MicroRNAs are regulators of gene expression implicated in cancer development. Using a prospective study, we addressed the possibility that microRNAs serve as biomarkers of breast cancer development. The study observed that upregulation of miRNA-513a-5p was associated with breast cancer risk.
Abstract
MicroRNAs (miRNAs) might be considered both predictors and players of cancer development. The aim of the present report was to investigate whether many years before the diagnosis of breast cancer miRNA expression is already disregulated. In order to test this hypothesis, we compared miRNAs extracted from leukocytes in healthy women who later developed breast cancer and in women who remain healthy during the whole 15-year follow-up time. Accordantly, we used a case–control study design nested in the hOrmone and Diet in the ETiology of breast cancer (ORDET) prospective cohort study addressing the possibility that miRNAs can serve as both early biomarkers and components of the hormonal etiological pathways leading to breast cancer development in premenopausal women. We compared leukocyte miRNA profiles of 191 incident premenopausal breast cancer cases and profiles of 191 women who remained healthy over a follow-up period of 20 years. The analysis identified 20 differentially expressed miRNAs in women candidate to develop breast cancer versus control women. The upregulated miRNAs, miR-513-a-5p, miR-513b-5p and miR-513c-5p were among the most significantly deregulated miRNAs. In multivariate analysis, miR-513a-5p upregulation was directly and statistically significant associated with breast cancer risk (OR = 1.69; 95% CI 1.08–2.64; P = 0.0293). In addition, the upregulation of miR-513-a-5p displayed the strongest direct association with serum progesterone and testosterone levels. The experimental data corroborated the inhibitory function of miR-513a-5p on progesterone receptor expression confirming that progesterone receptor is a target of miR-513a-5p. The identification of upregulated miR-513a-5p with its oncogenic potential further validates the use of miRNAs as long-term biomarker of breast cancer risk.
Introduction
Recent advances in high-throughput technology for gene expression led to the discovery that most human transcriptional regulatory units are non-coding RNAs (1). These RNA molecules do not encode proteins but have important structural, catalytic or regulatory functions, including regulation of gene expression involved in important functions. One such non-coding RNAs, microRNAs (miRNAs), are small RNA molecules (on average 22 nucleotides in length) that are involved in the modulation of, among others, hormone and metabolic pathways through post-transcriptional gene silencing (2,3). MicroRNAs may play a role in cancer progression functioning as oncogenes or tumor suppressors (4–6). At the same time, miRNAs have been recently indicated as non-invasive biomarkers for early detection of cancer (7). In a previous report from our group, we observed differences in leukocytes miRNA expression between healthy postmenopausal women who later became affected with breast cancer and in women who did not develop the disease during the same 20-year follow-up period (8). This provides strong evidence to the role of leukocyte miRNAs as early, long-term biomarkers of breast cancer.
The present investigation aimed to study miRNAs not only as biomarkers of breast cancer risk, this time in premenopausal women, but also to characterize their functional activity to unravel their specific role in breast cancer development. Previously conducted prospective cohort studies, including the hOrmone and Diet in the ETiology of breast cancer (ORDET) study, observed that endogenous sex steroids were directly (estrogens and androgens) or inversely (progesterone) associated with risk of breast cancer (9–14). Thus, we expected to observe that the early disregulated miRNAs had specific function in modulating the steroid hormone synthesis and/or metabolism associated with risk or protection of breast cancer.
To test this working hypothesis, we compared leukocyte miRNA profiles of healthy premenopausal women who subsequently became affected with breast cancer with women who remained healthy.
The present investigation was conducted in two succeeding steps: (i) a case–control study nested in the ORDET prospective cohort over a follow-up period of 20 years; (ii) an experimental study to validate the observational study results. As a part of the first step, we have validated the ORDET study findings in a different cohort, the METABRIC study (15) where molecular profile of tumor and non-tumor tissue samples collected from 1375 breast cancer patients have been extensively characterized.
Materials and methods
Study design and population
The study has been conducted in the context of the ORDET prospective cohort study.
The ORDET cohort was established in Northern Italy between June 1987 and June 1992, in which 10786 healthy women aged 35–69 years were enrolled (16). They were all residents of the Varese province, an area covered by the population-based Lombardy Cancer Registry (17). They had heard about the study through the media, at public meetings and volunteered to participate. At recruitment, we measured anthropometric variables and collected demographic information and blood samples. Because the study’s original focus was on endogenous hormones in relation to breast cancer risk, we also applied stringent inclusion criteria and highly standardized conditions on the collection of biologic samples.
Information on cancer outcomes available from the Lombardy Cancer Registry has been linked to the ORDET cohort to identify incident breast cancer cases up to December 31, 2006 (18) for a median follow-up time of 15.4 years. The ORDET file was also linked to the Varese residents’ file to check participants’ vital status. Participants were censored at the time of cancer diagnosis, death or loss to follow-up, whichever came first. Case subjects were women who developed breast cancer after their recruitment into the ORDET cohort and before the end of the follow-up. We randomly chose one control for each case, from appropriate risk sets consisting of all cohort members who satisfied the matching criteria. Matching characteristics were age (±3 years) at enrolment and date of recruitment (±180 days). An incidence density sampling protocol for control selection was used, such that controls could include subjects who became a case later, while each control subject could also be sampled more than once (19). Selecting controls with incidence density sampling provides important advantages such as the possibility to obtain a direct estimate of the rate ratio and to calculate unbiased estimates where biases can always occur due to differential loss to follow-up among the exposed versus unexposed controls (20).
Women were classified as premenopausal if they had at least one menstruation in the 6 months prior to recruitment. The analysis included 191 incident invasive breast cancer cases in premenopausal status at recruitment and 191 matched control subjects.
This study was approved by the Ethical Review Board of the National Cancer Institute of Milan (Italy).
Biomarker stability and blood collection
A relevant feature of miRNAs is their remarkable stability, which indicates their use as biomarkers in a number of different tissues (21,22). In this study, we evaluated the miRNA expression profile of leukocytes derived from buffy coats collected at recruitment.
Blood samples were drawn after overnight fasting between 7:30 AM and 9:00 AM from each premenopausal woman. Women were timed to be collected between the 20th and 24th day of their menstrual cycle (i.e. during the mid luteal phase). For further verification of their luteal phase during blood collection, women were given a postcard to report the date of the subsequent bleeding after the blood drawing (13). Blood samples from each breast cancer case and related control were handled identically and assayed together on the same day and in the same run. All samples were taken out of the freezer simultaneously and sent to laboratory in the same parcel on dry ice. They were stored at –80°C until the bioassays included in the present report. Laboratory personnel were blinded to case–control status.
