Abstract
Functional disruptions of susceptibility genes by large genomic structure variant (SV) deletions in germlines are known to be associated with cancer risk. However, few studies have been conducted to systematically search for SV deletions in breast cancer susceptibility genes. We analysed deep (> 30x) whole-genome sequencing (WGS) data generated in blood samples from 128 breast cancer patients of Asian and European descent with either a strong family history of breast cancer or early cancer onset disease. To identify SV deletions in known or suspected breast cancer susceptibility genes, we used multiple SV calling tools including Genome STRiP, Delly, Manta, BreakDancer and Pindel. SV deletions were detected by at least three of these bioinformatics tools in five genes. Specifically, we identified heterozygous deletions covering a fraction of the coding regions of BRCA1 (with approximately 80kb in two patients), and TP53 genes (with ∼1.6 kb in two patients), and of intronic regions (∼1 kb) of the PALB2 (one patient), PTEN (three patients) and RAD51C genes (one patient). We confirmed the presence of these deletions using real-time quantitative PCR (qPCR). Our study identified novel SV deletions in breast cancer susceptibility genes and the identification of such SV deletions may improve clinical testing.
Introduction
Over the past few decades, multiple breast cancer susceptibility genes have been identified. Those reliably established include BRCA1, BRCA2, ATM, TP53, CHEK2, PALB2, CDH1, STK11, NF1 and PTEN (1–10). Previous studies have focused primarily on evaluating protein truncating variants identified as small insertion/deletion (Indel) frameshift, nonsense or splice site variants, or potentially pathogenic missense variants in these genes. Structure variants (SVs) typically involved in DNA fragments range from a few hundred bases to a million bases. SVs may have multiple functional consequences, influencing gene sequence, gene expression, or post-transcriptional regulation when they remove gene structures and non-coding regulatory elements, thus leading to an abnormal phenotype with a distinct clinical manifestation (11). SVs involve several classes including large deletions, duplications, translocations or inversions; here we concentrate specifically on large deletions (‘SV deletions’). SV deletions have been identified as one of the major sources of genetic variation in the human genome, contributing to a variety of complex diseases and cancers (12–16). In our recent study conducted among Chinese women, we found that an SV deletion in APOBEC3A/B genes is associated with an increased risk of breast cancer (17). This deletion results in a fusion gene, which increases the stability of the APOBEC3A protein, leading to elevated mutation rates in breast cells (18). It has now been demonstrated that somatic mutation signatures driven by APOBEC enzymes are one of the most important signatures in human cancers (18–22). A second breast cancer susceptibility locus on 2q35 may also be mediated through a large deletion (23). In this study, we searched for SV deletions in breast cancer susceptibility genes using deep whole-genome sequencing (WGS) data.
Results
Blood DNA samples from 128 breast cancer patients, 107 of which were European and 21 of which were of East Asian ancestry were subjected to WGS. The WGS data had an average sequencing depth of 37.8X and an average mapping rate of 98.0%. We applied five different SV calling tools including Genome STRiP (24), Delly (25), Manta (26), BreakDancer (27), and Pindel (28) to discover SV deletions. We evaluated SV deletions identified in the regions of 20 known, or suspected, breast cancer susceptibility genes including BRCA1, BRCA2, ATM, TP53, CHEK2, PALB2, CDH1, STK11, PTEN, PIK3CA, NF1, BRIP1, RAD51C, BARD1, PTEN, FANCM, RAD50, FAM175A, XRCC2, FANCC and RAD51D (1–10). We identified SV deletions in five genes including BRCA1, TP53, PALB2, PTEN and RAD51C by at least three SV calling tools: Genome STRiP tool, Delly, and Manta (Supplementary Material, Data 1, Table 1). Specifically, all of these SV deletions were detected in high concordance by these three tools (Supplementary Material, Data 1). No patient carried variants in more than one gene (Table 1).
Table 1.
