Abstract
Background
The mechanism underlying the interaction between host plant and host-selective toxin (HST)-producing Alternaria alternata during infection is of particular interest for sustainable crop production. Alternaria blotch of apple (Malus × domestica Borkh.) caused by A. alternata apple pathotype is a major disease particularly in East Asia, which is the largest producer of apples globally. A single dominant gene, Alt, controls the susceptibility of the apple cultivar ‘Delicious’ to Alternaria blotch. In this study, we fine mapped the Alt locus and characterized three potential candidate genes.
Results
We used 797 F1 individuals derived from 15 crosses between apple accessions susceptible (Alt/alt) and resistant (alt/alt) to Alternaria blotch to construct physical and genetic maps of the Alt locus located on the top of chromosome 11. Susceptible accessions were derived from ‘Delicious.’ To fine map the Alt locus, we constructed a BAC library of ‘Starking Delicious,’ a sport of ‘Delicious,’ and used graphical genotyping to delimit the Alt locus to a region of 43 kb. Three genes predicted within the candidate Alt region were potentially involved in plant defense response, among which the gene encoding a coiled coil-nucleotide binding-leucine rich repeat (CC-NB-LRR) type disease resistance protein was the most promising. Moreover, a 12-bp insertion was uniquely identified in the 5′ untranslated region of the Alt-associated allele of this gene, the presence or absence of which co-segregated with the susceptibility or resistance to A. alternata apple pathotype, respectively, among 43 tested cultivars including old ones and founders of modern apple breeding.
Conclusion
A disease resistance protein has been suggested as a determinant of susceptibility/resistance to HST-producing A. alternata for the first time. Our finding provides new insight into the mechanism of HST-mediated disease control used by A. alternata against host plants.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1737-7) contains supplementary material, which is available to authorized users.
Keywords: Alternaria alternata, Bacterial artificial chromosome library, Disease resistance, Host-selective toxin, Guard model
Background
Plant–pathogen interactions are of major interest for sustainable crop production with minimal use of chemicals. While most Alternaria species are saprophytic fungi that exist in soil or on decaying tissues [1, 2], host-selective toxin (HST) producing Alternaria alternata has acquired pathogenic traits and cause disease in a wide range of host plants. HST-producing A. alternata has seven pathogenic variants (pathotypes), each producing different HSTs specific for genera belonging to Rosaceae (Malus, Pyrus, Fragaria) and Rutaceae (Citrus) as well as Solanaceae species (tomato [Solanum lycopersicum] and tobacco [Nicotiana tobacum]) [3, 4]. HSTs are low molecular weight secondary metabolites that are toxic only to the host of the fungus producing the toxin but do not affect other plants.
While fungal pathogenicity has been extensively studied, the susceptibility of hosts to fungal infection remains poorly understood. Chemical structures have been elucidated for six of the seven HSTs, and the HST biosynthetic genes have been cloned, with the exception of the tobacco pathotype [3, 4]. Moreover, complementary experiments using A. alternata mutants that vary in their ability to produce HST have revealed the mechanism underlying its pathogenicity against host plants [5–7]. While the primary target sites of HSTs have been identified [3, 4], genes that determine host susceptibility have been identified only in rough lemon (Citrus jambhiri Lush.) and tomato [8, 9]. Therefore, elucidation of the mechanism underlying host susceptibility/resistance is needed to obtain a comprehensive understanding of the relationship between plant hosts and HST-producing A. alternata.
A. alternata apple pathotype (previously known as A. mali Roberts.) produces AM-toxin [10–12] and causes Alternaria blotch in apple (Malus × domestica Borkh.). AM-toxin has three related molecular species, AM-toxins I, II, and III, among which AM-toxin I is the most abundant. AM-toxin biosynthetic genes, AMT1, AMT2, AMT3, and AMT4, have been cloned and are clustered together in a small conditionally dispensable chromosome with many transposon-like sequences [6]. Alternaria blotch affects apple orchards in East Asia, which surpasses North America, and Europe as the region with the highest production of apples [13, 14]. It is characterized by the appearance of circular brown or blackish spots on leaves in late spring to early summer leading to defoliation. This results in reduction of photosynthesis and deterioration of fruit quality, leading to significant losses in apple production.
Although the level of susceptibility of apple cultivars to Alternaria blotch varies, the cutivars ‘Delicious,’ ‘Indo,’ and their sports and descendants are particularly susceptible [15]. Susceptibility to Alternaria blotch is controlled by the dominant gene Alt, which is mapped to chromosome 11 of ‘Starking Delicious’ (SD), a sport of ‘Delicious’ [16, 17]. Resistant cultivars are of the genotypes alt/alt, and susceptible cultivars are either Alt/alt or Alt/Alt although cultivars homozygous for Alt have not been identified [16]. Several simple sequence repeat (SSR) markers have been previously identified as flanking the Alt locus, which serve as good starting points for the positional cloning of Alt. In this study, we performed fine mapping of Alt using F1 progenies derived from crosses between apple accessions resistant and susceptible to Alternaria blotch. All susceptible accessions used in this study are derivatives of ‘Delicious.’ A bacterial artificial chromosome (BAC) library of ‘SD’ was constructed for fine mapping the Alt locus. Furthermore, a PCR-based genotyping marker was designed for scoring the resistance/susceptible phenotype of apple cultivars.
Results
Fine mapping of Alt
For fine mapping the Alt locus with 797 F1 plants (Table 1, Additional file 1: Figure S1), we first applied a marker enrichment approach for the genetic map of chr 11 constructed from two reciprocal full-sibs of ‘SD’ and ‘Jonathan.’ Forty-five SSR markers described previously [17] were tested, of which 25 markers were polymorphic in ‘SD’ and were subjected to linkage analysis (Additional file 2: Table S1).
Table 1.