Laboratory methods
RNA extraction, labeling and microarray hybridization
Leucocytes were lysed in 1 ml of TRIZOL Reagent, a lysis reagent from Ambion, according to the manufacturer’s instructions. The concentration and purity of total RNA were assessed using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies). Total RNA (100 ng) was labeled, hybridized to human microRNA Microarray V2 (Agilent Technologies), and scanned with Agilent DNA Microarray Scanner (P/N G2565BA) according to the manufacturer’s instructions. Feature Extraction Software (Version 10.5) was used for data extraction from raw microarray image files using the microRNA_105_Dec08 FE protocol. This miRNA Agilent expression profile was submitted to the Gene Expression Omnibus (GEO) with the accession number GSE54470. Minimum information about a microarray gene experiment (MIAME) guidelines were followed as instructions. Furthermore, representative RNA preparations were evaluated for integrity using the 2100 Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies; data not shown). We also assessed the expression of miRNA-223 in randomly selected samples by Northern blot analysis. miR223 is highly specific for hematopoietic cells and constitutes a regulator of myelopoiesis (23). The blot was hybridized with a [32P]gATP-radiolabeled LNA oligonucleotide complementary to miRNA-223 sequence. The specificity and strength of hybridization of ORDET RNA samples was as good as that of human promyelocytic HL60 cells treated with retinoic acid (10-6 mol/l), a known inducer of miRNA-223.
Hormone assay
Stability and reliability of the ORDET collection method for sex steroids in premenopausal women have been previously described (13). Control of analytic error was based on the inclusion of two standard samples.
All samples were assayed in duplicate, by using commercially available kits following the manufacturer’s instructions. Plasma sex steroid measurements (testosterone, estradiol and progesterone) were conducted by Centro Medico Diagnostico Emilia (Bologna, Italy). For testosterone and progesterone we used Coat-A-Count procedure, a solid-phase radioimmunoassay (Diagnostic Products Corporation, Los Angeles, CA) and for estradiol, Orion Diagnostica SPECTRIA Sensitive RIA test, a coated-tube radioimmunoassay (Orion Diagnostica Oy, Espoo, Finland). Quality control was done at three concentrations for total testosterone and at four concentrations for total estradiol and progesterone. Intra- and inter-assay coefficients of variation were, respectively, 4.2–4.6% and 8.0–9.1% for total testosterone; 4.3–8.6% and 6.0–15.2% for free testosterone; 2.2–7.0% and 6.8–13.8% for DHEAS; 6.3–8.7% and 5.3–10.6% for progesterone; 2.7–3.5% and 4.4–9.6% for SHBG; 1.5–2.7% and 2.4–4.8%. In each batch, quality-control samples were evaluated in quadruplicate. Average between-batch coefficients of variation for high and low concentrations were 7.4 and 16.4% for estradiol, 8.7 and 18.5% for total testosterone and 5.3 and 10.6% for progesterone. Serum levels of progesterone were compatible with ovulatory cycles, ranging between 5.3 and 21.5 ng/ml in control subjects and between 4.8 and 20.8 ng/ml in incident breast cancer cases; 6.3–8.7% and 5.3–10.6% for progesterone and 6.3– 8.7% and 5.3–10.6% for progesterone.
Microarray data analysis
Data were verified and extracted by the Agilent Extraction 10.7.3.1 software and analyzed using an inhouse built routines by Matlab (The MathWorks Inc.). Background-subtracted signal of 851 human miRNA assays was used in the study. All arrays were quantile normalized, assuming that all samples were measured and analyzed under the same condition, enforcing all the arrays to assume the same mean distribution. The Pearson coefficient was calculated to assess the correlation between technical replicates of some randomly chosen samples.
We fitted a linear model to the expression values for each miRNA, to assess the significance of differential expression between case and control. In addition, we used empirical Bayes methods implemented in the Linear Models for MicroArrays (LIMMA) package to construct moderated t statistics and incorporated the statistical tools to adjust for the multiplicity of the tests. The Benjamini method (24) was used to control for false discovery. We considered the linear model including the matched case–control study design, the case–control status and the error term.
Statistical methods
Data pre-processing and differential expression analysis were done using the Bioconductor AgiMicroRna package (25). The total gene signal (TGS) provided by the Agilent Feature Extraction image analysis software was used as the quantitative measure of miRNA expression. The TGS provided by the Agilent Feature Extraction image analysis software was used as the quantitative measure of miRNA expression. We set all negative TGS values to 0.5 before log transformation, so that the log ratios are shrunk toward zero at lower intensities. The miRNA expression data (i.e. TGS) were quantile normalized before determining differential expression. The data were analyzed using the R software package. For differential expression analysis, the AgiMicroRna package incorporates the linear model with matched pair features from the Bioconductor LIMMA package (26). The LIMMA approach fits a linear model to the expression value for each miRNA to assess the significance of differential expression between different experimental conditions. In addition, the method uses empirical Bayes methods (27) to construct moderated t statistics and incorporates statistical tools to adjust for multiple testing. The Benjamini method (24) was used to control for false discovery rate (FDR), and we ranked the miRNAs according to FDR. We considered the top-ranked 20 miRNAs and investigated the upregulated and downregulated miRNAs identified from premenopausal samples. At first, each object is assigned to its own cluster and then the algorithm proceeds iteratively. At each stage, the two most similar clusters are combined to form a larger cluster, continuing until there is just a single cluster. At each stage, distances between clusters are computed by the Lance–Williams dissimilarity update formula. Details about the clustering algorithm are given in the book by Kaufman and Rousseeuw (28). This clustering method partitions the dataset into clusters, in which similar miRNA expression patterns are assigned to the same cluster.
Several breast cancer risk factors were used in a univariate and multivariate logistic regression models to predict positive or negative z-scores of normalized signal intensity of specific miRNAs. Confidence intervals were set at 95%. In the regression models the variables with a P value less than 0.05 were included in a multivariate model. Variables with a P value less than 0.1 in the multivariate model were considered significant.
All regression analyses were performed by Matlab (The MathWorks).
Target prediction was assessed by using several prediction software included in the web server tool MirWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). Prediction was considered reliable if confirmed by at least three different software. Predicted targets were used for pathway analysis.
In order to validate the prediction reported in the ORDET patients, we performed a validation analysis in the patients from a different cohort, the METABRIC cohort (15), a cohort of 1359 molecularly well characterized breast cancer patients and carried out a functional analyses separating the patient low and high expression levels of the miRNAs selected. In particular, we conducted a correlation analysis on matched miRNA\mRNA samples from METABRIC cohort.
Cell cultures and transfection
Human luminal breast cancer cell lines MCF-7, T47D, ZR-75-1 and BT-474 were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% (v/v) fetal bovine serum; cells were grown at 37°C in a balanced air humidified incubator with 5% CO2. All fresh cell lines were purchased from ATCC that has authenticated them by STR genotyping with Promega PowerPlex® 1.2 system and the Applied Biosystems Genotyper 2.0 software for analysis of the amplicons. The cells were maintained in culture no more than six passages. All the cell lines have been tested by PCR/IF for Mycoplasma presence.
For mature miR-513a-5p expression, we used mirVana miRNA Mimic Negative Control #1 (ThermoFisher) and miR-513a-5p mirVana miRNA Mimic (ThermoFisher) at final concentration of 5 nM.
Cells were transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
During hormone treatments, cells were serum-starved for 12–16 h in Dulbecco's modified Eagle's medium (Invitrogen), and then treatments were performed in Dulbecco's modified Eagle's medium with 0.5% bovine serum albumin.
