Animals
All mouse experiments were performed in accordance with relevant guidelines and regulations. Animal protocols were approved by Harvard IACUC. Adult (3-month-old) mice from the inbred strain C57BL/6NTac (Taconic) were used.
Published data
Known zone indices of 82 ORs were downloaded from Supplementary Table 1 of (Miyamichi et al. 2005) and converted to modern gene names through the Mouse Genome Informatics (Supplementary Table 4), among which 77 were sufficiently detected in our experiment.
RNA-Seq experiments
Small pieces were isolated by forceps from dissected MOE. Total RNA was extracted with an RNeasy Mini Kit (Qiagen) and a TissueLyser II (Qiagen) (25 Hz for 2 min, invert, and another 2 min, with 5 mm stainless steel beads), quantified by a NanoDrop 2000 (Thermo), and assessed by an RNA 6000 Pico Kit on a Bioanalyzer 2100 (Agilent).
From each piece, we extracted 0.1–1.0 ug of total RNA (Supplementary Table 1), compared to 10–20 ug from a typical whole MOE.
Libraries were prepared with a TruSeq RNA Sample Prep Kit v2 (Illumina) with poly(A) selection, quantified by a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) and a High Sensitivity DNA Analysis Kit on a Bioanalyzer 2100 (Agilent), and sequenced on a HiSeq 2500 (Illumina).
Data analysis
RNA-Seq reads were mapped to the mouse reference genome GRCm38.p5 (downloaded from the GENCODE M12 release) with tophat v2.0.11 (with default parameters) (Kim et al. 2013). Expression levels were quantified with cufflinks v2.2.1 (with parameters “-u --max-bundle-frags 100000000”) (Trapnell et al. 2010) based on the comprehensive gene annotation (ALL) from the above GENCODE release. Restricting quantification to reads that have the highest mapping quality (of 50) had no impact on the results. Transcripts per million (TPM) values were calculated after removing genes that have a gene type beginning with “Mt_”, “miRNA”, “rRNA”, “scRNA”, “snRNA”, “snoRNA” “sRNA”, “scaRNA”, or “vaultRNA”. Gene descriptions were downloaded from Ensembl BioMart (mouse genes GRCm38.p5). OR genes (1417 genes) were defined as having “Olfr” in the gene name (1419 entries, 4 pairs of which refer to the same genes: ENSMUSG00000109148 and ENSMUSG00000074985 are both Olfr1452-ps1, ENSMUSG00000109806 and ENSMUSG00000073919 are both Olfr663, ENSMUSG00000109020 and ENSMUSG00000061501 are both Olfr197, and ENSMUSG00000063732 and ENSMUSG00000110991 are both Olfr908) or having “olfactory receptor” in the gene description (2 additional genes, “Olrf445-ps1” and “OR4P4”). Pseudogenes (5580 genes, 189 of which are ORs) were defined as having “pseudogene” in the gene type.
For spatial inference, expression levels (in TPMs) were normalized such that the average is 1 for each gene across the 12 MOE pieces. Two-dimensional t-SNE was performed with its MATLAB implementation on pair-wise Euclidean distances between normalized expression values (the “tsne_d” function, with a default perplexity of 30). Below is a step-by-step guide for the inference procedure:
1. FPKM values of each gene were loaded from the output of cufflinks.
2. Certain genes were removed (see above).
3. For each tissue piece, TPM values were calculated by dividing each FPKM value by the total FPKM value in that piece and then multiplying by 106.
4. Only genes with an average TPM of at least 0.5 across the 12 tissue pieces were retained (“sufficiently detected”).
5. For each gene, TPM values were normalized by its summed TPM across the 12 tissue pieces (yielding its “normalized expression values”).
6. Assignment of OR genes to the unusual location of Olfr459: after performing t-SNE on all retained OR genes, the 22 ORs in the smaller cluster were assigned to the unusual location.
7. Assignment of non-OR genes to the unusual location of Olfr459: a non-OR gene was potentially in the unusual location if the Euclidean distance between its normalized expression values (a 12-dimensional vector) and that of Olfr459 was ≤4.