F1 progenies used for fine mapping of Alt
| Maternal parenta | Paternal parent | Abbreviation | No. of individuals | No. of recombinantsb |
|---|---|---|---|---|
| Starking Delicious | Jonathan | SDJ | 57 | 0 |
| Jonathan | Starking Delicious | JSD | 57 | 3 |
| Sansa | Starking Delicious | P1 | 39 | 2 |
| Starking Delicious | Sansa | P2 | 46 | 2 |
| Sansa | Redgold | P3 | 36 | 2 |
| Jonathan | Redgold | P4 | 40 | 3 |
| Redgold | Jonathan | P5 | 27 | 0 |
| Gala | Morioka 61 | P6 | 50 | 2 |
| Sansa | Morioka 61 | P7 | 57 | 2 |
| Morioka 61 | 5–6393 | P8 | 3 | 0 |
| 4–547 | Morioka 61 | P9 | 26 | 0 |
| 4–161 | 5–5102 | P10 | 81 | 1 |
| Sensyu | Morioka 61 | P11 | 86 | 1 |
| Morioka 59 | 5–3645 | P12 | 38 | 0 |
| Morioka 61 | Silken | P13 | 154 | 5 |
| Total | 797 | 23 (2.9%) |
aApples shown with an underline are susceptible (Alt/alt) to Alternaria blotch. Morioka 59: Kitakami × Hatsuaki, Morioka61: Tsugaru × Kitakami, 5–6393: Akane × 4–23 (Fuji × [Golden Delicious × Indo]), 4–547: Fuji × Hatsuaki, 4–161: Hatsuaki × Starking Delicious, 5–3645: Sansa × Tsugaru, 5–5102: Tsugaru × Fuji. A graphical illustration of the pedigree of susceptible apples is also shown in Additional file 1: Figure S1
bIndividuals in which recombination occurred between Mdo.chr11.27 and Mdo.chr11.44
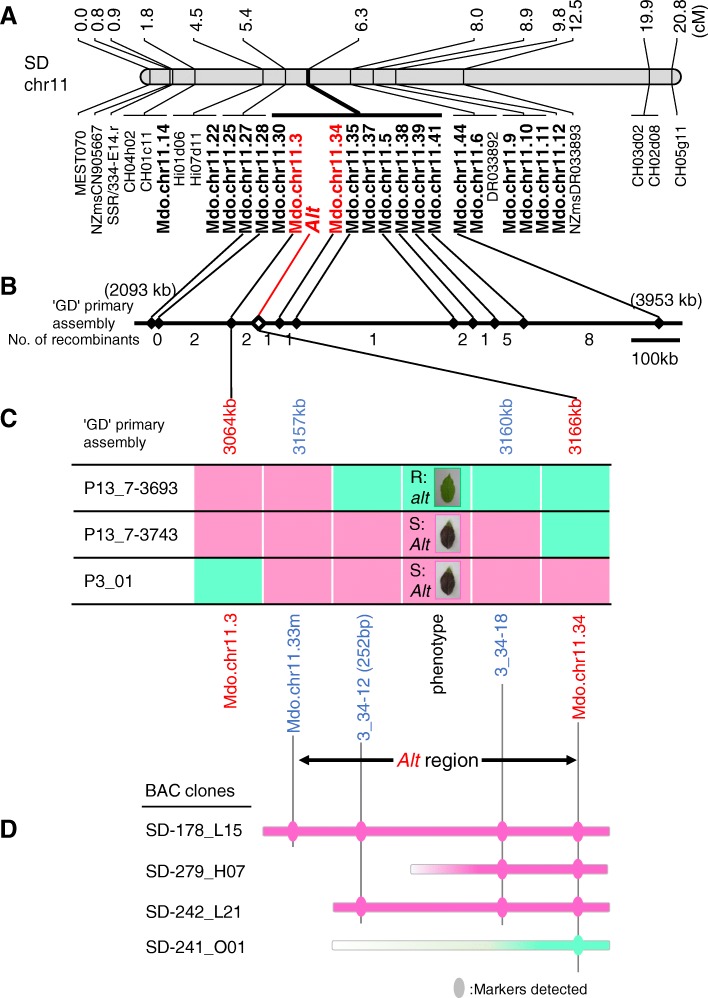
Of the 25 SSR markers, 21 mapped to chr 11. Of these 21 SSRs, 9 co-segregated with the Alt locus (Fig. 1a). The SSR markers, Mdo.chr11.28 and Mdo.chr11.44 flanked the Alt locus on either side. However, because Mdo.chr11.28 was less polymorphic, Mdo.chr11.27 was used together with Mdo.chr11.44 for further fine mapping of the Alt locus. Among 683 F1 individuals, 20 plants showing recombination between Mdo.chr11.27 and Mdo.chr11.44 were identified (Table 1 and Additional file 3: Table S2). Of these 20 recombinants plus an additional three recombinants derived from ‘SD’ and ‘Jonathan’ reciprocal full-sibs, 9 and 14 plants were identified as susceptible and resistant to Alternaria blotch, respectively (Table 1). None of the plants exhibited moderate resistance. Exploiting the phenotypic and the genetic linkage data, the Alt region was represented as a graphical genotype (Fig. 1b and Additional file 3: Table S2). The candidate region of Alt was identified between Mdo.chr11.3 and Mdo.chr11.34, spanning a physical distance of 102 kb according to the ‘Golden Delicious’ genome version 1.0 primary assembly. Three plants showing recombination between Alt and either Mdo.chr11.3 or Mdo.chr11.34 were identified.
Fig. 1.
Fine mapping of the Alt locus. a Marker-enriched initial map of ‘Starking Delicious’ (SD) constructed from 114 individuals of reciprocal crosses between ‘SD’ and ‘Jonathan.’ Simple sequence repeat (SSR) markers developed in this study are indicated in bold. Chr: chromosome. b Detailed physical map of Alt with genetic information using 797 F1 individuals derived from 15 crosses. The physical position of each marker is based on the ‘Golden Delicious’ genome version 1.0 primary assembly. Graphical representation of the genotype is also shown in Additional file 3: Table S2. c Final graphical genotype of the sequence flanking the Alt locus. Only three key recombinants are shown. The candidate region of Alt is indicated using double-headed arrow. Green and pink segments indicate Alt- and alt-associated regions, respectively. d Screening of bacterial artificial chromosome (BAC) clones of ‘SD’
To narrow down the candidate region further, we designed a new SSR marker Mdo.chr11.33 m by redesigning the primer sequence of Mdo.chr11.33 which was designed from the region between Mdo.chr11.3 and Mdo.chr11.34 and did not amplify in the marker enrichment approach and even developed two additional SSR markers (3_34–12 and 3_34–18) between Mdo.chr11.3 and Mdo.chr11.34 (Table 2). To identify Alt-associated alleles, we initially mapped these markers to the ‘SD’ map (data not shown). For markers Mdo.chr11.33 m and 3_34–18, 267-bp and 299-bp fragments were PCR amplified, respectively, both of which represented the Alt-associated alleles. For the marker 3_34–12, two fragments (288 and 252 bp) representing the Alt-associated alleles were PCR amplified, suggesting that primer pairs of 3_34–12 amplified two tightly linked loci (3_34–12 [288 bp] and 3_34–12 [252 bp]). We tested these SSR markers on 23 recombinants and identified that 3_34–12 (252 bp) and 3_34–18 co-segregated with Alt (Fig. 1c). Mdo.chr11.33 m was located between Alt and Mdo.chr11.3. The 3_34–12 (288 bp) locus mapped to a genomic location between Mdo.chr11.3 and Mdo.chr11.28, outside the candidate Alt region (Additional file 3: Table S2). Overall, fine mapping delimited Alt to a 9-kb region between Mdo.chr11.33 m and Mdo.chr11.34 according to the ‘Golden Delicious’ genome version 1.0 primary assembly.
Table 2.
Novel SSR markers developed in this study from apple contigs
| Marker | Primer sequence (5′ → 3′) | Origin of apple contig | Contig start position (bp) | SSR start position in contig (bp) | Motif type | Copy number |
|---|---|---|---|---|---|---|
| Mdo.chr11.33 m | F: GTTCGATCGGGGTGAAAGT R: CCCCATCCATTTACCCTACC |
MDC021160.220 | 3,150,224 | 6672 | GA | 15.5 |
| 3_34–12 | F: CCAATTGAAGACCTCCCAAA R: CCAGGAAAAGGACGCTACTG |
MDC004702.449 | 3,152,413 | 4269 | TAA | 5.7 |
| 3_34–18 | F: GAATCCCGAACTGAACCAAA R: GCTAAAATTTGGGCTTTAGGC |
MDC001844.206 | 3,160,267 | 223 | AT | 13 |
Identification of BAC clones spanning the Alt region
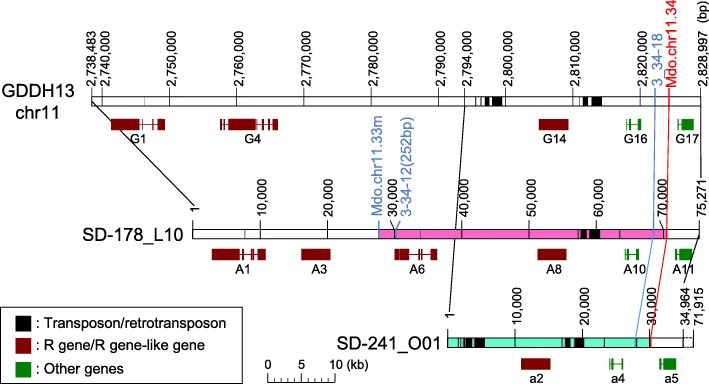
To identify genes in the candidate region of Alt, we constructed a BAC library of ‘SD’ with an average insert size of 180 kb and a total of 49,920 BAC clones at the first trial. Thus, the size of the BAC library is approximately 8985 Mb, which is approximately 11 times the size of the apple genome. However, clones harboring each of the four SSR markers (Mdo.chr11.3, Mdo.chr11.34, 3_34–12, and Mdo.chr11.33 m) could not be identified. Therefore, we performed the second trial with shorter fragments than the first trial and then developed an additional 61,056 BAC clones although their average insert size was not verified. Eventually, four BAC clones (SD-178_L15, SD-279_H07, SD-242_L21, and SD-241_O01) harboring at least one SSR marker were identified (Fig. 1d). The BAC clones, SD-178_L15, SD-279_H07, SD-242_L21 were identified as Alt-associated clones, whereas SD-241_O01 was identified as the alt-associated clone. Only one Alt-associated clone covered the complete candidate region (SD-178_L15), whereas the span of the alt-associated clone (SD-241_O01) was unconfirmed. Both SD-178_L15 and SD-241_O01 were shotgun sequenced and determined to span a length of 75,271 and 71,915 bp, respectively (Fig. 2). The Alt candidate region spanned approximately 43 kb, which was located within the range of 27–70 kb from the 5′-end of SD-178_L15. The BAC clone SD-241_O01 harbored Mdo.chr11.34 and only the forward primer of 3_34–18 at 30,344 bp and 28,015 bp from the 5′-end, respectively, but did not harbor the other two markers, 3_34–12 and Mdo.chr11.33 m.
