Abstract
In multicellular organisms, glycosylation regulates various developmental signaling pathways including the Notch pathway. One of the O-linked glycans added to epidermal growth factor-like (EGF) repeats in animal proteins including the Notch receptors is the xylose–xylose–glucose-O oligosaccharide. Drosophila glucoside xylosyltransferase (Gxylt) Shams negatively regulates Notch signaling in specific contexts. Since Shams adds the first xylose residue to O-glucose, its loss-of-function phenotype could be due to the loss of the first xylose, the second xylose or both. To examine the contribution of the second xylose residues to Drosophila Notch signaling, we have performed biochemical and genetic analysis on CG11388, which is the Drosophila homolog of human xyloside xylosyltransferase 1 (XXYLT1). Experiments in S2 cells indicated that similar to human XXYLT1, CG11388 can add the second xylose to xylose–glucose-O glycans. Flies lacking both copies of CG11388 (Xxylt) are viable and fertile and do not show gross phenotypes indicative of altered Notch signaling. However, genetic interaction experiments show that in sensitized genetic backgrounds with decreased or increased Notch pathway components, loss of Xxylt promotes Delta-mediated activation of Notch. Unexpectedly, we find that in such sensitized backgrounds, even loss of one copy of the fly Gxylt shams enhances Delta-mediated Notch activation. Taken together, these data indicate that while the first xylose plays a key role in tuning the Delta-mediated Notch signaling in Drosophila, the second xylose has a fine-tuning role only revealed in sensitized genetic backgrounds.
Keywords: developmental biology, Drosophila, Notch signaling, xylose, xylosyltransferases
Introduction
Notch signaling is a highly conserved cell to cell communication pathway which regulates numerous aspects of embryonic development and adult tissue homeostasis in metazoans (Artavanis-Tsakonas and Muskavitch 2010). Canonical Notch signaling is mediated by the activation of Notch receptors by Delta and Serrate/Jagged families of ligands and is modulated by multiple post-translational modifications, including phosphorylation, acetylation, ubiquitination and glycosylation (Siebel and Lendahl 2017). Mutations in Notch pathway components cause various human diseases, including developmental abnormalities and cancer (Allenspach et al. 2002; Talora et al. 2008; Penton et al. 2012). Notably, loss of one copy of some Notch pathway receptors and ligands can cause significant phenotypes in animal model organisms and human patients, indicating that a number of developmental processes are highly sensitive to perturbations in the Notch pathway activity (Penton et al. 2012; Masek and Andersson 2017). Moreover, even in the absence of mutations in the core components of the Notch pathway, dysregulation of Notch signaling has been implicated in various other diseases, including several forms of cancer and non-neoplastic diseases of the heart, lung, muscle and the liver (Siebel and Lendahl 2017). Therefore, identification and characterization of modulators of the Notch pathway has the potential to shed light on the pathophysiology of Notch-related diseases and might establish new paradigms for therapeutic manipulation of this pathway.
Notch sugar modifications affect Notch signaling (Stanley and Okajima 2010; Takeuchi and Haltiwanger 2014; Haltom and Jafar-Nejad 2015). One such modification is O-glucosylation, which refers to the addition of an O-linked glucose residue by the enzyme protein O-glucosyltransferase 1 (Poglut1) onto a specific consensus sequence found on EGF repeats in Notch receptors and a number of other animal proteins (Hase et al. 1988; Acar et al. 2008). Drosophila Poglut1, which is encoded by rumi, promotes Notch activation by adding O-glucose to multiple EGF repeats across the Notch extracellular domain (Acar et al. 2008; Leonardi et al. 2011; Perdigoto et al. 2011). Loss of O-glucose glycans from Drosophila Notch impairs Notch cleavage without affecting Notch-ligand binding (Acar et al. 2008; Leonardi et al. 2011).
O-glucosylated EGF repeats can be extended to a trisaccharide by addition of two xylose residues (Hase et al. 1988; Acar et al. 2008). In mammals, two glucoside xylosyltransferases (GXYLT1 and GXYLT2) are reported to add the first xylose to O-glucosylated EGF repeats (Sethi et al. 2010). Drosophila has only one GXYLT homolog, Shams, which negatively regulates fly Notch signaling in specific contexts by adding xylose residues to a subset of Notch EGF repeats (Lee et al. 2013). Moreover, transgenic overexpression of human GXYLT1 impairs Notch signaling in the fly wing (Lee et al. 2013). Genetic, cell aggregation and ligand-binding assays indicate that loss of the first xylose residue from O-glucosylated EGF16-20 specifically enhances the activation of Drosophila Notch by the Delta ligand (Lee et al. 2017). Since Shams is the only GXYLT in flies, loss of Shams results in the loss of both xylose residues from Notch EGF repeats. Therefore, it is possible that the enhancement of Notch signaling in shams mutant flies is at least in part due to the loss of the second xylose.
In mammals, a type II transmembrane protein called xyloside xylosyltransferase 1 (XXYLT1) adds a second xylose residue to the first xylose to form xylose–xylose–glucose-O trisaccharides on Notch EGF repeats (Sethi et al. 2012). Transgenic overexpression of human XXYLT1 inhibits Notch signaling in flies (Lee et al. 2013). Moreover, human XXYLT1 is frequently amplified in several cancer types associated with decreased Notch signaling (Cerami et al. 2012; Yu et al. 2015). Together, these observations suggest that the second xylose might play an inhibitory role in Notch signaling. However, a loss-of-function analysis has not been performed on mammalian or fly XXYLT1.
Based on the protein domain structure and percent amino acid identity, Drosophila CG11388 is the only fly protein showing a high degree of homology with human XXYLT1. Here, we report that CG11388 possesses xyloside xylosyltransferase activity towards Notch EGF repeats, but shows EGF repeat specificity, unlike the human enzyme. CG11388-null flies are viable and do no show characteristic phenotypes of altered Notch signaling. However, loss of CG11388 promotes Delta-mediated signaling in sensitized genetic backgrounds. In addition, genetic interaction studies suggest that loss of shams (Gxylt) modulates Drosophila Notch signaling in a dosage-sensitive manner. Altogether, our data indicate that the enhanced Notch signaling observed in shams mutants is primarily caused by the loss of first xylose residue from O-glucose glycans on Notch EGF16-20, and that the second xylose can modulate Notch signaling when the level of Notch or Delta is limiting.
Results
Drosophila CG11388 encodes a xyloside xylosyltransferase (Xxylt), adding the second xylose to xylose–glucose-O disaccharides on specific Notch EGF repeats
Mutations in the fly glucoside xylosyltransferase Shams result in the loss of both xylose residues from xylose–xylose–glucose-O glycans on Notch and cause a mild gain of Notch signaling in flies (Lee et al. 2013). To understand the contribution of each xylose residue to the observed phenotypes, we decided to compare the phenotypes of shams mutants with those of the fly xyloside xylosyltransferase responsible for the addition of the second xylose to these glycans. It has previously been shown that recombinantly expressed Drosophila Notch in S2 cells harbors limited xylosylation, with a low amount of the full trisaccharide on a limited number of EGF repeats, mostly in the EGF16-20 region (Lee et al. 2013; Harvey et al. 2016). While this indicates that Xxylt activity exists in Drosophila, it is not known whether CG11388, which is the fly homolog of the human XXYLT1, has Xxylt activity.