8. Creation of the standard curve: For each value of x from 1 to 5 with an increment of 0.05, all OR genes with known zone indices between x ± 0.5 were selected. A 12-dimensional vector y was calculated by averaging the normalized expression values across the selected genes. In this way, a 12-dimensional standard curve y = f(x) was created, which mapped each zone index x to its expected (smoothed) normalized expression values across the 12 tissue pieces.
9. Inference of zone indices: For each gene, the Euclidean distance between its normalized expression values (a 12-dimensional vector) and each point on the standard curve (y = f(x) for each x from 1 to 5 with an increment of 0.05) was calculated. The zone index was inferred as the value x that led to the shortest distance.
10. Filtering for non-OR genes: a non-OR gene was potentially spatially regulated along the DV axis if (1) its average TPM was at least one across the 12 tissue pieces, (2) its coefficient of variation (CV) was at least 0.5 across the 12 pieces, and (3) its shortest distance was ≤2 (or ≤1.5 for the more stringent list) from the standard curve.
In the text, the average difference in zone indices given a genomic separation was calculated from all OR pairs with log10 genomic separations within ±0.1 of the desired value, regardless of directionality (upstream or downstream).
Results
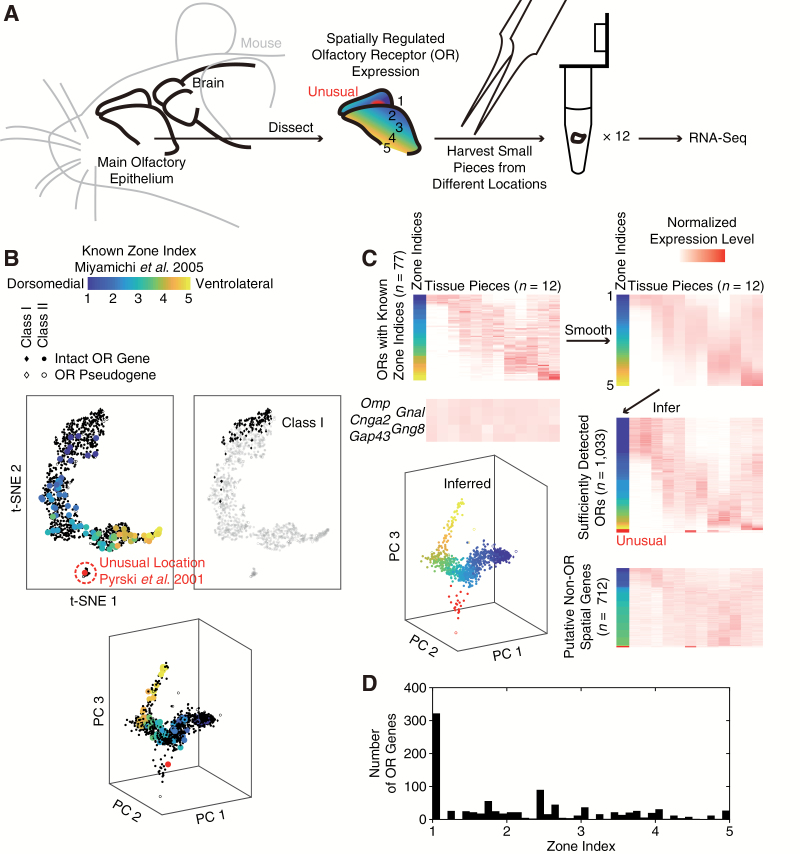
To investigate the spatial pattern of gene expression in the mouse MOE, we sequenced mRNA from 12 isolated MOE pieces throughout the DV axis of both the septum and the turbinates with an average of 12.5 million single-end 100-bp reads per piece (min = 4.9 million, max = 21.0 million) (Supplementary Tables 1–3). In mice, the expression region of each OR assumes a complex shape that approximates a cylindrical shell, concentric to each other (Ressler et al. 1993). Following a previous study (Miyamichi et al. 2005), we quantified the DV location of each OR by its “zone index”, where a zone index of 1 represents the most dorsomedial region of the MOE, a zone index of 5 (previously known as “zone 4b”) represents the most ventrolateral, and the zone index of each OR is not necessarily an integer (Figure 1A). The geometry is further complicated by an unusual, medial location of Olfr459 (also known as OR-Z6 or MOR120-1, orthologous to human OR9A2) (Pyrski et al. 2001) (red region in Figure 1A). We tackle the problem of complex geometry by isolating 12 very small tissue pieces across the MOE, each of which would contain only a small portion of the entire DV axis. In this way, the expression pattern of each OR across the 12 MOE pieces would reflect its location along the DV axis. Indeed, normalized expression levels of the 1033 OR genes that were sufficiently detected [defined as having an average of at least 0.5 transcripts per million (TPM)] showed a prominent pattern of differential expression across the 12 pieces, as visualized by t-distributed stochastic neighbor embedding (t-SNE) (Maaten and Hinton 2008) and principal component analysis (PCA) (black points in Figure 1B). This pattern reflected spatial expression along the DV axis, because on the t-SNE and PCA plots a small subset of 78 ORs of known zone indices (Pyrski et al. 2001; Miyamichi et al. 2005) were distributed according to their zone indices (colored points in Figure 1B) (Supplementary Table 4). In particular, on the t-SNE plot, Olfr459 of the unusual location resided in a small, separate cluster at the bottom, while the other 77 ORs with zone indices of 1–5 followed a continuous “trajectory” along the larger cluster.