Fig. 2.
Genetic characterization of the Alt region. Partial chromosome 11 of GDDH13, and BAC clones SD-178_L15 (Alt associated) and SD-241_O01 (alt associated) are shown. Genes predicted using FGNESH are indicated
In silico gene identification and characterization
Sequences of the BAC clones SD-178_L15 and SD-241_O01 and the partial sequence of GDDH13, a new genome sequence derived from a ‘Golden Delicious’ double haploid line [18], were examined for the presence of genes, transposable elements, and other features. The structure and location of transposable elements were clearly distinct between SD-178_L15, SD-241_O01, and GDDH13 (Fig. 2). We identified 11 (A1–11), 12 (a1–12), and 17 (G1–17) predicted ORFs in SD-178_L15, SD-241_O01, and GDDH13 (Table 3 and Fig. 2). Protein sequences of several predicted ORFs showed high similarity to those of genes involved in plant defense responses (Table 3). InterProScan identified a leucine-rich repeat (LRR) domain, a major component of disease resistance (R) genes, in four, three, and three ORFs of SD-178_L15, SD-241_O01, and GDDH13, respectively (Table 4 and Fig. 2). RIN4, a pathogenic type III effector avirulence factor Avr cleavage site, was also identified in A10, a4, and G16 ORFs. The Alt candidate region of SD-178_L15 included five predicted ORFs (A6–10), of which three showed characteristic features of R genes and two represented retrotransposon-like sequences and were discarded from the analysis.
Table 3.
ORFs predicted in this study from BAC clones and partial genomic sequence
| BAC clone/genome | ORF | Transcription start (bp) | Transcription end (bp) | Deduced amino acid length (aa) | Strand | Homologous protein (Species) | E-value | Accession no. |
|---|---|---|---|---|---|---|---|---|
| SD-178_L15 | A1 | 2061 | 11,998 | 1679 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008384572.2 |
| A2 | 15,932 | 13,969 | 203 | – | hypothetical protein DOTSEDRAFT_19432 (Dothistroma septosporum NZE10) | 0.24 | EME48939.1 | |
| A3 | 15,987 | 20,894 | 1411 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Pyrus × bretschneideri) | 0.0 | XP_018507989.1 | |
| A4 | 25,096 | 26,511 | 214 | + | PREDICTED: uncharacterized protein LOC103409892 (Malus domestica) | 5.00E-132 | XP_008346905.1 | |
| A5 | 27,488 | 28,157 | 40 | + | None | |||
| A6 | 29,534 | 36,522 | 835 | + | PREDICTED: putative disease resistance protein RGA3 isoform X2 (Pyrus × bretschneideri) | 0.0 | XP_009379454.1 | |
| A7 | 43,448 | 45,613 | 168 | + | Hypothetical protein COA78_37210 (Blastopirellula sp.) | 0.47 | PHR87371.1 | |
| A8 | 50,640 | 55,659 | 1405 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008366183.1 | |
| A9 | 55,769 | 57,610 | 296 | + | Uncharacterized protein LOC110755101 (Prunus avium) | 1.00E-104 | XP_021811957.1 | |
| A10 | 63,542 | 66,456 | 174 | + | PREDICTED: uncharacterized protein LOC103967858 isoform X1 (Pyrus × bretschneideri) | 2.00E-37 | XP_009379452.1 | |
| A11 | 71,752 | 74,786 | 650 | + | PREDICTED: WEB family protein At5g55860 (Malus domestica) | 0.0 | XP_008384579.2 | |
| SD-241_O01 | a1 | Not identified | 3754 | 222 | + | PREDICTED: uncharacterized protein LOC108170510 (Malus domestica) | 7.E-63 | XP_017181004.1 |
| a2 | 10,358 | 15,380 | 1405 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008366183.1 | |
| a3 | 15,499 | 17,345 | 322 | + | Uncharacterized protein LOC110755101 (Prunus avium) | 5E-132 | XP_021811957.1 | |
| a4 | 23,336 | 26,622 | 115 | + | PREDICTED: uncharacterized protein LOC103967858 isoform X1 (Pyrus × bretschneideri) | 1.00E-36 | XP_009379452.1 | |
| a5 | 31,018 | 34,478 | 650 | + | PREDICTED: WEB family protein At5g55860 (Malus domestica) | 0.0 | XP_008384579.2 | |
| a6 | 34,684 | 38,521 | 473 | + | PREDICTED: uncharacterized protein LOC103447176 (Malus domestica) | 0.0 | XP_008384578.1 | |
| a7 | 44,893 | 39,592 | 324 | + | PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1-like (Pyrus × bretschneideri) | 0.0 | XP_009367226.1 | |
| a8 | 51,499 | 52,557 | 69 | – | PREDICTED: kinesin-like protein KIF1C (Lates calcarifer) | 0.11 | XP_018537717.1 | |
| a9 | 59,979 | 54,964 | 609 | + | PREDICTED: serine/threonine-protein kinase D6PK-like (Pyrus × bretschneideri) | 0.0 | XP_009367228.1 | |
| a10 | 60,552 | 63,449 | 152 | – | Pre-rRNA-processing protein TSR2-like (Prunus avium) | 3.00E-66 | XP_021818064.1 | |
| a11 | 69,641 | 64,724 | 353 | – | PREDICTED: DNA-directed RNA polymerases I and III subunit rpac1-like (Malus domestica) | 3.00E-164 | XP_008349490.1 | |
| a12 | 69,866 | Not Identified | 325 | – | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008384580.1 | |
| GDDH13 Chr 11 partial | G1 | 2,740,584 | 2,749,771 | 1689 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008384572.2 |
| G2 | 2,754,217 | 2,752,730 | 125 | – | Putative reverse transcriptase family member (Malus domestica) | 2.00E-36 | CBL94165.1 | |
| G3 | 2,754,290 | 2,757,403 | 577 | – | Putative COBL7 (COBRA-LIKE 7) (Malus domestica) | 0.0 | CBL94184.1 | |
| G4 | 2,757,523 | 2,766,112 | 1770 | + | PREDICTED: putative disease resistance protein RGA3 isoform X2 (Pyrus × bretschneideri) | 0.0 | XP_009379454.1 | |
| G5 | 2,769,661 | 2,774,105 | 1049 | + | T4.15 (Malus × robusta) | 0.0 | CCH50976.1 | |
| G6 | 2,774,818 | 2,774,245 | 133 | – | T4.14 (Malus × robusta) | 3.00E-89 | CCH50975.1 | |
| G7 | 2,774,858 | 2,778,564 | 786 | + | Hypothetical protein (Malus domestica) | 3.00E-149 | AEJ72571.1 | |
| G8 | 2,778,594 | 2,779,821 | 161 | + | PREDICTED: uncharacterized protein LOC103420450 (Malus domestica) | 8.00E-11 | XP_008356739.1 | |
| G9 | 2,781,742 | 2,780,939 | 49 | – | None | |||
| G10 | 2,781,835 | 2,787,073 | 592 | + | Putative COBL7 (COBRA-LIKE 7) (Malus domestica) | 1.00E-145 | CBL94184.1 | |
| G11 | 2,787,274 | 2,788,139 | 225 | + | T4.14 (Malus × robusta) | 2.00E-144 | CCH50975.1 | |
| G12 | 2,791,526 | 2,788,282 | 1416 | – | T4.15 (Malus × robusta) | 0.0 | CCH50976.1 | |
| G13 | 2,794,998 | 2,799,499 | 430 | + | PREDICTED: uncharacterized protein LOC103438444 (Malus domestica) | 3.00E-135 | XP_017188663.1 | |
| G14 | 2,804,362 | 2,809,384 | 1405 | + | PREDICTED: putative disease resistance RPP13-like protein 1 (Malus domestica) | 0.0 | XP_008366183.1 | |
| G15 | 2,809,494 | 2,812,371 | 338 | + | Uncharacterized protein LOC110755101 (Prunus avium) | 6.00E-100 | XP_021811957.1 | |
| G16 | 2,817,267 | 2,820,181 | 174 | + | PREDICTED: uncharacterized protein LOC103967858 isoform X1 (Pyrus × bretschneideri) | 2.00E-37 | XP_009379452.1 | |
| G17 | 2,825,063 | 2,828,512 | 650 | + | PREDICTED: WEB family protein At5g55860 (Malus domestica) | 0.0 | XP_008384579.2 | |
Table 4.
