Abstract
Genomic islands (GIs) that are associated with microbial adaptations and carry sequence patterns different from that of the host are sporadically distributed among closely related species. This bias can dominate the signal of interest in GI detection. However, variations still exist among the segments of the host, although no uniform standard exists regarding the best methods of discriminating GIs from the rest of the genome in terms of compositional bias. In the present work, we proposed a robust software, MTGIpick, which used regions with pattern bias showing multiscale difference levels to identify GIs from the host. MTGIpick can identify GIs from a single genome without annotated information of genomes or prior knowledge from other data sets. When real biological data were used, MTGIpick demonstrated better performance than existing methods, as well as revealed potential GIs with accurate sizes missed by existing methods because of a uniform standard. Software and supplementary are freely available at http://bioinfo.zstu.edu.cn/MTGI or https://github.com/bioinfo0706/MTGIpick.
Keywords: genomic island detection, genomic signature, feature selection, multiscale statistical test, boundary detection
Introduction
Bacteria have greatly diversified over billions of years as a result of their adaption to a wide range of environments. One of the major contributors to adaptability of bacteria is horizontal gene transfer (HGT). An HGT event was first reported in 1990 when Hacker et al. [1] found that a few clusters of virus genes present in several Escherichia coli genomes are absent in their close relatives; these gene clusters are referred to as pathogenicity islands (PAIs). At least a dozen types of PAIs, such as ‘secretion islands’, antimicrobial ‘resistance islands’ and ‘metabolic islands’, have been detected thereafter. Genomic island (GI) was then used as a more general term to refer to a cluster of 10–200 kb long genes acquired through horizontal transfer. Typically, these horizontally transferred regions are first denoted as GIs until further inspection of their gene function provides basis for the use of a more specific term [2].
The importance of GIs should not be underestimated in the genomic era. Given a newly sequenced genome, researchers usually intend to find some genomic regions that differentiate an organism from other species or strains. By comparing related taxa, one can possibly discern GIs that encode functions related to complex changes in ecological niche [3]. For example, GIs are responsible for type III secretion systems, iron uptake functions, toxin and adhesion secretion, which augment the ability of pathogens to survive within a host and thus cause diseases [4, 5]. Some researchers have reported that pathogenicity can be modulated with the help of selective loss or regain of specific GIs [6, 7], and PAIs can be spontaneously excised from a chromosome at detectable rates, resulting in distinct pathogenic phenotypes [8, 9]. In addition, GIs apparently confer many other adaptations to bacteria, including metal resistance, antimicrobial resistance and secondary metabolic properties, which are of environmental or industrial interest [5]. Therefore, identification of GIs in different genomes has become of great interest in studies on microbial evolution and function.
With the help of large-scale comparative genomics, researchers have found that GIs are characterized by varying sequence composition, flanking direct repeats and presence of mobility and transfer RNA (tRNA) genes. Exploring and using these features in turn can lead to better GI detection [3, 10–12]. GIs are sporadically distributed among closely related species, and they carry some phyletic patterns that differ from the host, allowing researchers to identify them by comparing the divergence of the 16S ribosomal RNAs or other orthologs among distantly related species [13]. Several alignment-based methods, such as basic local alignment method [14] and whole-genome alignment [15], have been developed to detect GIs. These tools rely on the observation that genomic regions that are not aligned across multiple genome alignment or uniquely aligned to a genome are more likely to be putative GIs compared with the conserved regions. For more complex cases, several methods to construct and apply multiple layers of large-scale genomic comparisons were reported. For example, MobilomeFINDER first finds shared tRNA genes among several related genomes and then uses Mauve, an alignment method, to search for GIs in the upstream and downstream regions of orthologous tRNA genes [16]. Given that GIs identified using this method are associated with disrupted tRNAs, GIs without tRNA genes as insertion sites will be missed. To address this problem, MOSAIC launched a method to identify strain-specific regions that were not necessarily inserted into a tRNA [17]. Unfortunately, inversions and translocations are often mistakenly identified as strain-specific regions. IslandPick is one of the most widely used tools for GI identification [18]. Given a genome, IslandPick first automatically selects suitable comparison genomes without any bias, and then Mauve is adopted to construct whole-genome alignments. To avoid duplication, IslandPick uses BLAST as secondary filter to recheck the regions aligned by Mauve. IslandPick has been integrated into Islandviewer website, where pre-computed data sets of GIs can be downloaded [19, 20]. In addition, comparative genomics method relies heavily on the genomes used in comparison and thus can be of limited use during annotation or when closely related genomes are unavailable. Even when more genomes are available, researchers will have to spend more effort on selecting genomes from the species of interest [21].
Apart from comparative genomics, composition-based methods are highly sensitive for GI detection. Given that GIs often exhibit a sequence composition that is significantly different from that of the host, a detection algorithm, to be efficient, must discriminate anomalous regions from the remainder of the genome in terms of compositional bias. In practice, composition-based methods are desirable because they allow rapid GI identification from a genome or from a sequence that is analysed without requiring additional genomes. The GC content and two to nine long oligonucleotides are widely used to describe sequence composition in GI detection [10, 22–25]. For example, PAI Finder calculates GC content anomalies and codon usage bias to detect GIs and further evaluates a candidate PAI only if the PAI-like region partly or entirely spans the GI [26]. PAI Finder has been integrated into the PAI database, where comprehensive information on all annotated PAIs and predicted ones in prokaryotic genomes can be downloaded [27, 28]. Hidden Markov models (HMMs) are also introduced to assist in removing or detecting anomalous regions containing compositional biases [22, 29–31]. For example, score-based identification of genomic islands using Hidden Markov Models (SIGI-HMM) constructs an HMM to remove ribosomal regions with biased codon usage [29, 30]. In addition, IslandPath-DIMOB [31] uses an HMMer to identify mobility genes [11] by searching each predicted gene against PFAM37 mobility gene profiles [32], whereas Alien_Hunter introduces a scoring system based on a flexible length of k-mers and refines the boundaries of the predicted GIs using an HMM [22]. Although these HMM-based methods demonstrate better performance in GI detection, they involve a relatively high number of parameters and heavy training calculation and thus longer computational time is required to detect GIs.
Instead of evaluating a cluster of genes, several researchers first split a genome into distinct overlapping or non-overlapping windows and then extract the compositional features of the genome [33–36]. To identify the compositionally distinct windows, they measure whether the difference between two windows is significant or not. To accomplish this goal, centroid identifies some windows as GIs because they are identified by distance values lying outside the other values [33]. However, one limitation of this method is that the signatures of the host are estimated based on all the windows without selection, resulting in some noise in the native information of the host. To overcome this problem, INDeGenIUS uses a sequence-clustering method to obtain a ‘major cluster’ to estimate the native signatures of the host [34]. However, measuring each oligonucleotide is not necessary, and simultaneous presence of a subset of oligonucleotides is generally viewed as strong evidence for a horizontal transfer. Thus, instead of selecting all possible tetranucleotides, SigHunt selects informative tetranucleotides from a range of organisms using the tetranucleotide quality score [36]. These window-based methods can provide a rapid GI prediction, although several problems still exist as described below:
Outcomes of the SigHunt depend on the additional related genomes used in selection. For example, inclusion of distant genomes that have extensive rearrangements renders selection of informative oligonucleotide difficult and can potentially lead to false-positive predictions. Moreover, SigHunt can be of limited use when closely related genomes are unavailable.