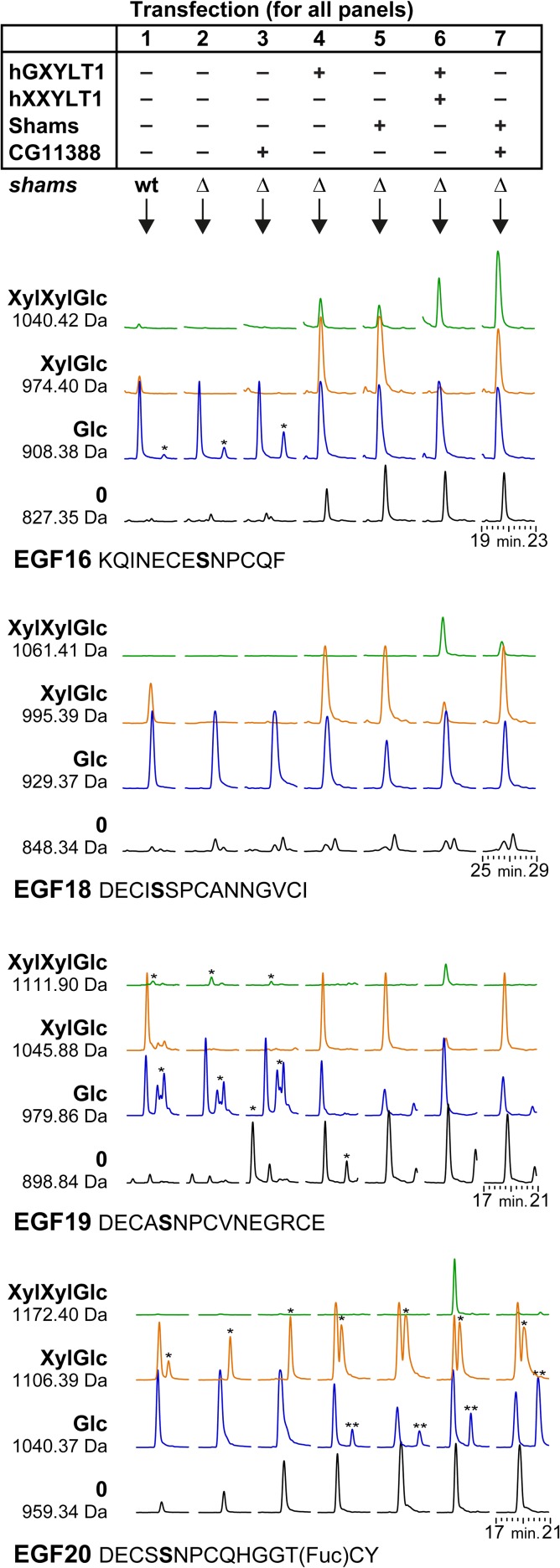
To evaluate CG11388 and to compare its activity with its human homolog, CG11388 and human XXYLT1 were overexpressed in Drosophila S2 cells and analyzed by peptide mass spectrometry of a co-expressed Notch EGF16-20 fragment. To avoid the endogenous glucoside xylosyltransferase activity, shams knock-out S2 cells were generated by CRISPR/Cas9 technique, as described in the Materials and methods section. In the background of a knock-out of the endogenous shams, overexpressed human GXYLT1 and Shams both showed robust activity (i.e., addition of first xylose to O-glucosylated EGF repeat) on the four EGF repeats 16, 18, 19 and 20 that could be analyzed using an AspN proteolytic digest of the Notch fragment (Figure 1). The observed activity of Shams and human GXYLT1 provided a high level of putative acceptor (i.e., xylose–glucose-O-EGF) for the second xylosyltransferase. We note that xylose–xylose–glucose-O trisaccharide was observed on EGF16 and less on EGF18 (hardly visible in Figure 1) even without overexpression of Xxylt, which is in accordance with previous observations (Lee et al. 2013; Harvey et al. 2016).
Fig. 1.
CG11388 functions as a xyloside xylosyltransferase on Drosophila Notch. A plasmid with His6-tagged EGF repeat 16 to 20 was transiently transfected into plain S2 cells, S2 cells lacking endogenous Shams (Δshams), or Δshams S2 cells co-transfected with expression vectors for the indicated human and fly proteins. AspN digested peptides from the purified protein were analyzed by mass spectrometry and revealed the glycosylation pattern of EGF16, 18, 19 and 20. Shown here are extracted ion chromatograms of expected masses of glycopeptides (all 2+). Black and blue chromatograms represent naked and O-glucosylated (monosaccharide) EGF repeats, respectively. Plain S2 cells (without any plasmid transfection) show limited xylosylation (column 1), which is completely abolished in Δshams cells (column 2). Both GXYLT1 and Shams (columns 4 and 5) formed xylose–glucose (XylGlc) and acted similarly on all four EGF repeats. Only EGF16 showed the relevant XylXylGlc peak, likely generated by the endogenous Xxylt. Simultaneous overexpression of human XXYLT1 or Drosophila CG11388 (columns 6 and 7) showed that the human enzyme was able to transfer a second xylose to all four EGF repeats, whereas the Drosophila enzyme only acted on EGF16 and to some extent on EGF18. Importantly, overexpression of CG11388 in Δshams S2 cells does not generate new glycopeptide peaks (column 3), confirming that CG11388 can only add the second xylose to xylose–glucose-O disaccharide. Of note, CG11388 showed relatively stronger activity on EGF16 compared to the human enzyme. For each peptide of each transfection condition, we set the highest peak as 100% and normalized the other peaks accordingly. For example, in EGF16 with hGXYLT and hXXYLT (column 6), the Glc peak is set at 100% and the other peaks are shown in proportion to this highest peak. Asterisks (*) mark nonspecific peaks that appeared within the extracted ion chromatograms because their m/z-values were similar to the search mass ± 0.3 Dalton. Double asterisks (**) mark peaks corresponding to the 1040.42 Da masses of EGF16 (XylXylGlc), which run close to the 1040.37 Da peaks of EGF20 (Glc). MS/MS spectra confirmed the identity of 1040.42, 1061.41 and 1172.40 peptides from the hGXYLT1 + hXXYLT1 sample (Supplementary data, Figures S1, S2 and S3).
Overexpression of human XXYLT1 together with GXYLT1 resulted in the appearance of xylose–xylose–glucose on all four analyzed EGF repeats. This suggests that the xylosyltransferase activity of the human enzyme is not limited to specific EGF repeats, as long as they harbor the consensus sequence. Moreover, the data indicate that there is no detection problem of peptides with trisaccharide modification.
Overexpression of Drosophila CG11388 together with Shams clearly increased the trisaccharide signal on EGF16 and EGF18 (Figure 1). Moreover, CG11388 overexpression was not able to extend the O-glucose saccharides in shams knock-out cells, confirming that the xylose–glucose-O disaccharide is required for the activity of CG11388 (Figure 1). These data indicate that similar to human XXYLT1, Drosophila CG11388 is a xylosyltransferase, capable of generating the trisaccharide structure. However, no second xylose was observed on EGF19 and EGF20 despite CG11388 overexpression. These observations suggest that unlike human XXYLT1, CG11388 shows specificity for certain EGF repeats, even when the enzyme is overexpressed. Human XXYLT1 was previously reported to show xylosyltransferase activity towards a synthetic xylose–glucose oligosaccharide acceptor lacking an EGF repeat backbone (Sethi et al. 2012). However, in agreement with our S2 cell assays, Drosophila CG11388 does not show any xylosyltransferase activity towards the same synthetic acceptor (data not shown). Together, these data establish that fly CG11388 is a xyloside xylosyltransferase, although its function is limited to specific EGF repeats.
Loss of Xxylt does not show impaired Notch signaling phenotypes in flies
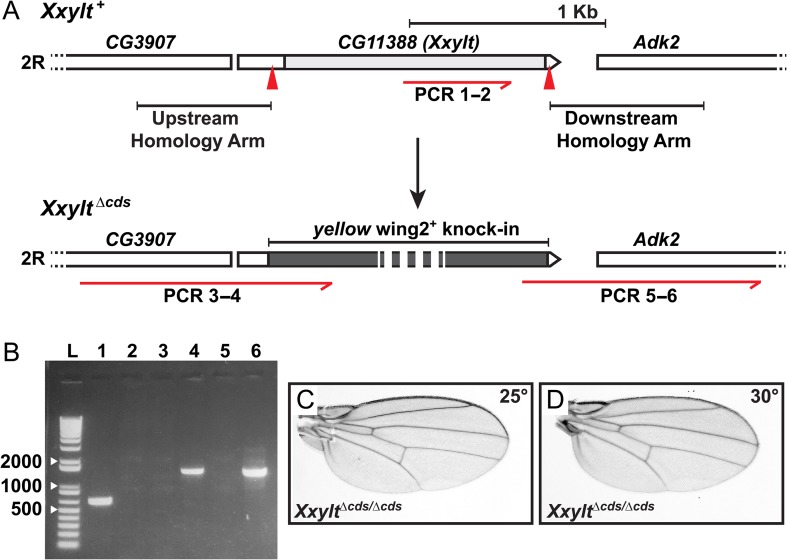
To examine the contribution of the second xylose residue of xylose–xylose–glucose-O trisaccharide on Notch EGF repeats to Drosophila Notch signaling, we utilized CRISPR/Cas9 genome engineering to generate a loss-of-function allele of CG11388, to which we will refer as Xxylt hereafter. In this allele (called XxyltΔcds) 87% of the gene region including the entire coding region was replaced by the yellow wing2+ marker (Li-Kroeger et al. 2018) (Figure 2A). Flies homozygous for this allele (XxyltΔcds/Δcds) are viable at temperatures ranging from 18°C to 30°C (Table I). Unlike the flies homozygous for the shamsΔ34 allele, which show loss of wing vein and head bristles (Lee et al. 2013), XxyltΔcds/Δcds flies do not exhibit any phenotypes suggestive of aberrant Notch signaling such as loss of wing vein (Figure 2C, D) or head bristles (data not shown) at these temperatures. Moreover, both male and female XxyltΔcds/Δcds flies are fertile. We conclude that Xxylt is not essential for embryonic and larval development and Notch signaling in flies.
