Abstract
Background
Porcine circovirus type 2 (PCV2) is a major emerging virus of porcine circovirus-associated disease (PCVAD), which has brought huge economic losses to the global pig industry. Pigs are well known as the natural reservoir of PCV2. Recently, many researchers have revealed PCV2 could infect many other mammals like mice, calves, minks, dogs and goats. In 2018, our laboratory has admitted six cases of raccoon dogs from Qinhuangdao city of China, which were characterized by inappetence, lethargy, depression, abortion, and sterility.
Results
At last, six raccoon dog-origin PCV2 strains were isolated in this study. Pairwise-sequence comparisons demonstrated that the six raccoon dog-origin PCV2 strains shared a nucleotide similarity of 92.1–99.8% among 40 PCV2 representative strains. Phylogenetic analysis indicated these PCV2 isolates belonged to Chinese epidemic genotypes PCV2b and PCV2d. And aborted or sterile symptom was significantly associated with PCV2 infection in raccoon dogs by the chi-square test (χ2 = 87.3, p < 0.001). The retrospective study revealed that raccoon dog-origin PCV2 strains shared 100% sequence similarity with the PCV2 stains isolated from pig farms around these raccoon dog farms, respectively.
Conclusion
In this study, the first supported evidence of PCV2 prevalence in raccoon dog farms of China was documented. PCV2 may be one of the most significant causative agents resulting in the reproductive failure of farmed raccoon dogs, implying that PCV2 could transmit from pigs to raccoon dogs. That indicated that PCV2 cross-species transmission will be a serious threat to China’s fur animal farming industry.
Keywords: Raccoon dogs, Porcine circovirus type 2, Phylogenetic analysis
Background
After 2010, China has increased to become one of the biggest fur animal farming countries. But poor feeding condition is still the serious adverse factor of restricting China’s fur animal farming industry [1]. Furthermore, the small-scale family breeding model is still the dominant breeding model of fur animals in China. Many cross-species diseases from other breeding animals could easily attack fur animals, especially those from surrounding pig farms [2–4]. Porcine circoviruses (PCV) belong to the Circovirus genus within the Circoviridae family, which are small non-enveloped single-strand circular DNA viruses. They have been firstly identified in 1982, and contain three types, including PCV1, PCV2 and PCV3 [5, 6].
PCV1 is known to have no association with clinical diseases [7]. PCV3 has recently been first identified in the USA in pigs with characteristics of cardiac and multisystemic inflammation with metagenomic sequencing [8, 9]. PCV2 has been recognized as one of the main agents responsible for PCV-associated disease (PCVAD) which has caused huge economic losses to the global pig industry [10, 11]. PCV2 has been currently classified into six major genotypes (PCV2a, PCV2b, PCV2c, PCV2d, PCV2e and PCV2f) [12–14], of which PCV2b and PCV2d genotypes are the predominant strains in China [15–17]. The complete genome of PCV2 ranges from 1766 or 1768 bp and encodes at least 11 predicted open reading frames (ORFs) [18, 19]. To date, PCV2 was reported to be discovered in many other reservoirs such as mice, calves, minks, dogs, foxes and goats, besides pigs as its natural reservoirs [4, 20–26]. In this study six PCV2 isolates were identified from farmed raccoon dogs with the symptom of reproductive failure. These findings suggested the possibility of PCV2 cross-species transmission. And this is the first time to report the genetic analysis of raccoon dog-origin PCV2 isolates.
Methods
Samples treatment
In 2018, our laboratory has admitted six cases of raccoon dogs from Qinhuangdao, China with clinical symptoms of inappetence, lethargy, depression, abortion, and sterility. After collecting raccoon dog samples, bacterial culture was immediately carried out. The lymph nodes and spleens of all these cases were cultured onto both blood agar plates and tryptic soy agar plates at 37 °C for 24–48 h under the aerobic and anaerobic conditions respectively. In addition, all the above tissues were aseptically collected and homogenized by TissueLyser II (QIAGEN, Germany), then centrifuged at 8000 g for 10 min for further study.
