Abstract
Approximately 90% of all cancer deaths arise from the metastatic spread of primary tumours. Of all the processes involved in carcinogenesis, local invasion and the formation of metastases are clinically the most relevant, but they are the least well understood at the molecular level. As a barrier to metastasis, cells normally undergo an apoptotic process known as ‘anoikis’, in circulation. The recent technological advances in the isolation and characterisation of rare circulating tumour cells (CTCs) will allow a better understanding of anoikis resistance. Detailed molecular and functional analyses of anoikis-resistant cells may provide insight into the biology of cancer metastasis and help identify novel targets for prevention of cancer dissemination. To uncover the molecular changes that govern the transition from a primary lung tumour to a secondary metastasis and specifically the mechanisms by which CTCs survive in circulation, we carried out whole genome sequencing (WGS) of normal lung, primary tumours and the corresponding brain metastases from five patients with progressive metastatic non-small-cell lung carcinoma. We also isolated CTCs from patients with metastatic cancer and subjected them to whole genome amplification and Sanger sequencing of genes of interest. While the primary tumours showed mutations in genes associated with cell adhesion and motility, brain metastases acquired mutations in adaptive, cytoprotective genes involved in response to cellular stress such as Keap-1, Nrf2 and P300, which are key players of the Keap1-Nrf2-ARE survival pathway. Nrf2 is a transcriptional factor that upon stress translocates into the nucleus, binds to the anti-oxidant response elements (ARE) and drives the expression of anti-oxidant genes. The identified mutations affect regulatory domains in all three proteins, suggesting a functional role in providing a survival advantage to CTCs in the peripheral blood allowing their dissemination to distant organs.
Introduction
It is estimated that metastasis is responsible for ~90% of cancer deaths (1). Brain metastases are among the most debilitating complications of many primary tumours. They are the most common type of intra-cranial neoplasm, occurring 5 to 10 times more frequently than primary tumours of the central nervous system (CNS) (2). Lung cancer is the main source of brain metastases; patients with non-small-cell lung carcinoma (NSCLC) develop CNS metastases in ~20–40% of cases and have a very poor prognosis (3).
Metastatic spread, a complex process initiated by the dissemination, seeding and engraftment of malignant cancer cells in sites distant to the primary tumour (4–6), is the leading cause of cancer-related death (7). It has been proposed that circulating tumour cell (CTC) populations in the blood of carcinoma patients contain cells with the clonal capacity to initiate metastatic growth in distant organs, thus behaving as metastasis-initiating cells(MICs) (6,8). In favour of this hypothesis, the presence of five or more CTCs (breast, prostate and lung cancer) (9–11) or three or more CTCs (colorectal cancer) (12) per 7.5 ml of peripheral blood is an indicator of decreased progression-free survival and overall survival in several types of carcinoma. In addition, the number of CTCs in patients with metastatic breast cancer appears to be a better indicator of overall survival than traditional radiologic imaging techniques (13).
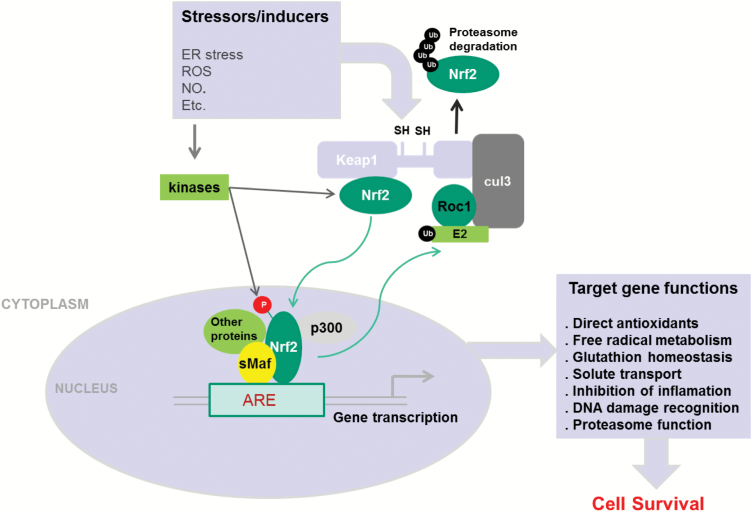
The mechanical and biochemical environment of the circulation is vastly different from the epithelial tissues in which metastatic carcinomas originate and represents a significant challenge for cancer cells to survive. Therefore, activation of survival mechanisms is a critical prerequisite to efficient metastasis formation. The Keap1-Nrf2-ARE pathway is the major regulator of cytoprotective responses to endogenous and exogenous stresses caused by reactive oxygen species (ROS) and electrophiles (14). The key signalling proteins within the pathway are the transcription factor such as nuclear factor erythroid 2-related factor 2 (Nrf2) that binds together with small Maf proteins to anti-oxidant response elements (ARE) in the regulatory regions of target genes, and Kelch ECH associating protein 1 (Keap1), a repressor protein that binds to Nrf2 and promotes its degradation by the ubiquitin proteasome pathway (15–17). As shown in Figure 1, under quiescent conditions, Nrf2 interacts with the actin-anchored protein Keap1 largely localised in the cytoplasm. This quenching interaction maintains low basal expression of Nrf2-regulated genes. However, upon recognition of stress signals imparted by oxidative and electrophilic molecules, Nrf2 is released from Keap1, escapes proteasomal degradation, translocates to the nucleus and transactivates the expression of several dozen cytoprotective genes that enhance cell survival such as NAD(P)H, quinone oxidoreductase 1 (NQO1), heme oxygenase 1 (HMOX1), glutamate-cysteine ligase (GCL) and glutathione S-transferases (GSTs) (18–31).