Fig. 2.
Loss of Xxylt does not show impaired Notch signaling phenotypes in flies. (A) Generation of a CG11388 loss-of-function allele using CRISPR/Cas9 engineering. Top panel shows schematic representation of the genomic region of second chromosome (2 R) containing CG11388 (Xxylt) and its neighboring genes. Light gray box indicates the coding sequence. Red arrowheads mark the sgRNA binding sites. Homology arms used for homologous recombination-mediated repair are shown upstream and downstream to the sgRNA binding sites. The red half arrow indicates a region of the CG11388 coding sequence used to confirm deletion by PCR. Bottom panel shows yellow wing2+ knock-in (dark gray box) and extended upstream and downstream homology regions used to confirm the knock-in. Numbers written under the red half arrows refer to the lanes on the PCR gel shown in panel B. (B) PCR amplification products for CG11388 internal (lanes 1 and 2), extended upstream homology region (lanes 3 and 4) and extended downstream homology region (lanes 5 and 6) from y w (control; lanes 1, 3 and 5) and XxyltΔcds/Δcds (lanes 2, 4 and 6) genomic DNA. Absence of PCR amplification product for CG11388 internal and presence of extended upstream and downstream homology regions confirmed the replacement of CG11388 with yellow wing2+. L marks the DNA ladder. (C and D) Wings from adult XxyltΔcds/Δcds flies raised at 25°C (C) and 30°C (D) show no sign of impaired Notch signaling.
Table I.
Xxylt Δcds/Δcds mutants are viable
| Genotype (XxyltΔcds/CyO x XxyltΔcds/CyO) | Temp | Observed(%) | Expected(%) | Chi-square |
|---|---|---|---|---|
| Heterozygous (XxyltΔcds/CyO) | 30°C | 121 (68.4%) | 118 (66.7%) | P = 0.733 |
| Homozygous (XxyltΔcds/Δcds) | 56 (31.6%) | 59 (33.3%) | ||
| Heterozygous (XxyltΔcds/CyO) | 25°C | 144 (67.6%) | 142 (66.7%) | P = 0.836 |
| Homozygous (XxyltΔcds/Δcds) | 69 (32.4%) | 71(33.3%) | ||
| Heterozygous (XxyltΔcds/CyO) | 18°C | 102 (68.9%) | 99 (66.7%) | P = 0.708 |
| Homozygous (XxyltΔcds/Δcds) | 46 (31.1%) | 49 (33.3%) |
Temp: temperature.
Notch haploinsufficient phenotypes are partially rescued by simultaneous loss of Xxylt
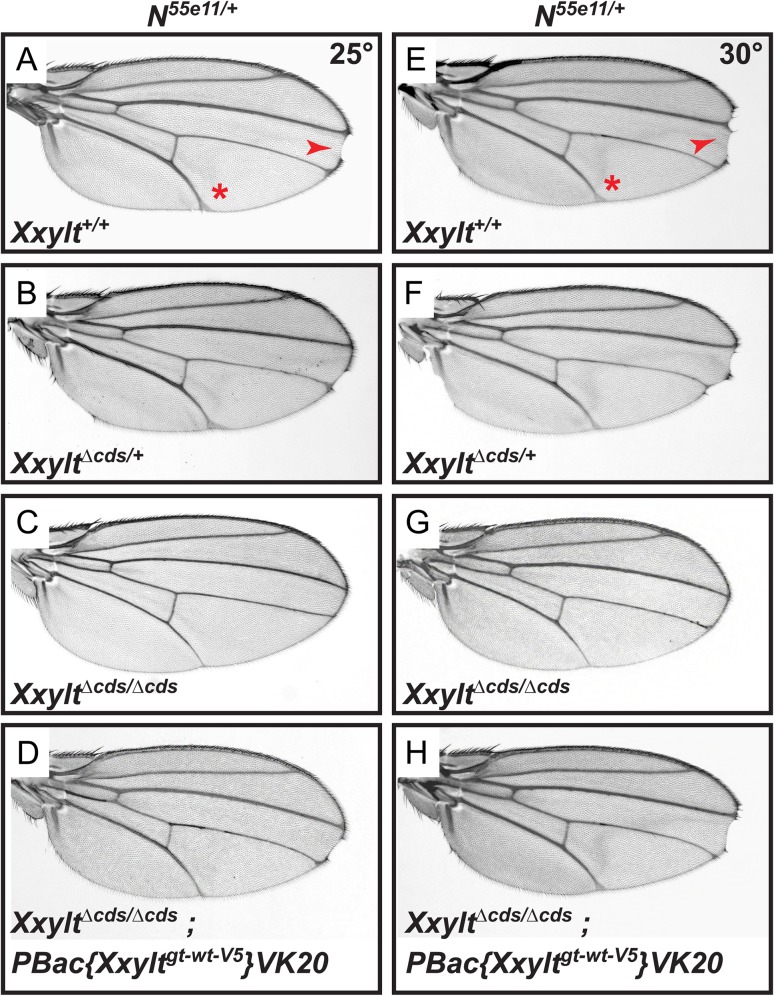
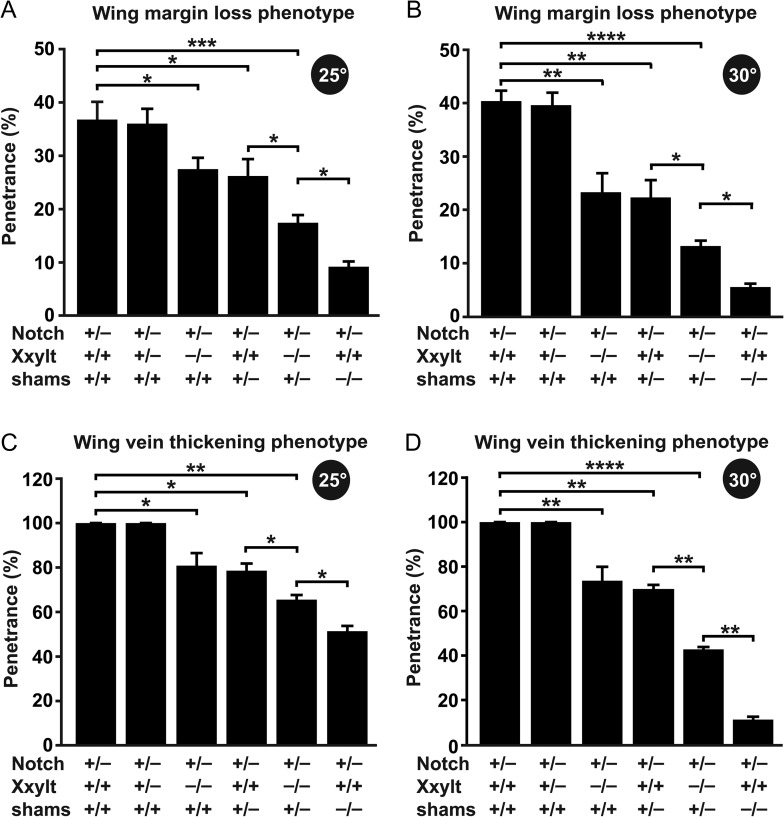
To examine if Xxylt shows any genetic interaction with Notch, we assessed the effect of loss of Xxylt on a Notch heterozygous background, which shows haploinsufficient phenotypes like loss of wing margin and wing vein thickening. At 25°C, N55e11/+ flies show a 39% penetrant wing margin loss and a 100% penetrant wing vein thickening phenotype (Figure 3A). Loss of one copy of Xxylt does not modify these phenotypes (Figure 3B). However, simultaneous loss of both copies of Xxylt in a N55e11/+ background partially rescues the wing margin loss (27% penetrant) and wing vein thickening (79% penetrant) (Figure 3C). Addition of one copy of an Xxylt genomic transgene reverts this rescue (37% and 100% penetrant wing margin loss and wing vein thickening, respectively; Figure 3D). This indicates that the partial rescue of N55e11/+ phenotypes in N55e11/+; XxyltΔcds/Δcds flies is because of loss of both copies of Xxylt. At 30°C, the penetrance of wing margin loss and wing vein thickening in N55e11/+ flies is 40% and 100%, respectively (Figure 3E). Loss of Xxylt shows a similar rescue of N55e11/+ phenotypes at this temperature, as evidenced by the 23% and 73% penetrance of wing margin loss and wing vein thickening, respectively, in N55e11/+; XxyltΔcds/Δcds animals (Figure 3G). Again, addition of an Xxylt genomic transgene reverts this rescue (38% and 100% penetrant wing margin loss and wing vein thickening, respectively; Figure 3H). Loss of one copy of Xxylt did not modify the penetrance of wing margin loss (39%) and wing vein thickening (100%) in N55e11/+ flies raised at 30°C (Figure 3F). These observations suggest that in a Notch heterozygous background, Xxylt negatively regulates Notch signaling in the Drosophila wing.