Viral genome extract and detection
Special detecting primers were synthesized to detect the common viruses infected raccoon dogs such as canine parvovirus (CPV), canine adenovirus (CAV), canine distemper virus (CDV), Pseudorabies virus (PRV), Hepatitis E Virus (HEV) and PCV2, as described previously [15, 27–31]. Viral DNA or RNA was extracted from each samples using a TIANamp virus DNA/RNA extraction kit (Tiangen, Beijing) following the manufacturer’s instructions.
RT-PCR was carried out to detect CDV and HEV with the PrimeScript One Step RT-PCR kit (TaKaRa, Dalian) following the manufacturer’s instructions. RT-PCR conditions comprised 50 °C for 30 min, 94 °C for 2 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s, ended with a final extension at 72 °C for 10 min. PCR was carried out to detect CAV, CPV, PRV, and PCV2 with Ex Taq DNA Polymerase (TaKaRa, Dalian) following the manufacturer’s instructions. The PCR conditions comprised 95 °C for 5 min, 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, ended with a final step at 72 °C for 10 min. RT-PCR and PCR products were run on a 1% agarose gel and imaged under ultraviolet light.
Viral isolation and indirect immunofluorescence assay (IFA)
Porcine Kidney Epithelial (PK-15) cells were purchased from American Type Culture Collection (ATCC CCL-33), and cells between passages 21 to 25 were used for experiments. The tissue samples of raccoon dogs were treated and cultured on PK-15 cells for three serial passages. Then the cell lysates were infected on PK-15 cells for IFA assay to detect PCV2, as described previously [26]. Finally, the IFA results were observed using inverted fluorescent microscope (Olympus IX73). The PCV2 strain Hebei2 (GenBank no. MG1825) preserved in our laboratory was infected with PK-15 cells as a positive control, while mock-infected PK-15 cells as a negative control.
Viral genome sequencing
The PCV2 genome sequences were amplified using a pair of special primers, as described previously [32]. PCR was performed using the following conditions: at 95 °C for 5 min, followed by 33 cycles of denaturation at 95 °C for 1 min, annealing at 54 °C for 1 min, and extension at 72 °C for 2 min, and ended with a final step at 72 °C for 10 min. PCR products of PCV2 fragments were purified with Gel Extraction Kit (Tiangen, Beijing) and subcloned into the pMD18-T vector (TaKaRa, Dalian). The recombinant plasmids were identified by enzyme digestion and sequenced using Sanger sequencing (Augct DNA-Syn Biotechnology, Beijing).
Multiple sequences comparison and phylogenetic analyses
The complete sequences of all PCV2 stains were assembled using the SeqMan v7.1.0 program (Lasergene, DNAStar, USA), and sequence homology analysis was performed using the MegAlign v7.1.0 program (Lasergene, DNAStar, USA). Phylogenetic tree was constructed using the neighbour-joining method of MEGA 7.0 software [33]. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history, and the neighbour-joining method was used to infer the evolutionary history [34, 35].
The retrospective and tracing investigations
Three hundred serum samples of raccoon dogs were collected from eight raccoon dog farms in Qinhuangdao, which included 150 samples with no symptoms and 150 samples with symptoms of abortion or sterility. A real-time SYBR green PCR was performed to quantify PCV2 in these serum samples, as described previously [36]. Cycling conditions were 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 59 °C for 10 s, and extension at 72 °C for 30 s. Fluorescence normalization and data analysis were performed using the ABI Prism 7500 (Applied Biosystems Inc., USA). A chi-square test was used to evaluate the association between aborted or sterile symptom and PCV2 infection in raccoon dogs.
For tracing the origin of the PCV2 stains isolated from raccoon dogs, 210 pig serum samples from 21 pig farms (10 samples per farm) around the six raccoon dog farms were collected and detected for PCV2 by PCR assay. One positive sample from every PCV2-positive farm was selected to amplify the PCV2 complete genome as above. PCR fragments of PCV2 were purified, cloned, sequenced, and assembled as above. Multiple sequence alignments were generated using the MegAlign v7.1.0 program. Sequence similarity analysis demonstrated the sequence distances between pig-origin PCV2 strains and raccoon dog-origin PCV2 strains.