Summary of SV deletions in breast cancer high/moderate penetrance genes
| Gene | BRCA1 | TP53 | PALB2 | PTEN | RAD51C |
|---|---|---|---|---|---|
| chr17: 41, 196, 311–41, 277, 500 | chr17: 7, 565, 097–7, 579, 937 | chr16: 23, 614, 482–23, 652, 678 | chr10: 89, 623, 194–89, 728, 532 | chr17: 56, 769, 962–56, 811, 692 | |
| Regiona | chr17: 41, 198, 000–41, 285, 000/41, 218, 000–41, 295, 000 | chr17: 7, 564, 612−7, 566, 205 | chr16: 23, 627, 238–23, 628, 395 | chr10: 89, 652, 820–89, 653, 726 | chr17: 56, 777, 849–56, 778, 979 |
| Annotation | Covering the completed gene deletion/Covering from the promoter to the 17th partial intron | Covering the whole last exon of one particular isoform | Covering the 3rd partial intron | Covering the 1st partial intron | Covering the 3rd partial Intron |
| Europeanb | 2 | 2 | 1 | 3 | 0 |
| Asianb | 0 | 0 | 0 | 0 | 1 |
Note: All deletions were heterozygous.
Two breakpoints for BRCA1 were inferred based on the analysis of read density, while other deletion breakpoints were inferred by the GenomeSTRiP tool (hg19).
Number of breast cancer patients carrying the deletion.
SV deletions in BRCA1
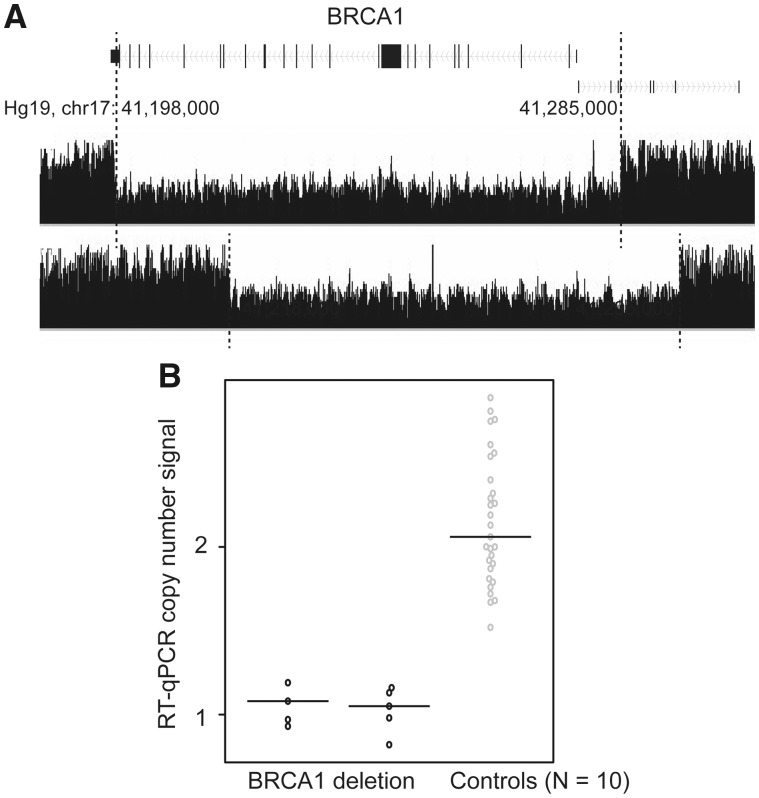
We identified two deletions involving BRCA1, both in a single patient of European descent. The deletions were approximately 87kb and 77 kb in length, and both covered almost all of the coding sequence. The breakpoints for them were located within 10kb of each other (Fig. 1A;Table 1). The deletions were validated using quantitative real-time polymerase chain reaction (qPCR) experiments in the two case samples, and the 10 control samples that were predicted to have no deletions (Fig. 1B and Materials and Methods).
Figure 1.
SV deletions in the coding regions of BRCA1 and qPCR validation. (A) SV deletions in the coding regions of BRCA1 were identified by multiple tools, including GenomeSTRiP, Delly, and Manta. The black track indicates the two patients carrying the SV deletion, which can be observed through the reduction of the read densities to half in the deleted regions, compared with the flanking undeleted regions (indicated by the dashed lines). (B) Results from qPCR validation. Deletions were observed among two patients predicted to carry the deletions based on the WGS data but not in 10 control samples (predicted to not be carrying the deletions based on WGS data or 1000 Genomes). Technique replications of qPCR result for each sample are represented by each dot.