Fig. 3.
Loss of Xxylt partially rescues Notch haploinsufficient phenotypes. (A–D) Animals were raised at 25°C. (A) N55e11/+ animals show wing vein thickening (asterisk) and margin defects (arrowhead). Penetrance of wing vein thickening and wing margin loss is 100% and 39%, respectively. (B) XxyltΔcds/+ does not suppress the N55e11/+ phenotypes. (C) XxyltΔcds/Δcds partially rescues the N55e11/+ phenotypes (penetrance of 79% for wing vein thickening and 27% for wing margin loss). (D) Providing a copy of wild-type Xxylt transgene (PBac{Xxyltgt-wt-V5}VK20) reverted the rescue of N55e11/+ phenotypes in an XxyltΔcds/Δcds background. (E–H) Animals raised at 30°C show a similar trend.
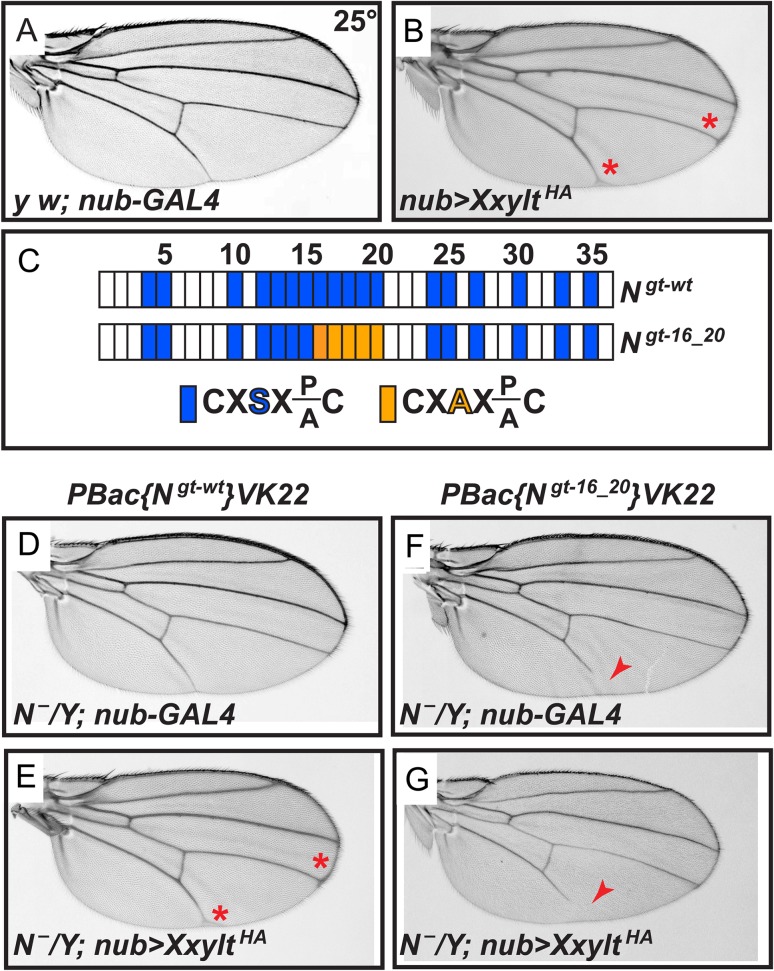
To further examine the role of fly Xxylt in Notch signaling, we performed overexpression studies. Overexpression of an HA-tagged version of Xxylt (XxyltHA) in the developing wing by nubbin-GAL4 results in wing vein thickening, suggestive of a mild Notch loss-of-function phenotype (Figure 4B; red asterisks). This favors a role for Xxylt towards negative regulation of Notch signaling. Previously, serine to alanine (S/A) mutations in O-glucosylation sites of EGF16-20 of Notch (Figure 4C) were reported to rescue the severe Notch loss-of-function phenotypes induced by overexpression of human XXYLT1 in fly wings (Lee et al. 2013). Similar genetic interaction studies show that S/A mutations in Notch EGF16-20 rescue the wing vein thickening phenotype induced by overexpression of XxyltHA (Figure 4D–G). These data indicate that Xxylt overexpression induces wing vein thickening by adding xylose residues to one or more EGF repeats in the EGF16-20 region, likely EGF16 and potentially EGF18 based on our mass spectrometry data shown in Figure 1 and previous reports (Lee et al. 2013; Harvey et al. 2016).
Fig. 4.
Overexpression of Xxylt causes a mild wing vein thickening phenotype by adding xylose to one or more EGF repeats in the Notch EGF16-20 region. (A) Control y w; nubbin-GAL4 (nub-GAL4) wings show normal wing veins. (B) Wing-specific overexpression of XxyltHA by nub-GAL4 shows mild wing vein thickening (red asterisks). (C) Schematic of the EGF repeats of wild-type and mutant Notch genomic transgenes. Blue boxes show EGF repeats with a consensus O-glucosylation site; orange boxes denote EGF repeats with a serine-to-alanine mutation in the O-glucosylation site, which prevents the addition of O-glucose and therefore both xylose residues. (D) N¯/Y; PBac{Ngt-wt}VK22/+; nub-GAL4 males exhibit no wing vein thickening. (E) N¯/Y; PBac{Ngt-wt}VK22/+; nub-GAL4 UAS-XxyltHA (nub>XxyltHA) males show mild thickening of wing veins (red asterisks). (F) N¯/Y; PBac{Ngt-16_20}VK22/+; nub-GAL4 males show a wing vein loss phenotype (red arrowhead). (G) N¯/Y; PBac{Ngt-16_20}VK22/+; nub>XxyltHA males do not show a wing vein thickening phenotype. In most cases (89%), the wing vein loss phenotype is not rescued by Xxylt overexpression (red arrowhead).
Xylosylation of the Notch receptor modulates Notch signaling in a dosage-sensitive manner
As mentioned above, a partial rescue of N55e11/+ phenotypes was observed upon loss of both copies of Xxylt, but loss of one copy of Xxylt showed no rescue. Moreover, homozygosity for the shamse01256 allele, which is a piggyBac transposon insertion in the locus, was previously shown to suppress the Notch haploinsufficient phenotypes in the wing (Lee et al. 2013). However, it is not known whether Notch haploinsufficient phenotypes are sensitive to the gene dosage of shams in the presence or absence of Xxylt. To address this question, we compared the penetrance/rescue of the Notch haploinsufficient phenotypes in different genetic backgrounds with loss of one or both copies of fly xylosyltransferases individually or together (i.e., XxyltΔcds/+, XxyltΔcds/Δcds, shamsΔ34/+, XxyltΔcds/Δcds; shamsΔ34/+ and shamsΔ34/Δ34). Quantification of the penetrance of wing margin loss (Figure 5A and B) and wing vein thickening (Figure 5C and D) phenotypes in the animals harboring different gene dosages of xylosyltransferases at 25°C and 30°C revealed a dosage-sensitive rescue of both phenotypes by loss of xylosylation. A comparable penetrance/rescue of N55e11/+ phenotypes is observed in XxyltΔcds/Δcds and shamsΔ34/+ backgrounds, while XxyltΔcds/Δcds; shamsΔ34/+ background shows an additive degree of rescue of N55e11/+ phenotypes. In agreement with the temperature sensitivity of the shams loss-of-function phenotypes (Lee et al. 2013), loss of xylosyltransferases appears to better rescue the N55e11/+ phenotypes at a higher temperature (Figure 5B and D as compared to A and C). These observations suggest that Drosophila Notch signaling is sensitive to the level of Notch xylosylation, especially when the level of Notch is limiting.