Results
Sample detecting
Bacterial culture revealed no common bacterium was observed on the plates cultivated with tissue samples of these raccoon dogs. The (RT)-PCR results proven all samples negative for CDV, CAV, CPV, HEV, and PRV using specific detecting primers to these viruses, but all samples positive for PCV2.
Virus isolation and identification
The tissue homogenates of raccoon dogs were subsequently inoculated on PK-15 cells. After three blind passages, no cytopathic effect (CPE) was observed. However, PCR detection showed all these passage cultures were PCV2-positive. Further IFA experiment demonstrated all PK-15 cells infected with these viral cultures could cross-react with anti-PCV2 Mab, implying that PCV2 strains were isolated from these raccoon dogs. At last, the complete sequences of raccoon dog-origin PCV2 isolates were designated as Rac-hb1~Rac-hb6 respectively.
Genome sequencing of raccoon dog-origin PCV2 strains
The fragments of PCV2 complete genome sequences were amplified as described previously [17]. After sequence assembling, the complete genome sequences of strains Rac-hb1~Rac-hb6 were obtained with all 1767 nt in length, and deposited into the GenBank database under accession numbers MH373555~MH373560 respectively.
Multiple sequences comparison and phylogenetic analyses
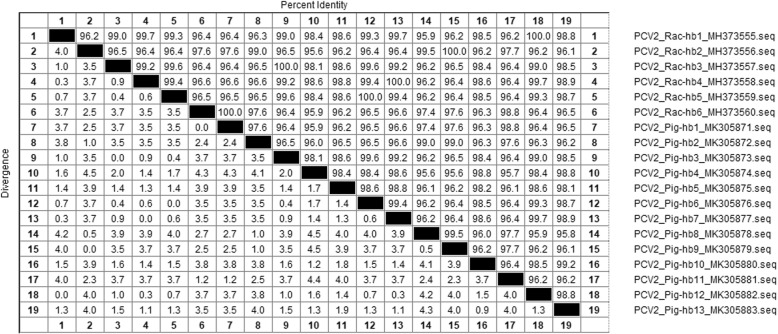
The complete genome sequences of six raccoon dog-origin PCV2 strains showed a high similarity varied from 92.1 to 99.8% compared with those of 40 PCV2 representative strains (Table 1) based on multiple sequences comparison analysis. Considering the phylogenetic analysis of PCV2 complete genome sequences, the six raccoon dog-origin PCV2 strains belong to the epidemic PCV2 strains of China (PCV2b and PCV2d) [15, 17, 37, 38]. Thereinto, strains Rac-hb1, Rac-hb3, Rac-hb4 and Rac-hb5 belong to PCV2b, and strains Rac-hb2 and Rac-hb6 belong to PCV2d (Fig. 1).
Table 1.