Figure 1.
Spatial-transcriptomic mapping of 1033 OR genes and 712 putative non-OR genes along the dorsomedial–ventrolateral axis of the mouse MOE. (A) We investigated the spatial pattern of gene expression by sequencing mRNA from 12 tissue pieces isolated across the mouse MOE. Each piece would contain only a small portion of the entire DV axis. (B) Normalized expression levels of the 1033 OR genes (diamonds for Class I ORs (Zhang and Firestein 2002) and dots for Class II) that were sufficiently detected showed a prominent pattern of spatial expression across the 12 pieces, as visualized by 78 ORs of known zones (colored points) (Miyamichi et al. 2005) via t-distributed stochastic neighbor embedding (t-SNE) and PCA. We assigned the 22 ORs in the smaller t-SNE cluster (red dashed circle) to the unusual location of Olfr459 (red dot). (C) We made a standard curve from a zone index of 1 to a zone index of 5 by smoothing the normalized expression levels of the 77 known ORs (top heatmaps), and inferred the zone indices of the 1033 ORs by finding the closest point on the standard curve (middle right heatmap). Results of the inference were visualized on a PCA plot. Putative non-OR genes that may exhibit spatial expression along the DV axis were selected based on expression levels, uniformity across the MOE pieces, and distance to the standard curve or to Olfr459 (bottom heatmap). In contrast, known marker genes (Omp, Gnal, Cnga2 for mature neurons, and Gng8, Gap43 for immature neurons) exhibited uniform expression (middle left heatmap). (D) Distribution of inferred zone indices of the 1033 OR genes. Consistent with a previous report (Miyamichi et al. 2005), zone indices were continuous along the DV axis except for the most dorsomedial region (left: zone index = 1).
We inferred the zone indices of 992 intact OR genes (81% of all 1228, based on the GENCODE annotation) and 41 OR pseudogenes (22% of all 189) using the observed expression patterns of 78 known ORs as standards. Among the 1033 ORs that were sufficiently detected, we began by assigning the 22 ORs in the smaller t-SNE cluster (Figure 1B) to the unusual location (Pyrski et al. 2001), showing for the first time that the unusual expression pattern of Olfr459 is not an isolated case. For the remaining ORs, which would have zone indices between 1 and 5, we first made a 12-dimensional standard curve from a zone index of 1 to a zone index of 5 by smoothing the normalized expression levels of the 77 known ORs (Miyamichi et al. 2005) with a half window size of ± 0.5 zone indices (top heatmaps in Figure 1C). The zone index of each OR was then inferred by finding the closest point (as measured by Euclidean distance) on the standard curve (middle heatmap and visualization on a PCA plot in Figure 1C) (Supplementary Table 5). Uncertainties in zonal inference could be estimated by leave-one-out cross-validation (LOOCV) on the 77 known ORs, yielding a root-mean-square error of 0.3 zone indices (min = 0.0, max = 1.1). Together, these ORs accounted for 99.4% of the total OR mRNA abundance (as measured by average TPMs) in the MOE. Consistent with a previous report (Miyamichi et al. 2005), zone indices were continuous along the DV axis except for the most dorsomedial region (zone index = 1) (Figure 1D).