Fig. 5.
Xylosylation of the Notch receptor modulates Notch signaling in a dosage-sensitive manner. (A–D) Graphs show the penetrance of wing margin loss (A and B) and wing vein thickening (C and D) phenotypes of Notch heterozygous animals upon loss of the indicated xylosyltransferases at 25°C (A and C) and 30°C (B and D). Error bars indicate standard deviations of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The Delta haploinsufficient phenotype in the wing is partially suppressed by simultaneous loss of Xxylt
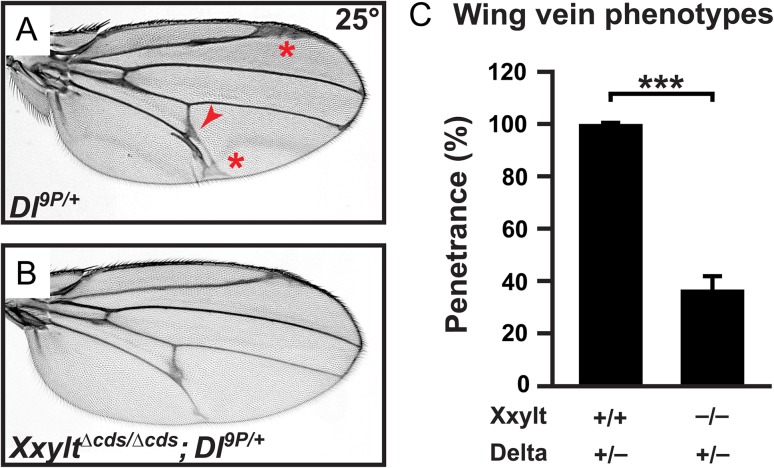
We recently reported that loss of shams specifically enhances Delta-mediated Notch signaling (Lee et al. 2017). In order to examine whether loss of Xxylt also affects Delta-mediated Notch signaling, we performed genetic interaction studies between Xxylt and Delta. Loss of one copy of Delta shows fully penetrant wing vein thickening and extra wing vein material phenotypes (Figure 6A, C), as reported previously (Vassin and Campos-Ortega 1987). Simultaneous loss of both copies of Xxylt partially rescues these phenotypes (Figure 6B, C). These data indicate that loss of the second xylose from xylose–xylose–glucose-O trisaccharides on Notch EGF repeats also contributes to modulation of Delta-mediated Notch signaling in shams mutants.
Fig. 6.
The Delta haploinsufficient phenotype in the wing is partially suppressed by simultaneous loss of Xxylt. All animals were raised at 25°C. (A) Adult wing from a Dl9P/+ animal. Note the wing vein thickening (red asterisks) and formation of extra wing vein material (red arrowhead). (B) XxyltΔcds/Δcds; Dl9P/+ adult wings show a partial suppression of the wing vein phenotypes present in Dl9P/+ mutants. (C) Graph showing the penetrance of Dl9P/+ mutant phenotypes in indicated genotypes. Error bar indicate standard deviation of three independent experiments. ***P < 0.001.
Loss of Xxylt enhances the trans-activation of Notch by ectopic Delta
The Notch receptor at the surface of the signal-receiving cells is trans-activated by the ligands expressed on the signal-sending cells (Bray 2006; Fortini 2009). In contrast, binding of Notch with ligands from the same cell leads to cis-inhibition of the signaling (Doherty et al. 1996; de Celis and Bray 1997; Micchelli et al. 1997; Jacobsen et al. 1998). A balance between trans-activation and cis-inhibition of Notch by ligands ensures optimal Notch singling in some contexts (Sprinzak et al. 2010; del Alamo et al. 2011). Our recent work showed that loss of the first xylose from O-glucosylated EGF repeats of Notch (in shamsΔ34/Δ34 mutants) enhances the trans-activation of Notch by ectopic Delta without affecting the cis-inhibition of Notch by ligands (Lee et al. 2017). Considering the partial rescue of the Delta (Dl) haploinsufficient phenotypes in XxyltΔcds/Δcds; Dl9P/+ flies, we aimed to examine the effects of loss of Xxylt on trans-activation and cis-inhibition of Notch by ectopic Delta. We used the GAL4-UAS system (Brand and Perrimon 1993) to overexpress Delta along the anterior–posterior boundary of the developing third instar wing imaginal discs.
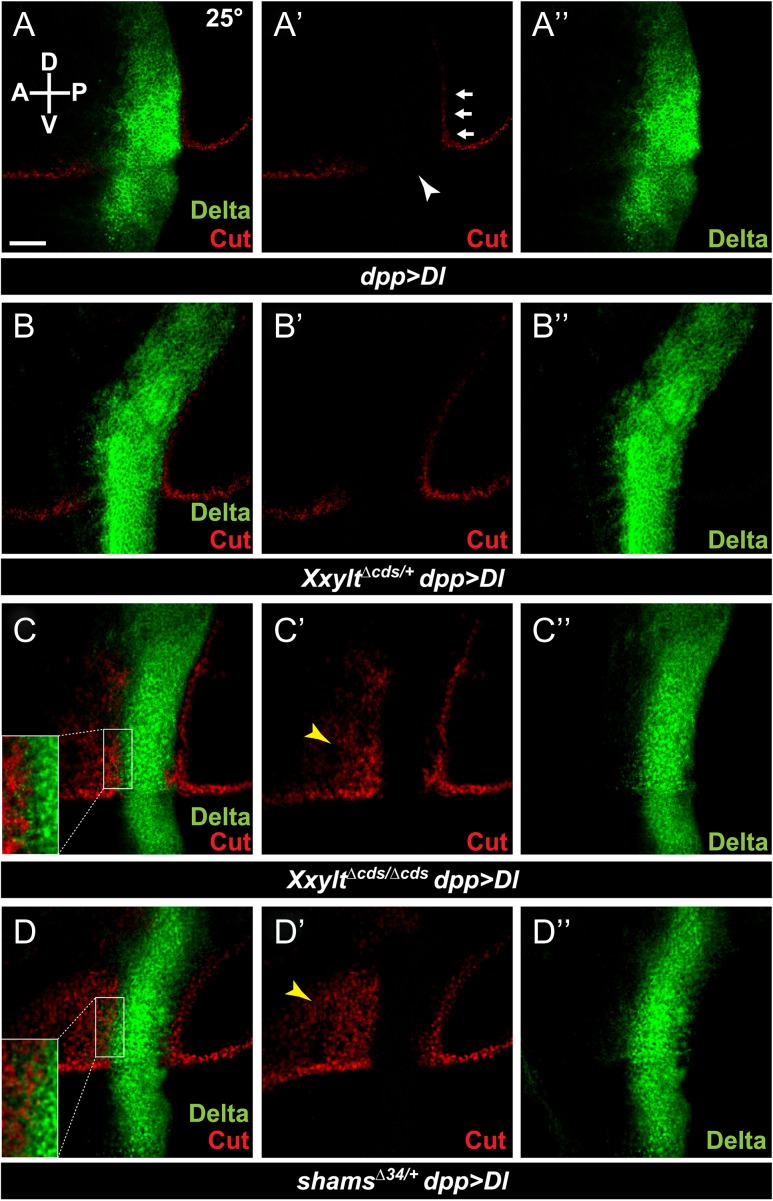
Third instar wing imaginal discs from genetic control animals (dpp-GAL4) show proper expression of the Notch downstream target Cut along the dorsal–ventral boundary, which generates the adult wing margin (Supplementary data, Figure S4). At 25°C, overexpression of Delta within the Dpp expression domain (dpp > Dl) results in loss of the Cut-positive cells at the dorsal–ventral boundary within the Dpp expression domain due to cis-inhibition (Figure 7A-A”, arrowhead in A’; n = 12). It also induces ectopic Cut-positive cells flanking the Dpp expression domain in the dorsal-posterior quadrant due to trans-activation (Figure 7A’, arrows; n = 12). Removing one copy of Xxylt does not affect the expression pattern of Cut in imaginal discs of the dpp > Dl animals (Figure 7B-B”, n = 11). However, loss of both copies of Xxylt results in ectopic activation of Cut in a significant portion of the dorsal-anterior quadrant (Figure 7C-C”, yellow arrowhead; n = 14). This suggests that loss of Xxylt enhances the trans-activation of Notch by Delta. For the most part, loss of Xxylt does not affect the loss of Cut within the Dpp expression domain, suggesting no effect on the cis-inhibition of Notch by Delta. At the border of the Dpp-expression domain, some cells seem to co-express Cut with Delta (Figure 7C-C”, inset in C). Of note, this phenomenon is only seen in cells that express low levels of Delta. This is compatible with a scenario in which loss of xylose promotes the trans-activation of Notch by neighboring cells expressing high levels of Delta, while the low levels of Delta expressed in the same cell are not sufficient to cis-inhibit Notch.