PCV2 representative strains used in this study
| Strain name | Accession no. | Source | Genotype | Strain name | Accession no. | Source | Genotype |
|---|---|---|---|---|---|---|---|
| Canada | AF055392 | pig | PCV2a | DK1990PMWSfree | EU148505 | pig | PCV2c |
| GX0841a | GQ359003 | pig | PCV2a | MiSD-2 | KP282146 | mink | PCV2d |
| MN500 | GQ404853 | Homo | PCV2a | TJ | AY181946 | pig | PCV2d |
| V2177/00 | KX352445 | dog | PCV2a | BJ0901b | GU001710 | pig | PCV2d |
| USA | AY699793 | pig | PCV2a | German | AY713470 | wild boar | PCV2d |
| SPA3 | AF201310 | pig | PCV2a | GXWM | EF675241 | pig | PCV2d |
| CL | HM038033 | pig | PCV2a | Buffalo2 | KM116514 | buffalo | PCV2d |
| HLJ1502 | KY940535 | pig | PCV2a | BDH | HM038017 | pig | PCV2d |
| France | AF055394 | pig | PCV2b | SD | AY181947 | pig | PCV2d |
| Ha08 | FJ804417 | calf | PCV2b | Goat2014–4 | KX894318 | goat | PCV2d |
| GX0841b | GQ359004 | pig | PCV2b | HZ0301 | AY510375 | pig | PCV2d |
| MN614 | GQ404852 | Homo | PCV2b | 09HeN | HQ395033 | pig | PCV2d |
| SD6 | DQ218421 | pig | PCV2b | GX0601 | EF524532 | pig | PCV2e |
| Ha10 | HQ231328 | calf | PCV2b | BJ0901a | GU001709 | pig | PCV2e |
| Buffalo1 | KM116513 | buffalo | PCV2b | XJ0901 | GU370063 | pig | PCV2e |
| JF | HM038022 | pig | PCV2b | Buffalo3 | KM116515 | buffalo | PCV2e |
| CC1 | JQ955679 | pig | PCV2b | ML-4 | LC004739 | pig | PCV2f |
| MiSD-1 | KP282147 | mink | PCV2b | MZ-6 | LC004751 | pig | PCV2f |
| DK1980PMWSfree | EU148503 | pig | PCV2c | Papuan08 | KT369069 | pig | PCV2f |
| DK1987PMWSfree | EU148504 | pig | PCV2c | Hebei2 | MG182436 | pig | PCV2b |
Fig. 1.
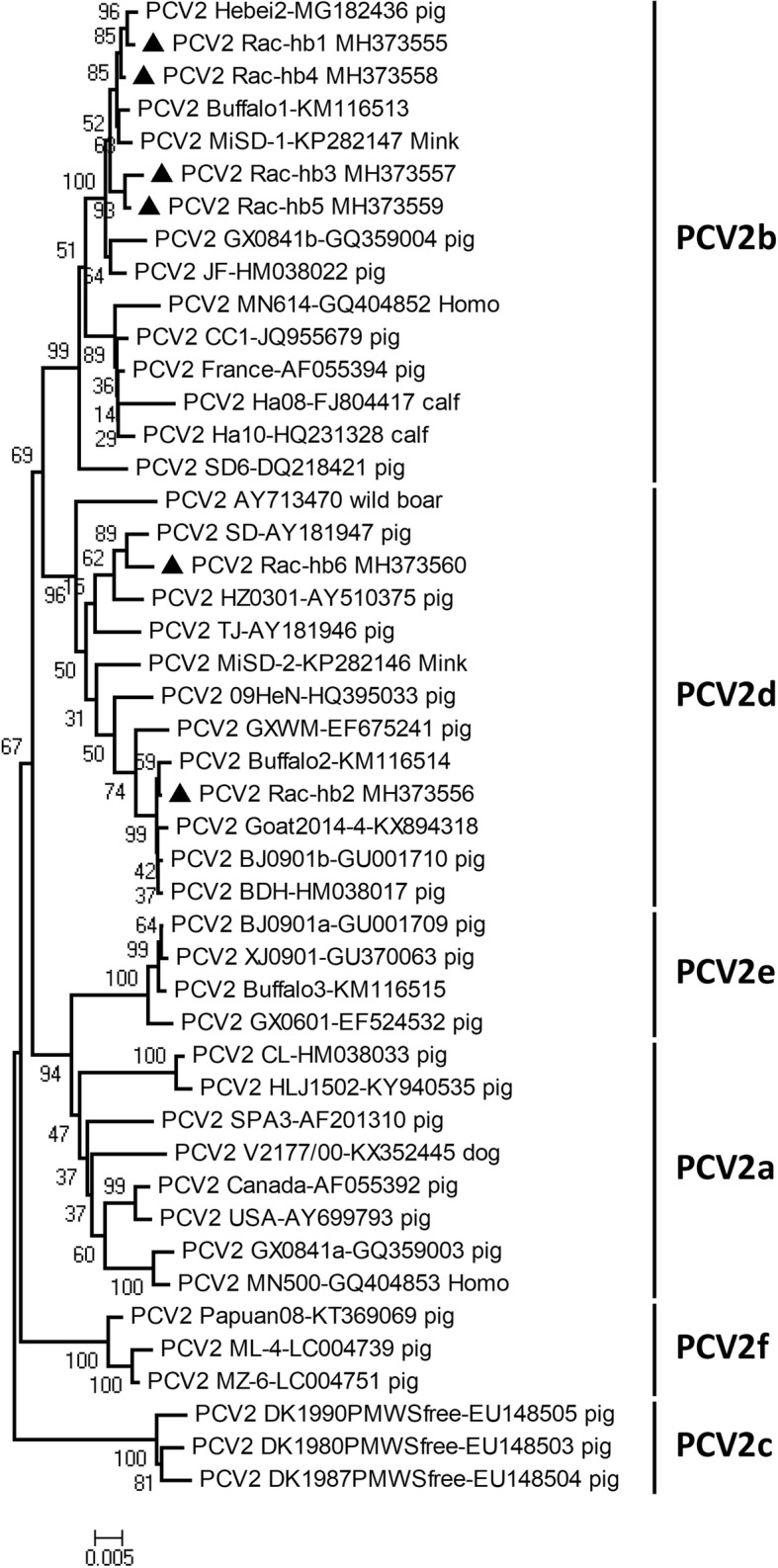