We further identified 666 intact non-OR genes and 46 non-OR pseudogenes that may exhibit spatial expression along the DV axis. The large number of annotated genes in the mouse genome necessitated stringent criteria. We removed genes whose expression was either low (average TPM < 1) or uniform across the 12 pieces (CV < 0.5). Among the remaining 3228 non-OR genes, putative spatially regulated genes were selected based on Euclidean distance to the aforementioned standard curve (699 candidate genes with distance ≤ 2) or to the unusual zone’s Olfr459 (13 genes with distance ≤ 4) (bottom heatmap in Figure 1C, Supplementary Table 6). This list included known zonal genes Acsm4 (also known as O-MACS) (Oka et al. 2003), Nqo1 (Gussing and Bohm 2004), Ncam2 (also known as OCAM) (Yoshihara et al. 1997), Foxg1 (Duggan et al. 2008), Eya2 (Tietjen et al. 2003), Msx1, Nrp2 (Norlin et al. 2001), Gstm5 (Whitby-Logan et al. 2004), and trace-amine-associated receptors (TAARs) (Liberles and Buck 2006). Novel genes included transcription factors (Six3, Yy2, Isl1, Prdm16, Tbx3/15, Bach2, Foxa1, Pitx1, Tcea3/l3/l5, Npas3/4, Dlx3, Twist1/2, E2f2/8, Zfp97/365/382/950), chemokines (Cxcl10/14, Ccl5/8), cytochromes (Ling et al. 2004) (Cyp2a4/2b10/2c44/2e1/7b1), and aldehyde dehydrogenases (Norlin et al. 2001) (Aldh1a7/3a1/3b1). The false discovery rate (FDR) could be estimated to be 51% by random permutation of MOE-piece labels. A more stringent list of 202 genes (distance ≤1.5 to the standard curve) was also created (Supplementary Table 7), with an estimated FDR of 18%. Note that the distribution of zone indices in our lists may not reflect the distribution of all spatially regulated non-OR genes, because our inference method might have different sensitivities in different regions and some non-OR genes may exhibit broader regions than those of OR genes. These genes provide promising candidates for the spatial regulation of OR expression and of other cellular characteristics, e.g. cilia lengths (Challis et al. 2015).
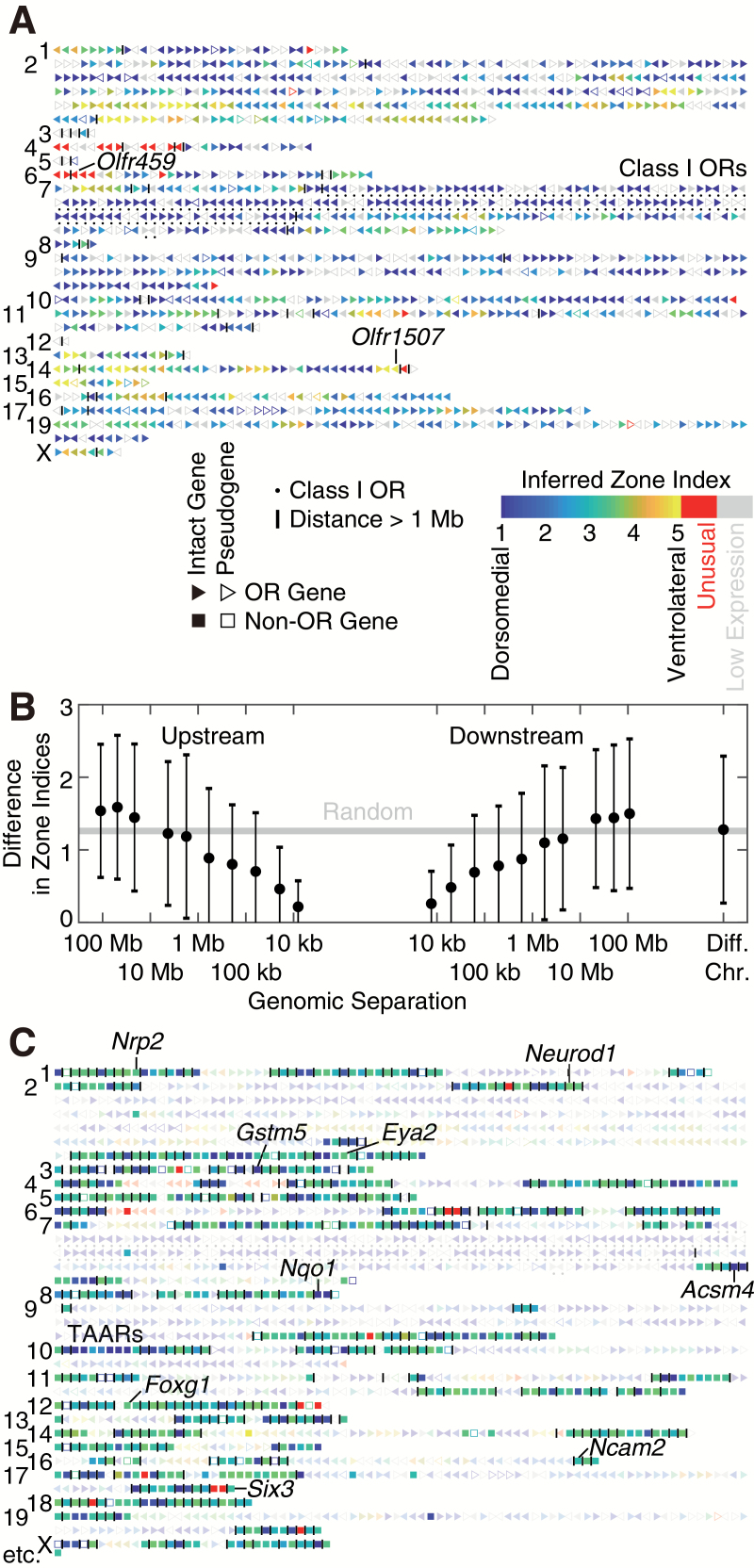