Atypical regions are frequently reported as GIs only in terms of the established threshold. If the compositional difference of a region is larger than the established threshold, then this region is deemed atypical. Given that different data sets can result in different thresholds, determining all of the GIs from different data sets solely on the basis of the established thresholds is therefore difficult. However, when atypical regions can be estimated by a standard statistical test, the efficiency of a detection method can be evaluated regardless of different data sets.
Iteration is virtually never used in GI identification. In most reports, the number of GIs from a one-step prediction is insufficient. Moreover, samples are nearly always considerably small to select optimal threshold values for various regions.
Most window-based methods predict GIs without refining the island boundaries. If the boundaries of the predicted GIs are further refined, then the validity and efficiency of the prediction are likely to be improved.
To address these problems, we reported herein a novel software called ‘MTGIpick’, the first multiscale statistical test for GI identification. For each region of a genome, we proposed an iteration of a small-scale t-test with large-scale feature selection (IST-LFS) to quantify compositional differences of a genome from that of a host rather than to calculate the distance or discrete interval accumulative score of each region. At the core of the IST-LFS method is a selection method for informative tetranucleotides using kurtosis and a highly sensitive measure based on a two-sample t-test. Unlike the predetermined thresholds and limited information from individual windows in the existing methods, we investigated the variability of genomic signatures and used multiscale segmentation algorithm (MSA) to identify large, multiwindow segments. After delineating these compositionally distinct segments, GIs were selected with respect to their enrichment scores. Finally, the boundaries of predicted GIs were further refined using Markovian Jensen–Shannon divergence (MJSD) and the GC-based segmentation method.
Materials and methods
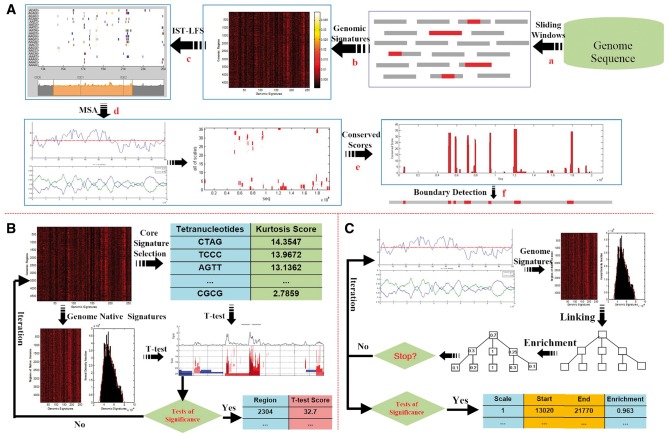
We now describe the framework for the robust GI identification using multiscale statistical testing. The steps are schematically illustrated in Figure 1A and are described as follows:
Figure 1.
Overview of the MTGIpick algorithm. (A) The workflow of the MTGIpick algorithm, with (a) split a genome into non-overlapping windows with a size of 1 kb; (b) extract genomic signatures represented as a heatmap; (c) score each window using IST-LFS; (d) identify large segments using MSA; (e) calculate conserved score of the predicted genomic islands; and (f) refine the boundaries of the GIs using the GC-MJSD method. (B) The workflow of the IST-LFS algorithm, in which signatures of the host are extracted using the CIWV, and core signatures are selected based on ordered kurtosis. During an iteration, we score each window using the two-sample t-test and select the windows whose scores are large enough to be considered to be statistically significant. (C) The workflow of the MSA algorithm. Starting from the IST-LFS scores and GC content, we select signatures of the host using the CIWV. During an iteration, we construct a continuous linear scale-space using a blurring strategy, score the enrichment of all the segments using the Z test and select the windows whose enrichment scores are large enough to be considered to be statistically significant.
Steps a–b: Split a genome into non-overlapping windows with a size of 1 kb and extract genomic signatures
To detect regions with distinct composition, one must extract features from each region of the genome. Several approaches have been proposed to extract genomic signatures within a given window. Different window sizes provide different information on DNA segments. In other words, each window size describes a different view of the genomic signatures: a longer window misses small details of local genomic signatures, whereas a shorter window preserves details of genomic signature, although it suffers greatly from clusters in different windows.
We split a genome into n non-overlapping windows of size 1 kb and calculated the frequencies of the tetranucleotides in each window as genomic signature.
Step c: Score each window using an IST-LFS
At a smaller scale, we proposed the use of IST-LFS to quantify the compositional differences of a region from the host (Figure 1B). The steps of the IST-LFS are described below:
- Extract the signatures of the host by using the confidence intervals on the windows' variances (CIWV). For each region, we calculated the variance s2 of the oligonucleotide frequencies and further estimated the confidence interval of their mean as follows:
where is the average of all of the window variances, is the standard variance of all of the windows’ variances, is a confidence level and N is the total number of regions. If variance of a region falls within the confidence interval, then the region is possibly considerably conserved and thus can be considered a region from the host.(1) - Calculate the kurtosis of each tetranucleotide across n windows and select the windows with a larger kurtosis as informative signatures. Kurtosis is formally defined as follows:
where is the sample mean and n is the total number of observations.(2) - Measure the divergence of the ith window from the host using two-sample t-test. For each informative signature , a t-test was used to determine whether the means of the two samples and are equal, and its P-value was calculated as follows:
where(3)
and ( and ) are the means (variances) of the informative signature from the eye regions surrounding the ith region and from the host , respectively, is the total number of selected regions of the host and is the total number of selected informative signatures. Summing all of the P-values of the informative signatures, we obtained the divergence of the ith region from the host as follows:(4) Select windows whose scores are sufficiently large to be considered statistically significant.
Delete the selected windows and update all windows of the genome; repeat Steps 2–4 until no window is found.
Step d: Identify segments using MSA
At a large scale, we investigated the variability of genomic signatures and used MSA to identify large, multiwindow segments (Figure 1C). MSA is described as follows:
- Create a scale space with S scales, where the first scale in the scale space (s = 1) is the score x(i) obtained using the IST-LFS method. The subsequent scales are obtained using a Gaussian window with increasing width. The standard deviation of this window for scale s is defined as follows:
The convolved signal at scale s is denoted by .(5) Choose a set of starting positions, which will serve as starting nodes of the segmentation tree. To reduce computational complexity, we only select these positions of the genomic signal where a differential signal intensity, i.e. or , exits.
- For each starting node at scale t (child), the best successor node at scale t + 1(parent) is sought within a limited domain. The potential successor nodes are selected on the basis of the affection to a given starting node at scale t. This affection is defined as follows:
where(6)
with and being the spatial positions of the child and the parent, respectively, and and being the scales of the child and parent levels, respectively. The candidate successor node with the highest affection value is selected to become the best successor node at scale t + 1. Divide the complete genomic signal at scale t + 1 into segments whose boundaries are the best successor nodes of the starting nodes at scale t.