Phylogenetic tree of the complete genome sequences of PCV2. The tree was constructed using MEGA 7.0 software and analyzed by neighbour-joining method using 1000 replicates on bootstrap analysis. Six raccoon dog-origin PCV2 strains isolated in this study were labeled with black triangles (▲)
The retrospective and tracing investigations of raccoon dog-origin PCV2 strains
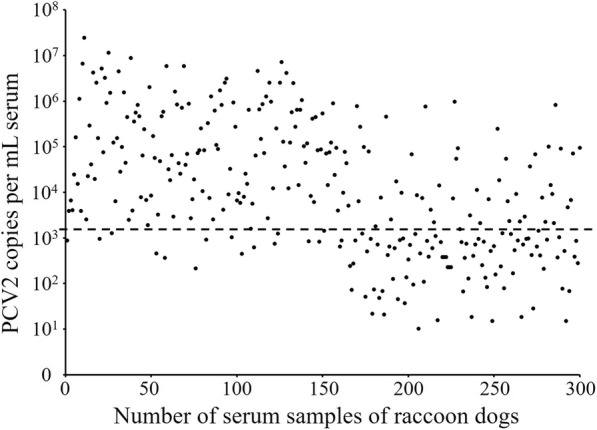
The retrospective epidemiological study on PCV2-infected raccoon dogs was performed to detect 300 serum samples collected from the raccoon dog farms in Qinhuangdao using real-time PCR. The results demonstrated the positive rate of PCV2 was 63.3% (190/300), including 89.3% (134/150) in diseased raccoon dogs with symptoms of abortion or sterility and 37.3% (56/150) in raccoon dogs with no symptoms (Fig. 2). There was a higher positive rate of PCV2 in the aborted or sterile raccoon dogs, while a lower positive rate of PCV2 in healthy raccoon dogs. The Chi-square test of significant difference (χ2 = 87.3, p < 0.001) revealed that aborted or sterile symptom was significantly associated with PCV2 infection in raccoon dogs.
Fig. 2.
Nucleotide sequence similarity (%) of the complete genome sequences between raccoon dog-origin PCV2 strains and pig-origin PCV2 strains
The tracing study revealed that 13 genome sequences of pig-origin PCV2 isolates were obtained and designated as Pig-hb1~Pig-hb13 (GenBank accession no. MK305871~MK305883). Sequence similarity analysis showed that the complete genomes of six pig-origin PCV2 strains (Pig-hb12, Pig-hb9, Pig-hb3, Pig-hb7, Pig-hb6, and Pig-hb1) were shared 100% nucleotide identity with those of strains Rac-hb1~Rac-hb6, respectively (Fig. 3).