SV deletions in TP53
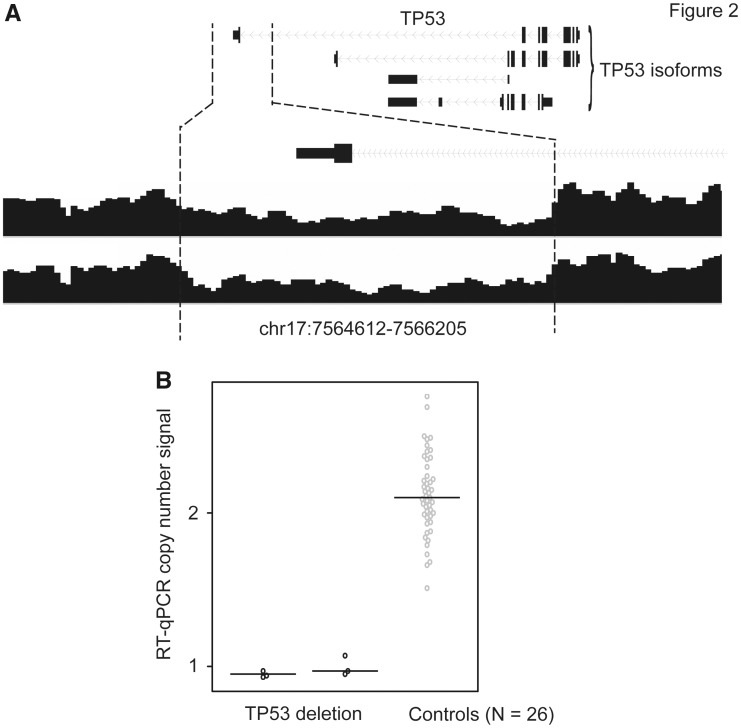
A heterozygous deletion in one TP53 isoform was observed in two patients of European descent. This deletion was approximately 1.6kb, covering the whole last exon and the 3’ untranslated region (UTR) of TP53 (Fig. 2A;Table 1). Using qPCR experiments, we confirmed that the deletion was present only in these two patients (Fig. 2B).
Figure 2.
SV deletion in the TP53 and qPCR validation. (A) An SV deletion in the last exon of the TP53 isoform was identified in two patients. (B) Results from qPCR validation for these two cases and 26 control samples.
SV deletions in the intronic regions of PTEN, PALB2 and RAD51C
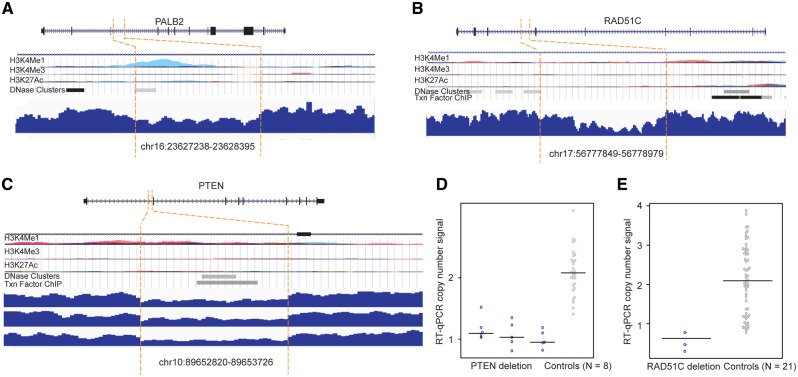
We found three small deletions (∼1 kb) in the intronic regions of PALB2, PTEN, and RAD51C (Table 1). The PALB2 deletion was observed in one patient and the PTEN deletion in three patients of European descent, while the RAD51 deletion was found in one patient of Asian descent. We evaluated the functional significance of the deletion regions by investigating the epigenetic data from the Encyclopedia of DNA Elements (ENCODE) (see Materials and Methods). The deletion regions for the PALB2 and PTEN genes were observed to have epigenetic signals with evidence of enhancer marker H3K4Me1 and DNase and/or ChIP-seq enriched peaks (Fig. 3A and B). No epigenetic signal was observed in the deletion region for the RAD51C gene (Fig. 3C).