- The observed intensity of a segment is simply the summed signal intensity in the segment and is denoted by X. The expected intensity of a segment follows a normal distribution with P = I/B. Herein, I and B are the total summed signal intensity (the IST-LFS score) and total background signal intensity (the GC score) across the complete genomic signal, respectively, and n is the summed intensity of the background signal of the segment under investigation. The enrichment and depletion of all the segments at scale t + 1 are calculated using standard enrichment tests (the Z-test). We picked a P-value threshold . The P-value threshold was converted into a Z score using the inverse error function. The probabilities were then solved using the following equations:
where . Thus, if the observed intensity of some segments is equal to or greater than the observed intensity X, they are expected to exhibit probability , which is the P-value. Given that np is the expected mean background intensity of the segment, its enrichment can be calculated by using the following equation:(7) (8) Select the segments whose enrichment scores are sufficiently large to be considered statistically significant.
Repeat Steps 3–6 until the given scale is achieved.
Step e: Calculate conserved score of each nucleotide according to the total number of appearances in the selected segments, from which GIs are detected with respect to their conserved scores
Step f: Refine the boundaries of the predicted GIs using the GC content bias and Markovian Jensen–Shannon divergence
Window-based methods usually select atypical windows as putative GIs without refining their boundaries. Herein, we proposed a simple method to refine the boundaries of predicted GIs based on GC content bias and Markovian Jensen–Shannon divergence (GC-MJSD) [21, 35]. Suppose that is a predicted GI whose start and end positions are and , respectively, then the proposed method allows users to search for its boundaries in regions upstream and downstream of from its start and end positions. The GC content bias describes the differences among the DNA fragments and captures some strong signatures for GI detection [37, 38]. To refine the start position of the GI, we segmented the sequence into several distinct regions according to the GC content bias and obtained a series of the breakpoints . For each breakpoint , we calculated the MJSD between the and using the following equation:
| (9) |
where and are the Markov entropies of the sub-sequences and , respectively, and is the Markov entropy of the sequence . The breakpoint showing the maximum MJSD value was then selected as start point:
| (10) |
Following the same method, we obtained the end point of :
| (11) |
For the parameter , 2 was set as the default value.
Results
Expected mer and window size in GI detection
In all window-based methods, different window sizes provide different information about k-mer counts. Each window size describes a different view of the genomic signatures: a longer window misses small details of local genomic signatures, whereas a shorter window preserves details of genomic signature, although it suffers greatly from clusters in different windows. To provide a robust view of the multilevel composition, appropriate levels of expectation should be determined first. Suppose that four nucleotides, A, C, T and G, occur in equal probabilities and occur independently of one another. Should we want to observe that a specific k-mer appears at least t times in a window of length n with a 95% chance, then by using binomial distribution, we obtain the following equation:
| 12 |
Given an expectation level, we can solve for the window length n that optimally summarizes the information of the given k-mer with the help of the above equation. However, what is truly needed in GI identification is an appropriate window length that optimally summarizes the genomic signatures. We initially investigated the relationship between these parameters and found that window size rises sharply with k-mer length or their expected count increases, especially starting from 7-mer (Supplementary Figure S1A and Table S1). Although no biological evidence exists for a minimum size of GIs, many methods typically use a minimum cut-off of 8 kb (34). In the case of a 7-mer, the minimum window size in which we expect k-mer to appear at least once is ∼50 kb (Supplementary Table S1), which is much larger than the minimum GI size. This finding suggests that k-mers that are ≥7 are not desirable for development of methods for GI detection.
GIs contain clusters of genes that are acquired by horizontal transfer, and detecting these genes in turn can lead to better prediction of GIs. To further optimize the k-mer length and window size, we analysed the length distribution of horizontally transferred genes. We collected 118 131 transferred genes (15–11 792 bp) in 479 prokaryotic genomes from the Horizontal Gene Transfer DataBase. We observed that the average length of these transferred genes is <1 kb in prokaryotic genomes (Supplementary Figure S1B). However, a 1 kb window only allows us to observe the maximum k-mer length of 4, with an expected count of at least 1. In other words, tetranucleotides can be the largest k-mers that are sufficiently sensitive for detection of HGTs.
Moreover, window size is associated with expected k-mer count. We subsequently investigated the window size based on the expected count. When 4-mer count increases by 10, the window size increases from 0.8 to 4 kb (Supplementary Figure S1C). To further understand the influence of the expected counts, we scored the genome sequences using the IST-LFS without iteration in which non-overlapping window sizes were used from 1 to 4.5 kb. These window sizes guarantee that the ‘at least’ the expected 4-mer counts ranges from 1 to 10. We performed this experiment on four genomes with different GI sizes. One genome is the Salmonella enterica serovar CT18, which contains 21 large known GIs with an average size of 29 kb, and the other genome are the three chromosomes of Aspergillus fumigatus, which contains 86 small known GIs whose average size is 5 kb. We noted that the area under curve (AUC) the receiver operating characteristic curve for A. fumigatus decreases as the window size increases, and the best window size is 1 kb (Supplementary Figure S1D). In contrast, the AUC scores for S. enterica serovar CT18 increase first and then decrease from 2 kb, with the best window size being 1.7 kb in which a 4-mer can be observed at least three times (Supplementary Figure S1D). All of these results suggest that a 4-mer with an expected count of at least four is sufficient to predict the GIs by detecting horizontally transferred gene. In this work, tetranucleotides within a non-overlapping window with a size of 1 kb were chosen to detect the GIs.
Comparison of window-based approaches to classify GIs/non-GIs
To evaluate the proposed method, we first used the proposed method to classify GIs/non-GIs constructed as a standard data set to evaluate GI predictors (Supplementary Data). We applied MTGIpick to classify GIs/non-GIs, where the IST-LFS was run with default parameters, using six iterations in the IST-LFS and 0.28 standard error in MSA. MTGIpick finally used 1 kb upstream/downstream of ‘raw’ GIs to refine the boundaries of predicted GIs. For comparison, the window-based methods centroid [33], INDeGenIUS [34] and SigHunt [36] were all run using default values on the same 118 chromosomes. For SigHunt and INDeGenIUS, we used the same significance test with a significance level of 0.05, which was used in our method to detect putative GIs. DIAS value of >5, which is used in SigHunt, does not work in our experiment because all of the tetranucleotides were used to calculate DIAS rather than the 16 selected tetranucleotides because of the lack of related genomes for core signature selection. The precision, recall and overall accuracy of each method were calculated at the nucleotide level (Supplementary Data).
We found that MTGIpick achieved the best performance, with an overall accuracy of 86.15%, whereas others demonstrated similar overall accuracies ranging from 81 to 84% (Supplementary Table S2). MTGIpick was the only prediction method that achieved a prediction of over 70% and a recall of over 45%, whereas the other methods lagged behind. Zisland Explorer achieved the best performance in terms of precision but at the cost of lower recall (25.49%) [39]. SigHunt did not achieve the expected prediction, and this finding is apparently caused by a large number (758) of predicted GIs and a relatively short average length (4670 bp) in SigHunt compared with those in the other methods (number: 277–522, average length: 13 146–30 352 bp). In addition, the chosen significance level possibly exerts influence on the performances of SigHunt and INDeGenIUS. To minimize the effects of this parameter, we further performed the same experiments with selected significance levels of 0.05–0.2. The overall accuracy of INDeGenIUS slightly increases, whereas that of SigHunt decreases as the significance level increases (Supplementary Figure S2).