All hormones were dissolved in 100% ethanol and used at a final concentration of 10 nM. The hormones used were progesterone (P4; Sigma) and 17b-estradiol (E2, Sigma).
MCF-7 cell line viability assay
Cell viability was evaluated using the ATPlite™ Luminescence Assay System and following the manufacturer’s instructions. Luminescence was read by the EnSpire® Multimode Plate Reader (PerkinElmer, Whitman, MA).
Total RNA extraction from cells and reverse transcriptase
Total RNA was extracted using the TRIZOL Reagents (GIBCO). One microgram of total RNA was reverse-transcribed at 37°C for 60 min in the presence of random hexamers and Moloney murine leukemia virus reverse transcriptase (Invitrogen). Specific oligonucleotides for the genes listed in Supplementary Table 2, available at Carcinogenesis Online, for RTq-PCR analyses. GAPDH, progesterone receptor (PR) and ATP1B1 genes expressions were measured by RTq-PCR using the Sybr Green assay (Applied Biosystems) on a StepOne instrument (Applied Biosystems).
Small amount of RNA (10 ng) was reverse-transcribed using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems). Reverse transcription was carried out in a final volume of 10 ul using ABI Prism 7000 Sequence Detection System (Applied Biosystems). The PCR Reactions were initiated with a 10-min incubation at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. RTq-PCR quantification of miR expression was performed using TaqMan MicroRNA® Assays (Applied Biosystems) according to the manufacturer’s protocol. RNU19 was used as endogenous control to normalize miR expression. All reactions were performed in duplicate.
The sequences of primers used for RTq-PCR were listed in Supplementary Table 6, available at Carcinogenesis Online.
Lysate preparation and immunoblotting analysis
Cells were lysed in buffer with 50 mM Tris–HCl pH 8, with 1% NP-40 (Igepal AC-630) 150 mM NaCl, 5 mM EDTA and fresh protease inhibitors. Extracts were sonicated for 10 s and centrifuged at 12000× rpm for 10 min to remove cell debris. Protein concentrations were determined by colorimetric assay (Bio-Rad).
Western blotting was performed using the following primary antibodies: mouse monoclonal anti-Gapdh (Santa Cruz Biotechnology), mouse monoclonal anti-PR (Santa Cruz Biotechnology), rabbit polyclonal anti-p21 (Santa Cruz Biotechnology) and rabbit polyclonal anti-PARP (Cell Signaling).
Secondary antibodies used were goat anti-mouse, goat anti-rabbit, conjugated to horseradish peroxidase (Amersham Biosciences).
Immunostained bands were detected by chemiluminescent method (Pierce).
Results
Leucocyte miRNAs differentially expressed in ORDET samples
The baseline characteristics did not differ between the 191 breast cancer cases and 191 controls: age at recruitment as well as age at menarche and age at first birth were not statistically associated to the case–control status as well as other reproductive, hormonal and life-style risk factors that could have represented confounders of the studied association (Supplementary Table 1, available at Carcinogenesis Online).
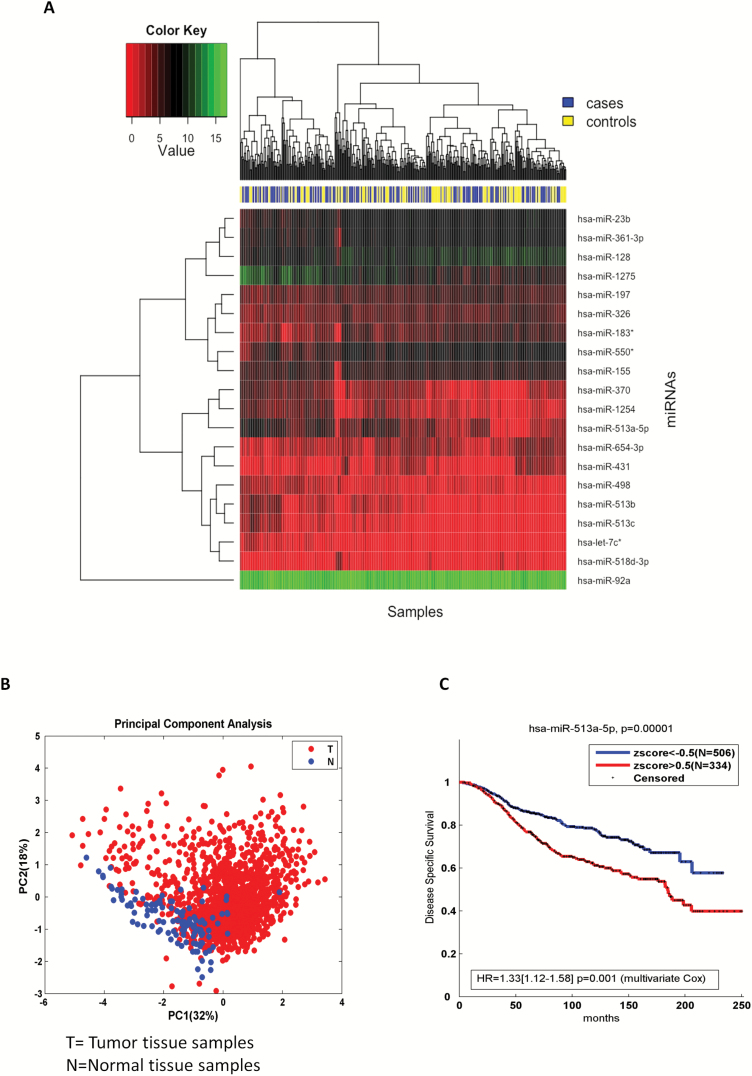
When we conducted class comparisons to identify differentially expressed miRNAs, we first performed a moderated t test (27) for each miRNA on all 382 premenopausal women. In our analysis, we included the top-ranked 20 miRNAs (ranked according to the FDR values): the difference in miRNA expression between cases and controls was balanced between up- and downregulation with 11 miRNAs downregulated and 9 upregulated (see Supplementary Table 2, available at Carcinogenesis Online). Among the upregulated miRNAs, miR-513c-5p was characterized by the lowest FDR (close to 8%), resulting in the most statistically significant differentially expressed miRNA. Among the same group of upregulated miRNAs, miR-513a-5p ranked second together with the downregulated miR-431, miR-550 and miR-23b (all at 13% FDR). An examination of miRNA expression of the 20 top-ranked miRNAs revealed that, among the upregulated miRNAs in candidate breast cancer versus control women, the first and second ranked miRNAs, miR-513c-5p and the miR-513a-5p together with miR-513b-5p belonged to a subfamily member of a miR-506-514 cluster located on the X chromosome, whose oncogenic role has been already well characterized in melanoma (29,30). Figure 1A shows the heatmap of the all the 20 top-ranked miRNA profiles following an unsupervised hierarchical clustering analysis. The heatmap revealed that the three miRNAs were also grouped together in one of the two largest clusters identified across the 20 top-ranked miRNAs. In order to better characterize the identified miRNA signature in breast cancer, we used data from the METABRIC cohort. As it is displayed in Principal Component Analysis (Figure 1B), the signature was moderately able to discriminate non-tumoral versus tumoral tissues. Moreover, in the METABRIC breast cancer patients higher values of miR-513a-5p in the tumor tissue were associated to a poor prognosis at univariate level as well as adjusting for clinical covariates, such as T,N, stage, age, hystotype, menopause status and ER, PR and HER2 status (Figure 1C).