Functional domains identified in predicted ORFs in the Alt region
| BAC clone/genome | Genea | Functional domains (InterPro) | InterPro ID | Match position (aa) |
|---|---|---|---|---|
| SD-178_L15 | A5 | NPa | ||
| A6 | RX-CC_like | CD14798 | 10..98 | |
| Leucine-rich repeat domain superfamily | IPR32675 | 106..563 | ||
| F-box-like domain superfamily | IPR001810 | 641..691 | ||
| A7 | NP | |||
| A8 | RX-CC_like | CD14798 | 33..136 | |
| P-loop containing nucleoside triphosphate hydrolase | IPR027417 | 160..425 | ||
| NB-ARC | IPR002182 | 173..451 | ||
| Winged helix-turn-helix DNA-binding domain superfamily | IPR011991 | 405..487 | ||
| Leucine-rich repeat domain superfamily | IPR32675 | 487..1393 | ||
| A9 | Gag-polypeptide of LTR copia-type | IPR029472 | 16..59 | |
| A10 | RIN4, pathogenic type III effector avirulence factor Avr cleavage site | IPR008700 | 12..47 | |
| SD-241_O01 | a1 | Gag-polypeptide of LTR copia-type | IPR029472 | 30..70 |
| a2 | RX-CC_like | CD14798 | 33..136 | |
| P-loop containing nucleoside triphosphate hydrolase | IPR027417 | 161..425 | ||
| NB-ARC | IPR002182 | 173..451 | ||
| Winged helix-turn-helix DNA-binding domain superfamily | IPR011991 | 405..490 | ||
| Leucine-rich repeat domain superfamily | IPR032675 | 487..1393 | ||
| a3 | Gag-polypeptide of LTR copia-type | IPR029472 | 16..59 | |
| a4 | RIN4, pathogenic type III effector avirulence factor Avr cleavage site | IPR008700 | 12..47 | |
| GDDH13 | G4 | RX-CC_like | CD14798 | 123..249 |
| P-loop containing nucleoside triphosphate hydrolase | IPR027417 | 275..544 | ||
| NB-ARC | IPR002182 | 304..570 | ||
| Winged helix-turn-helix DNA-binding domain superfamily | IPR011991 | 524..608 | ||
| Leucine-rich repeat domain superfamily | IPR032675 | 985..1428 | ||
| Receptor L-domain superfamily | IPR036941 | 1310..1427 | ||
| RIN4, pathogenic type III effector avirulence factor Avr cleavage site | IPR008700 | 1439..1474 | ||
| F-box-like domain superfamily | IPR036047 | 1532..1586 | ||
| F-box domain | IPR001810 | 1530..1570 | ||
| G5 | Endonuclease/exonuclease/phosphatase superfamily | IPR036691 | 110..358 | |
| SWR1-complex protein 5/craniofacial development protein | IPR027124 | 112..406 | ||
| Reverse transcriptase domain | IPR000477 | 621..867 | ||
| G6 | NP | |||
| G7 | NP | |||
| G8 | NP | |||
| G9 | NP | |||
| G10 | NP | |||
| G11 | NP | |||
| G12 | Endonuclease/exonuclease/phosphatase superfamily | IPR036691 | 94..300 | |
| SWR1-complex protein 5/craniofacial development protein | IPR027124 | 57..78 | ||
| SWR1-complex protein 5/craniofacial development protein | IPR027124 | 97..298 | ||
| G13 | Gag-polypeptide of LTR copia-type | IPR029472 | 30..70 | |
| GAG-pre-integrase domain | IPR025724 | 268..328 | ||
| G14 | RX-CC_like | CD14798 | 33..136 | |
| P-loop containing nucleoside triphosphate hydrolase | IPR027417 | 161..425 | ||
| NB-ARC | IPR002182 | 173..451 | ||
| Winged helix-turn-helix DNA-binding domain superfamily | IPR011991 | 405..487 | ||
| Leucine-rich repeat domain superfamily | IPR032675 | 487..1393 | ||
| G15 | Gag-polypeptide of LTR copia-type | IPR029472 | 16..59 | |
| G16 | RIN4, pathogenic type III effector avirulence factor Avr cleavage site | IPR008700 | 12..47 | |
aNP none predicted
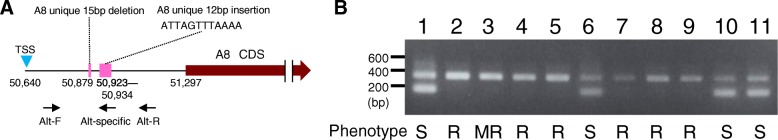
Within the Alt candidate region, the A6 ORF was uniquely present on SD-178_L15, and was absent from SD-241_O01, and GDDH13. The A6 ORF showed high similarity to coiled coil-nucleotide binding-LRR (CC-NB-LRR) type proteins; however, it lacked the NB domain (Table 4 and Additional file 4: Figure S2). It showed the highest similarity (62%) to the predicted ORF MD11G1030400 located on 2.656 Mb of chr 11 of the GDDH13 genome. The A8, a2, and G14 ORFs showed high similarity to CC-NB-LRR class R proteins, clearly indicating that they belong to this category of protein. They represented highly conserved 1405 amino acids (> 99% similarity) encoding CC, NB, and LRR domains (Table 4) and were predicted by InterPro to be extracellular proteins (data not shown). The A8 ORF harbored several unique sequence variations, including several amino acid substitutions in the CC and NB domains (Additional file 5: Figure S3), and a 12-bp insertion and a 15-bp deletion in the 5′ untranslated region (UTR) (Fig. 3). The A10, a4, and G16 ORFs harbored RIN4, a pathogenic type III effector avirulence factor Avr cleavage site domain (Table 4). The amino acid sequences of A10 and G16 were identical, and their promoter sequences 2 kb upstream of the transcription start site were also identical, except for 1-bp mismatch. However, the predicted amino acid sequence of a4, especially at the C-terminus, differed significantly from that of A10 and G16 (Additional file 6: Figure S4).
Fig. 3.
DNA marker for the detection of the Alt-associated allele of A8 ORF. a Schematic representation of the A8 ORF unique 12-bp insertion structure and primers for its detection. b Gel image showing PCR products amplified from the genomic DNA of apple cultivars with three primers “Alt-F,” “Alt-R,” and “Alt-specific.” Different lanes of the gel represent different apple cultivars: 1, ‘Starking Delicious’; 2, ‘Jonathan’; 3, ‘Golden Delicious’; 4, ‘Ralls Janet’; 5, ‘Worcester Pearmain’; 6, ‘Indo’; 7, ‘Cox’s Orange Pippin’; 8, ‘McIntosh’; 9, P13_7–3693; 10, P13_7–3743; and 11, P3_01. S = susceptible, R = resistant, MR = moderately resistant. TSS: transcription start site, CDS: coding sequences
DNA marker for Alt
The unique insertion in the 5′ UTR of the A8 ORF was used to develop a PCR-based genotyping marker for the Alt region (Fig. 3). Accessions carrying the insertion were susceptible (score > 2.5) to infection with the AKI-3 isolate of A. alternata apple pathotype (Additional file 7: Table S3 and Fig. 3).