Fig. 7.
Loss of Xxylt enhances the activation of Notch by ectopic Delta. (A–D) Third instar wing imaginal discs from larvae raised at 25°C. Staining with α-Cut and α-Delta antibodies are shown in red and green, respectively. Anterior is to the left, and dorsal is up. Scale bar in (A) is 50 μm and applies to all panels. (A-A”) Expression of Cut upon Delta overexpression using dpp-GAL4 (dpp > Dl) in a wild-type background. Arrows and the arrowhead indicate trans-activation and cis-inhibition, respectively. (B-B”) Loss of one copy of Xxylt does not alter the Cut expression pattern in a dpp > Dl wing disc. (C-C”) In an XxyltΔcds/Δcds background, dpp > Dl induces ectopic activation of Cut anterior to the Dpp domain (yellow arrowhead). (D-D”) In a shams heterozygous background, dpp > Dl results in ectopic activation of cut comparable to dpp > Dl in an XxyltΔcds/Δcds background (yellow arrowhead). Note that in both C and D, some cells at the anterior border of the dpp expression domain co-express Cut with low levels of Delta, likely because the Delta level in these cells is not sufficient to cis-inhibit Notch (insets).
We also examined the effect of one copy loss of shams on trans-activation of Notch by dpp > Dl. In shamsΔ34/+ animals, a significant ectopic activation of Cut by dpp > Dl is observed in dorsal-anterior quadrant of the wing imaginal discs (Figure 7D-D”, yellow arrowhead; n = 13). A similar overlap between the expression of Cut and low levels of Delta is seen in some cells (Figure 7>D-D”, inset in D). Of note, ectopic activation of Cut by dpp > Dl is comparable in shamsΔ34/+ and XxyltΔcds/Δcds backgrounds (Figure 7C’ and D’), but is much weaker than the strong and broad ectopic Cut activation in the dorsal-anterior quadrant induced by dpp > Dl in shamsΔ34/Δ34 animals (Lee et al. 2017). These observations match the above-mentioned genetic interaction studies between Notch55e11 and xylosyltransferases, and suggest that decreasing Notch xylosylation enhances the trans-activation of Notch by Delta in a dosage-sensitive manner.
Discussion
Xylose–xylose–glucose-O glycans and their shorter versions (xylose–glucose-O and glucose-O) are found on EGF repeats with a CXSX(P/A)C consensus sequence (Hase et al. 1988; Acar et al. 2008; Fernandez-Valdivia et al. 2011; Rana et al. 2011; Lee et al. 2013; Haltom et al. 2014; Ramkumar et al. 2015; Harvey et al. 2016). Previously, mass spectrometric analysis of the Drosophila Notch showed that all 18 Notch EGF repeats with this consensus sequence are efficiently modified with O-glucose, but xylosylation is only observed in a subset of Notch EGF repeats, mostly in the EGF14-20 region (Acar et al. 2008; Lee et al. 2013; Harvey et al. 2016). Furthermore, loss of function analysis of the Drosophila enzymes that add the glucose and the first xylose to EGF repeats (Poglut1/Rumi and Gxylt/Shams), combined with in vivo structure–function analysis of Notch showed that glucose and xylose residues play opposite roles in Drosophila Notch signaling. Specifically, multiple O-glucose residues across the Notch extracellular domain function in redundant and additive manners to promote Notch signaling in all contexts studied so far (Leonardi et al. 2011). In contrast, xylose residues on O-glucosylated EGF16-20 of Notch inhibit Notch signaling in specific contexts (Lee et al. 2013, 2017). Here we report the generation and characterization of a null allele of the fly xyloside xylosyltransferase (Xxylt/CG11388). In agreement with the limited presence of the second xylose on Notch EGF repeats (Lee et al. 2013; Harvey et al. 2016), Xxylt mutant flies are viable and fertile and do not show any gross morphological changes suggestive of altered Notch signaling. These observations indicate that in wild-type flies, the second xylose on EGF repeats is dispensable for animal development and Notch signaling, and that the shams phenotypes are caused by the loss of the first xylose.
A number of Notch pathway mutants exhibit haploinsufficient phenotypes in animal models and human patients (Masek and Andersson 2017). Moreover, both decreased and increased Notch signaling have been causally linked to cancer and other human diseases (Allenspach et al. 2002; Siebel and Lendahl 2017). Accordingly, it is of interest and potential clinical importance to know whether mutations in a given gene can modify the Notch pathway activity in sensitized backgrounds with decreased or increased Notch signaling. Our genetic interaction studies indicate that loss of the second xylose residue in xylose–xylose–glucose-O trisaccharide enhances Notch signaling in contexts where the gene-dosages of Notch or Delta are altered. Moreover, in such sensitized backgrounds, even loss of one copy of shams (Gxylt) is sufficient to enhance Notch signaling. Therefore, when the level of Notch or Delta is decreased, loss or even a decrease in Notch xylosylation results in partial to near complete restoration of signaling. It has been reported that loss of one copy of Poglut1/Rumi partially suppresses the haploinsufficient phenotypes of the Notch ligand Jag1 in a mouse model of Alagille syndrome (Thakurdas et al. 2016). Therefore, the sensitivity of the Jag1+/− phenotypes to the gene dosage of Poglut1 might be explained by a concomitant decrease in the xylosylation of Notch and/or JAG1.
Although O-glucosylation occurs at high stoichiometry at all 18 Rumi/Poglut1 targets in Drosophila Notch, elongation with xylose is limited to a subset of Notch EGF repeats (Lee et al. 2013; Harvey et al. 2016). This might in part be explained by the presence of two Gxylt enzymes in mammals (GXYLT1 and GXYLT2) but only one in flies (Shams) (Sethi et al. 2010; Lee et al. 2013). Nevertheless, semi-quantitative analysis of mass spectrometry data on Drosophila Notch indicates that even on those EGF repeats that are modified by Shams, the second xylose is found at a very low stoichiometry (Harvey et al. 2016). Given that Xxylt overexpression in S2 cells can efficiently add a second xylose to EGF16 (Figure 1), the low stoichiometry of the second xylose on EGF16 in wild-type cells (Figure 1) might be due to low expression level of endogenous Xxylt. In contrast, our S2 cell overexpression experiments strongly suggest that the limited distribution of the second xylose on Drosophila Notch is not caused by low endogenous levels of the Drosophila Xxylt, but is due to site specificity of this enzyme. This might also explain why the Notch loss-of-function phenotype (i.e., wing vein thickening) upon overexpression of the fly Xxylt (Figure 4) is much weaker than the phenotypes observed upon overexpression of human XXYLT1 using the same GAL4 driver (Lee et al. 2013).
The Notch pathway was originally found to have an oncogenic effect in T-cell leukemia (Ellisen et al. 1991; Weng et al. 2004). However, later work showed that loss-of-function mutations in Notch pathway components can lead to other types of cancer (Agrawal et al. 2011; Klinakis et al. 2011; Wang et al. 2011; Siebel and Lendahl 2017). Notably, analysis of publically available cancer genomics data in cBioPortal (Cerami et al. 2012) revealed that the human XXYLT1 gene is frequently amplified in a number of cancer types, some of which are associated with loss of Notch signaling (Yu et al. 2015). Therefore, it is tempting to speculate that the role of xylose in mammalian Notch signaling is similar to its role in flies shown here and previously (Lee et al. 2013, 2017). It remains to be seen whether xylosylation of Notch negatively regulates mammalian Notch signaling, and whether Notch xylosyltransferases can be used as therapeutic targets for modulation of Notch signaling.