Figure 1.
A general scheme for the induction of gene expression through the Keap1-Nrf2-ARE signalling pathway (adapted from Kensler et al. (18)). Nrf2-regulated genes are activated as a result of exposure to endogenous or exogenous molecules. This leads to disruption of Nrf2 interaction with Keap1 leading to stabilisation of Nrf2 protein and its nuclear accumulation. Nrf2 interacts with multiple partners in the nucleus allowing for transactivation of ARE-responsive genes. Induction of these genes results in resistance of cells to environmental stresses.
It is noteworthy that many cancers have been found to up-regulate Nrf2. This is thought to provide an advantage for the cancer cells to evade apoptosis, proliferate and even metastasise when exposed to ROS (15–17). Several mechanisms by which Nrf2 signalling pathway is constitutively activated in various cancers have been described: (i) Somatic mutations in Keap1 or the Keap1-binding domain of Nrf2 disrupting their interaction; (ii) epigenetic silencing of Keap1 expression leading to defective repression of Nrf2; (c) accumulation of disruptor proteins such as p62 leading to dissociation of the Keap1-Nrf2 complex; (d) transcriptional induction of Nrf2 by oncogenic K-Ras, B-Raf and c-Myc; and (e) post-translational modification of Keap1 cysteines by succinylation (32–38). Constitutively, abundant Nrf2 protein causes increased expression of genes involved in drug metabolism, thereby increasing the resistance to chemotherapeutic drugs and radiotherapy (39). In addition, high Nrf2 protein level is associated with poor prognosis in cancer (40). Overactive Nrf2 also affects cell proliferation by directing glucose and glutamine towards anabolic pathways augmenting purine synthesis and influencing the pentose phosphate pathway to promote cell proliferation (41).
P300 was cloned based on its interaction with the adenovirus-transforming protein E1A (42,43). P300 and CBP (CREB-binding protein) share a high degree of homology and possess intrinsic histone acetyl-transferase activity. They are both believed to serve as transcription coactivators by acetylating core histones to facilitate chromatin decondensation and recruiting basic RNA polymerase machinery (44,45). Recent findings have shown that many nonhistone proteins, particularly transcription factors, are substrates for P300/CBP, which greatly expands the possible mechanisms of P300/CBP in transcriptional activation (46,47). Nrf2 has been found to be acetylated by P300/CBP, and this acetylation augments promoter-specific DNA binding of the protein (48). However, it is not known whether this acetylation affects Nrf2-Keap1 interaction. Interestingly, acetylation of p53 at multiple lysine residues is an indispensable event that destabilises the p53-Mdm2 interaction and enables the p53-mediated stress response (49).
Materials and Methods
Patient samples and blood collection
In the original study, matching samples from five NSCLC patients were obtained from the University of Cincinnati Tumor Bank. Genomic DNA was extracted, and 3 µg per sample was subjected to whole genome sequencing (WGS; Macrogen, Rockville, MD). For the follow-up study for CTC isolation, 10 metastatic breast, colon and melanoma cancer patients, who were older than 18 years of age, were enrolled in The University of Cincinnati Institutional Review Board (IRB)–approved protocol. All patients gave their informed consent to participate in the study. Blood samples were collected before and during treatment. Peripheral blood (7.8–17.7 ml) was collected in BD Vacutainer tubes (BD Biosciences, San Jose, CA) for CTC isolation and processed within 4 h of blood collection.
Sample processing for negative depletion and enrichment of CTCs
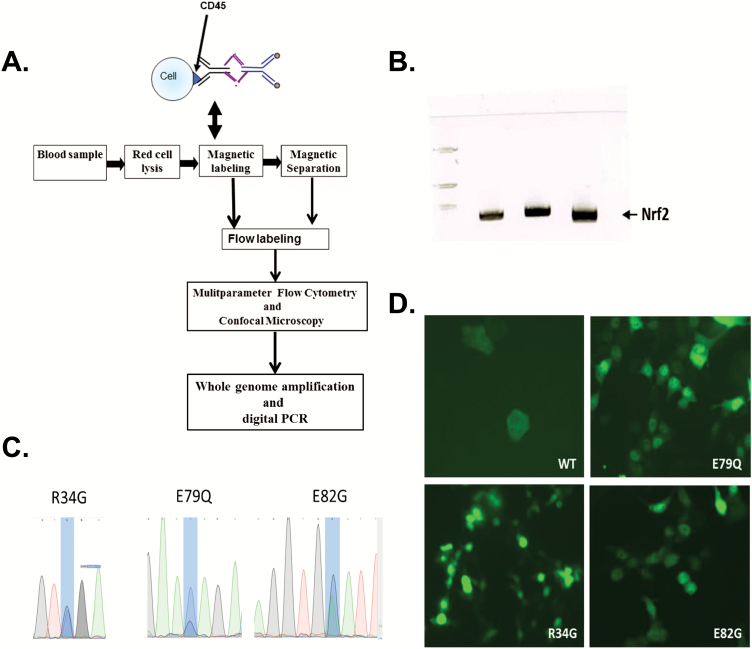
Blood samples (7.8–17.7 ml) were subjected to red blood cells lysis using hypotonic solution of ammonium chloride. Samples were kept at ambient temperature and processed using a negative depletion immunomagnetic methodology that we have described previously (50–53). Briefly, blood samples were centrifuged to isolate plasma and blood cells, and then subjected to an RBC lysis step and labelled with anti-CD45 tetrameric antibody complex (STEMCELL Technologies, Vancouver, BC, Canada). The magnetic nanoparticles were added and incubated with the cell suspension to allow for coupling to CD45-positive cells which were run through the quadrupole magnetic separation sorter system. Cells were then counted and labelled for further analysis.