In addition, we performed the same experiments using MTGIpick with 3-mer, 4-mer and 5-mer, and discussed the effects of the size of k-mer. The overall accuracies of MTGIpick with 3-mer, 4-mer and 5-mer are 85.43, 86.15 and 85.6%, respectively, which also confirmed that tetranucleotides (4-mers) are more sensitive for detection of genomic islands in the proposed detection method.
Comparison with the tools in IslandViewer for GI identification
Islandviewer [19, 20] is an integrated interface for computational identification and visualization of GIs and is a combination of a comparative genomics method, namely, IslandPick [18], and two HMM-based methods, namely, SIGI-HMM [29, 30] and IslandPath-DIMOB [31]. To examine the proposed method, we identified the GIs of the available genomes by using IslandViewer and further compared the results with those obtained using IslandPick, SIGI-HMM and IslandPath-DIMOB. Over 3000 additional publicly available complete genomes have been pre-computed for GIs; we selected some of these genomes in which at least 20 kb of the DNA segments were predicted to be GIs by at least two of the three methods, and at least 40% of the total DNA were predicted to be GIs by any of the three methods [21]. Considering these requirements, we selected 20 genomes from IslandViewer and used the proposed method (MTGIpick) to predict the GIs (Supplementary Table S3); the IST-LFS was run in default parameters and 0.3 standard error in MSA. Finally, MTGIpick used 1 kb upstream/downstream of ‘raw’ GIs to refine the boundaries of predicted GIs.
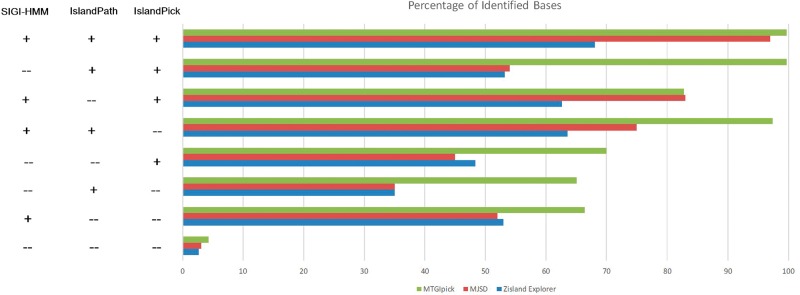
MTGIpick successfully identified the GI regions (99.6% of the bases) that were previously predicted by all of the three methods (Figure 2), and the accuracy of MTGIpick was ∼>3% than that of the MJSD [21] and >31.6% than that of Zisland Explorer [39]. For the regions identified by two of the three previously mentioned methods (in other words, the regions missed by SIGI, IslandPick or IslandPath but were detected by the two other methods), 82.8–99.7% (average of 93.3%) of the bases were identified as GIs by MTGIpick (Figure 2). In contrast, the regions identified by MJSD were between 54 and 83% (average of 74%), and the regions identified by Zisland Explorer were between 53 and 63% (average of 60%). For GIs identified by one of the three previous methods, 65.1–70% of them (average of 67.1%) were detected by MTGIpick (Figure 2), whereas 35–54% of them (average of 44%) were detected by MJSD and 35–53% of them (average of 45%) were detected by Zisland Explorer. In addition, low amount of DNA was classified as GIs that were deemed native by the three previous methods (Figure 2), although the misclassification of native DNA as GIs is slightly higher in our method than in MJSD (1.2%) and Zisland Explorer (1.6%). These results demonstrate that nearly all of the GIs (predicted by at least two of the three methods) and 67.1% of the GIs detected by one of the three methods can be identified correctly by the proposed method MTGIpick. Thus, these consistent and robust detection results are sufficient to warrant its use in general detection of GIs.
Figure 2.
Performance of the MTGIpick, MJSD and Zisland Explorer in predicting the identified genomic islands by IslandPick (IsPick), SIGI-HMM (SIGI) and IslandPath-DIMOB (IsPath). The accuracy in identifying islands reported by one, two or all three of the above-mentioned methods is assessed by obtaining the percentage of island nucleotides that are correctly labelled as genomic islands by the proposed MTGIpick method.
Identification of horizontal transfers in assembled/unassembled genomes
To test the proposed method on real biological data, we first selected the assembled genomic sequences of A. fumigatus whose 189 horizontal transfers were annotated and their locations are known [40]. Examination of those horizontal transfers reveals that the average length is ∼5 kb, which is less than the minimum GI size of 8 kb but much larger than the average length (<1 kb) of horizontally transferred genes in most prokaryotic genomes. This examination enabled us to cross-check the proposed method (MTGIpick) and the other methods. We first scored each window by using all of the evaluated methods with default settings, and we sorted the windows in descending order according to their scores. The AUC was calculated based on the selected top 10–25% of the windows. MTGIpick outperforms the other methods (Table 1). In the top 10%, the Alien_Hunter and INDeGenIUS methods both perform well in identifying horizontal transfers, whereas SigHunt outperforms them when the top percentage is >15% (Table 1). Moreover, all of the methods did not perform as well as expected in the above two experiments, and their AUC values reflect that challenges in identifying small horizontal transfers still exist because of the weak atypical characters of these genes.
Table 1.
Performance of the MTGIpick, SigHunt, INDeGeNIUS, centroid and Alien_Hunter on the detection of 189 horizontal transfers identified in A. fumigatus, in which the AUC is calculated based on the top 10–25% of the ordered windows
| Method | Percentage of the windows with the top 10–25% of scores |
|||
|---|---|---|---|---|
| 10 | 15 | 20 | 25 | |
| Alien_Hunter | 0.5814 | 0.5413 | 0.5418 | 0.5654 |
| Centroid | 0.5373 | 0.6717 | 0.6638 | 0.6832 |
| INDeGenIUS | 0.5868 | 0.6338 | 0.666 | 0.6699 |
| SigHunt | 0.5648 | 0.699 | 0.7262 | 0.7376 |
| MTGIpick | 0.6537 | 0.7621 | 0.7847 | 0.7952 |
The bold value indicates the best among the values.
We subsequently examined these methods on Cryptosporidium [41] and Galdieria sulphuraria [42]. While the genomic sequences of G. sulphuraria were not yet assembled into chromosomes, limited numbers of horizontal transfers (24 horizontal transfers identified in the 7 chromosomes of Cryptosporidium and 79 horizontal transfers identified in the 18 chromosomes of G. sulphuraria) were identified, and small horizontal transfers (average length of 1.8 and 1.3 kb) render the identification of horizontal transfers more challenging. The sequences of the unassembled genome are concatenated into a single sequence as input of MTGIpick for GI prediction [36]. In contrast to the experiment on A. fumigatus, we evaluated these methods by counting the established horizontal transfers covering >50% of the top 10–25% of the ranked windows (Table 2). In G. sulphuraria, MTGIpick identifies 50 of the 79 previously identified horizontal transfers using the top 25% of the ranked windows followed by SigHunt, which identifies 44 horizontal transfers, whereas the others lag behind. For Cryptosporidium, MTGIpick can recognize 16 of the 24 identified horizontal transfers using the top 25% followed by Alien_Hunter, which identifies 12 established horizontal transfers. The same result holds for the top 10–20%. These results demonstrate that MTGIpick is still efficient in detecting small horizontal transfers regardless of whether the genome is assembled or not.