Fig. 3.
PCV2 copies per mL serum of raccoon dogs followed by real-time PCR on the quantification of PCV2 viral DNA. The lower detection limit (2.2 × 103 copies) is generated based on the standard curve (the broken line)
Discussion
PCV2 is the key agents responsible for PCVAD in pigs, including several clinical syndromes such as postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephropathy syndrome (PDNS), porcine respiratory disease complex (PRDC) and PCV2-associated reproductive failure [39, 40]. Recent studies have suggested that PCV2 was detected in non-porcine species with no symptoms or mild diarrhea [4, 21, 24, 25, 41]. Surprisingly, we firstly detected and isolated PCV2 from raccoon dogs with reproductive failure. Phylogenic analysis indicated that these raccoon dog-origin PCV2 isolates were confirmed to belong to the genotypes PCV2b and PCV2d, which were the dominating PCV2 genotypes in current China [15, 17, 37, 38].
The retrospective investigation has revealed that aborted or sterile symptom was significantly associated with PCV2 infection in raccoon dogs. This indicated that PCV2 may be one of the most significant causative agents resulting in the reproductive failure of farmed raccoon dogs. However, further animal experiment needs to be validate the effect on PCV2 infection in raccoon dogs during oestrus.
The tracing study has demonstrated these raccoon dog-origin PCV2 strains originates from pig farms around raccoon dog farms. Previous studies have also revealed that PCV2 could be detected and isolated from minks and foxes in China, implying that PCV2 becomes an increasing threat to China’s fur animal farming industry [4, 26]. And horizontal transmission especially contact between animals proved to be the main route of PCV2 transmission [42]. What’s more, the small-scale family breeding model of fur animals in China, mixed-breeded with other different livestock, has increased the risk of PCV2 cross-species transmission. Therefore, in order to prevent PCV2 cross-species transmission, very strong biosecurity barriers should be built between fur-animal farms and pig farms.
Conclusion
In summary, this study has provide first evidence of PCV2 prevalence in raccoon dogs in China. The genetic analysis and epidemiological investigation of raccoon dog-origin PCV2 stains in this research will help enrich the data of PCV2 cross-species transmission. Much attention should be paid to the cross-species infectious mechanism of PCV2 and the commercial PCV2 vaccine for raccoon dogs.
Acknowledgements
The specific anti-PCV2 monoclonal antibody (Mab) was kindly provided from Dr. Xiangdong Li (worked at National Research Center for Veterinary Medicine).
Funding
This work was supported by Hebei Provincial Modern Agro-industry Technology System of China (HBCT2018110207), the National Natural Sciences Foundation of China (31502187), the Scientific Research Foundation of Hebei Normal University of Science and Technology (2018YB017), and the Key Research and Development Program of Hebei Province (18226619D).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Authors’ contributions
TS, PR and ZM designed research; TS, JH, SX and SZ performed research; TS and ZM wrote the first draft of the manuscript; RZ, WH, MT, WL, CW, QF and HR contributed to modify the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All the samples was collected under the permission from of raccoon dog farmers in Qinhuangdao, China. This study did not include animal experiments.
Consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tao Song, Email: songtaoer@126.com.
Jianxiang Hao, Email: 2713068641@qq.com.
Ran Zhang, Email: zhangranvet@163.com.
Menghu Tang, Email: 2710252509@qq.com.
Wenao Li, Email: 642120608@qq.com.
Weirong Hui, Email: 18733521926@163.com.
Qiyuan Fu, Email: fuqiyuanyl@163.com.
Chunfang Wang, Email: 459747294@qq.com.
Shuyang Xin, Email: 836849024@qq.com.
Shoucong Zhang, Email: 1007292179@qq.com.
Ping Rui, Email: rp1969@126.com.