Multi-parameter flow cytometry
Sample aliquots were taken prior to and after magnetic labelling for setting up gating controls for multi-parametric flow cytometry (FCM). The enriched sample after separation was split into two fractions, one of which was stained using a multistep sequential labelling protocol for FCM. Briefly, the FCM sample was labelled for surface markers, fixed with 4% paraformaldehyde to stabilise the surface staining, permeabilised with 0.1% Triton X-100 and stained for intracellular proteins. A BD LSR II flow cytometer or a BD FACSAria II cell sorter (BD Biosciences) equipped with three excitation lasers (405, 488 and 633 nm) were used for initial FCM analysis. Both automatic and manual compensation were evaluated for each patient sample. During the study period, a BD FACSAria III cell sorter was acquired. The FACSAria III is equipped with 355-, 488-, 561- and 635-nm lasers, which allows for fluorescein isothiocyanate and phycoerythrin dyes to be excited with different lasers (488 and 561 nm) incident to the cell at different locations in the stream, thereby significantly reducing the need for compensation. To determine whether the exclusion of non-viable cells, determined with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (L34957; Life Technologies, Carlsbad, CA), would change the staining distribution of cells, selected patient samples for this assay were assessed using the FACSAria III system. All gates were determined on the basis of unstained and single-colour controls of each patient sample, control samples from healthy volunteers or buffy coats. Because at most two colours were excited by a single laser (one dye per laser on the FACSAria III), this gating strategy was equivalent to the fluorescence minus one approach. For the samples using the viability assay, the initial step included using a side scatter width setting to exclude obvious doublets.
Detection of Nrf2 mutations in CTCs
To detect Nrf2 mutations, genomic DNA from CTCs was whole genome amplified using REPLI-g Single Cell Kit (Qiagen). The amplified DNA was used to amplify different regions of NRF2 using the following primer sets:
Fragment 1: NRF2 Forward F1: TTGACATACTTTGG AGGCAAGA and NRF2 Reverse R1: ACTGGTTTCTGACTGG ATGTG
Fragment 2: NRF2 Forward F2: TTCAGATGCCACAGTCAACA and NRF2 Reverse R2: GCCAAGTAGTGTGTCTCCATAG
Fragment 3: NRF2 Forward F3: GTACAACCCTTGTCACC ATCTC and NRF2 Reverse R3: CAATTCTGAGCAGCCA CTTTATTC.
Droplet digital polymerase chain reaction
To develop a droplet digital polymerase chain reaction (ddPCR) assay for detection of NRF2 mutations, we used the following custom primers and probes (Life Technologies) to detect the R34G on Nrf2:
Forward primer: TTGATTGACATACTTTGGAGGCAAGA, reverse primer: CTCTTTCCGTCGCTGACTGA and wild-type NRF2 probe: TCAAATACTTCTCGACTTACT and mutant R34G probe: CAAATACTTCTCCACTTACT.
The assay was first validated using wild-type NRF2 plasmid construct titrated with decreasing amounts of R34G NRF2 plasmid mutant. Before directly analysing the CTC-amplified whole genome for NRF2 R34G, we also validated the ddPCR assay using the same NRF2 PCR product from whole genome-amplified CTC DNA that showed R34G mutation by Sanger sequencing.
The ddPCR reaction mix contained 2xTaqMan Master Mix, 40× custom primers/probes, a stabiliser and DNA template. We first generated droplets using Raindance system and a chip source, and then subjected the droplets to the following thermal cycling programme: 10 min at 95°C followed by 45 cycles of 10 s at 95°C, and 1 min at 60°C, then 10°C hold. Droplet reactions were then transferred to Raindance sense for fluorescence quantification. ddPCR data were analysed using Raindance Software version II.
Data analysis
Data were processed utilising our previously published analysis pipeline (54,55). Briefly, the pipeline is tailored to the detection of copy number variation (CNV) and large (>200 bp) structural variations (SVs): deletions, insertions, inversions and translocations. While single-nucleotide polymorphisms and small insertions/deletions (indels) are detectable using the pipeline, we will limit our discussion to relevant point mutations. BAM files underwent an indel realignment and base quality recalibration process using the Genome Analysis Tool Kit (GATK) (56). CNV events were detected using Control-FREEC (57), and SVs were called using BreakDancer (58) with a minimal Map Q = 30 and the presence of at least five non-redundant read pairs defining an SV event. Structural variant calls were confirmed through visualisation using Integrative Genomics Viewer (IGV) (59). In addition, tandem duplication events were detected by ITX using BreakDancer.
Generation of Nrf2 mutant constructs
pcDNA3-EGFP-C4-Nrf2 plasmid that expresses an EGFP-Nrf2 fusion protein was obtained from Addgene (Cambridge, MA). All the mutations were introduced by using Q5 Site-Directed Mutagenesis Kit (New England Biolabs). Following introduction of diverse somatic mutations in the Nrf2 cDNA, NRF2 plasmids were transfected into MDA231 cells, and localisation of the GFP-Nrf2 wild-type and mutant proteins was visualised by fluorescence.