Table 2.
Number of established horizontal transfers which are covered by > 50% of the top 10–25% of the windows from MTGIpick, SigHunt, INDeGeNIUS, centroid and Alien_Hunter on red algae Galdieria and chromalveolates Cryptosporidium
| Organism | Previously established GIs | Method | Percentage of the windows with decent scores |
|||
|---|---|---|---|---|---|---|
| 10 | 15 | 20 | 25 | |||
| Red algae Galdieria | 79 | Alien_Hunter | 12 | 16 | 24 | 30 |
| Centroid | 10 | 13 | 16 | 21 | ||
| INDeGenIUS | 7 | 12 | 17 | 21 | ||
| SigHunt | 15 | 25 | 31 | 44 | ||
| MTGIpick | 25 | 34 | 42 | 50 | ||
| Chromalveolates Cryptosporidium | 24 | Alien_Hunter | 6 | 9 | 12 | 12 |
| Centroid | 2 | 3 | 6 | 7 | ||
| INDeGenIUS | 2 | 2 | 2 | 6 | ||
| SigHunt | 3 | 4 | 5 | 6 | ||
| MTGIpick | 8 | 11 | 14 | 16 | ||
The bold value indicates the best among the values.
GI identification in the L-data set
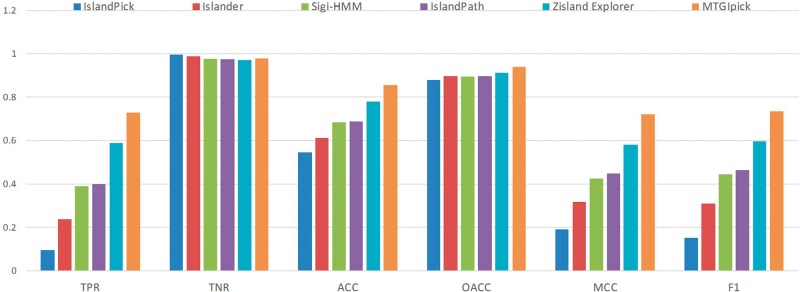
To further assess the proposed method MTGIpick, we used the proposed method to identify the genomic islands in the L-data set constructed by Wei et al. [39]. They collected the genomic islands in 11 genomes identified using a genome-wide comparative approach from published literature (Supplementary Data). We applied MTGIpick to identify GIs in the L-data set, where the IST-LFS was run with 0.2–0.4 standard error 0.03–0.07, using four to seven iterations in the IST-LFS and 0.2–0.4 standard error in MSA. MTGIpick finally used 10 kb upstream and 2–4 kb downstream of ‘raw’ GIs to refine the boundaries of predicted GIs. We also compared our results with those of the methods IslandPick [18], Islander [43], SIGI-HMM [30] and IslandPath-DIMOB [31] from Supplementary Table S4 in the article [39]. As for Zisland Explorer, we downloaded the predicted genomic islands in 11 genomes from Zisland Explorer and calculated the sensitivity (TPR), specificity (TNR), overall accuracy (OACC), accuracy (ACC), F1 and Matthews correlation coefficient (MCC) that were defined in the article [39] (Figure 3).
Figure 3.
Comparison of the TPR, TNR, OACC, ACC, F1 and MCC of IslandPick, Islander, SIGI-HMM, IslandPath-DIMOB, Zisland Explorer and MTGIpick on the L-data set.
For the six methods compared using the L-data set, MTGIpick was the only prediction method that achieved a TPR of >73% and an ACC of >85%, whereas the other methods lagged behind. This finding suggests that the proposed method MTGIpick was able to detect more true genomic islands in L-data set (Figure 3 and Supplementary Table S4). In addition, we found that MTGIpick achieved the best performance in terms of the F1 score and MCC. To be specific, the F1 score and MCC of MTGIpick were ∼>14% than those of the second-best tool, Zisland Explorer [39]. These results demonstrate that MTGIpick is efficient in detecting genomic islands in L-data set and has the best TPR/TNR balance and TPR/precision balance.
GI identification in S. enterica serovar typhi CT18
We subsequently examined the performances of MTGIpick when detecting large GIs in genuine genomes. Herein, we attempted to analyse the S. enterica serovar typhi CT18 genome, whose GIs have been explored extensively [22, 44]. A total of 17 PAIs have been annotated in Salmonella genomes, and 13 of these are speculated to be present and active in S. enterica serovar typhi CT18 [22]. In addition, this strain contains multiple bacteriophage insertions and two other islands that were not previously identified [45, 46], resulting in 21 large regions that are confirmed to be foreign origin [21].
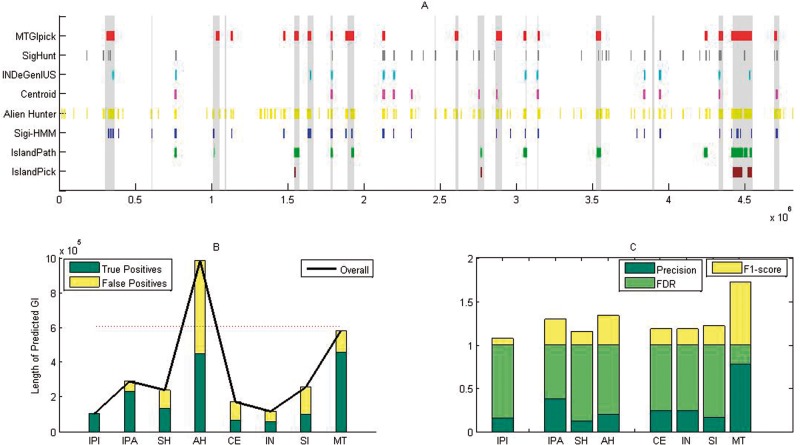
We applied MTGIpick to identify the GIs in the S. enterica serovar typhi CT18 genome, where IST-LFS was run in default parameters, and 0.31 standard error and 25 scale in MSA were used. Finally, MTGIpick used 6 kb upstream/downstream of ‘raw’ GIs to refine the boundaries of the predicted GIs. For comparison, we also used six composition-based approaches (SIGI-HMM [30], Alien_Hunter [22], centroid [30], IslandPath-DIMOB [31], INDeGenIUS [34] and SigHunt [36]), as well as a comparative genomics method (IslandPick [18]); these methods are highly accurate in GI prediction. All of the evaluated methods were run in default values. We used the same significance test at a significance level of 0.05 in our method to identify putative GIs based on their scores. Figure 4A illustrates the results of various prediction methods when detecting 21 known GIs in S. enterica serovar typhi CT18 [22]. A large number of putative regions were detected by Alien_Hunter, which predicts the longest GIs (Figure 4B). Although Alien_Hunter detected 451 of the 605 kb of DNA encoded by established islands, the number of false positives was extremely high (Figure 4B). Therefore, Alien_Hunter always demonstrates the best recall at the cost of low precision and accuracy (Figure 4C and Supplementary Tables S5–6).