Hai Ren, Email: hairen1982@163.com.
Zengjun Ma, Email: mzj0712@hevttc.edu.cn.
References
- 1.Li G, Bao K, Zhang X, Si F, Yan S. Review on the development of special economic animal breeding industry in China. J Agr (China) 2018;8(01):140–144. [Google Scholar]
- 2.Jin HL, Gao SM, Liu Y, Zhang SF, Hu RL. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch Virol. 2016;161(2):445–448. doi: 10.1007/s00705-015-2659-9. [DOI] [PubMed] [Google Scholar]
- 3.Marcaccini A, Lopez Pena M, Quiroga MI, Bermudez R, Nieto JM, Aleman N. Pseudorabies virus infection in mink: a host-specific pathogenesis. Vet Immunol Immunop. 2008;124(3):264–273. doi: 10.1016/j.vetimm.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Wang GS, Sun N, Tian FL, Wen YJ, Xu C, Li J, et al. Genetic analysis of porcine circovirus type 2 from dead minks. J Gen Virol. 2016;97(9):2316–2322. doi: 10.1099/jgv.0.000529. [DOI] [PubMed] [Google Scholar]
- 5.Zhai SL, Chen SN, Xu ZH, Tang MH, Wang FG, Li XJ, et al. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J. 2014;11(1):88. doi: 10.1186/1743-422X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72(6):5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986;91(3–4):271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 8.Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91(1):e01879–e01816. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13(1):184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng XJ. Spread like a wildfire--the omnipresence of porcine circovirus type 2 (PCV2) and its ever-expanding association with diseases in pigs. Virus Res. 2012;164(1–2):1–3. doi: 10.1016/j.virusres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Kim HH, Park SI, Hyun BH, Park SJ, Jeong YJ, Shin DJ, et al. Genetic diversity of porcine circovirus type 2 in Korean pigs with postweaning multisystemic wasting syndrome during 2005-2007. J Vet Med Sci. 2009;71(3):349–353. doi: 10.1292/jvms.71.349. [DOI] [PubMed] [Google Scholar]
- 12.Harmon KM, Gauger PC, Zhang J, Pineyro PE, Dunn DD, Chriswell AJ. Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc. 2015;3(6). [DOI] [PMC free article] [PubMed]
- 13.Franzo G, Cortey M, Olvera A, Novosel D, Castro AM, Biagini P, et al. Revisiting the taxonomical classification of porcine circovirus type 2 (PCV2): still a real challenge. Virol J. 2015;12:131. doi: 10.1186/s12985-015-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao F, Mi S, Luo Q, Guo H, Tu C, Zhu G, et al. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound Emerg Dis. 2018;65(2):432–440. doi: 10.1111/tbed.12721. [DOI] [PubMed] [Google Scholar]
- 15.Jiang CG, Wang G, Tu YB, Liu YG, Wang SJ, Cai XH, et al. Genetic analysis of porcine circovirus type 2 in China. Arch Virol. 2017;162(9):2715–2726. doi: 10.1007/s00705-017-3414-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, Guo X, Ge X, Wang Z, Chen Y, Cha Z, et al. Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Res. 2009;145(1):151–156. doi: 10.1016/j.virusres.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Yang S, Yin S, Shang Y, Liu B, Yuan L, Zafar Khan MU, et al. Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transbound Emerg Dis. 2018;65(2):e383–ee92. doi: 10.1111/tbed.12768. [DOI] [PubMed] [Google Scholar]
- 18.Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J. 2010;7(1):273. doi: 10.1186/1743-422X-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol. 2013;87(3):1420–1429. doi: 10.1128/JVI.01443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorincz M, Csagola A, Biksi I, Szeredi L, Dan A, Tuboly T. Detection of porcine circovirus in rodents - short communication. Acta Vet Hung. 2010;58(2):265–268. doi: 10.1556/AVet.58.2010.2.12. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, et al. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84(4):1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Shan T, Soji OB, Alam MM, Kunz TH, Zaidi SZ, et al. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol. 2011;92:768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kappe EC, Halami MY, Schade B, Alex M, Hoffmann D, Gangl A, et al. Bone marrow depletion with haemorrhagic diathesis in calves in Germany: characterization of the disease and preliminary investigations on its aetiology. Berl Munch Tierarztl Wochenschr. 2010;123(1–2):31–41. [PubMed] [Google Scholar]