Figure 1.
(A) Heatmap of top ranked 20 miRNAs from premenopausal samples. The heatmap provides insight into the data structure for each microRNA and samples. We used red and green colors for defining low and high expression values. The blue represents the cases and yellow represents the controls. The clustering methods partitioned the dataset into three clusters identified on the left side of the figure. Each miRNA is listed on the right side. (B) Principal component analysis of the miRNAs signature from METABRIC breast cancer patient sample analysis. (C) Breast cancer survival data based on miR-513a-5p low and high expression. Higher values of miR-513a-5p were associated to a poor prognosis from Kaplan–Meier method and multivariate Cox proportional-hazards regression. Several clinical variables, such as T, N, stage, age, hystotype, menopause status and ER, PR, HER2 receptors, were included in the multivariate model.
In Table 1, we describe the most significant cancer predicted pathways targeted by miR-513a-5p, miR-513b-5p and miR-513c-5p. It is worth noting that the pathways identified by the three differentially upregulated miRNAs were very similar and related to both breast cancer and more general cancer development (e.g. ErbB, mTOR, TGFb and Wnt pathways) (31–35). We then also observed that the three miRNAs targeted pathways also related to insulin and other endocrine/metabolic pathways recognized in the ORDET cohort, as well as in other studies as pathways involved in breast cancer development (10,32,36). We repeated the prediction miR-513a-5p pathways analysis using data from the METABRIC study. First we identified putative targets of miR-513a-5p predicted in at least three softwares using miRWALK2.0 prediction tool. Then we split patients basing on their zscores as defined in KM curves. The Spearman’s correlation coefficient was used to select negative correlated gene targets separately in the two subgroups of patients. ConsensusPathDB (37) was interrogated for a pathway analysis of the identified genes.
Table 1.
Cancer predicted pathways targeted by miR-513a-5p, miR-513b-5p and miR-513c-5p listed among the nine differentially upregulated miRNAs
| microRNA | Most significant predicted pathways | P value |
|---|---|---|
| hsa-miR-513a-5p | Pathways in cancer | 3.3E–08 |
| Wnt signalling pathway | 1.1E–07 | |
| Hedgehog signalling pathway | 1.7E–04 | |
| ErbB signalling pathway | 1.7E–04 | |
| Insulin signalling pathway | 2.6E–04 | |
| mTOR signalling pathway | 1.5E–03 | |
| Chemokine signalling pathway | 2.8E–03 | |
| TGF beta signalling pathway | 2.5E–02 | |
| Metabolic pathways | 4.1E–02 | |
| hsa-miR-513b-5p | Pathways in cancer | 1.5E–11 |
| Wnt signalling pathway | 5.4E–08 | |
| ErbB signalling pathway | 5.9E–07 | |
| Insulin signalling pathway | 6.4E–05 | |
| MAPK signalling pathway | 1.3E–03 | |
| mTOR signalling pathway | 4.3E–03 | |
| Hedgehog signalling pathway | 4.7E–03 | |
| Metabolic pathways | 4.7E–02 | |
| Cell cycle | 3.9E–02 | |
| Chemokine signalling pathway | 4.1E–02 | |
| TGF beta signalling pathway | 4.1E–02 | |
| hsa-miR-513c-5p | Wnt signalling pathway | 9.7E–08 |
| Pathways in cancer | 2.9E–07 | |
| MAPK signalling pathway | 7.3E–05 | |
| Hedgehog signalling pathway | 2.7E–04 | |
| ErbB signalling pathway | 3.8E–04 | |
| Insulin signalling pathway | 2.1E–03 | |
| mTOR signalling pathway | 6.4E–03 | |
| TGF beta signalling pathway | 3.5E–02 |
Results of this analysis indicated that miR-513a-5p also in that cohort modulates the activation of those pathways observed in the ORDET cohort and associated to cancer development (Supplementary Tables 3 and 4, available at Carcinogenesis Online).
To identify and quantify the potential association between the 20 top-ranked miRNAs with breast cancer risk, we performed first an univariate and then a fully adjusted logistic regression analysis on all the considered miRNAs. We observed that among all the miRNAs, only the miR-513a-5p differential expression was directly and significantly associated with risk of breast cancer. The univariate and the multivariate odds ratios were similar ranging between 1.62 (95% CI 1.08–2.43; P = 0.018) to 1.69 (95% CI 1.08–2.64; P = 0.0293) adjusted for age, age at menarche, age at first pregnancy, BMI, smoking, alcohol intake, fasting glucose, IGF-1, insulin, progesterone, estradiol and total testosterone as breast cancer risk factors earlier reported in the ORDET cohort.
Previous reports from the ORDET cohort indicated serum sex steroids were implicated in breast cancer development (11,13), thus, we examined the association between progesterone, serum total testosterone and estradiol and each single miRNA expression first at univariate and then at multivariate level. We excluded from this analysis the let-7c* as it has been define a dubious miRNA (38). While both serum progesterone and testosterone were not associated with breast cancer risk in the present set of the ORDET cases and controls [progesterone OR = 0.99 (95% CI 0.64–1.57); testosterone OR = 1.10 (95% CI 0.71–1.71)], at univariate level, miR-513a-5p showed the strongest association with serum levels of progesterone at borderline statistical significance (β coeff. = 0.39; P = 0.07) (Supplementary Table 5, available at Carcinogenesis Online). Subsequently, we performed a multivariate analysis of the top-ranked miRNAs in association with Progesterone (Table 2) with the inclusion of the same potential confounders used for the odds ratio assessment (Univariate analysis results are included in Supplementary Table 5, available at Carcinogenesis Online). Interestingly, we observed that, again, in both univariate and multivariate analysis, miR-513a-5p was the only miRNA associated with progesterone level and this time the positive association was stronger and reached the statistical significance (β coeff. = 0.55; P = 0.04). When subsequently, we examined the association between miR-513a-5p and each single covariate, adjusting the point estimates for all the other variables, the data showed that miR-513a-5p upregulation was significantly and independently associated with testosterone and progesterone serum levels (Table 3) (Box plots are also included as Supplementary Figure 1A and B, available at Carcinogenesis Online).
Table 2.