Figure 4.
Performance of the proposed MTGIpick (MT), SIGI-HMM (SH), Alien_Hunter (AH), centroid (CE), IslandPath-DIMOB (IPA), INDeGenIUS (IN), SigHunt (SI) and IslandPick (IPI) on the detection of genomic islands in S. enterica serovar typhi CT18. (A) Predicted GIs found by all of the methods, and the known genomic islands are shown as vertical bars. (B) Overall length of the predicted genomic islands, true positives and false positives of all of the evaluated methods at the nucleotide level. (C) Precision, false discovery rate (FDR) and F1 score of all of the evaluated methods at the island level, in which the precision, false-positive rate and F1 score are calculated based on the number of known GIs, which are covered by > 50% of the results of the prediction method.
In contrast, the comparative genomics IslandPick only discovered six putative GIs with extremely low false-positive results and consequently achieved the best precision according to the number of overlapping nucleotides between the predicted and annotated GIs. However, the limitation of this method is that it reports high false positives and suffers greatly from having the lowest recall, as well as in predicting small GIs. To further measure the prediction power at the GI level, we calculated the precision, false positives and F1 score based on the number of the known GIs, which are covered by >50% of the results of the prediction method (Supplementary Data). As expected, there exists only one known GI whose half region was predicted by IslandPick, resulting in the lowest F1 score (Figure 4C and Supplementary Tables S5–6).
For MTGIpick, 18 genomic regions were detected as putative GIs showing the largest average lengths among those identified by all other prediction methods (Supplementary Table S5). Among the 582 232 nucleotides in the predicted GIs, ∼80% of them are located in the published GIs. Similar to Alien_Hunter, MTGIpick achieved a good true-positive rate but with low false-positive rate (Figure 4B). We then examined the known GIs, >50% of which was covered by the proposed method, and found that the 14 annotated GIs were largely overlapping in the predicted results, leading to the highest precision and the highest F1 score among all of the evaluated methods (Figure 4C and Supplementary Table S6).
This comprehensive comparison further indicates that IslandPick (comparative genomics) is reliable but misses a large number of GIs, resulting in high false-negative results. Although Alien_Hunter is sensitive, it suffers greatly from the clutter in different regions, resulting in high false-positive results (Figure 4B). The window-based methods, namely, centroid [33], INDeGenIUS [34] and SigHunt [36], discover many putative extremely small GIs, leading to low true-positive and high false-positive results (Figure 4B). Thus, these results provide compelling evidence that the proposed MTGIpick method is superior in identifying GIs.
Figure 4A also shows the sizes of the predicted GI obtained by MTGIpick are more accurate than those from other methods. To be specific, we listed all of the start and end positions of the known genomic islands and predicted genomic islands from IslandPick [18], SIGI-HMM [30], Alien_Hunter [22], centroid [30], IslandPath-DIMOB [31], INDeGenIUS [34], SigHunt [36], Zisland Explorer [39] and MTGIpick (Supplementary Table S7). It is easy to find that ∼80% of the predicted GIs from MTGIpick are located in the published GIs, leading to the high accurate sizes. In addition, the window-based methods centroid, INDeGenIUS and SigHunt identify some windows as GIs without refining the island boundaries, and thus the size of their predicted genomic islands is an integer multiple of the window size (Supplementary Table S7). However, MTGIpick refined the boundaries of predicted GIs using MJSD and the GC-based segmentation method and therefore can detect the exact boundaries of the genomic islands.
Method efficiency
One obvious benefit of the proposed method is the utilization of small patterns (tetranucleotides) rather than using large ones. SigHunt has the same purpose, although it predicts small putative GIs, leading to low true positives and high false positives (Figure 4). MTGIpick uses the multiscale statistical test to improve GI prediction. It uses simple kurtosis to select informative tetranucleotides from a single genome instead of a range of organisms, as well as uses t-test to measure the divergence between two regions rather than computing DIAS based on density distribution, which leads to an improvement of MTGIpick speed. For example, MTGIpick took about 6 min to complete GI prediction of S. enterica serovar typhi CT18, whereas IslandPick, Alien_Hunter and INDeGenIUS algorithms require 11, 26 and 47 min, respectively. SIGI-HMM, centroid, IslandPath-DIMOB and SigHunt run this prediction under 1 min, and Zisland Explorer took about 2 min. Therefore, the computational efficiency of the proposed MTGIpick is higher than those of IslandPick, Alien_Hunter and INDeGenIUS, but less efficient than SIGI-HMM, centroid, Zisland Explorer, IslandPath-DIMOB and SigHunt.
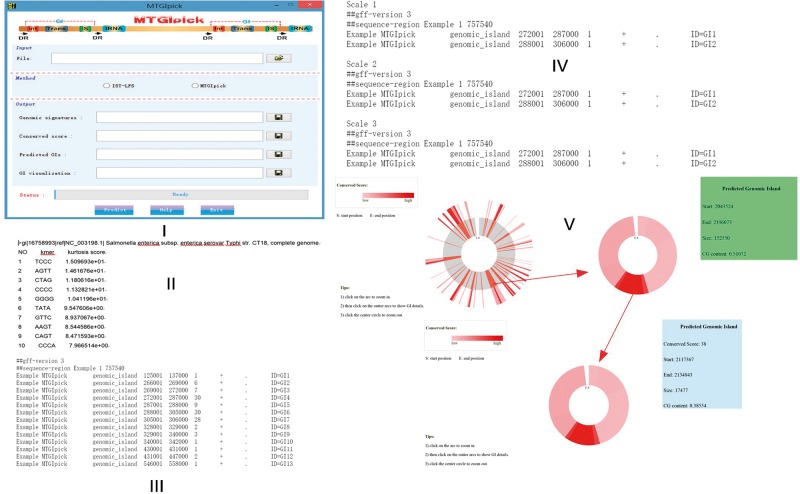
Software and application
We provided an online service and a software of the MTGIpick tool with graphical user interface to run locally (Figure 5I). MTGIpick has been compiled and tested under Sun Java interpreter and Matlab. MTGIpick can be used in Windows- and Linux-based platforms. Java Virtual Machine and MATLAB Compiler Runtime are required for MTGIpick setup on your platform. However, we strongly advise the use of openjdk instead of the Oracle version of java virtual machine when working in Linux-based machines as the Oracle version may result in some exceptions during the analyses. MTGIpick is an open-source software, and it is available at http://bioinfo.zstu.edu.cn/MTGI or https://github.com/bioinfo0706/MTGIpick.
Figure 5.
MTGIpick software and applications.
The input format of the MTGIpick follows the standard FASTA format and multiple DNA sequences are supported. There will be an upload progress bar to monitor upload progress when clicking the button to upload a file. If the input file you uploaded contains at least two sequences, a dialog box appears to tell you to select predicting each sequence separately or assembling and predicting to process the input file. This software consists of two prediction methods: IST-LFS and MTGIpick. IST-LFS is a proposed small-scale t-test with large-scale feature selection, and it is efficient at detecting HGTs or GIs with small sizes.