Materials and methods
Drosophila strains and genetics
The following strains were used in this study: (1) y w, (2) y w; L/CyO; D/TM3,Sb1, (3) w; nocSco/CyO; TM3, Sb1/TM6, (4) dpp-GAL4, (5) Dl9P/TM6, Tb1, (6) y w N54l9FRT19A/FM7, B1, (7) N55e11/FM7c, B1 (Bloomington Drosophila Stock Center), (8) shamsΔ34/TM6, Tb1 (Lee et al. 2013), (9) PBac{Ngt-wt}VK22, (10) PBac{Ngt-16_20}VK22 (Leonardi et al. 2011), (11) w; UAS-Dl (Wang and Struhl 2004), (12) nubbin-GAL4 (Georg Halder), (13) UASattB-CG11388-HA-VK20 (UAS-XxyltHA), (14) PBac{Xxyltgt-wt-V5}VK20, (15) XxyltΔcds/CyO, (16) XxyltΔcds/CyO; dpp-GAL4/TM6, Tb1, (17) dpp-GAL4 shamsΔ34/TM6, Tb1, (18) XxyltΔcds/CyO; UAS-Dl (this study).
All crosses were performed on standard media and incubated at the indicated temperatures. To examine the effect of Notch mutations on Xxylt overexpression in the wing, B+, Sb+ male progeny were scored from nubbin-GAL4 UASattB-CG11388-HA/TM3, Sb1 males crossed to y w N5419FRT19A/FM7, B1; PBac{Ngt-wt}VK22/+ or N54l9/FM7, B1; PBac{Ngt-16_20}VK22/+ females.
Biochemical characterization of CG11388
The complete open reading frames of CG11388 (sequence identical to NCBI NP_611871.2) and Shams (NP_001097911 with Y108F and I255V) were amplified from Drosophila cDNA and cloned in the HindIII/EcoRI sites of pIB (ThermoFisher). The open reading frames of human GXYLT1 and XXYL1 (Sethi et al. 2010, 2012) were cloned in the same vector. A Notch fragment containing amino acid 638 to 832 (numbering as in NP_476859) was cloned in AvaI digested pMTBip/V5His (ThermoFisher). The secreted Notch fragment contains EGF repeats 16 to 20 and a C-terminal V5-His6 tag.
Δshams S2 cells were generated using CRISPR/Cas9 vector pAc-sgRNA-Cas9 (gift from Ji-Long Liu; Addgene plasmid # 49330) (Bassett et al. 2014) with oligo sequence GAGACCACCACGATGTAAAG. Single clones were selected with puromycin in 96 well plates with un-transfected S2 cells as feeder cells using a co-transfected plasmid with the puromycin resistance gene. The clone used in this study contains a homozygous 8 bp deletion in shams, resulting in a frameshift after amino acid 47 of the protein.
Plasmids were transiently expressed in S2 cells (WT or Δshams) using Polyethylenimine (PEI, MW 40,000, Polysciences) transfection. S2 cells were grown in Xpress medium (Lonza) in suspension culture flasks at 24°C with 30 rpm shaking. Ten-milliliter cultures (in a T75 flask) were transfected with 20 μg plasmid/100 μg PEI in 2 mL of 20 mM HEPES buffer pH 7.4. Cells were transfected with 10 μg of Notch EGF16-20 combined with 5 μg of hGXYLT1 and hXXYL1 each or Shams and Xxylt each. Empty vector was used to reach 20 μg for the samples with only one or no xylosyltransferase. On the next day, cells were induced with 0.2 mM CuSO4 after a medium exchange. After 3 additional days of culture, the cell culture supernatant was collected, filtered (0.2 μm) and supplemented with 5 mL of 500 mM NaCl, 20 mM TRIS/HCl pH 7.5. Imidazole was added to a total concentration of 20 mM. Using 1 mL HisTrap HF columns (GE Healthcare) and a linear gradient to 500 mM imidazole in 7 mL, the His-tagged Notch fragment was purified. The peak fraction was concentrated using acetone precipitation and separated by SDS-PAGE. In-gel digestion using AspN and further analysis by mass spectrometry on a Waters ESI Q-TOF Ultima coupled to a nanoACQUITY was carried out essentially as described (Shcherbakova et al. 2017). Extracted ion chromatograms were generated of expected masses of peptides of EGF16, EGF18, EGF19 and EGF20 without glycosylation and the putative glycosylated forms. Fragmentation MS/MS analyses confirmed the identity of the peptides shown in Figure 1 (see Supplementary data, Figures S1–S3).
Generation of Xxylt overexpression and genomic rescue transgenes
To generate a V5-tagged genomic rescue transgene for Xxylt, the following primer pairs were used in an overlap-extension PCR program (Bryksin and Matsumura 2010) to insert a V5 tag after the first methionine of CG11388 and to flank the genomic region with AscI and NotI restriction enzymes (restriction sites and the sequences encoding the V5 tag are in uppercase):
5′-CG11388-AscI: ggtGGCGCGCCatggggtcgtcttcta
3′-CG11388-V5: GCCCAGCAGGGGGTTGGGGATGGGCTTGCCcatttcgaaatcagcggagga
5′-CG11388-V5: CCCAACCCCCTGCTGGGCCTGGACTCCACCgccaagaacagctttaagact
3′-CG11388-NotI: tcgGCGGCCGCgttaactacgtatgctcg
The PCR product was cloned into the pSC-B vector (Stratagene). AscI and NotI were used to transfer the CG11388-V5 genomic region from pSC-B to attB-P[acman]-ApR vector (Venken et al. 2006).
To generate an overexpression transgene for Xxylt, the following primers were used to amplify the CG11388 coding region (restriction sites are in uppercase):
5′-CG11388-BglII: tatccAGATCTgatttcgaaatggccaagaacagc
3′-CG11388-XbaI: ctataTCTAGAttatttattctggaattcgcgtattacagttg
BglII and XbaI were used to clone the CG11388 coding region in a modified version of pUASTattB (Bischof et al. 2007) which contains an HA-tag after the ATG.
After verifying the sequence, ϕC31-mediated transgenesis was used to integrate the transgenes into the VK20 locus on the third chromosome (Venken et al. 2006).
Generation of the XxyltΔcds allele using CRISPR/Cas9 genome engineering
CRISPR/Cas9 gene editing utilizing homology directed repair (HDR) was used to create a loss-of-function allele for Xxylt (CG11388) by replacing 87% of the gene region including the entire coding sequence (CDS) with yellow wing2+, a 2.9 kb swappable insertion cassette containing a dominant marker that expresses the yellow gene product within wings of Drosophila melanogaster (Li-Kroeger et al. 2018). Briefly, homology arms were PCR amplified from genomic DNA with Q5 polymerase (NEB) using the following primers (5′-3′). The BsaI recognition and restriction sites are in uppercase:
Upstream Forward-ctctctCGTCTCtGACCagagcagcttgtcctgggagttgg
Upstream Reverse-acacacCGTCTCgATCCgcaacagtacgtcggcgtcaacg
Downstream Forward-tctctCGTCTCtTTCCtataggaatattaaattaattaaatgcagggtatttttatttgg
Downstream Reverse-acacacCGTCTCgTATAgagaccagtttgccggcatcc
The amplified products were run on an agarose gel and purified with the QIAquick Gel Extraction Kit (Qiagen). The homology arms, the pBH donor vector and the yellow wing2+ dominant marker cassette vector (Li-Kroeger et al. 2018) were combined by Golden Gate assembly (Engler et al. 2008) using established methods (Housden and Perrimon 2016). The resulting reaction products were transformed into Stbl2 Chemically Competent Cells (ThermoFisher), and plated overnight under kanamycin selection. Colonies were cultured for 24 h at 30°C and DNA was prepared by miniprep. The entire homology arm sequence and the adjacent yellow wing2+ marker were verified prior to injection.
Two sgRNA constructs, one upstream (designated 5′, sense sequence CGCCGACGTACTGTTGCAAT) and one downstream (designated 3′, sense sequence AGAATAAATAAAATTGTTAT) of the CDS were cloned into pCFD3 as described previously (Port et al. 2014). Sense and antisense oligos containing the 20 bp guide target sequences were annealed and phosphorylated with T4 Polynucleotide Kinase (NEB), then inserted between BbsI sites in the pCFD3 vector. Ligation products were transformed into TOP10 Competent Cells (ThermoFisher), and plated overnight. Colonies were cultured, DNA prepared by miniprep, and sequences verified prior to injection. A mix of 25 ng/μL of each sgRNA and 150 ng/μL donor DNA was injected into isogenized fly embryos of the genotype y1M{nos-Cas9.P}ZH-2A w*; isogenic (II) by GenetiVision (Houston, USA). Injected founders were then crossed to y¯ flies and resultant offspring screened for the presence of yellow+ wing. For this experiment, 450 embryos were injected producing 16 fertile crosses. One vial of the 16 produced two offspring with yellow+ wings, from which balanced lines were established. PCR analysis followed by Sanger sequencing was performed on genomic DNA of the positive strains to confirm that the coding sequence was correctly replaced with the donor construct.