Results
Several pathways are differentially mutated in the brain metastases compared with primary lung tumours
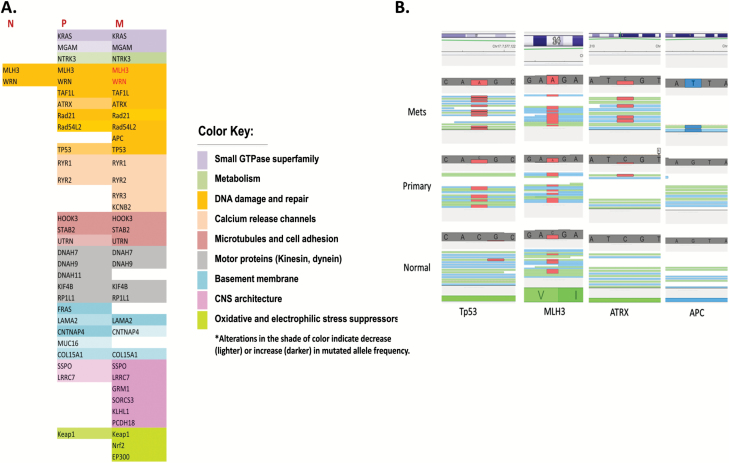
To better understand the mechanism(s) by which metastatic spread occurs, we carried out WGS of normal lung tissue, primary tumours and the corresponding metastases from five NSCLC patients. Genomic DNA was extracted from the tissue, sheared into ~500 bp fragments and used to make libraries. The libraries were subjected to pair-end sequencing with Illumina HiSeq 2500 2 × 100-bp reads, and data were analysed using our data analysis pipeline (54,55). Data analysis revealed enrichment in mutations affecting genes involved in multiple pathways (Figure 2A). The DNA damage and repair pathway was the most mutated, and the mutated genes were involved either in DNA damage checkpoint or in DNA repair. Some genes are mutated in both primary and metastatic tumours; others are mutated in the metastasis only while some genes lost heterozygosity in the metastasis. Tp53, Rad54L2 and ATRX are mutated in the primary tumour, but the mutations were highly enriched in the metastasis. The APC gene was only mutated in the metastasis while genes such as WRN and MLH3 were heterozygous in the normal tissue but lost heterozygosity in the metastases (Figure 2A and B). Other genes involved in metabolism such as NTRK3 or are part of the small GTPase superfamily such KRAS were also mutated in primary tumours but were enriched in the metastasis (Figure 2A). Other affected pathways include microtubules and cell adhesion, motor proteins, calcium release channels and basement membrane proteins. Interestingly, six genes involved in the CNS architecture were found to be mutated mostly in the metastases, indicating a potential role in establishing the tumours in the brain. More important is our finding that mutations in the Keap1-Nrf2-ARE cell survival pathway were present in 80% of the patients and were either specific to or enriched in the metastases.
Figure 2.
Somatic mutations identified in the primary NSCLC tumours and their corresponding metastases. (A) A table listing mutations detected in all the five patients and were categorised into different functional groups. An increase in colour intensity indicates enrichment in mutant population as indicated by an increase in the number of reads from the sequencing data. N, normal; P, primary; M, metastasis. Low-intensity colour corresponds to a low number of supporting reads for the mutation (indicative of small allele frequency) and the high-intensity colour corresponds to a higher number of supporting reads (indicative of high allele frequency). (B) Mutations detected in some DNA damage and repair genes from four patients (Tp53, MLH3, ATRX and APC). The red colour indicates a mutation to an adenine base (A), and the blue colour indicates a mutation to a thymine base (T). Genes that lost heterozygosity are in red.
The majority of the sequenced tumour genomes carry a mutation in the Keap1-Nrf2-ARE pathway
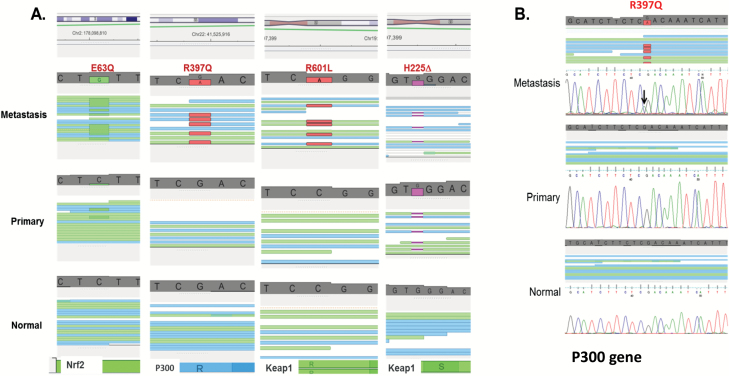
Four of the sequenced patients presented mutations in the Keap1-Nrf2-ARE pathway. IGV (59) data in Figure 3A show for example that the mutation 63Q (in DNA: C to G mutation) in Nrf2 is not detectable in the normal tissue, barely present in the primary tumour but highly enriched in the metastasis. The R397Q (G to A mutation) in P300 and R601L (C to A mutation) in Keap1 are absent in both normal tissue and primary tumours but are highly detectable in the metastasis, indicating a potential role of these mutations in the metastatic spread of lung tumours to the brain. Besides single-nucleotide mutations, these genes can also be inactivated by truncation. For example, the Keap1 gene is truncated in one of the patients as a result of a deletion (H225∆). This deletion is present in both the primary and metastatic tumours.
Figure 3.
Mutations in Keap1, Nrf2 and P300 detected by whole genome sequencing. (A) Visualisation of the mutations in four patients using the genomic browser IGV shows mutations in metastatic cells in Keap1, Nrf2 and P300. The mutated bases are flagged in green for glycine (G) or red (A) and the single-nucleotide deletion in pink. All mutations are supported by a large number of reads and were confirmed by Sanger sequencing. (B) Sanger sequencing confirming the WGS sequencing data. Sanger sequencing shows the presence of the G to A mutation in P300 gene.
To confirm the presence of these mutations, DNA extracted from normal, primary and metastatic tissues was subject to Sanger sequencing that confirmed our next-generation sequencing results. Sanger sequencing results for P300-R397Q (G to A mutation) are shown in Figure 3B and confirm our NGS data.