- 24.Herbst W, Willems H. Detection of virus particles resembling circovirus and porcine circovirus 2a (PCV2a) sequences in feces of dogs. Res Vet Sci. 2017;115:51–53. doi: 10.1016/j.rvsc.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Li W, Xu X, Wang W, He K, Fan H. Phylogenetic analysis of two goat-origin PCV2 isolates in China. Gene. 2018;651:57–61. doi: 10.1016/j.gene.2018.01.095. [DOI] [PubMed] [Google Scholar]
- 26.Song T, Zhang S, Hao J, Xin S, Hui W, Tang M, et al. First detection and genetic analysis of fox-origin porcine circovirus type 2. Transbound Emerg Dis. 2019;66(1):1–6. doi: 10.1111/tbed.13004. [DOI] [PubMed] [Google Scholar]
- 27.Walker D, Fee SA, Hartley G, Learmount J, O'Hagan MJ, Meredith AL, et al. Serological and molecular epidemiology of canine adenovirus type 1 in red foxes (Vulpes vulpes) in the United Kingdom. Sci Rep. 2016;6:36051. doi: 10.1038/srep36051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandi S, Kumar M. Canine parvovirus: current perspective. Indian J Virol. 2010;21(1):31–44. doi: 10.1007/s13337-010-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan J, Zeng X, Zhang G, Wu Q, Niu J, Sun B, et al. Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013-2014. J Vet Sci. 2016;17(3):369–375. doi: 10.4142/jvs.2016.17.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H, Su S, Deng S, Gu H, Ji F, Wang L, et al. The prevalence of hepatitis E virus infections among swine, swine farmers and the general population in Guangdong Province, China. PLoS One. 2014;9(2):e88106. doi: 10.1371/journal.pone.0088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Shan F, Zhou X, Li B, Zhai JQ, Zou SZ, et al. Outbreak and genotyping of canine distemper virus in captive Siberian tigers and red pandas. Sci Rep. 2017;7(1):8132. doi: 10.1038/s41598-017-08462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Wang X, Ma T, Feng Z, Li Y, Jiang P. Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes. 2010;40(2):244–251. doi: 10.1007/s11262-009-0438-y. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.McIntosh KA, Tumber A, Harding JC, Krakowka S, Ellis JA, Hill JE. Development and validation of a SYBR green real-time PCR for the quantification of porcine circovirus type 2 in serum, buffy coat, feces, and multiple tissues. Vet Microbiol. 2009;133(1–2):23–33. doi: 10.1016/j.vetmic.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Yuan W, Guo H, Ma Z, Song Q, Wang X, et al. Prevalence and genetic variation of porcine circovirus type 2 in Hebei, China from 2004 to 2014. Gene. 2016;586(2):222–227. doi: 10.1016/j.gene.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Ge X, Wang F, Guo X, Yang H. Porcine circovirus type 2 and its associated diseases in China. Virus Res. 2012;164(1–2):100–106. doi: 10.1016/j.virusres.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Investig. 2007;19(6):591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 40.Segales J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012;164(1–2):10–19. doi: 10.1016/j.virusres.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Zhai SL, He DS, Qi WB, Chen SN, Deng SF, Hu J, et al. Complete genome characterization and phylogenetic analysis of three distinct buffalo-origin PCV2 isolates from China. Infect Genet Evol. 2014;28:278–282. doi: 10.1016/j.meegid.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Rose N, Opriessnig T, Grasland B, Jestin A. Epidemiology and transmission of porcine circovirus type 2 (PCV2) Virus Res. 2012;164(1–2):78–89. doi: 10.1016/j.virusres.2011.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.