Figure 3.
SV deletions in the intronic regions of PTEN, PALB2 and RAD51C and qPCR validation. (A–C) SV deletions in the intronic regions of PALB2, PTEN, and RAD51C. At the top of each panel: epigenetic landscape of SV deletion. From top to bottom, RefSeq genes; layered H3K4Me1, H3K4Me3, and H3K27Ac histone modifications; DNase clusters; clustered ChIP-seq binding sites; The signals of different layered histone modifications from the same ENCODE cell line are shown in the same color. (D) qPCR results for the deletion in PTEN. (E) qPCR results for the deletion in RAD51C.
We performed qPCR in the breast cancer patients carrying these SV deletions and in the controls without these deletions to technically validate our findings. The PTEN deletion was fully validated in three patients and eight controls (Fig. 3D). The deletion in RAD51C was confirmed in the patient sample, but also found in some control samples (Fig. 3E). We could not perform qPCR validation for the deletion in the PALB2 gene due to the lack of a DNA sample in the patient carrying the deletion.
Discussion
To our knowledge, this is the first study to investigate germline SV deletions in known breast cancer susceptibility genes using deep WGS technology. We found multiple SV deletions in these genes. In particular, two patients carrying SV deletions in the BRCA1 gene showed definitive evidence that SVs disrupted gene function by removing the gene almost entirely, and thus contributed to breast cancer susceptibility. The deletions we detected in this study in TP53, PTEN and PALB2 may directly or indirectly affect gene function by removing either coding regions or functional elements, likely contributing to breast cancer susceptibility as well. In particular, the TP53 function may be affected by the disruption of protein translation or by the dysregulation of post-transcriptional levels of mRNA stability, as the deletion involved the last exon of the gene. These SV deletions could be strong candidates for clinical testing to identify women with high-risk for breast cancer.
Previous family-based studies have revealed many potential pathogenic single nucleotide coding variants or small Indel in penetrance genes, including BRCA1, BRCA2, TP53 and others (1–10). In regards to them, SVs have been believed to be a major source of variation in the human genome, and may contribute to a variety of diseases (11,13,29,30). However, SVs remain uninvestigated due to their being more challenging to discover than single nucleotide variants. The analysis of WGS data provides an unprecedented opportunity to capture SVs in high confidence and to provide novel resources for the discovery of genetic variation in breast cancer.
We compared the identified SV deletions with deletions in public SV databases, including the Database of Genomic variants (DGV) (31), the 1000 Genomes project (29), the Exome Aggregation Consortium (ExAC), and the Mills_and_1000G deletions (see Materials and Methods). We did not find the PALB2 deletion identified in our study overlapped in any public database. For the two deletions in BRCA1, we observed highly overlapped deletions with ∼90% of reciprocal overlap (RO), which were presented in the ExAC and DGV databases (Supplementary Material, Data 2). The other two deletions in the TP53 and PTEN genes have been reported in European descendants, with allele frequency (AF) being 0.01 and 0.03, respectively, in the 1000 Genomes project. The deletion in the RAD51C gene was only reported in the DGV. All of these three genes showed RO > 0.95. However, since some deletions (i.e. PTEN, TP53, and RAD51C) were also observed in normal subjects, a thorough validation study with increased sample size for these candidate SV deletions would be required to further confirm the deletions for breast cancer susceptibility. Previously, we reported a strong association between a deletion in the APOBEC3A/B gene and breast cancer. Here, we investigated this deletion based on the WGS data (hg19, chr22: 39, 358, 340–39, 388, 452). We found that 10 of the 21 Asian descendants included in this study carried the APOBEC3A/B deletion, including nine heterozygous and one homozygous deletion. These data were fully consistent with the results based on the qPCR experiments performed in our previous work (32,33). In European descendants, 22 of 107 breast cancer patients carried the APOBEC3A/B deletion, including 19 heterozygous and three homozygous deletions. The frequency distribution in European descendants was similar to our previous qPCR results (34). The findings actually supported our discovery of SVs with high reliability.