Discussion
In the present study, we fine mapped the Alt locus to a 43 kb region on the top of chr 11 using BAC clones (Figs. 1 and 2) and predicted three ORFs as potential candidates underlying the susceptibility or resistance response to the A. alternata apple pathotype. One of these predicted ORFs encodes a typical R protein.
Genes controlling Alternaria blotch susceptibility
An SSR marker (CH05g07) linked to a dominant gene conferring susceptibility to the A. alternata apple pathotype at a distance of 5.6 cM has been previously reported in ‘Golden Delicious’ [19]. However, because CH05g07 is located on two different chr (12 and 14) [20], the genomic position of the gene identified by Li et al. [19] possibly differs from that of the Alt locus mapped in the present study (Fig. 1). This inconsistency may be due to differences in genes responsible for the reaction to pathogen isolate attack or to the experimental conditions used. It may be that the virulence of A. alternata in the same cultivar differs between China and Japan [15, 21]. For example, ‘Golden Delicious’ has been defined as susceptible to Alternaria blotch in Chinese studies [19] but as resistant [15, 22, 23] or moderately resistant [24] in Japanese studies. The finding that application of HST at high concentrations to resistant cultivars induces susceptibility [25] suggests that pathogens used in those studies differ in their ability to produce HST; the Chinese one might produce AM-toxin in great quantities than the Japanese one. Therefore, it is important to characterize the virulence of the pathogens studied. Moreover, the long incubation time (7 days) used for the phenotypic evaluation of genotypes by Li et al. [19] compared with that used by Abe et al. [15] may raise other resistance/susceptibility systems controlled by genes that are not identical to the one identified in the present study. The mechanisms underlying these inconsistencies need to be investigated in future studies.
Candidate gene identification
No alt-associated BAC clone was found to completely span the Alt region, suggesting that the nature of the sequence or chromosomal structure of this region inhibited its cloning. One possibility could be that the region surrounding the Alt candidate region contains many HindIII recognition sites, and this impeded the formation of long fractions we harvested at the first trial. The in silico ORF prediction revealed several R genes in BAC clones and the GDDH13 partial genome sequence. This result is consistent with previous observations of the CC-NB-LRR gene cluster at the top of chr 11 [26]. Although the predicted ORFs were not verified using RT-PCR, we predicted two ORFs encoding R proteins (A6 and A8) and one encoding a defense response-related protein (A10) within the candidate region (Table 3 and Fig. 2). Amino acid sequences of both A6 and A8 showed high similarity to CC-NB-LRR class R proteins; however, because A6 lacked the NB-ARC domain that is typical and critical for R protein function in this class of genes [27], A6 is probably a pseudogene. The absence of A6 allele in SD-241_O01 and GDDH13 could be explained by duplication of NB-LRR gene. In the vicinity of A6, the non-coding nucleotide sequence of SD-178_L10 is very different from those of GDDH13 and SD-241_O01 (data not shown); there is almost no similarity between SD-178_L10 and the other two, suggesting ectopic recombination and/or gene translocation, which are the driving forces behind enlargement of the resistance gene analogous (RGA) family, including NB-LRR genes, in the plant genome. The fact that A6 showed the highest similarity to the MD11G1030400 located approximately 100 kb from A6 is in line with the findings of a previous study [28]; NB-LRR pseudogenes are often present within 100 kb of the bonafide NB-LRR gene. Therefore, it could be possible that A6 was generated by gene duplication and is in the process of defunctionalizing.
Because the predicted ORFs A8, a2, and G14 were considered to be CC-NB-LRR class R genes, the encoded proteins could recognize an effector directly or indirectly, thus inducing a hypersensitive response (HR). It was suggested that several amino acid substitutions in the CC and NB domains could be associated with changes in their functions. The high conservation of amino acid sequences of these ORFs (> 99% similarity) suggests A8 to be a functional R gene, and unique polymorphisms in its 5′ UTR imply the distinct expression profile of the Alt-associated allele, although the future research is necessary to collect evidence supporting this theory.
The ORFs, A10, a4, and G16 were predicted to encode RIN4, a pathogenic type III effector avirulence factor Avr cleavage site (Table 4). In Arabidopsis thaliana, RIN4 mediates HR against Pseudomonas syringae and is associated with the well-studied plant–pathogen relationship called the “guard model” [29–31]. The R proteins, RPM1 and RPS2 (i.e., guards), monitor RIN4 (i.e., guardee) invariability. The attack by pathogenic effectors secreted by P. syringae modifies RIN4, thereby activating RPM1 and RPS2 and inducing HR. A similar mechanism could exist in apple, where A10, a candidate of Alt, would function as a guardee protein. However, because sequences of A10 and G16 predicted proteins and promoters were highly conserved, it suggests that there are no functional and expression level differences between them and therefore do not appear to be plausible candidates for the Alt locus.
Consequently, among the three predicted in the SD-178_L15 BAC clone, A8 appears to be the most promising candidate for the Alt locus. These findings also suggest the elicitor activity of AM-toxin. Although the suppressor effect of AM-toxin is well documented [32], the elicitor activity of AM-toxin has been described for the first time in this study.
Predicted mechanism underlying susceptibility control
Despite counter evidence, HR is thought as a plants’ defense mechanism against invading pathogens whereby the pathogen is contained within the dead tissue by inducing programmed cell death in areas surrounding the site of infection [33]. In typical relationships between plants and fungal pathogens mediated by R genes, resistance is usually dominantly inherited as a consequence of gene-to-gene interaction. By contrast, the relationship between apple and A. alternata apple pathotype exhibits an opposite trend, whereby Alternaria blotch susceptibility is dominantly inherited [16]. This is similar to the relationship between A. thaliana and Cochliobolus victoriae [34]. C. victoriae is a necrotrophic fungus that produces the HST victorin that affects A. thaliana. The susceptibility of A. thaliana to C. victoriae is dominantly inherited [35], which can be explained on the basis of the guard model [36, 37]. Lorang et al. [38] identified the susceptibility gene, LOV1 that encodes a CC-NB-LRR class R protein. LOV1 guards TRXh5, a target of victorin. The attack by victorin on TRXh5 increases free LOV1, resulting in LOV1-mediated HR. Thus, C. victoriae exploits the plants’ defense response for its own pathogenesis [34]. Because the present study also suggested the involvement of R genes in the infection by HST-producing A. alternata, it is possible that the relationship between apple and HST-producing A. alternata is similar to that observed between A. thaliana and C. victoriae.
Tabira et al. [39] indicated that cell death is not necessary for the infection of apple by A. alternata apple pathotype, suggesting that a certain step of HR induction is sufficient for A. alternata infection. It is noteworthy that A. alternata, a saprophyte, mimics biotrophic pathogens and elicits a resistance response to exploit the hosts’ defense system for its invasion. However, details of the molecular mechanisms underlying the infection of apple by A. alternata apple pathotype need to be elucidated.
Comprehensive understanding of the relationship between A. alternata and its host
The relationship between HST-producing A. alternata and host plants among the Rosaceae is similar. For example, black spot disease of Japanese pear and that of strawberry (F. × ananassa Duch.) is caused by the Japanese pear and strawberry pathotypes of A. alternata, respectively [3, 4]. It has been shown that the susceptibility of host plants to the disease is dominantly inherited, similar to the Alternaria blotch in apple [16, 40, 41]. HSTs produced by these pathotypes target the plasma membrane of the host cells [4]. In Japanese pear, the susceptibility gene A has been mapped to pear chr 11 between the markers Mdo.chr11.28 and Mdo.chr11.34 [17], thus perfectly corresponding to the Alt candidate region in apple; the chromosomal location of the susceptibility gene in strawberry is unknown. The A. alternata apple pathotype has recently been shown to cause black spot disease in the European pear (P. communis L.) [42]. Although the inheritance pattern has not been studied, AM-toxin induces veinal necrosis in leaves of specific European pear cultivars, such as ‘Le Lectier’ and ‘General Leclerc’ but not in ‘Bartlett’ [42], suggesting the existence of a similar HST-producing A. alternata – host relationship in the European pear. Based on these findings, we hypothesize that A. alternata exploits plant defense systems to express pathogenicity against the Rosaceae family, and that genes of Roseaceae hosts involved in susceptibility to A. alternata may be functionally conserved.