Distribution of zone indices along the mouse genome provides insights into the mechanism of spatial regulation. Within each OR gene cluster, inferred zone indices typically vary gradually along the chromosome, although drastic changes between adjacent ORs occasionally occur (Figure 2A). On average, 2 ORs differed by an average of 0.3, 0.8, 0.9, or 1.3 zone indices, respectively, when they are separated by 10, 100 kb, 1 Mb, or 2 different chromosomes (Figure 2B). This hints to a model where the pattern of chromatin silencing morphs continuously along the DV axis, in each region of the MOE exposing only a subset of ORs—namely, ORs with corresponding zone indices—to some universal machinery of transcriptional activation. This model is consistent with a recent observation that removal of H3K9-mediated heterochromatic silencing obliterated spatial expression of ORs, among other changes in OR expression (Lyons et al. 2014). In addition, genomic locations of drastic spatial differences may harbor important regulatory sites for OR silencing. Although most non-OR candidates reside in separate genomic locations (Figure 2C), several are embedded in OR clusters and share DV locations with nearby ORs, likely a byproduct of chromatin organization.
Figure 2.
Distribution of zone indices across the mouse genome. (A) Spatial information for all 1417 annotated ORs (triangles, with ▶ denoting the plus strand and ◀ denoting the minus strand). Typically, inferred zone indices vary gradually along the chromosome, although drastic differences between adjacent ORs can be occasionally seen. Numbers on the left denote chromosomes. Grey denotes insufficient detection because of low expression levels. OR clusters are separated by vertical lines. (B) Nearby ORs tend to have similar zone indices. All pairs of ORs from the canonical regions (zone indices between 1 and 5) were grouped into bins according to their log10 genomic separation (in base pairs, upstream or downstream) with a bin size of 0.5. Bins with fewer than 10 OR pairs were excluded. Each black dot denotes the average difference in zone indices and the average log10 genomic separation in each bin. Error bar denotes standard deviation (SD); standard error of the mean (SEM) is smaller than the size of the dot. The gray line (“random”) denotes the expected average difference in zones if ORs are randomly permuted, which equals to the average pairwise difference between all pairs of ORs. (C) Spatial information for 712 non-OR candidates (squares), displayed together with ORs (triangles, faded for visual clarity).