The sample size of our current study is relatively small, and thus many SV deletions remain to be discovered. Although we performed functional annotation using epigenomic data including TFs ChIP-seq data, DNase I hypersensitive sites, and histone modification markers from the ENCODE data, in vitro assays are needed to characterize the functions of these SV deletions. The current study focuses on identifying SV deletions, but other types of SVs should be investigated in the future, including short insertions and deletions (indels), insertion, inversion and tandem duplications.
Materials and Methods
Study populations
This study included 128 breast cancer patients from the Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium. Of these patients, there were 107 breast cancer patients of European descent with a family history, or early onset case, of cancer. These patients were part of the Nurse Health Study (N = 30) (35) and the SEARCH breast cancer study (N = 77) (36). Also included were 21 Chinese women diagnosed with triple-negative breast cancer from the Shanghai Breast Cancer Genetics Study (37).
Data processing
The raw sequencing reads with pair-end of a length of 100 bp were generated using HiSeq 2000 from Illumina (target: average 30X coverage). The bioinformatics analysis started with the sequencing fastq files, which was provided by the Illumina sequencing center. The sequencing reads for each of the 128 samples were mapped to the human reference genome (hg19) using the Burrows-Wheeler Aligner BWA program (version 0.7.9a) (38). After the alignment, duplicate reads were removed using Picard MarkDuplicates (http://picard.sourceforge.net/). The remaining aligned reads were further processed for local realignment using the GATK package following best-practice recommendations (39). The final BAM file for each sample was generated for subsequent SV tool calling analysis.
SV deletions calling
We applied multiple SV calling tools including Genome STRiP tool v2 (24), Delly2 (25), Manta (26), BreakDancer-1.1 (27), and Pindel 0.2.5 (28) to discover SV deletions based on the BAM files from the 128 breast cancer patients. Using the Genome STRiP tool, we performed the initial SV deletion discovery using the ‘SVDiscovery’ script. We then filtered those SVs following the criteria from the tutorial. The genotype for each identified SV was then generated across all samples using ‘SVGenotyper’ script. We further applied Script ‘RedundancyAnnotator’ to remove redundant SV deletions with similar coordinates. We applied the SV calling Delly tool with the default parameters. Poor SV deletions from the generated BCF file were further filtered using the ‘Delly filter’ function. We applied the Manta tool including two python scripts, configManta.py and runWorkflow.py, to call SV deletions for each sample. We then filtered SVs deletions through VCFtools in the VCF file with a ‘PASS’ flag and merged all files for downstream analysis. We applied both BreakDancer and Pindel tools with the default parameters to call SV deletions for each sample. Only the SV deletions with >7 supporting reads detected in at least one sample were retained. For the SV deletions identified by each of the above five tools, we removed those located in telomere and centromere regions and only analysed the ones with deletion sizes ranging from 50bp to 1Mb. To remove redundant SVs with similar coordinates identified by each tool, we developed an in-house PERL script to merge any SV deletions having RO of at least 50% into single SV deletions by averaging their start and end breakpoints.
Comparison of identified SV deletions with public SV databases
To compare the SV deletions identified in this study with those from SV public databases, we downloaded a total of 68 818 SVs from the 1000 Genomes project (29), 392 583 SVs from the DGV (31), 15 734 SVs from the ExAC (40) and 1 274 580 Indels from the Mills_and_1000G (ftp://ftp.broadinstitute.org/bundle/). The intersection of SV deletions identified in the study and the above databases were analysed using the ‘bedtools intersect’ function.
Functional annotation of SV deletions
To systematically annotate SV deletions involved in gene coding regions, we downloaded the human transcriptome annotation Gencode version 17 (hg19) from the GENCODE browser (http://www.gencodegenes.org/releases/17.html). We assigned each SV deletion a gene body region, including the coding, exonic, and intronic regions of genes, using the function ‘GeneOverlapAnnotator’ from the GenomeSTRiP package. Functional annotation for SVs deletions in noncoding regions was also accessed through the UCSC Genome Browser (https://genome.ucsc.edu/cgi-bin/hgGateway). The epigenetic landscape of histone markers H3K4Me1, H3K4Me3, H3K27Ac, DNase I hypersensitive sites and transcription factor chromatin immunoprecipitation with sequencing (ChIP-seq) binding sites were also examined through layered histone tracks on all available ENCODE cell lines from the UCSC Genome Browser.