Multivariate analysis of miRNA signature and progesterone
| miRNA | β | P value |
|---|---|---|
| hsa-miR-23b | 0.28 | 0.3 |
| hsa-miR-361-3p | 0.2 | 0.43 |
| hsa-miR-128 | 0 | 0.98 |
| hsa-miR-1275 | 0.34 | 0.2 |
| hsa-miR-197 | 0.31 | 0.23 |
| hsa-miR-326 | −0.1 | 0.71 |
| hsa-miR-183* | −0.05 | 0.85 |
| hsa-miR-550* | 0.19 | 0.49 |
| hsa-miR-155 | 0.27 | 0.33 |
| hsa-miR-370 | 0.31 | 0.25 |
| hsa-miR-1254 | 0.31 | 0.25 |
| hsa-miR-513a-5p | 0.55 | 0.04 |
| hsa-miR-513b | 0.25 | 0.37 |
| hsa-miR-513c | 0.04 | 0.9 |
| hsa-miR-654-3p | –0.01 | 0.98 |
| hsa-miR-431 | 0.1 | 0.7 |
| hsa-miR-498 | −0.2 | 0.46 |
| hsa-miR-518d-3p | −0.56 | 0.1 |
| hsa-miR-92a | −0.1 | 0.69 |
β values represent the expected change in log odds for a one unit of progesterone. High and low values were defined by positive and negative Z-scores progesterone was adjusted for BMI, age menarche, insulin, igf1, tts, glucose, age, full-term pregnancy, alcohol, age first birth, smoke and estradiol.
Table 3.
Multivariate analysis of miR-513a-5p
| β | P value | |
|---|---|---|
| BMI | −0.26 | 0.33 |
| Age menarche | −0.26 | 0.29 |
| Insulin | −0.09 | 0.72 |
| IGF1 | −0.24 | 0.35 |
| TTS | 0.69 | 0.01 |
| PG | 0.55 | 0.04 |
| E2 | −0.4 | 0.14 |
| Glucose | −0.19 | 0.47 |
| Age | −0.14 | 0.59 |
| Full-term pregnancy | 0.31 | 0.23 |
| Alcohol | 0.35 | 0.16 |
| Age first birth | 0.2 | 0.42 |
| Smoke | 0.32 | 0.21 |
β values represent the expected change in log odds for a one unit of the variable. High and low values were defined by positive and negative Z-scores. Each variable was adjusted for the others.
In order to understand whether miR-513a-5p expression was modulated in different histological subgroups, considering the unavailability of the breast cancer tissue samples of the ORDET study, we conducted the analysis within the METABRIC cohort. In that cohort we observed higher miRNA513a-5p expression level in HER2 and a lower expression in the luminal A subgroup (Supplementary Figure 2A, available at Carcinogenesis Online). Furthermore, we did not observe any statistically significant difference across menopausal status (Supplementary Figure 2B, available at Carcinogenesis Online).
At the end of this phase of analysis, we observed that:
a) MiR-513a-5p expression was directly and significantly associated with almost 70% increase in breast cancer risk;
b) MiR-513a-5p expression was directly and significantly associated with progesterone and testosterone serum levels
We then conducted two different in vitro experiments to clarify the role of miR-513a-5p as simultaneous risk factor for breast cancer and endocrine modulator.
MiR-513a-5p impinges on progesterone receptor protein expression in breast cancer cells
Recent data from experimental studies showed that miR-513a-5p reduces the expression of PR through its direct binding to the 3′UTR of PR mRNA (39). The human PR gene holds a long 3′UTR sequence, about 13 kb in length, containing several putative miRNA target sites, thereby suggesting an important involvement of this region in the control of PR translation (40).
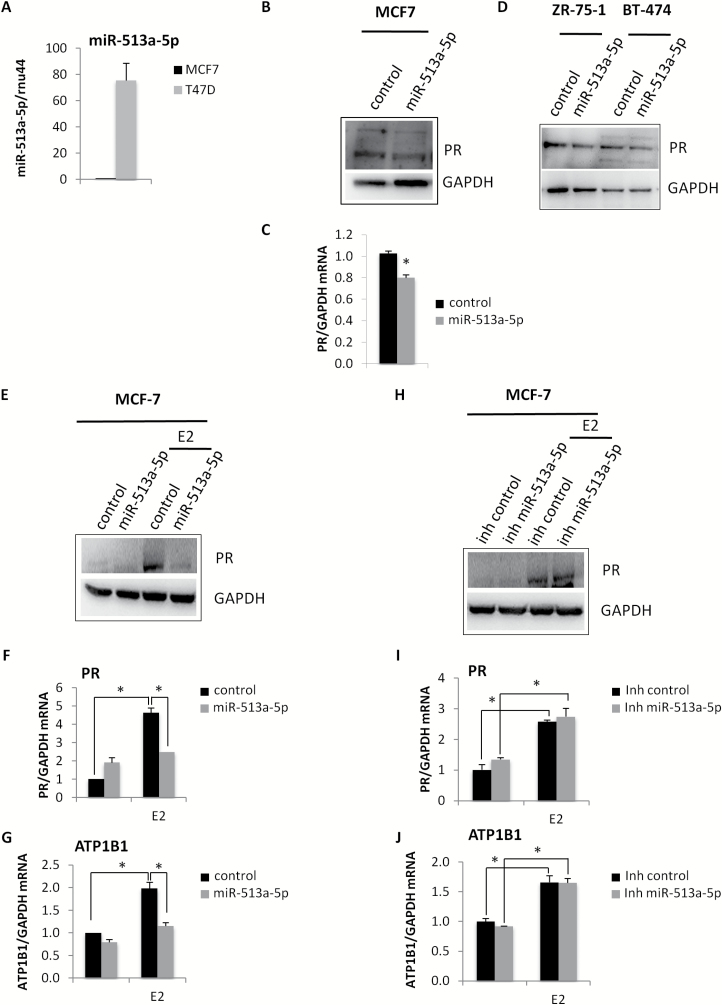
Due to the previously reported evidences about miR-513a-5p in the regulation of PR levels, we aimed to confirm this direct effect in our cellular model (39). In particular, we initially analysed the expression levels of miR-513a-5p in two luminal breast cancer cell lines, MCF7 and T47D (ER+ and PR+) (Figure 2A), that constitutively expressed high levels of PR. MCF-7 cell line, which presented the lowest levels of miR-513a-5p, has been chosen as cellular system in which to perform ectopical expression of miR-513a-5p. As shown in Figure 2B and C, the over-expression of miR-513a-5p determined a significant reduction in PR protein and mRNA levels compared to the relative control. This inhibitory effect on PR protein expression levels occurred also in two additional luminal breast cancer cell lines, ZR-75-1 and BT-474 (Figure 2D). To better investigate the direct effect of miR-513a-5p on PR expression levels, we treated cells with estradiol (E2), that induces the transcription of PR gene. As it has been shown in Figure 2E and F, in a cellular context characterized by higher levels of PR, the inhibitory effect of miR-513a-5p on PR mRNA and protein levels was more pronounced. This also pairs with the striking reduction of mRNA levels of ATP1B1, a transcriptional target of PR (Figure 2G). Conversely, the induction of PR and ATP1B1 was similar to the control cells in E2 treated MCF-7 cells depleted for the miR-513a-5p expression (Figure 2H–J). Taken together, these findings contribute to establish PR as a direct target of miR-513a-5p in breast cancer cells.
Figure 2.