Discussion
Our results provide by far the most complete spatial map of mouse ORs in the MOE, covering an order of magnitude more OR genes than state-of-the-art results obtained by traditional in situ hybridization (Miyamichi et al. 2005). Future work includes improving spatial resolution and inferring lowly expressed ORs by deeper sequencing of more MOE pieces, improving spatial inference by allowing variable kernel sizes and investigating expression patterns within the most dorsomedial region, mapping out the second stage of the olfactory map by applying the same spatial-transcriptomic approach to the olfactory bulb, and comparing spatial expression between mouse and human.
Supplementary material
Supplementary material can be found at Chemical Senses online.
Funding
This work was supported by a Transformative Research Award (grant number R01 EB010244) and a Director’s Pioneer Award (grant number DP1 CA186693) from the National Institutes of Health to X.S.X. L.T. was supported by an Howard Hughes Medical Institute International Student Research Fellowship.
Conflict of interests
The authors declare that they have no conflict of interest.
Acknowledgements
The authors thank Stavros Lomvardas and Kevin Monahan (Columbia University), Chenghang Zong (Baylor College of Medicine), Catherine Dulac (Harvard University), Yang Shi (Boston Children’s Hospital), Stephen Liberles, Qian Li, Sean Megason, and Tim Mitchison (Harvard Medical School) for helpful discussion. Raw sequencing data were deposited at the National Center for Biotechnology Information with accession number SRP127539 at the following link: http://www.ncbi.nlm.nih.gov/sra/SRP127539.
References
- Challis RC, Tian H, Wang J, He J, Jiang J, Chen X, Yin W, Connelly T, Ma L, Yu CR, et al. 2015. An olfactory cilia pattern in the mammalian nose ensures high sensitivity to odors. Curr Biol. 25:2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. [DOI] [PubMed] [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. 2008. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 28:5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, et al. 2017. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell. 20:817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Bear DM, Lassance JM, Bloom ML, Tsukahara T, Pashkovski SL, Masuda FK, Nowlan AC, Kirchner R, Hoekstra HE, et al. 2016. A family of non-GPCR chemosensors defines an alternative logic for mammalian olfaction. Cell. 165:1734–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussing F, Bohm S. 2004. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci. 19:2511–2518. [DOI] [PubMed] [Google Scholar]
- Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, Pachter L, Trapnell C, Buck LB. 2015. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 350:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz LF, Saraiva LR, Kuang D, Yoon KH, Buck LB. 2014. Olfactory receptor patterning in a higher primate. J Neurosci. 34:12241–12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW. 2014. The olfactory transcriptomes of mice. PLoS Genet. 10:e1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X, Nakahara TS, Lilue J, Jiang Y, Trimmer C, Souza MA, Netto PH, Ikegami K, Murphy NR, Kusma M, et al. 2017. Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated. Elife. 6:e21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H. 2015. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat Neurosci. 18:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanageswaran N, Demond M, Nagel M, Schreiner BS, Baumgart S, Scholz P, Altmüller J, Becker C, Doerner JF, Conrad H, et al. 2015. Deep sequencing of the murine olfactory receptor neuron transcriptome. PLoS One. 10:e0113170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keydar I, Ben-Asher E, Feldmesser E, Nativ N, Oshimoto A, Restrepo D, Matsunami H, Chien MS, Pinto JM, Gilad Y, et al. 2013. General olfactory sensitivity database (GOSdb): candidate genes and their genomic variations. Human Mutat. 34:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. 2006. A second class of chemosensory receptors in the olfactory epithelium. Nature. 442:645–650. [DOI] [PubMed] [Google Scholar]