SV deletions genotyping via qPCR
SV deletion genotyping was performed in a duplex real-time qPCR reaction with TaqMan® copy number variant (CNV) assays for each of the target genes BRCA1 (Assay ID: Hs01865955_cn), PTEN (Assay ID: Hs05141067_cn), TP53 (Custom-designed assay ID: TP53R_CCBJXVN) and RAD51C (Custom-designed assay ID: RAD51Cv2_CC20TN6), and the reference gene RPPH1 (4403328 with VIC fluorophore) (Applied Biosystems, Foster City, CA), using 2× TaqMan® genotyping Master Mix (Applied Biosystems, Foster City, CA), and 10 ng of genomic DNA. The 10-µl reactions were run in technical triplicates using the Applied Biosystems 7900HT Fast Real-Time PCR System under standard conditions. Samples with predicted not carrying deletions (CN = 2) based on our WGS data or based on the 1000 Genomes project were included as positive controls. Distilled and deionized water was included as a negative control on each PCR plate. To determine the copy number (CN) for each SV deletion, the qPCR data were analysed using CopyCaller® software v2.0 (Applied Biosystems). This program performs a comparative qPCR cycle at threshold (Ct) relative quantification on the real-time data to determine the CN with the formula CN = 2 × 2-ΔΔCt (41), where ΔΔCt = (Ct reference genesample − Ct target genesample) -(Ct reference genecalibrator − Ct target genecalibrator). After the ΔΔCt was calculated, the genotype for each deletion was determined: homozygote deletion (CN < 0.10), heterozygote deletion (0.8 < CN < 1.2), and no deletion (1.8 < CN < 2.5). If the CN value was not within the above ranges, those DNA samples were re-genotyped into triplicates. All qPCR assays were performed by a single lab staff member (J. Shi), and all CN calls were conducted by two independent staff members.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank Jie Wu and Regina Courtney for laboratory assistance. The authors also thank Nancy Kennedy and Marshal Younger for assistance with editing and manuscript preparation. The data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
Conflict of Interest statement. None declared.
Funding
U19CA14806, R01CA158473, Cancer Research UK (CR-UK), Cancer Research UK (C1287/A16563).
References
- 1. Apostolou P., Fostira F. (2013) Hereditary breast cancer: the era of new susceptibility genes. Biomed. Res. Int., 2013, 747318.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan M.H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. (2012) Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer. Res., 18, 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H.. et al. (2010) Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet., 42, 410–414. [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez K.D., Noltner K.A., Buzin C.H., Gu D., Wen-Fong C.Y., Nguyen V.Q., Han J.H., Lowstuter K., Longmate J., Sommer S.S.. et al. (2009) Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J. Clin. Oncol., 27, 1250–1256. [DOI] [PubMed] [Google Scholar]
- 5. Stratton M.R., Rahman N. (2008) The emerging landscape of breast cancer susceptibility. Nat. Genet., 40, 17–22. [DOI] [PubMed] [Google Scholar]
- 6. Pujana M.A., Han J.D., Starita L.M., Stevens K.N., Tewari M., Ahn J.S., Rennert G., Moreno V., Kirchhoff T., Gold B.. et al. (2007) Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet., 39, 1338–1349. [DOI] [PubMed] [Google Scholar]
- 7. Rahman N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A., Reid S., Spanova K., Barfoot R., Chagtai T.. et al. (2007) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet., 39, 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K.. et al. (2006) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet., 38, 1239–1241. [DOI] [PubMed] [Google Scholar]
- 9. Renwick A., Thompson D., Seal S., Kelly P., Chagtai T., Ahmed M., North B., Jayatilake H., Barfoot R., Spanova K.. et al. (2006) ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet., 38, 873–875. [DOI] [PubMed] [Google Scholar]
- 10. Pharoah P.D., Guilford P., Caldas C. and International Gastric Cancer Linkage, C. (2001) Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology, 121, 1348–1353. [DOI] [PubMed] [Google Scholar]
- 11. Weischenfeldt J., Symmons O., Spitz F., Korbel J.O. (2013) Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet., 14, 125–138. [DOI] [PubMed] [Google Scholar]
- 12. Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D.. et al. (2005) Fine-scale structural variation of the human genome. Nat. Genet., 37, 727–732. [DOI] [PubMed] [Google Scholar]