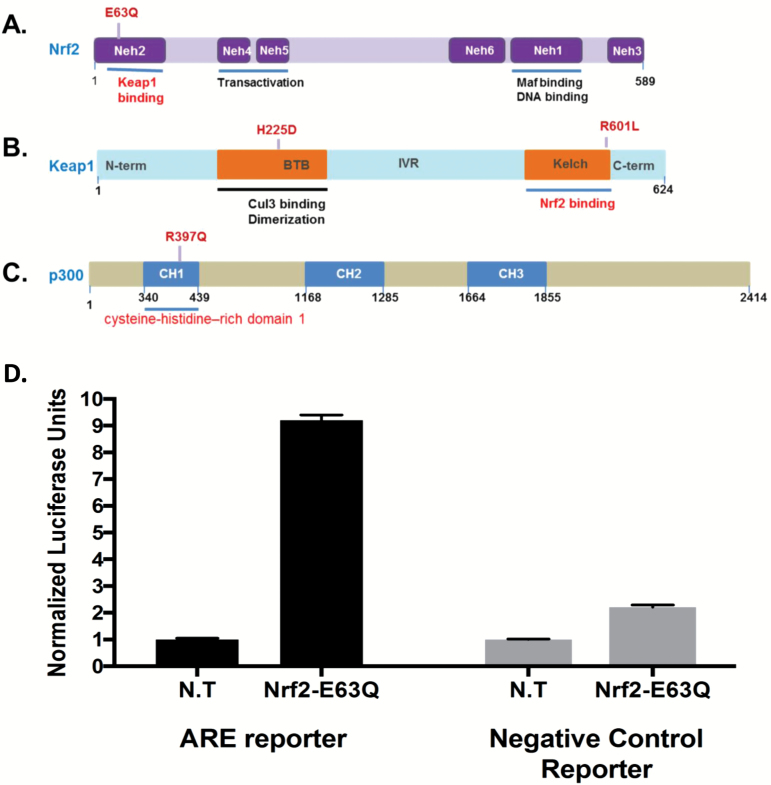
The detected mutations affect functional domains on the Keap1, Nrf2 and P300 proteins
The mutations detected in the three genes affect functional domains that are involved in the interaction between Keap1 and Nrf2 or the transcriptional adaptor zinc-finger domain 1 in P300 that may be important for the protein’s ability to acetylate Nrf2 and increase its transcriptional activity (Figure 4A–C). Indeed, E63Q mutation in Nrf2 affects the Nh2 domain involved in Keap1 binding and the R601L mutation in Keap1 affects the Kelch domain which is also involved in Nrf2 binding while the single-nucleotide deletion in Keap1 leads to a truncated protein that lacks the Nrf2 binding domain. This loss of interaction between Keap1 and Nrf2 would lead to Nrf2 translocation to the nucleus leading to transcription activation of genes involved in stress response and cell survival. To assess whether the identified mutations do indeed affect Nrf2 activity, we used an ARE-luciferase reporter assay (Qiagen) designed for monitoring the activity of the Nrf2 protein in cultured cells. The Nrf2-responsive luciferase construct encodes the firefly luciferase reporter gene under the control of a minimal (m)CMV promoter and tandem repeats of the ARE transcriptional response element. The number of response elements and the intervening sequence between these response elements has been experimentally optimised to maximise the signal to noise ratio. This construct monitors both increases and decreases in the transcriptional activity of Nrf1 and Nrf2, and therefore the activity of the anti-oxidant response pathway. Normalised luciferase activity was measured in cells expressing an inducible ARE-luciferase construct transfected with either an NRF2-E63Q mutant or left untransfected. As a control, we also used cells expressing a non-inducible luciferase construct and that were transfected with either an NRF2-E63Q mutant or left untransfected. Figure 4D shows that Nrf2 E63Q was able to induce ARE-luciferase activity ~8-fold higher than control cells not transfected with Nrf2 E63Q mutant construct and that the non-inducible Luciferase construct failed to show a comparable activity whether the cells were transfected or not with NRF2 E63Q mutant.
Figure 4.
Domain architecture of Keap1, Nrf2 and P300 and the effect of the mutations on Nrf2 activity. (A) Nrf2 consists of 589 amino acids and has six evolutionarily highly conserved domains, Neh1-6. Neh2 contains ETGE and DLG motifs, which are required for the interaction with Keap1. (B) Keap1 consists of 624 amino acid residues and has five domains. The two protein–protein interaction motifs, the BTB domain and the Kelch domain, are separated by the intervening region (IVR). The Kelch domain and the C-terminal region mediate the interaction with Neh2 on Nrf2. (C) P300 consists of 2414 amino acid residues and has three domains. CH1, cysteine-histidine–rich (CH) region 1 also known as TAZ1, transcriptional adaptor zinc-finger domain 1; The RING-PHD segment also known as the CH2 region and the ZZ-TAZ2 domains as the CH3 region. (D) Measurement of NRF2-E63Q activity using an Nrf2 pathway-responsive ARE-luciferase reporter. Error bars were calculated based on three replicates.
Mutations in Nrf2 are also found in CTCs of metastatic cancer patients
To extend our study to other metastatic cancer types beyond lung tumours, we collected blood from 10 patients with metastatic melanoma, breast or colon cancer. Blood samples were subjected to RBCs lysis, and CTCs were isolated using a combination of magnetic nanoparticles CD45 depletion and positive EpCAM-based FCM sorting. Genomic DNA isolated from the enriched CD45-EpCAM− and CD45-EpCAM+ CTC populations was then used for whole genome amplification (Figure 5A). We first used three sets of primers to amplify three distinct regions of NRF2 from the amplified whole genome (Figure 5B) and subjected these PCR products to Sanger sequencing. We found mutations in neh2 domain but not in other regions of NRF2, and the mutations that we found were among cancer-related mutations reported by the Catalogue Of Somatic Mutations In Cancer (COSMIC) database. Two patients showed a single R34G somatic mutation, while two others had E79Q and E82G mutation in the genomic DNA of CTCs (Figure 5C).