A dialog box appears to tell you to select a way to download the results once your project is complete. There are two ways to download the results: download the results by clicking save button and find the results in the same directory where the input file is stored. The outputs of the MTGIpick consist of genomic signatures (Figure 5II), predicted GIs of total scales (Figure 5III), predicted GIs of each scale (Figure 5IV) and GIs visualization (Figure 5V). Output are Zip files whose names are created by the input file name. If the input file contains at least two sequences, each Zip file contains all of the results for all the sequences.
MTGIpick provides a new interactive genome visualization tool (Figure 5V), which uses zoomable sunburst and sunburst partition to represent predicted GIs with conserved score along the whole genome. MTGIpick has generated a number of HTML files in the same directory where the input file is stored, and you can open them directly and view the predicted GIs with conserved scores. Orange regions in the first circle represent GIs predicted by MTGIpick method at all the scales. You can click on the arc to zoom in, click on the outer to show GI details and click the circle to zoom out.
Discussion
An integrated strategy for bottom-up and top-down approaches
Top-down and bottom-up approaches, which are strategies for information processing, have been both widely used in GI prediction. Bottom-up methods usually detect a few of the constituent genes as sufficiently atypical to be deemed foreign and thus their predicted GIs consist of a large number of small predicted fragments. To circumvent the problems in bottom-up approaches, a top-down method was proposed to detect GIs by splitting a genome into successively smaller regions by using a recursive segmentation procedure [21]. Motivated by these approaches, we attempted to adopt an integrated strategy, where the bottom-up method (IST-LFS) is used to calculate the score of each small region deviating from the host. With the aid of the top-down method (GC-based segmentation), we further split the predicted large segments into optimal distinct segments and then identified the GIs. Thus, the proposed method can be regarded as a specific way to combine both the top-down and bottom-up approaches for GI prediction.
Integrated strategy for local and global testing
Previous approaches usually execute global testing to detect GIs and focus on whether local signatures of a region are significantly different from the host. However, genomic signatures at different scales exhibit different genomic characteristics: at a large scale, the local genomic signature is poor and it misses small details to detect more ancient GI insertions, resulting in false-negative predictions of GIs; at a small scale, details of genomic signature are preserved, although GI detection suffers greatly from clutter in different regions and can result in false-positive predictions. Herein, we proposed a multiscale statistical testing method, MTGIpick, to explore the multiscale genomic signatures. In IST-LFS, we used small-scale t-testing with large-scale feature selection to quantify the compositional differences from the host genome. In contrast, MSA used a large-scale statistical testing to identify some multiwindow segments. As expected, MTGIpick performs better in identifying GIs (Figure 4). This work is the first to use multiscale statistical testing to improve GI prediction, and the resulting new insights can be used to develop more powerful prediction methods.
Complementarity of the existing prediction methods
Window-based methods
Window-based methods are capable and versatile tools to detect GIs despite their high false-negative and false-positive rates. They attempt to enhance their discriminative power by selecting core signatures. These approaches have achieved promising results but are limited by the use of related sequenced genomes. The proposed method does not replace the existing window-based approaches; rather, they provide a novel method for host signature extraction and core signature selection to overcome their inherent weakness and thus should be used along with existing methods. Moreover, the window-based methods select consecutive atypical windows as ‘raw’ GIs without refining their boundaries. To address this problem, we proposed a simple method to detect the boundaries of the ‘raw’ GIs, and this proposed method can also be merged with the window-based methods.
Annotation-based methods or comparative genomics
Direct/inverted repeats, tRNA/tmRNA and mobility genes, in addition to sequence composition features, are widely used by annotation-based methods to identify GIs. One of their limitations is that they require fully annotated genomes and sometimes fail to identify GIs that are devoid of flanking features. Moreover, comparative genomics are often the most reliable methods to detect laterally acquired genes, although the success of such methods clearly depends on the breadth and depth of the sequence database. In contrast, the proposed method requires only a single genome sequence analysed without any annotation information and offers a better performance than comparative genomics (Figure 4). Therefore, the proposed method will complement the annotation-based methods or comparative genomics when fully annotated genomes and closely related genomes are lacking.
HMM-based methods
HMM was constructed and applied to remove or detect anomalous regions in GI detection. For example, Alien_Hunter introduces an HMM to refine the boundaries of predicted GIs [22]. Although these methods achieve good performance in GI detection, they involve a relatively high number of parameters and training calculations; thus, longer time is required for GI detection with a risk of overtraining. To address the same problem, the proposed method provides a rapid and accurate method to detect the boundary of ‘raw’ GIs and thus can also be merged with existing HMM-based methods for GI detection.
Current limitations of the proposed method
Although competitive performance of the proposed method has been achieved, this method is not a universal solution to detect all GIs or horizontal transfers in different organisms. The genomic signatures in the proposed method are limited because the method relies on observation of different tetranucleotides. As shown by our experiments using simulated or real data sets, tetranucleotides are not always sufficiently strong and credible for GI detection and thus may result in false-negative predictions of GIs. For example, some random islands or small GIs do not provide sufficient oligonucleotide patterns from the host genome, making their detection difficult. In addition, a phenomenon where some GIs originated from a species with similar oligonucleotide patterns limits the ability of the proposed method to detect the GIs. As more signatures are added into the proposed model, the accuracy of GI prediction improves.
Key Points
MTGIpick is a software that uses the first multiscale statistical test to detect genomic islands.
We proposed an IST-LFS to quantify compositional differences of a genome from that of a host.
We investigated the variability of genomic signatures and used MSA to identify multiwindow segments.
MTGIpick can refine the boundaries of predicted GIs using MJSD and the GC-based segmentation method.
MTGIpick can identify GIs from a single genome without annotated information of genomes or prior knowledge from other data sets.
Supplementary Material
Acknowledgments
The authors thank Prof. Kamil S. Jaron, Jiri C. Moravec and Natalia Martinkova for providing data sets and technical help for SigHunt; Prof. Sharmila S. Mande for providing the software of centroid and INDeGenIUS; and Prof. Wei Wen for providing the technical help and prediction results of other tools on L-data set.
Funding
The National Natural Science Foundation of China (grant number 61370015), the National Basic Research Program of China (grant number 2012CB316503), Zhejiang Provincial Natural Science Foundation of China (grant number LY14F020046), Public Projects of Zhejiang Province (grant number 2015C33141) and 521 Talent Cultivation Plan of Zhejiang Sci-Tech University. Funding for open access charge: National Natural Science Foundation of China. The NIH/NIAID (grant number AI116610 to M.Q.Z.).
Qi Dai is a professor at the College of Life Sciences, Zhejiang Sci-Tech University, and is a visiting research scientist at Center for Systems Biology, University of Texas at Dallas.
Chaohui Bao is a PhD candidate at the College of Life Sciences, Zhejiang Sci-Tech University, and her main research interests include genomic island prediction and evolution study.
Yabing Hai is a PhD candidate at the College of Life Sciences, Zhejiang Sci-Tech University, and focuses on statistical methods and computing tools to advance genomic research.
Sheng Ma is a PhD candidate at the College of Life Sciences, Zhejiang Sci-Tech University, and his research interests include protein structure prediction and machine learning.
Tao Zhou is a PhD candidate at the College of Life Sciences, Zhejiang Sci-Tech University, and focuses on genomic island prediction and software design.