- 13. Alkan C., Coe B.P., Eichler E.E. (2011) Genome structural variation discovery and genotyping. Nat. Rev. Genet., 12, 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pang A.W., MacDonald J.R., Pinto D., Wei J., Rafiq M.A., Conrad D.F., Park H., Hurles M.E., Lee C., Venter J.C.. et al. (2010) Towards a comprehensive structural variation map of an individual human genome. Genome Biol., 11, R52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L.. et al. (2007) Paired-end mapping reveals extensive structural variation in the human genome. Science, 318, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. (2004) Detection of large-scale variation in the human genome. Nat. Genet., 36, 949–951. [DOI] [PubMed] [Google Scholar]
- 17. Long J., Delahanty R.J., Li G., Gao Y.T., Lu W., Cai Q., Xiang Y.B., Li C., Ji B.T., Zheng Y.. et al. (2013) A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst., 105, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caval V., Suspene R., Shapira M., Vartanian J.P., Wain-Hobson S. (2014) A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat. Commun., 5, 5129.. [DOI] [PubMed] [Google Scholar]
- 19. Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E.. et al. (2014) Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet., 46, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burns M.B., Temiz N.A., Harris R.S. (2013) Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet., 45, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burns M.B., Leonard B., Harris R.S. (2015) APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J., 38, 102–110. [DOI] [PubMed] [Google Scholar]
- 22. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L.. et al. (2013) Signatures of mutational processes in human cancer. Nature, 3, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wyszynski A., Hong C.C., Lam K., Michailidou K., Lytle C., Yao S., Zhang Y., Bolla M.K., Wang Q., Dennis J.. et al. (2016) An intergenic risk locus containing an enhancer deletion in 2q35 modulates breast cancer risk by deregulating IGFBP5 expression. Hum. Mol. Genet., 25, 3863–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Handsaker R.E., Van Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., McCarroll S.A. (2015) Large multiallelic copy number variations in humans. Nat. Genet., 47, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O. (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics, 28, i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Kallberg M., Cox A.J., Kruglyak S., Saunders C.T. (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics, 32, 1220–1222. [DOI] [PubMed] [Google Scholar]
- 27. Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P.. et al. (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods, 6, 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics, 25, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Hsi-Yang Fritz M.. et al. (2015) An integrated map of structural variation in 2, 504 human genomes. Nature, 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haraksingh R.R., Snyder M.P. (2013) Impacts of variation in the human genome on gene regulation. J. Mol. Biol., 425, 3970–3977. [DOI] [PubMed] [Google Scholar]
- 31. MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. (2014) The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res., 42, D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G.. et al. (2013) An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet., 45, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Delahanty R., Guo X., Zheng W., Long J. (2015) Integrative genomic analysis reveals functional diversification of APOBEC gene family in breast cancer. Hum. Genomics, 9, 34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xuan D., Li G., Cai Q., Deming-Halverson S., Shrubsole M.J., Shu X.O., Kelley M.C., Zheng W., Long J. (2013) APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis, 34, 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tworoger S.S., Eliassen A.H., Sluss P., Hankinson S.E. (2007) A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J. Clin. Oncol., 25, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 36. Lesueur F., Pharoah P.D., Laing S., Ahmed S., Jordan C., Smith P.L., Luben R., Wareham N.J., Easton D.F., Dunning A.M.. et al. (2005) Allelic association of the human homologue of the mouse modifier Ptprj with breast cancer. Hum. Mol. Genet., 14, 2349–2356. [DOI] [PubMed] [Google Scholar]
- 37. Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L.. et al. (2009) Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet., 41, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ruderfer D.M., Hamamsy T., Lek M., Karczewski K.J., Kavanagh D., Samocha K.E., Exome Aggregation C., Daly M.J., MacArthur D.G., Fromer M.. et al. (2016) Patterns of genic intolerance of rare copy number variation in 59, 898 human exomes. Nat. Genet., 48, 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.