Figure 5.
Detection of Nrf2 mutations in CTCs and nuclear localisation of Nrf2 mutant proteins. (A) Enrichment and analysis methodology used in this study. (B) Neh2 domain of Nrf2 is amplified from CTCs. (C) Sanger sequencing of the amplified fragment and detection of the somatic mutations. (D) Nuclear localisation of wild-type and mutant GFP-Nrf2 proteins.
To check whether these mutations can affect the interaction between Nrf2 and Keap1, we generated lentiviral constructs with Nrf2 gene either wild type or carrying the detected mutations. Figure 5D shows that while the Nrf2 wild type is mainly cytoplasmic, introduction of the R34G, E79Q or E82G mutations led to stabilisation and nuclear localisation of Nrf2 protein. This finding confirms that the detected mutations are important for Nrf2 translocation to the nucleus and suggest that these mutations may play a role in the metastatic spread of the cancer cells.
Quantification of Nrf2 mutations in CTCs from metastatic cancer patients
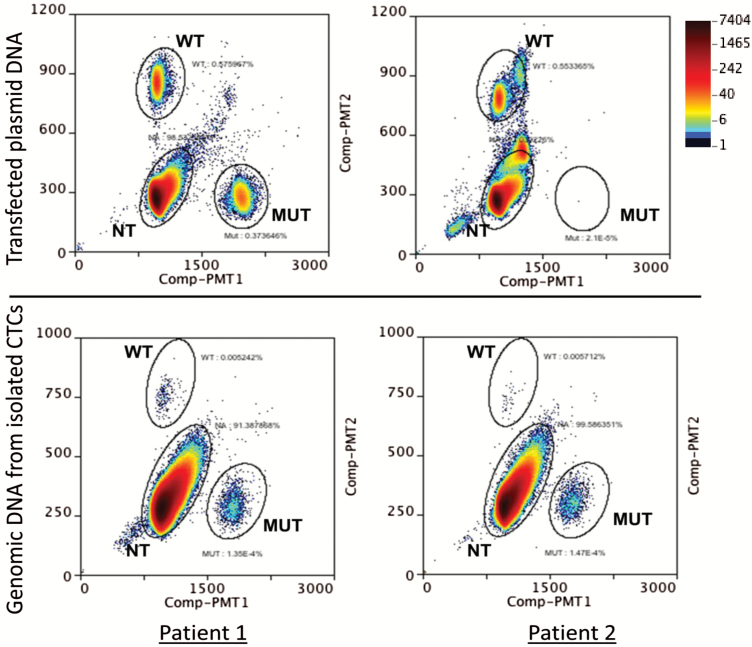
To detect Nrf2 mutations in circulation in patients with metastatic cancer, CTCs were isolated from peripheral blood (7.8–17.7 ml) from 10 metastatic breast, colon and melanoma cancer patients. Genomic DNA from CTCs was extracted and subjected to ddPCR using specific probes and primers described in Materials and Methods. The assay was first validated using wild-type and mutant R34G NRF2 plasmid constructs (Figure 6, upper panel). Once the probes were validated using the plasmid constructs, the wild-type and mutant probes were tested in the genomic DNA extracted from CTCs from multiple metastatic cancer patients. As shown in Figure 6, lower panel, mutant R34G and wild-type NRF2 gene are detected in the isolated CTCs and suggest that detection of these mutations in circulation may serve as a biomarker to predict metastatic progression of cancer.
Figure 6.
Quantification of Nrf2 mutations in circulation. Droplet-based digital PCR was used to quantify the expression of mutant Nrf2 gene in CTCs. CTCs were isolated using the protocol described in Figure 5A. Genomic DNA was extracted and the mutations were quantified using specific probes that detect either wild-type or mutant alleles. Upper panel shows ddPCR quantification of plasmid DNA carrying either wild-type and R34G mutant DNA (left) or wild type only (right). The lower panel shows quantification of mutant DNA in CTCs from two patients with metastatic cancer. NT indicates droplets that contain no genomic DNA but probes only.
Discussion
The most deadly aspect of cancer is its ability to spread or metastasise. To metastasise, a cancer cell must break away from its tumour, invade either the circulatory or lymph system, which will carry it to a new location, and establish itself in the new site. The body has many safeguards to prevent cells from surviving in circulation and reach distant organs, yet many cancer cells have the ability to overcome these safeguards. One such safeguard is anoikis, a form of programmed cell death that is induced by anchorage-dependent cells detaching from the surrounding extracellular matrix (60). The vast majority of cells released into circulation will undergo cell death and only a very small fraction will survive. It is therefore very important to understand the mechanism(s) by which cancer cells have mutated to circumvent these defences and freely travel to distant sites.
To decipher the molecular mechanisms underlying the transition from a primary to a metastatic tumour, we carried out WGS of normal lung, primary tumours and the corresponding metastases from five patients with progressive metastatic NSCLC and found that genes that belong to several key pathways are mutated, or the mutated form enriched in the metastatic tissue. Of great importance is our finding that metastases acquired mutations in adaptive, cytoprotective genes involved in response to cellular stress mainly Keap-1, Nrf2 and P300, which are key players of the Keap1-Nrf2-ARE survival pathway. The high occurrence of mutations in this survival pathway suggests that it may play a role in cell survival in circulation. The identified mutations induce transcriptional activation of Nrf2 leading to higher transcription of anti-oxidant and detoxification enzymes possibly providing a survival advantage to CTCs in circulation and allowing their dissemination to distant organs. Experiments are underway to establish a correlation between activation of the Keap1-Nrf2-ARE pathway and metastasis in a longitudinal study of a large cohort of metastatic cancer patients.