- Ling G, Gu J, Genter MB, Zhuo X, Ding X. 2004. Regulation of cytochrome P450 gene expression in the olfactory mucosa. Chem Biol Interact. 147:247–258. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Magklara A, Goh T, Sampath SC, Schaefer A, Schotta G, Lomvardas S. 2014. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 9:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten LVD, Hinton G. 2008. Visualizing data using t-SNE. J Mach Learn Res. 9:2579–2605. [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. 2011. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 145:555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. 2005. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 25:3586–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Homma H, Tamura H, Matsui M, Mikoshiba K. 1996. Zonal distribution of sulfotransferase for phenol in olfactory sustentacular cells. EMBO J. 15:2050–2055. [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. 1996. Visualizing an olfactory sensory map. Cell. 87:675–686. [DOI] [PubMed] [Google Scholar]
- Norlin EM, Alenius M, Gussing F, Hägglund M, Vedin V, Bohm S. 2001. Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol Cell Neurosci. 18:283–295. [DOI] [PubMed] [Google Scholar]
- Oka Y, Kobayakawa K, Nishizumi H, Miyamichi K, Hirose S, Tsuboi A, Sakano H. 2003. O-MACS, a novel member of the medium-chain acyl-CoA synthetase family, specifically expressed in the olfactory epithelium in a zone-specific manner. Eur J Biochem. 270:1995–2004. [DOI] [PubMed] [Google Scholar]
- Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, et al. 2012. Promoter architecture of mouse olfactory receptor genes. Genome Res. 22:486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrski M, Xu Z, Walters E, Gilbert DJ, Jenkins NA, Copeland NG, Margolis FL. 2001. The OMP-lacZ transgene mimics the unusual expression pattern of OR-Z6, a new odorant receptor gene on mouse chromosome 6: implication for locus-dependent gene expression. J Neurosci. 21:4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 73:597–609. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. 1994. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 79:1245–1255. [DOI] [PubMed] [Google Scholar]
- Saraiva LR, Ibarra-Soria X, Khan M, Omura M, Scialdone A, Mombaerts P, Marioni JC, Logan DW. 2015. Hierarchical deconstruction of mouse olfactory sensory neurons: from whole mucosa to single-cell RNA-seq. Sci Rep. 5:18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P, Kalbe B, Jansen F, Altmueller J, Becker C, Mohrhardt J, Schreiner B, Gisselmann G, Hatt H, Osterloh S. 2016. Transcriptome analysis of murine olfactory sensory neurons during development using single cell RNA-Seq. Chem Senses. 41:313–323. [DOI] [PubMed] [Google Scholar]
- Shiao MS, Chang AY, Liao BY, Ching YH, Lu MY, Chen SM, Li WH. 2012. Transcriptomes of mouse olfactory epithelium reveal sexual differences in odorant detection. Genome Biol Evol. 4:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Li Q, Xie XS. 2015. Olfactory sensory neurons transiently express multiple olfactory receptors during development. Mol Syst Biol. 11:844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. 2003. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 38:161–175. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel J, Dulac CG. 2005. Single-cell transcriptional profiles and spatial patterning of the mammalian olfactory epithelium. Int J Dev Biol. 49:201–207. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall LB, Axel R. 1994. Topographic organization of sensory projections to the olfactory bulb. Cell. 79:981–991. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 74:309–318. [DOI] [PubMed] [Google Scholar]
- Vedin V, Molander M, Bohm S, Berghard A. 2009. Regional differences in olfactory epithelial homeostasis in the adult mouse. J Comp Neurol. 513:375–384. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 96:725–736. [DOI] [PubMed] [Google Scholar]
- Von Der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Dal Col J, Kan C, Carleton A, Rodriguez I. 2015. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat Neurosci. 18:1455. [DOI] [PubMed] [Google Scholar]
- Weth F, Nadler W, Korsching S. 1996. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci USA. 93:13321–13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby-Logan GK, Weech M, Walters E. 2004. Zonal expression and activity of glutathione S-transferase enzymes in the mouse olfactory mucosa. Brain Res. 995:151–157. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. 1997. OCAM: a new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 17:5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. 2002. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 5:124–133. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zhang X, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. 2004. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci USA. 101:14168–14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.