Marker-assisted breeding of Alternaria blotch-resistant apple
Using three primers (Alt-F, Alt-R, and Alt-specific), we were able to score the Alt genotype of modern breeding founders and old cultivars, indicating the utility of this marker for the breeding of Alternaria blotch-resistant apple. Co-segregation of the 12-bp insertion with susceptible phenotype suggests single origin of susceptibility to Alternaria blotch and sufficient linkage disequilibrium between the insertion and causal polymorphism(s) of Alt. However, it did not discriminate moderately resistant cultivars such as ‘Golden Delicious’ from resistant cultivars, which needs to be investigated in future research.
Conclusion
A CC-NB-LRR class R protein was identified as a promising candidate gene for Alt. This is the first study that identified a candidate gene involved in HR induction for the control of susceptibility/resistance to HST-producing A. alternata diseases. This finding provides new insights into the relationship between HST-producing A. alternata and host plants. This information will be useful in the development of better disease control strategies and will also improve our understanding of the co-evolution of plant defense mechanisms and fungal pathogenicity. Most importantly, the DNA marker developed in this study serves as a tool for marker-assisted breeding of Alternaria blotch-resistant apple.
Methods
Plant materials and DNA extraction
A total of 797 F1 plants derived from 15 crosses between Alternaria blotch-susceptible (Alt/alt) and -resistant (alt/alt) apple accessions were used in this study (Table 1). This included 114 F1 plants derived from reciprocal crosses between ‘SD’ and ‘Jonathan’ that have been previously assessed for their resistance to Alternaria blotch [43]. The susceptible accessions used in this study were derived from the common founder, ‘Delicious’ (susceptible, Alt/alt; Additional file 1: Figure S1). Moreover, eight founders of modern world apple cultivars and 35 old world apple cultivars described previously [15] were also included in this study (Additional file 7: Table S3). All apple genotypes were grown and maintained at the Apple Research Station, Institute of Fruit Tree and Tea Science, NARO, Morioka, Japan.
Genomic DNA was isolated from of all F1 progenies and their parents. Briefly, 100 mg of young leaves were ground in liquid nitrogen and incubated with 1 ml of an isolation buffer (10% PEG 6000, 100 mM Tris-HCl [pH 8.0], 350 mM sorbitol, and 50 mM EDTA [pH 8.0]) at 50 °C for 30 min. Genomic DNA was extracted using the DNA extraction device, PI-50α (Kurabo, Osaka, Japan) in accordance with the manufacturer’s instructions.
Evaluation of Alternaria blotch resistance
The monoconidial isolate of A. alternata apple pathotype, AKI-3 was used for the evaluation of Alternaria blotch resistance. Inoculation was performed using the detached-leaf method as described previously [15]. Briefly, five, second or third youngest leaves, were removed from the growing shoots of each plant and coated with a suspension of 2 × 105 conidia of the AKI-3 isolate using a mist sprayer. The inoculated leaves were incubated at 20 °C for 48 h in the dark in a plastic chamber. The resistance level of each leaf was scored on a scale of 0 (no visible symptoms) to 5 (almost complete necrosis of the whole leaf) as described previously [15]. To determine the resistance level of a genotype, resistance scores of all leaves belonging to the same genotype were averaged. Scoring of the resistance level of genotypes was slightly modified from Abe et al. [15]; a genotype was categorized as resistant: mean score ≤ 0.5, moderately resistant: mean score 0.5–2.5, and susceptible: mean score > 2.5.
SSR markers and linkage analysis
SSR makers (Mdo.chr11.1–11.44) developed previously for the fine mapping of the A gene, responsible for the susceptibility of Japanese pear (Pyrus pyrifolia Nakai) to black spot disease [17], were used for initial marker enrichment and fine mapping the Alt locus (Additional file 2: Table S1). These markers were developed from contigs spanning 2.5–4.0 Mb of chr 11 of the apple genome version 1.0 primary assembly [44] corresponding to the Alt location. Moreover, novel SSR markers were developed from the same assembly. Batch Primer3 [45] was used to identify SSRs (Table 2) and for primer design. PCR amplification and detection of these markers were performed as described previously [46]. Linkage analysis and genetic map construction were performed using JoinMap version 4.1 [47]. Genetic distances between markers were calculated using a pseudo-testcross mapping strategy [48] by applying the regression mapping algorithm and the Kosambi’s map function. A minimum LOD score of 10.0 was used to establish the linkage groups.
BAC library construction and shotgun sequencing
A BAC library of ‘SD’ was constructed as described previously [49]. Briefly, nuclear DNA isolated from ‘SD’ leaves was digested with HindIII restriction endonuclease. DNA fragments 100–180 kb in size at the first trial and slightly shorter than that at the second trial were ligated into the BAC vector pIndigoBAC-5 (Epicentre, Illumina), followed by transformation into Escherichia coli, ElectroMAX DH10B cells (Invitrogen, Life Technologies). Bacterial colonies were picked, transferred to LB medium dispensed to 384-well plates, and stored at − 80 °C.
To identify clones spanning the Alt region, four SSR markers (Mdo.chr11.3, Mdo.chr11.34, 3_34–12, and Mdo.chr11.33 m) were used to screen the BAC clones. PCR-amplified fragment length was used for the identification of Alt- or alt-associated clones. Because the alt-associated alleles of 3_34–12 and Mdo.chr11.33 m were not identified, these markers were able to detect only Alt-associated clones. DNA of selected BAC clones was randomly sheared and shotgun sequenced using the ABI 3730xl sequencer (Applied Biosystems, Life Technologies) with universal forward and reverse primers and the dye-terminator method. Shotgun sequences were assembled using PHRED and PHRAP software packages [50, 51]. To fill any gaps between assembled BAC contigs, additional shotgun sequencing was performed using other methods, such as dye-primer and transposon-tag sequencing [52].
In silico gene prediction and characterization
Two BAC clones (SD-178_L15 and SD-241_O01) and partial sequence of a doubled haploid line of ‘Golden Delicious’ (GDDH13) [18], trimmed as to correspond to SD-178_L15, were subjected to in silico analysis. Nucleotide sequences of transposable elements identified using RepeatMasker (http://repeatmasker.org) were substituted as Ns prior to predicting open reading frames (ORFs). The ORF prediction was performed using FGENESH [53] with the algorithm for dicot plants. Predicted ORFs were queried against the nonredundant (nr) protein database of the National Center for Biotechnology Information (NCBI; https://blast.ncbi.nlm.nih.gov/) using BLASTP and searched for conserved domains using InterProScan [54]. Protein alignments were performed using the online versions of Needle and ClustalW available at the European Molecular Biology Laboratory (EMBL; http://www.ebi.ac.uk/Tools/psa/) and the DNA Data Bank of Japan (DDBJ; http://clustalw.ddbj.nig.ac.jp/index.php?lang=en), respectively.
DNA marker for Alt
Three primers, “Alt-F” (5′-ATGTGTTTTATCCATCCAATTACG-3′), “Alt-R” (5′-AAGTTCAAATCTGACTCCGCTTA-3′), and “Alt-specific” (5′-GCCAGGGAGACTAAATTTTAAACTAAT-3′), were designed based on the sequence of the BAC clone SD-178_L15. PCRs were conducted with all three primers using the GoTaq Hot Start Master Mix (Promega, Madison, WI, USA). PCR conditions included an initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min.