MiR-513a-5p impairs the expression of PR in luminal breast cancer cell lines. (A) qRT-PCR analysis of miR-513a-5p expression levels in MCF-7 and T47D cell lines. (B, C) Western blot analysis and qRT-PCR analysis of PR expression levels in MCF7 cells transiently transfected with miR-513a-5p mimic or control. (D) Western blot analysis of PR protein expression in ZR-75-1 and BT-474 breast cancer cell lines transiently transfected with miR-513a-5p mimic or control. (E) Western blot analysis of PR protein expression in serum-starved MCF-7 cells treated for 48 h with 10 nM E2 upon miR-513a-5p over-expression. (F, G) qRT-PCR analysis of PR and ATP1B1 expression levels in serum-starved MCF-7 cells treated for 48h with 10nM E2 upon miR-513a-5p over-expression. (H) Western-blot analysis of PR protein expression in serum-starved MCF-7 cells treated for 48 h with 10 nM E2 upon miR-513a-5p inhibition (inh miR-513a-5p). (I, J) qRT-PCR analysis of PR and ATP1B1 expression levels in serum-starved MCF-7 cells treated for 48 h with 10 nM E2 upon miR-513a-5p inhibition (inh miR-513a-5p). *P < 0.05.
In order to further characterize miR-513a-5p function in breast cancer, we performed in silico analysis for miR-513a-5p target genes. Among the list of the putative targets, EGFR and ERBB2 resulted to be the more interesting, in terms of number of software that predicted them and pathways involved in breast cancer (Supplementary Figure 3A, available at Carcinogenesis Online). To verify the binding of miR-513a-5p on EGFR and ERBB2 3′UTRs, we evaluated their protein expression levels in ZR-75-1 breast cancer cells upon miR-513a-5p expression. As shown in Supplementary Figure 3B, available at Carcinogenesis Online, we observed a significant reduction in EGFR and ERBB2 protein levels, indicating an inhibitory effect of miR-513a-5p on these two receptors whose involvement in breast cancer progression is well established. Moreover, to assess the ability of miR-513a-5p to bind to EGFR 3′UTR we performed luciferase assays using a reporter construct with the full-length 3′UTR of EGFR. As shown in Supplementary Figure 3C, available at Carcinogenesis Online, miR-513a-5p significantly reduced the relative luciferase activity of the reporter, indicating a direct inhibitory effect of miR-513a-5p on EGFR mRNA translation.
MiR-513a-5p renders breast cancer cells more resistant to starvation stress
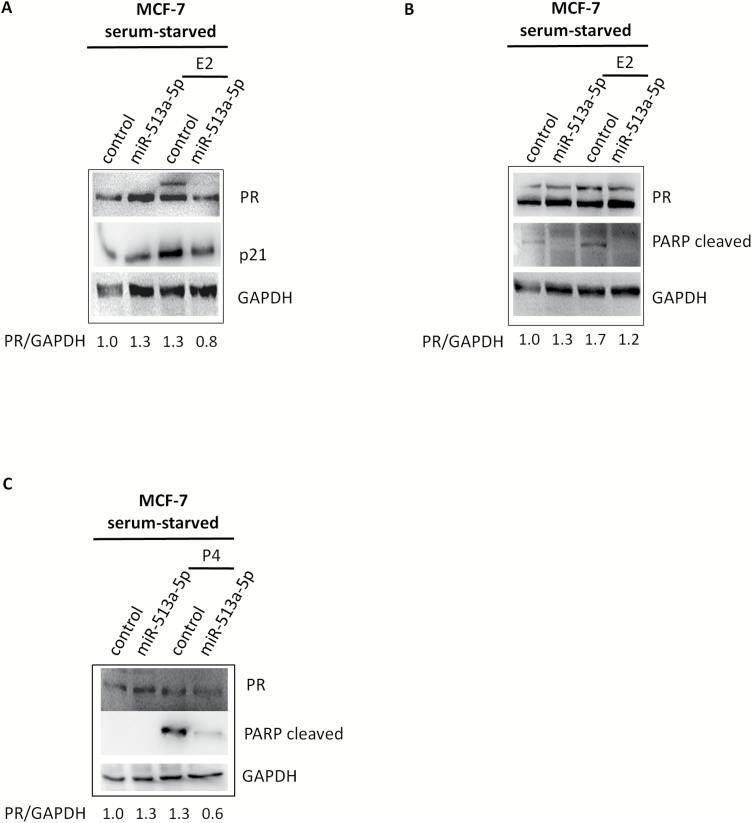
There is still scarce evidence of the functional effects of miR-513a-5p in breast cancer. To this purpose we aimed to test the effect induced either by miR-513a-5p ectopic expression or its depletion in MCF7 cells. No effect on cell viability was observed under standard cell culture condition (Supplementary Figure 4A and B, available at Carcinogenesis Online). Strikingly miR-513a-5p impacted the cell response upon serum-starvation. MCF7 underwent growth arrest and apoptosis upon treatment with E2 or progesterone in condition of serum starvation, which was typical cellular response to stress induced by deprivation of nutrients (41,42). This led to the increase of p21 and cleaved PARP protein expression, respectively (Figure 3A–C). MiR-513a-5p ectopic expression increased cell resistance to serum starvation. This also led to lower p21 and cleaved PARP protein levels, when compared with control cells (Figure 3A–C).
Figure 3.
MiR-513a-5p increases MCF-7 cells resistance to serum-starvation stress. (A, B) Western blot analysis of PR, p21 and cleaved PARP proteins expression in serum-starved MCF-7 cells treated for 48 h with 10 nM E2 upon miR-513a-5p over-expression. (C) Western blot analysis of PR and cleaved PARP proteins expression in serum-starved MCF-7 cells treated for 48 h with 10 nM P4 upon miR-513a-5p over-expression.
Upon E2 or progesterone treatment, we found an induction of PR expression (Figure 3A–C upper panels lane 3). Ectopic expression of miR-513a-5p mimic significantly reduced the expression of PR (Figure 3A–C upper panels lane 4).
In serum starvation condition, being it a stress condition, we observed a reduction in PR protein levels upon miR-513a-5p over-expression only upon stimulation of PR expression (Figure 3A–C). It is possible that in serum starvation conditions miR-513a-5p is not able to affect the basal levels of PR.
These findings originally highlight a putative oncogenic role of miR-513a-5p in breast cancer cells.
At the end of this phase of experimental studies, we understood that:
Progesterone receptor is a direct target of miR-513a-5p
The experimental data have pointed out a specific putative oncogenic function of miR-513a-5p corroborating its role as potential breast cancer risk factor.
Discussion
The present study has identified miR-513a-5p as prediagnostic miRNA that, when upregulated is able to reduce PR levels and potentially to decrease the biological function of progesterone. At the same time, the significant upregulation of miR-513a-5p in women candidate to develop breast cancer many years before the on-set of the disease, makes this miRNA an early, long-term biomarker of breast cancer risk in premenopausal women. The two combined observations provide evidence that reduction in progesterone biological activity may represent a risk factor for breast cancer.