Dissections, staining, image acquisition and processing
Standard methods were used for dissection and staining. Antibodies used were mouse α-Cut 1:500 (2B10; Developmental Studies Hybridoma Bank) (Blochlinger et al. 1990), guinea pig α-Delta 1:3000 (Huppert et al. 1997), goat α-mouse-Cy3 1:500, donkey α-guinea pig-Cy3 1:500 (Jackson ImmunoResearch Laboratories). Zeiss Axioscope-A1 and Nikon Ci-L upright microscopes were used to image adult wings. Confocal images were obtained using a Leica TCS-SP5 microscope and processed with Amira5.2.2. Images were processed with Adobe Photoshop CS5; Figures were assembled in Adobe Illustrator CS5.
Statistics
Statistical analysis was performed using the Prism software (GraphPad version 6.0, San Diego, CA, USA). One-way and two-way ANOVA were used, followed by post-hoc tests Dunnett and Bonferroni tests, respectively. To examine the Mendelian ratio, Chi-square test was used. Statistical significance level was ascribed as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Supplementary Material
Abbreviations
- EGF
epidermal growth factor-like
- GXYLT
glucoside xylosyltransferase
- Poglut1
protein O-glucosyltransferase
- XXYLT
xyloside xylosyltransferase
Funding
We acknowledge support from the NIH/NIGMS (R01GM084135 to H.J.N.), Mizutani Foundation for Glycoscience (grant #110071 to H.J.N.) and DFG (Deutsche Forschungsgemeinschaft; grant BA4091/4-1 to H.B.).
Acknowledgements
We thank Hugo Bellen for sharing an unpublished method with us and for his support; Yi-Dong Li for generating the CG11388 rescue transgene; Bloomington Drosophila Stock Center (NIH P40OD018537), the Developmental Studies Hybridoma Bank, Georg Halder, Marc Muskavitch and Gary Struhl for reagents; the Microscopy Core of the BCM IDDRC (1U54HD083092; the Eunice Kennedy Shriver NICHD), and the BCM Integrated Microscopy Core (supported by NCI-CA125123, NIDDK-56338-13/15, CPRIT-RP150578 and John S. Dunn Gulf Coast Consortium for Chemical Genomics).
Conflict of interest statement
None declared.
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. 2008. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 132:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333:1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allenspach EJ, Maillard I, Aster JC, Pear WS. 2002. Notch signaling in cancer. Cancer Biol Ther. 1:466–476. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MA. 2010. Notch: The past, the present, and the future. Curr Top Dev Biol. 92:1–29. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. 2014. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open. 3:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jan LY, Jan YN. 1990. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4:1322–1331. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Bray SJ. 2006. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. [DOI] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques. 48:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Bray S. 1997. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 124:3241–3251. [DOI] [PubMed] [Google Scholar]
- del Alamo D, Rouault H, Schweisguth F. 2011. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 21:R40–R47. [DOI] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. 1996. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 10:421–434. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 66:649–661. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H. 2011. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 138:1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. 2009. Notch signaling: The core pathway and its posttranslational regulation. Dev Cell. 16:633–647. [DOI] [PubMed] [Google Scholar]
- Haltom AR, Jafar-Nejad H. 2015. The multiple roles of epidermal growth factor repeat O-glycans in animal development. Glycobiology. 25:1027–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltom AR, Lee TV, Harvey BM, Leonardi J, Chen YJ, Hong Y, Haltiwanger RS, Jafar-Nejad H. 2014. The protein O-glucosyltransferase Rumi modifies eyes shut to promote rhabdomere separation in Drosophila. PLoS Genet. 10:e1004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey BM, Rana NA, Moss H, Leonardi J, Jafar-Nejad H, Haltiwanger RS. 2016. Mapping sites of O-glycosylation and fringe elongation on Drosophila Notch. J Biol Chem. 291:16348–16360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase S, Kawabata S, Nishimura H, Takeya H, Sueyoshi T, Miyata T, Iwanaga S, Takao T, Shimonishi Y, Ikenaka T. 1988. A new trisaccharide sugar chain linked to a serine residue in bovine blood coagulation factors VII and IX. J Biochem (Tokyo). 104:867–868. [DOI] [PubMed] [Google Scholar]
- Housden BE, Perrimon N. 2016. Design and generation of donor constructs for genome engineering in Drosophila. Cold Spring Harb Protoc. 2016, 10.1101/pdbprot090787. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Jacobsen TL, Muskavitch MA. 1997. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 124:3283–3291. [DOI] [PubMed] [Google Scholar]
- Jacobsen TL, Brennan K, Arias AM, Muskavitch MA. 1998. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 125:4531–4540. [DOI] [PubMed] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E et al. 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Pandey A, Jafar-Nejad H. 2017. Xylosylation of the Notch receptor preserves the balance between its activation by trans-Delta and inhibition by cis-ligands in Drosophila. PLoS Genet. 13:e1006723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Sethi MK, Leonardi J, Rana NA, Buettner FF, Haltiwanger RS, Bakker H, Jafar-Nejad H. 2013. Negative regulation of notch signaling by xylose. PLoS Genet. 9:e1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Fernandez-Valdivia R, Li YD, Simcox AA, Jafar-Nejad H. 2011. Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development. 138:3569–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Kroeger D, Kanca O, Lee PT, Cowan S, Lee MT, Jaiswal M, Salazar JL, He Y, Zuo Z, Bellen HJ. 2018. An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. Elife. 7:e38709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek J, Andersson ER. 2017. The developmental biology of genetic Notch disorders. Development. 144:1743–1763. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 124:1485–1495. [DOI] [PubMed] [Google Scholar]
- Penton AL, Leonard LD, Spinner NB. 2012. Notch signaling in human development and disease. Semin Cell Dev Biol. 23:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto CN, Schweisguth F, Bardin AJ. 2011. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 138:4585–4595. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 111:E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkumar N, Harvey BM, Lee JD, Alcorn HL, Silva-Gagliardi NF, McGlade CJ, Bestor TH, Wijnholds J, Haltiwanger RS, Anderson KV. 2015. Protein O-glucosyltransferase 1 (POGLUT1) promotes mouse gastrulation through modification of the apical polarity protein CRUMBS2. PLoS Genet. 11:e1005551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS. 2011. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 286:31623–31637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. 2012. Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J Biol Chem. 287:2739–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. 2010. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J Biol Chem. 285:1582–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova A, Tiemann B, Buettner FF, Bakker H. 2017. Distinct C-mannosylation of netrin receptor thrombospondin type 1 repeats by mammalian DPY19L1 and DPY19L3. Proc Natl Acad Sci USA. 114:2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C, Lendahl U. 2017. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 97:1235–1294. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. 2010. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 465:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Okajima T. 2010. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 92:131–164. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Haltiwanger RS. 2014. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 453:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C, Campese AF, Bellavia D, Felli MP, Vacca A, Gulino A, Screpanti I. 2008. Notch signaling and diseases: An evolutionary journey from a simple beginning to complex outcomes. Biochim Biophys Acta. 1782:489–497. [DOI] [PubMed] [Google Scholar]
- Thakurdas SM, Lopez MF, Kakuda S, Fernandez-Valdivia R, Zarrin-Khameh N, Haltiwanger RS, Jafar-Nejad H. 2016. Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi). Hepatology. 63:550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin H, Campos-Ortega JA. 1987. Genetic analysis of delta, a neurogenic gene of Drosophila melanogaster. Genetics. 116:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. 2006. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 314:1747–1751. [DOI] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L et al. 2011. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 108:17761–17766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Struhl G. 2004. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 131:5367–5380. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271. [DOI] [PubMed] [Google Scholar]
- Yu H, Takeuchi M, LeBarron J, Kantharia J, London E, Bakker H, Haltiwanger RS, Li H, Takeuchi H. 2015. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat Chem Biol. 11:847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.