Our finding that the Keap1-Nrf2-ARE pathway is mutated in the majority of NSCLC patients that metastasised to the brain and in CTCs isolated from metastatic cancer patients supports our hypothesis that mutations in this pathway may indeed provide a survival advantage to these cells and help them reach distant sites. The finding also indicates that therapies that target this pathway to prevent Nrf2 activation may be useful to sensitise CTCs to anoikis and prevent these cells from dissemination to distant sites.
While this is an attractive approach to prevent metastasis, targeting Nrf2 is a double-edged sword. While there are strong opinions that further pharmacological development of drugs that enhance Nrf2 activity should be pursued for preventing not only cancer but also many other diseases in which oxidative and inflammatory stress are crucial for pathogenesis (61–63), many new drugs that activate Nrf2 are now in clinical trials. Other recent genetic analyses show that mutations in Nrf2 or Keap1 are found in some cancers; and that these mutations enhance Nrf2 activity and are associated with resistance to standard chemotherapy and poor survival from cancer (64–66). Therefore, it has also been suggested that inhibitors of Nrf2 should be developed for the treatment of cancer.
Our current data suggest a role for mutations in Keap1-Nrf2-ARE pathway in the ability of CTCs to survive the harsh environment in circulation and reach distant organs. While these findings still need to be confirmed using larger longitudinal studies in patients with metastatic disease, it provides some preliminary evidence that targeting this pathway may eliminate CTCs and prevent tumour metastases.
Funding
Funding for this project was provided by Gromada Foundation, award number (G401471-62624129981) and by the University of Cincinnati, award number (D100271-F100607), the UC Brain Tumor Center, award number (F101559-2015) and the National Center for Advancing Translational Sciences of the National Institutes of Health, award number 1UL1TR001425-01 (E.M.B.). The funding agencies had no involvement in the design of the experiments or in data interpretation.
Conflict of interest statement: None declared.
References
- 1. Chaffer C. L. and Weinberg R. A (2011) A perspective on cancer cell metastasis. Science, 331, 1559–1564. [DOI] [PubMed] [Google Scholar]
- 2. Eichler A. F., Chung E., Kodack D. P., Loeffler J. S., Fukumura D. and Jain R. K (2011) The biology of brain metastases-translation to new therapies. Nat. Rev. Clin. Oncol., 8, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Popper H. H. (2016) Progression and metastasis of lung cancer. Cancer Metastasis Rev., 35, 75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim M. Y., Oskarsson T., Acharyya S., Nguyen D. X., Zhang X. H., Norton L. and Massagué J (2009) Tumor self-seeding by circulating cancer cells. Cell, 139, 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen D. X., Bos P. D. and Massagué J (2009) Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer, 9, 274–284. [DOI] [PubMed] [Google Scholar]
- 6. Pantel K., Alix-Panabières C. and Riethdorf S (2009) Cancer micrometastases. Nat. Rev. Clin. Oncol., 6, 339–351. [DOI] [PubMed] [Google Scholar]
- 7. Jemal A., Bray F., Center M. M., Ferlay J., Ward E. and Forman D (2011) Global cancer statistics. CA Cancer J. Clin., 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 8. Yu M., Stott S., Toner M., Maheswaran S. and Haber D. A (2011) Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol., 192, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krebs M. G., Sloane R., Priest L. et al. (2011) Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol., 29, 1556–1563. [DOI] [PubMed] [Google Scholar]
- 10. Cohen S. J., Punt C. J., Iannotti N. et al. (2009) Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol., 20, 1223–1229. [DOI] [PubMed] [Google Scholar]
- 11. de Bono J. S., Scher H. I., Montgomery R. B. et al. (2008) Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res., 14, 6302–6309. [DOI] [PubMed] [Google Scholar]
- 12. Cristofanilli M., Budd G. T., Ellis M. J. et al. (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med., 351, 781–791. [DOI] [PubMed] [Google Scholar]
- 13. Budd G. T., Cristofanilli M., Ellis M. J. et al. (2006) Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin. Cancer Res., 12, 6403–6409. [DOI] [PubMed] [Google Scholar]
- 14. Zhang D. D. (2006) Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev., 38, 769–789. [DOI] [PubMed] [Google Scholar]
- 15. Homma S., Ishii Y., Morishima Y. et al. (2009) Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res., 15, 3423–3432. [DOI] [PubMed] [Google Scholar]
- 16. Singh A., Misra V., Thimmulappa R. K. et al. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med., 3, e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Padmanabhan B., Tong K. I., Ohta T. et al. (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell, 21, 689–700. [DOI] [PubMed] [Google Scholar]
- 18. Kensler T. W., Wakabayashi N. and Biswal S (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol., 47, 89–116. [DOI] [PubMed] [Google Scholar]
- 19. Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D. and Yamamoto M (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev., 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cullinan S. B., Gordan J. D., Jin J., Harper J. W. and Diehl J. A (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol., 24, 8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furukawa M. and Xiong Y (2005) BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol., 25, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K. and Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol., 24, 7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang D. D., Lo S. C., Cross J. V., Templeton D. J. and Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol., 24, 10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong K. I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S. and Yamamoto M (2007) Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell Biol., 27, 7511–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W. and Hannink M (2005) Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem., 280, 30091–30099. [DOI] [PubMed] [Google Scholar]
- 26. Itoh K., Chiba T., Takahashi S. et al. (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun., 236, 313–322. [DOI] [PubMed] [Google Scholar]
- 27. Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A. and Yamamoto M (2001) Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells, 6, 857–868. [DOI] [PubMed] [Google Scholar]