Additional files
Figure S1. Pedigree of susceptible apple accessions used in this study. (PDF 50 kb)
Table S1. Mapping of SSR markers used in marker enrichment analysis. (PDF 30 kb)
Table S2. Graphical illustration of genotypes showing recombination between Alt and flanking markers. (PDF 38 kb)
Figure S2. Pairwise alignment of A6, a predicted ORF in the Alt region and XP_009379454.1, the gene most similar to A6, as revealed by a database search. (PDF 49 kb)
Figure S3. Amino acid sequence alignment of predicted candidate genes of Alt, A8, a2, and G14. (PDF 37 kb)
Figure S4. Amino acid sequence alignment of predicted candidate genes of Alt, A10, a4, and G16. (PDF 36 kb)
Table S3. Presence of 12-bp insertion among founders and old cultivars. (PDF 50 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics-based Technology for Agricultural Innovation, HOR-2001). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The nucleotide sequences of BAC clones are available in DDBJ as LC360748 and LC360749 for SD-178_L10 and SD-241_O01, respectively.
Authors’ contributions
SM, ST, TY and KA conceived and designed the experiments. SM, KO, YA, TS and KA performed disease-resistance assessments of plant materials. SM and ST performed molecular marker development and detection. SM, ST, YK, HF, JW, HK and TY performed experiments involving the BAC library. SM and ST analyzed the data. SM drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shigeki Moriya, Email: moriyas@affrc.go.jp.
Shingo Terakami, Email: terakami@affrc.go.jp.
Kazuma Okada, Email: okak@affrc.go.jp.
Taku Shimizu, Email: takushimizu@affrc.go.jp.
Yoshihiko Adachi, Email: nagoya@affrc.go.jp.
Yuichi Katayose, Email: katayose@affrc.go.jp.
Jianzhon Wu, Email: jzwu@affrc.go.jp.
Hiroyuki Kanamori, Email: kanamo@affrc.go.jp.
Toshiya Yamamoto, Email: toshiya@affrc.go.jp.
Kazuyuki Abe, Email: kazuyuki@affrc.go.jp.
References
- 1.Rotem J. The genus Alternaria–biology, epidemiology and pathogenicity. St Paul: APS press; 1994. [Google Scholar]
- 2.Thomma BPHJ. Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol. 2003;4:225–236. doi: 10.1046/j.1364-3703.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- 3.Akimitsu K, Tsuge T, Kodama M, Yamamoto M, Otani H. Alternaria host-selective toxins: determinant factors of plant disease. J Gen Plant Pathol. 2014;80:109–122. doi: 10.1007/s10327-013-0498-7. [DOI] [Google Scholar]
- 4.Tsuge T, Harimoto Y, Akimitsu K, Ohtani K, Kodama M, Akagi Y, et al. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol Rev. 2013;37:44–66. doi: 10.1111/j.1574-6976.2012.00350.x. [DOI] [PubMed] [Google Scholar]
- 5.Johnson LJ, Johnson RD, Akamatsu H, Salamiah A, Otani H, Kohmoto K, et al. Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr Genet. 2001;40:65–72. doi: 10.1007/s002940100233. [DOI] [PubMed] [Google Scholar]
- 6.Harimoto Y, Hatta R, Kodama M, Yamamoto M, Otani H, Tsuge T. Expression profiles of genes encoded by the supernumerary chromosome controlling AM-toxin biosynthesis and pathogenicity in the apple pathotype of Alternaria alternata. Mol Plant-Microbe Interact. 2007;20:1463–1476. doi: 10.1094/MPMI-20-12-1463. [DOI] [PubMed] [Google Scholar]
- 7.Harimoto Y, Tanaka T, Kodama M, Yamamoto M, Otani H, Tsuge T. Multiple copies of AMT2 are prerequisite for the apple pathotype of Alternaria alternata to produce enough AM-toxin for expressing pathogenicity. J Gen Plant Pathol. 2008;74:222–229. doi: 10.1007/s10327-008-0089-1. [DOI] [Google Scholar]
- 8.Ohtani K, Yamamoto M, Akimitsu K. Sensitivity to Alternaria alternata toxin in citrus because of altered mitochondrial RNA processing. Proc Natl Acad Sci U S A. 2002;99:2439–2444. doi: 10.1073/pnas.042448499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandwagt BF, Mesbah LA, Takken FLW, Laurent PL, Kneppers TJA, Hille J, et al. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. Sp lycopersici toxins and fumonisin B1. Proc Natl Acad Sci U S A. 2000;97:4961–4966. doi: 10.1073/pnas.97.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuno T, Ishita Y, Sawai K, Matsumoto T. Characterization of Alternariolide, a host-specific toxin produced by Alternaria mali Roberts. Chem Lett. 1974;3:635–638. doi: 10.1246/cl.1974.635. [DOI] [Google Scholar]
- 11.Ueno T, Nakashima T, Hayashi Y, Fukami H. Structures of AM-toxin I and II, host specific phytotoxic metabolites produced by Alternaria mali. Agric Biol Chem. 1975;39:1115–1122. [Google Scholar]
- 12.Ueno T, Nakashima T, Umemoto M, Fukami H, Lee SN, Izumiya N. Mass spectrometry of Alternaria mali toxins and related cyclodepsipeptides. Biol Mass Spectrom. 1977;4:134–142. doi: 10.1002/bms.1200040303. [DOI] [PubMed] [Google Scholar]
- 13.Sawamura K. Compendium of apple and pear diseases. In: Jones AL, Aldwinkle HS, editors. Alternaria blotch. St. Paul: APS Press; 1990. pp. 24–25. [Google Scholar]
- 14.Filajdic N, Sutton TB. Identification and distribution of Alternaria mali on apples in North Carolina and susceptibility of different varieties of apples to Alternaria blotch. Plant Dis. 1991;75:1045–1048. doi: 10.1094/PD-75-1045. [DOI] [Google Scholar]
- 15.Abe K, Iwanami H, Kotoda N, Moriya S, Takahashi S. Evaluation of apple genotypes and Malus species for resistance to Alternaria blotch caused by Alternaria alternata apple pathotype using detached-leaf method. Plant Breed. 2010;129:208–218. doi: 10.1111/j.1439-0523.2009.01672.x. [DOI] [Google Scholar]
- 16.Saito K, Takeda K. Genetic analysis of resistance to Alternaria blotch in apple. Japan J Breed. 1984;34:197–209. doi: 10.1270/jsbbs1951.34.197. [DOI] [Google Scholar]
- 17.Terakami S, Moriya S, Adachi Y, Kunihisa M, Nishitani C, Saito T, et al. Fine mapping of the gene for susceptibility to black spot disease in Japanese pear (Pyrus pyrifolia Nakai) Breed Sci. 2016;66:271–280. doi: 10.1270/jsbbs.66.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daccord M, Celton JM, Linsmith G, Becker C, Choisne N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;48:1099–1106. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhang LY, Zhang Z, Cong PH, Cheng ZM. A simple sequence repeat marker linked to the susceptibility of apple to Alternaria blotch caused by Alternaria alternata apple pathotype. J Am Soc Hortic Sci. 2011;136:109–115. doi: 10.21273/JASHS.136.2.109. [DOI] [Google Scholar]
- 20.Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, et al. Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree Genet Genomes. 2006;2:202–224. doi: 10.1007/s11295-006-0045-1. [DOI] [Google Scholar]
- 21.Li Y, Hirst PM, Wan Y, Liu Y, Zhou Q, Gao H. Resistance to Marssonina coronaria and Alternaria alternata apple pathotype in the major apple cultivars and rootstocks used in China. HortScience. 2012;47:1241–1244. doi: 10.21273/HORTSCI.47.9.1241. [DOI] [Google Scholar]
- 22.Tsuchiya S, Yoshida Y, Haniuda T. Studies on the disease resistance in apple I investigation on the resistance to Alternaria blotch in apple varieties and hybrids. Bull Hort Res Sta Ser C. 1967;5:9–19. [Google Scholar]
- 23.Tsuyama H, Kodama Y, Nakazawa N. Histological studies on the different varietal reaction in Alternaria leaf spot disease of apples caused by Alternaria mali rob. Bull Fac Agric Iwate Univ. 1971;10:217–231. [Google Scholar]