Cong Wang is a master student at the College of Life Sciences, Zhejiang Sci-Tech University, and his research interests include computing tools in genomic research and software design.
Yunfei Wang, PhD, is a research scientist at the Center for Systems Biology, University of Texas at Dallas, and focuses on genetics, cancer research and systems biology study.
Wenwen Huo is a PhD candidate at the Center for Systems Biology, University of Texas at Dallas, and uses genomic and biochemical approaches to study antibiotic resistance in pathogenic bacteria.
Xiaoqing Liu, PhD, is an assistant professor at the College of Sciences, Hangzhou Dianzi University, and his research interests include statistical methods in genomic research.
Yuhua Yao, PhD, is a professor at the College of Life Sciences, Zhejiang Sci-Tech University, and focuses on genomic comparison, alignment-free comparison and classification.
Zhenyu Xuan is an assistant professor at the Center for Systems Biology, University of Texas at Dallas, and his research focuses on analysing cancer genomics data
Min Chen is an assistant professor at the Department of Mathematical Sciences, University of Texas at Dallas, and his research focuses on Statistical Genomics and Bioinformatics.
Michael Q. Zhang is a director, Center for Systems Biology, University of Texas at Dallas, and a guest professor at the Center for Systems Biology, Tsinghua University.
References
- 1. Hacker J, Bender L, Ott M, et al. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog 1990;8:213–25. [DOI] [PubMed] [Google Scholar]
- 2. Hacker J, Kaper JB.. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 2000;54:641–79. [DOI] [PubMed] [Google Scholar]
- 3. Nakamura Y, Itoh T, Matsuda H, Gojobori T.. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 2004;36:760–6. [DOI] [PubMed] [Google Scholar]
- 4. Gal-Mor O, Finlay BB.. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 2006;8:1707–19. [DOI] [PubMed] [Google Scholar]
- 5. Dobrindt U, Hochhut B, Hentschel U, et al. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2004;2:414–24. [DOI] [PubMed] [Google Scholar]
- 6. Lawrence JG. Common themes in the genome strategies of pathogens. Curr Opin Genet Dev 2005;15:584–8. [DOI] [PubMed] [Google Scholar]
- 7. Manson JM, Gilmore MS.. Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation. Mol Microbiol 2006;61:555–9. [DOI] [PubMed] [Google Scholar]
- 8. Middendorf B, Hochhut B, Leipold K, et al. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol 2004;186:3086–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finlay BB, Falkow S.. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev 1997;61:136–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol 2001;9:335–43. [DOI] [PubMed] [Google Scholar]
- 11. Hsiao WW, Ung K, Aeschliman D, et al. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet 2005;1:e62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vernikos GS, Parkhill J.. Resolving the structural features of genomic islands: a machine learning approach. Genome Res 2008;18:331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ragan MA. Detection of lateral gene transfer among microbial genomes. Curr Opin Genet Dev 2001;11:620–6. [DOI] [PubMed] [Google Scholar]
- 14. Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darling ACE, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004;14:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ou HY, Chen LL, Lonnen J, et al. A novel strategy for the identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Nucleic Acids Res 2006;34:e3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiapello H, Bourgait I, Sourivong F, et al. Systematic determination of the mosaic structure of bacterial genomes: species backbone versus strain-specific loops. BMC Bioinformatics 2005;6:171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langille MG, Hsiao WWL, Brinkman FSL.. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 2008;9:329.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langille MG, Brinkman FS.. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 2009;25:664–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dhillon BK, Chiu TA, Laird MR, et al. IslandViewer update: Improved genomic island discovery and visualization. Nucleic Acids Res 2013;41:W129–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aaron JA, Rajeev K, Azad AR, et al. Detection of genomic islands via segmental genome heterogeneity. Nucleic Acids Res 2009;37:5255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vernikos GS, Parkhill J.. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 2006;22:2196–203. [DOI] [PubMed] [Google Scholar]
- 23. Karlin S, Mrazek J, Campbell AM.. Codon usages in different gene classes of the Escherichia coli genome. Mol Microbiol 1998;29:1341–55. [DOI] [PubMed] [Google Scholar]
- 24. Sandberg R, Winberg G, Branden CI, et al. Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome. Res 2001;11:1404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsirigos A, Rigoutsos I.. A new computational method for the detection of horizontal gene transfer events. Nucleic Acids Res 2005;33:922–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoon SH, Hur GC, Kang HY, et al. A computational approach for identifying pathogenicity islands in prokaryotic genomes. BMC Bioinformatics 2005;6:184.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoon SH, Park YK, Lee S, et al. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic Acids Res 2007;35:D395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoon SH, Park YK, Kim JF.. PAIDB v2.0: exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res 2014;43:D624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merkl R. SIGI: score-based identification of genomic islands. BMC Bioinformatics 2014;5:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waack S, Keller O, Asper R, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 2006;7:142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsiao W, Wan I, Jones SJ, et al. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 2003;19:418–20. [DOI] [PubMed] [Google Scholar]
- 32. Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res 2008;36:D281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajan I, Aravamuthan S, Mande SS.. Identification of compositionally distinct regions in genomes using the centroid method. Bioinformatics 2007;23:2672–7. [DOI] [PubMed] [Google Scholar]
- 34. Shrivastava S, Reddy CV, Mande SS.. INDeGenIUS, a new method for high-throughput identification of specialized functional islands in completely sequenced organisms. J Biosci 2010;35:351–64. [DOI] [PubMed] [Google Scholar]
- 35. Azad RK, Lawrence JG.. Towards more robust methods of alien gene detection. Nucleic Acids Res 2011;39:e56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaron KS, Moravec JC, Martinkova N.. SigHunt: horizontal gene transfer finder optimized for eukaryotic genomes. Bioinformatics 2014;30:1081–6. [DOI] [PubMed] [Google Scholar]
- 37. Tu Q, Ding D.. Detecting pathogenicity islands and anomalous gene clusters by iterative discriminant analysis. FEMS Microbiol Lett 2003;221:269–75. [DOI] [PubMed] [Google Scholar]
- 38. Pundhir S, Vijayvargiya H, Kumar A.. PredictBias: a server for the identification of genomic and pathogenicity islands in prokaryotes. In Silico Biol 2008;8:223–34. [PubMed] [Google Scholar]
- 39. Wei W, Gao F, Du MZ, et al. Zisland Explorer: detect genomic islands by combining homogeneity and heterogeneity properties. Brief Bioinform 2016, in press. doi: 10.1093/bib/bbw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mallet LV, Becq J, Deschavanne P.. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genomics 2010;11:171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang J, Mullapudi N, Lancto CA, et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol 2004;5:R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schonknecht G, Chen WH, Ternes CM, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 2013;339:1207–10. [DOI] [PubMed] [Google Scholar]
- 43. Hudson CM, Lau BY, Williams KP.. Islander: a database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res 2015;43:D48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vernikos GS, Thomson NR, Parkhill J.. Genetic flux over time in the Salmonella lineage. Genome Biol 2007;8:R100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kingsley RA, van AK, Kramer N, et al. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun 2000;68:2720–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kingsley RA, Humphries AD, Weening EH, et al. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 2003;71:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.