- 28. Katsuoka F., Motohashi H., Ishii T., Aburatani H., Engel J. D. and Yamamoto M (2005) Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell Biol., 25, 8044–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nioi P., McMahon M., Itoh K., Yamamoto M. and Hayes J. D (2003) Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J., 374, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen G., Hebbar V., Nair S. et al. (2004) Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem., 279, 23052–23060. [DOI] [PubMed] [Google Scholar]
- 31. Zhu M. and Fahl W. E (2001) Functional characterization of transcription regulators that interact with the electrophile response element. Biochem. Biophys. Res. Commun., 289, 212–219. [DOI] [PubMed] [Google Scholar]
- 32. Taguchi K., Motohashi H. and Yamamoto M (2011) Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells, 16, 123–140. [DOI] [PubMed] [Google Scholar]
- 33. Sporn M. B. and Liby K. T (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer, 12, 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeNicola G. M., Karreth F. A., Humpton T. J. et al. (2011) Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature, 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanada N., Takahata T., Zhou Q. et al. (2012) Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer, 12, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang P., Singh A., Yegnasubramanian S., Esopi D., Kombairaju P., Bodas M., Wu H., Bova S. G. and Biswal S (2010) Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther., 9, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adam J., Hatipoglu E., O’Flaherty L. et al. (2011) Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell, 20, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ooi A., Wong J. C., Petillo D. et al. (2011) An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell, 20, 511–523. [DOI] [PubMed] [Google Scholar]
- 39. Kansanen E., Kuosmanen S. M., Leinonen H. and Levonen A. L (2013) The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol., 1, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma Q. and He X (2012) Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol. Rev., 64, 1055–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M. and Motohashi H (2012) Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell, 22, 66–79. [DOI] [PubMed] [Google Scholar]
- 42. Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R. and Goodman R. H (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature, 365, 855–859. [DOI] [PubMed] [Google Scholar]
- 43. Eckner R., Ewen M. E., Newsome D., Gerdes M., DeCaprio J. A., Lawrence J. B. and Livingston D. M (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- 44. Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H. and Nakatani Y (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 45. Roth S. Y., Denu J. M. and Allis C. D (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- 46. Glozak M. A., Sengupta N., Zhang X. and Seto E (2005) Acetylation and deacetylation of non-histone proteins. Gene, 363, 15–23. [DOI] [PubMed] [Google Scholar]
- 47. Yang X. J. and Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell, 31, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun Z., Chin Y. E. and Zhang D. D (2009) Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell Biol., 29, 2658–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang Y., Zhao W., Chen Y., Zhao Y. and Gu W (2008) Acetylation is indispensable for p53 activation. Cell, 133, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lara O., Tong X., Zborowski M. and Chalmers J. J (2004) Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp. Hematol., 32, 891–904. [DOI] [PubMed] [Google Scholar]
- 51. Yang L., Lang J. C., Balasubramanian P., Jatana K. R., Schuller D., Agrawal A., Zborowski M. and Chalmers J. J (2009) Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol. Bioeng., 102, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Balasubramanian P., Yang L., Lang J. C., Jatana K. R., Schuller D., Agrawal A., Zborowski M. and Chalmers J. J (2009) Confocal images of circulating tumor cells obtained using a methodology and technology that removes normal cells. Mol. Pharm., 6, 1402–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu Y., Deighan C. J., Miller B. L., Balasubramanian P., Lustberg M. B., Zborowski M. and Chalmers J. J (2013) Isolation and analysis of rare cells in the blood of cancer patients using a negative depletion methodology. Methods, 64, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Furgason J. M., Li W., Milholland B. et al. (2014) Whole genome sequencing of glioblastoma multiforme identifies multiple structural variations involved in EGFR activation. Mutagenesis, 29, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Furgason J. M., Koncar R. F., Michelhaugh S. K., Sarkar F. H., Mittal S., Sloan A. E., Barnholtz-Sloan J. S. and Bahassi e. l. M (2015) Whole genome sequence analysis links chromothripsis to EGFR, MDM2, MDM4, and CDK4 amplification in glioblastoma. Oncoscience, 2, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DePristo M. A., Banks E., Poplin R. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O. and Barillot E (2012) Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics, 28, 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen K., Wallis J. W., McLellan M. D. et al. (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods, 6, 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G. and Mesirov J. P (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frisch S. M. and Screaton R. A (2001) Anoikis mechanisms. Curr. Opin. Cell Biol., 13, 555–562. [DOI] [PubMed] [Google Scholar]
- 61. Calkins M. J., Johnson D. A., Townsend J. A. et al. (2009) The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid. Redox Signal., 11, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kwak M. K. and Kensler T. W (2010) Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol., 244, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boutten A., Goven D., Artaud-Macari E., Boczkowski J. and Bonay M (2011) NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol. Med., 17, 363–371. [DOI] [PubMed] [Google Scholar]
- 64. Wang X. J., Sun Z., Villeneuve N. F. et al. (2008) Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis, 29, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Taguchi K., Motohashi H. and Yamamoto M (2011) Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells, 16, 123–140. [DOI] [PubMed] [Google Scholar]
- 66. Yamadori T., Ishii Y., Homma S. et al. (2012) Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene, 31, 4768–4777. [DOI] [PubMed] [Google Scholar]