- 24.Sawamura K, Yanase H. Studies on spotted disease of apples 2. On Alternaria sp., causal organism of Alternaria blotch and Japanese name of the disease. Bull Hort Res Stn Series. 1963;1:77–94. [Google Scholar]
- 25.Kohmoto K, Taniguchi T, Nishimura S. Correlation between the susceptibility on apple cultivars to Alternaria mali and their sensitivity to AM-toxin I. Ann Phytopathol Soc Japan. 1977;43:65–68. doi: 10.3186/jjphytopath.43.65. [DOI] [Google Scholar]
- 26.Perazzolli M, Malacarne G, Baldo A, Righetti L, Bailey A, Fontana P, et al. Characterization of resistance gene analogues (RGAs) in apple (Malus x domestica Borkh.) and their evolutionary history of the Rosaceae family. PLoS One. 2014;9:e83844. doi: 10.1371/journal.pone.0083844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW. Plant disease resistance genes: current status and future directions. Physiol Mol Plant Pathol. 2012;78:51–65. doi: 10.1016/j.pmpp.2012.01.002. [DOI] [Google Scholar]
- 28.Marone D, Russo MA, Laido G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci. 2013;14:7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/S0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 30.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/S0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 31.Mackay D, Holt BF, III, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2003;108:743–754. doi: 10.1016/S0092-8674(02)00661-X. [DOI] [PubMed] [Google Scholar]
- 32.Zhu LM, Ni WC, Liu S, Cai BH, Xing H, Wang SH. Transcriptomics analysis of apple leaves in response to Alternaria alternata apple pathotype infection. Front Plant Sci. 2017;8:22. doi: 10.3389/fpls.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolpert TJ, Lorang JM. Victoria blight, defense turned upside down. Physiol Mol Plant Pathol. 2016;95:8–13. doi: 10.1016/j.pmpp.2016.03.006. [DOI] [Google Scholar]
- 35.Lorang JM, Carkaci-Salli N, Wolpert TJ. Identification and characterization of victorin sensitivity in Arabidopsis thaliana. Mol Plant Microbe Interac. 2004;17:577–582. doi: 10.1094/MPMI.2004.17.6.577. [DOI] [PubMed] [Google Scholar]
- 36.Sweat TA, Wolpert TJ. Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis thaliana. Plant Cell. 2007;19:673–687. doi: 10.1105/tpc.106.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC. Tricking the guard: exploiting plant defense for disease susceptibility. Science. 2012;338:659–662. doi: 10.1126/science.1226743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorang JM, Sweat TA, Wolpert TJ. Plant disease susceptibility conferred by a “resistance” gene. Proc Natl Acad Sci U S A. 2007;104:14861–14866. doi: 10.1073/pnas.0702572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabira H, Otani H, Shimomura N, Kodama M, Kohmoto K, Nishimura S. Light-induced insensitivity of apple and Japanese pear leaves to AM-toxin from Alternaria alternata apple pathotype. Ann Phytopathol Soc Jpn. 1989;55:567–578. doi: 10.3186/jjphytopath.55.567. [DOI] [Google Scholar]
- 40.Kozaki I. Black spot disease resistance in Japanese pear. I. Heredity of the disease resistance. Bull Hort Res Stn Japan A. 1973;12:17–27. [Google Scholar]
- 41.Yamamoto M, Namiki F, Nishimura S, Kohmoto K. Studies on host-specific AF-toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry cultivar Morioka-16. Ann Phytopath Soc Japan. 1985;51:530–35.
- 42.Tanahashi M, Nakano T, Akamatsu H, Kodama M, Otani H, Osaki-Oka K. Alternaria alternata apple pathotype (A. mali) causes black spot of European pear. Eur J Plant Pathol. 2016;145:787–795. doi: 10.1007/s10658-016-0866-1. [DOI] [Google Scholar]
- 43.Moriya S, Terakami S, Iwanami H, Haji T, Okada K, Yamamoto T, et al. Genetic mapping and marker-assisted selection of the gene conferring susceptibility to Alternaria blotch caused by Alternaria alternata apple pathotype in apple. Acta Hort. 2013;976:555–560. doi: 10.17660/ActaHortic.2013.976.79. [DOI] [Google Scholar]
- 44.Velasco Riccardo, Zharkikh Andrey, Affourtit Jason, Dhingra Amit, Cestaro Alessandro, Kalyanaraman Ananth, Fontana Paolo, Bhatnagar Satish K, Troggio Michela, Pruss Dmitry, Salvi Silvio, Pindo Massimo, Baldi Paolo, Castelletti Sara, Cavaiuolo Marina, Coppola Giuseppina, Costa Fabrizio, Cova Valentina, Dal Ri Antonio, Goremykin Vadim, Komjanc Matteo, Longhi Sara, Magnago Pierluigi, Malacarne Giulia, Malnoy Mickael, Micheletti Diego, Moretto Marco, Perazzolli Michele, Si-Ammour Azeddine, Vezzulli Silvia, Zini Elena, Eldredge Glenn, Fitzgerald Lisa M, Gutin Natalia, Lanchbury Jerry, Macalma Teresita, Mitchell Jeff T, Reid Julia, Wardell Bryan, Kodira Chinnappa, Chen Zhoutao, Desany Brian, Niazi Faheem, Palmer Melinda, Koepke Tyson, Jiwan Derick, Schaeffer Scott, Krishnan Vandhana, Wu Changjun, Chu Vu T, King Stephen T, Vick Jessica, Tao Quanzhou, Mraz Amy, Stormo Aimee, Stormo Keith, Bogden Robert, Ederle Davide, Stella Alessandra, Vecchietti Alberto, Kater Martin M, Masiero Simona, Lasserre Pauline, Lespinasse Yves, Allan Andrew C, Bus Vincent, Chagné David, Crowhurst Ross N, Gleave Andrew P, Lavezzo Enrico, Fawcett Jeffrey A, Proost Sebastian, Rouzé Pierre, Sterck Lieven, Toppo Stefano, Lazzari Barbara, Hellens Roger P, Durel Charles-Eric, Gutin Alexander, Bumgarner Roger E, Gardiner Susan E, Skolnick Mark, Egholm Michael, Van de Peer Yves, Salamini Francesco, Viola Roberto. The genome of the domesticated apple (Malus × domestica Borkh.) Nature Genetics. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 45.You FM, Huo N, Qiang GY, Luo MC, Ma Y, Hane D, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriya S, Okada K, Haji T, Yamamoto T, Abe K. Fine mapping of Co, a gene controlling columnar growth habit located on apple (Malus × domestica Borkh.) linkage group 10. Plant Breed. 2012;131:641–647. doi: 10.1111/j.1439-0523.2012.01985.x. [DOI] [Google Scholar]
- 47.van Ooijen JW (2006) JoinMap 4 Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
- 48.Grattapaglia D, Sederoff R. Genetic-linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137(Suppl 4):1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baba T, Katagiri S, Tanoue H, Tanaka R, Chiden Y, Saji S, et al. Construction and characterization of rice genomic libraries: PAC library of Japonica variety, Nipponbare and BAC library of Indica variety, Kasalath. Bull Natl Inst Agrobiol Resour. 2000;14:41–52. [Google Scholar]
- 50.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 51.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- 53.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pedigree of susceptible apple accessions used in this study. (PDF 50 kb)
Table S1. Mapping of SSR markers used in marker enrichment analysis. (PDF 30 kb)
Table S2. Graphical illustration of genotypes showing recombination between Alt and flanking markers. (PDF 38 kb)
Figure S2. Pairwise alignment of A6, a predicted ORF in the Alt region and XP_009379454.1, the gene most similar to A6, as revealed by a database search. (PDF 49 kb)
Figure S3. Amino acid sequence alignment of predicted candidate genes of Alt, A8, a2, and G14. (PDF 37 kb)
Figure S4. Amino acid sequence alignment of predicted candidate genes of Alt, A10, a4, and G16. (PDF 36 kb)
Table S3. Presence of 12-bp insertion among founders and old cultivars. (PDF 50 kb)
Data Availability Statement
The nucleotide sequences of BAC clones are available in DDBJ as LC360748 and LC360749 for SD-178_L10 and SD-241_O01, respectively.