We assessed miRNAs in leukocytes: the high blood stability of miRNAs, their resistance to RNA degradation and their reproducible detection make miRNAs suitable biomarker candidates (43). The relation between leukocytes intervention and inflammation in tumor development has been identified since the beginning of the 19th century. However, only in the last 10 years, experimental and clinical studies have identified inflammation as a major player in tumorigenesis, and some of the key basic mechanisms have been clarified (44). Inflammation responsibility in different aspects of tumorigenesis is now accepted, and it is now clear that an inflammatory macro- as well as micro-environment are very relevant components of cancer development (45). Chronic inflammation can be induced by endocrine and metabolic cancer risk factors (i.e. obesity, altered glucose metabolism, etc) and by environmental exposures (i.e. exposure to asbestos, pollutants). These same pro-inflammatory factors have been reflected into specific deregulation of miRNA in peripheral leukocytes (46). It has been indicated that an inflammatory microenvironment can lead to a raise in mutation rates together with amplifying the proliferation of transformed cells. As matter of fact, inflammatory cells may produce reactive oxygen species (ROS) and reactive nitrogen intermediates inducing DNA damage and genomic instability (47).Thus, in our study, leukocytes could in fact represent expression of that chronic inflammation status leading many years later to breast cancer development. Progesterone shows to have both anti-inflammatory activity and to inhibit oncogenic pathways (48). Thus, results of our study are in agreement with the evidence produced by leukocytes: low expression of PR could represent an ‘intracrine’ facilitating condition for cancer development.
As mentioned above, the role of progesterone in the natural history of breast cancer has been a scientific unresolved issue in particular for premenopausal women. In these women, the role of progesterone in breast cancer development has been challenged by different sources of variability such as the variability of progesterone serum concentrations over the menstrual cycle and the variability of cycle length within and between individuals (14). However, in spite of these limitations, the European Prospective Investigation on Cancer and Nutrition (EPIC), which is one of the largest prospective cohort studies on cancer aetiology, investigated the association between sex steroid hormones and breast cancer occurrence in premenopausal women (12). The study observed a statistically significant inverse relationship between serum levels of progesterone and breast cancer incidence. The study replicated the results of the first 7-year follow-up of the ORDET cohort study (11). An additional analysis conducted on a new follow-up of the ORDET cohort did not observe any risk associated with progesterone serum levels (13). The last published report on progesterone and breast cancer risk described the pooled analysis of the seven largest prospective cohort studies on hormones and breast cancer risk (10) also confirmed the lack of association between progesterone levels and risk of breast cancer.
When we looked at the PR as main outcome of the miR-513a-5p functional activity, the scientific literature included mainly studies describing the correlation between PR expression and different miRNAs, but only few evidences about direct effect of miRNAs on PR expression. In particular, in human breast cancer cells MCF-7 it has been demonstrated that PR expression was also negatively regulated by the direct binding of miRNA-181 and miRNA-26a on its 3′UTR (49). Recently Cochrane and coworkers, reported a feedback loop mechanism through which PR regulates its own levels by promoting the expression of miR-513a-5p that in turns targets the PR 3′UTR (39). In particular the authors observed that in luminal breast cancer cells, upon progestin treatments, there was an increase in miR-513a-5p expression levels and a concomitant reduction in the protein levels of PR (39). This reduction was partially due to the direct binding of miR-513-5p on three binding sites located in PR 3′UTR (39). In our study we observed that miR-513a-5p determined the reduction in PR levels, in particular upon E2-mediated stimulation of PR gene transcription, impinging on PR signaling pathway and concomitantly a significant induction of resistance to starvation stress in breast cancer cell lines, supporting a direct potential oncogenic role for miR-513a-5p. The association between miR-513a-5p and elevated levels of serum progesterone observed in our cohort is in agreement with the positive effect of progestin treatments on miR-513a-5p expression levels described by Cochrane et al. It may also represent a potential positive feed-back mechanism to compensate for the reduction in PR at the beginning of a process that later will create the condition for breast cancer development.
The association between miR-513a-5p with elevated levels of serum testosterone represents a new finding to be subsequently explored. Elevated levels of testosterone have been consistently found associated to breast cancer development in different studies, in pre- and postmenopausal women and in different populations. The association of miR-513a-5p upregulation with elevated circulating testosterone levels may delineate a risk profile to be better characterized and validated in different prospective cohort studies.
Limitations of this investigation warrant consideration. The adopted study design was powerful to infer on the etiological role of miRNAs on breast cancer risk definition and the follow-up was crucially long to adapt the study results to the natural history of the disease. However, we recognized that our inference were done on blood sample collected only once in the ORDET participants. Nevertheless, miRNAs are characterized by small intra-individual variability (50) showing that miRNAs have great potential as reliable blood biomarkers of early signs of disease development. We are also aware that in the first set of bioinformatics analysis FDRs differentiating the level of expression for the 20 top-ranked miRNAs were not characterized by a high level of statistical difference. In cancer prevention, which usually concerns with healthy individuals and very long follow-up, one should expect to observe relatively weak statistical significant differences especially in dealing with serum samples and not with tissue biopsies. In these studies, based on thousands of molecular variables, bioinformatic analysis and related applied statistics may provide general orientation for further investigations with the application of different strategies to validate the study results. Because of that, we have validated the findings from the ORDET cohort through both the use of a different cohort study, the METABRIC study, where we repeated part of the bioinformatic and biostatistical analyses and the specific design of experimental studies. Thus, the confirmatory results observed by these two different contexts validated the ORDET outcomes beyond the level of the initial statistical significance.
In summary, results of the present study may impinge different aspects of breast cancer development. The identification of miR-513a-5p with its oncogenic potential further validates both the use of miRNAs as long-term biomarker of breast cancer risk and the use of miRNAs to identify the pathways implicated in early breast cancer development. In this particular case, the reduction in PR level exerted by the upregulated miR-513a-5p may shed new light on the effect of natural progesterone in breast cancer etiology.
Funding
This work was supported by Department of Defense Grant W81 XWH 04 1 0195 and USA National Cancer Institute Grant CA98344.
Conflict of Interest Statement: None declared.
Supplementary Material
Acknowledgements
Dr. Muti wishes to thank the Arcelormittal Dofasco Company for the support of her research endeavor. We are indebted to the 10786 ORDET participants and 1,359 METABRIC participants. We would also like to thank Dr. P. Crosignani, Andrea Micheli, Paolo Contiero and Giovanna Tagliabue and the staff of the Varese Cancer Registry for technical assistance; Drs. G. Bolelli and F. Franceschetti for conducting sex steroid assays. Contribution of EPIGEN Flagship Project (13/05/R/42) to Dr. Blandino is greatly appreciated. The authors thank Dr. Oreste Segatto for kindly providing with 3′UTR-EGFR-plasmid for luciferase reporter assay.
Abbreviations
- FDR
false discovery rate
- miRNAs
microRNAs
- PR
progesterone receptor
- TGS
total gene signal
References
- 1. Birney E., et al. (2007)Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fletcher C.E., et al. (2014)Interplay between steroid signalling and microRNAs: implications for hormone-dependent cancers. Endocr. Relat. Cancer, 21, R409–R429. [DOI] [PubMed] [Google Scholar]
- 3. Filipowicz W., et al. (2008)Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?Nat. Rev. Genet., 9, 102–114. [DOI] [PubMed] [Google Scholar]
- 4. Li M.H., et al. (2015)Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol. Sin., 36, 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Acunzo M., et al. (2015)MicroRNA and cancer–a brief overview. Adv. Biol. Regul., 57, 1–9. [DOI] [PubMed] [Google Scholar]
- 6. Donzelli S., et al. (2016)MicroRNAs: non-coding fine tuners of receptor tyrosine kinase signalling in cancer. Semin. Cell Dev. Biol., 50, 133–142. [DOI] [PubMed] [Google Scholar]
- 7. Liu H. (2012)MicroRNAs in breast cancer initiation and progression. Cell. Mol. Life Sci., 69, 3587–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muti P. (2014)Is progesterone a neutral or protective factor for breast cancer?Nat. Rev. Cancer, 14, 146. [DOI] [PubMed] [Google Scholar]
- 9. Endogenous Hormones and Breast Cancer Collaborative Group et al. (2011)Circulating sex hormones and breast cancer risk factors in postmenopausal women: reanalysis of 13 studies. Br. J. Cancer, 105, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Endogenous Hormones and Breast Cancer Collaborative Group et al. (2013)Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol., 14, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Micheli A., et al. (2004)Endogenous sex hormones and subsequent breast cancer in premenopausal women. Int. J. Cancer, 112, 312–318. [DOI] [PubMed] [Google Scholar]
- 12. Kaaks R., et al. (2005)Serum sex steroids in premenopausal women and breast cancer risk within the European Prospective Investigation into Cancer and Nutrition (EPIC). J. Natl. Cancer Inst., 97, 755–765. [DOI] [PubMed] [Google Scholar]
- 13. Schernhammer E., et al. (2013)Sex hormones and breast cancer development in premenopausal women: the ORDET cohort study. Breast Cancer Res., 18, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muti P. (2014)Is progesterone a neutral or protective factor for breast cancer?Nat. Rev. Cancer, 14, 146. [DOI] [PubMed] [Google Scholar]
- 15. Dvinge H., et al. (2013)The shaping and functional consequences of the microRNA landscape in breast cancer. Nature, 497, 378–382. [DOI] [PubMed] [Google Scholar]
- 16. Muti P., et al. (1988)ORDET–prospective study on hormones, diet and breast cancer: feasibility studies and long-term quality control. Steroids, 52, 395–396. [DOI] [PubMed] [Google Scholar]
- 17. Parkin D.M., et al. (1997)Cancer Incidence in Five Continents, vol. 7. IARC, Lyon, France. [Google Scholar]
- 18. Contiero P., et al. (2008)Comparison with manual registration reveals satisfactory completeness and efficiency of a computerized cancer registration system. J. Biomed. Inform., 41, 24–32. [DOI] [PubMed] [Google Scholar]
- 19. Miettinen O. (1976)Estimability and estimation in case-referent studies. Am. J. Epidemiol., 103, 226–235. [DOI] [PubMed] [Google Scholar]
- 20. Lubin J.H., et al. (1984)Biased selection of controls for case-control analyses of cohort studies. Biometrics, 40, 63–75. [PubMed] [Google Scholar]
- 21. Cortez M.A., et al. (2011)MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol., 8, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strano S., et al. (2015)What biomarkers (if any) for precise medicine?Aging, 7, 533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fazi F., et al. (2005)A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell, 123, 819–831. [DOI] [PubMed] [Google Scholar]
- 24. Benjamini Y., et al. (2001)Controlling the false discovery rate in behavior genetics research. Behav. Brain Res., 125, 279–284. [DOI] [PubMed] [Google Scholar]
- 25. López-Romero P., et al. (2010)Processing of agilent microRNA array data. BMC Res. Notes, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smyth G.K. (2005)Limma: linear models for microarray data. In: Gentleman R. (ed.) Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, NY, pp. 397–420. [Google Scholar]
- 27. Smyth G.K., et al. (2003)Normalization of cDNA microarray data. Methods, 31, 265–273. [DOI] [PubMed] [Google Scholar]
- 28. Kaufman L., et al. (1990)Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons, Inc, New Jersey. [Google Scholar]
- 29. Streicher K.L., et al. (2012)A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene, 31, 1558–1570. [DOI] [PubMed] [Google Scholar]
- 30. Sun Z., et al. (2013)Functional divergence of the rapidly evolving miR-513 subfamily in primates. BMC Evol. Biol., 13, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bacus S.S., et al. (2000)Expression of erbB receptors and their ligands in breast cancer: implications to biological behavior and therapeutic response. Breast Dis., 11, 63–75. [DOI] [PubMed] [Google Scholar]
- 32. Vucenik I., et al. (2012)Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann. N. Y. Acad. Sci., 1271, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. ten Dijke P., et al. (2004)New insights into TGF-beta-Smad signalling. Trends Biochem. Sci., 29, 265–273. [DOI] [PubMed] [Google Scholar]
- 34. Zhuang Z., et al. (2013)LKB1 inhibits breast cancer partially through repressing the Hedgehog signaling pathway. PLoS One, 8, e67431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santilli G., et al. (2011)Breast cancer-initiating cells: insights into novel treatment strategies. Cancers (Basel), 3, 1405–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agnoli C., et al. (2010)Metabolic syndrome and postmenopausal breast cancer in the ORDET cohort: a nested case-control study. Nutr. Metab. Cardiovasc. Dis., 20, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamburov A., et al. (2013)The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res., 41(Database issue), D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozomara A., et al. (2014)miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res., 42(Database issue), D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cochrane D.R., et al. (2012)Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol. Cell. Endocrinol., 355, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yue X., et al. (2010)Transcriptional regulation by small RNAs at sequences downstream from 3’ gene termini. Nat. Chem. Biol., 6, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song R.X., et al. (2001)Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J. Natl. Cancer Inst., 93, 1714–1723. [DOI] [PubMed] [Google Scholar]
- 42. Ansquer Y., et al. (2005)Progesterone induces BRCA1 mRNA decrease, cell cycle alterations and apoptosis in the MCF7 breast cancer cell line. Anticancer Res., 25, 243–248. [PubMed] [Google Scholar]
- 43. Waldman S.A., et al. (2007)Translating microRNA discovery into clinical biomarkers in cancer. JAMA, 297, 1923–1925. [DOI] [PubMed] [Google Scholar]
- 44. Grivennikov S.I., et al. (2010)Immunity, inflammation, and cancer. Cell, 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mantovani A., et al. (2008)Cancer-related inflammation. Nature, 454, 436–444. [DOI] [PubMed] [Google Scholar]
- 46. Radom-Aizik S., et al. (2010)Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. J. Appl. Physiol., 109, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hussain S.P., et al. (2003)Radical causes of cancer. Nat. Rev. Cancer, 3, 276–285. [DOI] [PubMed] [Google Scholar]
- 48. Lee J.H., et al. (2012)Progesterone suppresses the mTOR pathway and promotes generation of induced regulatory T cells with increased stability. Eur. J. Immunol., 42, 2683–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maillot G., et al. (2009)Widespread estrogen-dependent repression of microRNAs involved in breast tumor cell growth. Cancer Res., 69, 8332–8340. [DOI] [PubMed] [Google Scholar]
- 50. Daniels S.I., et al. (2014)Improving power to detect changes in blood miRNA expression by accounting for sources of variability in experimental designs. Cancer Epidemiol. Biomarkers Prev., 23, 2658–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.