Abstract
Background
Enterocytozoon bieneusi is a unicellular microsporidian fungal pathogen that infects a broad range of animal hosts, including wild and domestic animals and humans. The infection burden of this parasite in wild animals in Korea is largely unknown. In this study, the occurrence and genotypes of E. bieneusi were investigated in wild animal populations in Korea.
Methods
A total of 157 fecal samples (97 from Korean water deer, 48 from raccoon dogs and 12 from other taxa) were collected from wild animals at five wildlife centers in Korea. Genomic DNA was extracted from the samples and screened by nested-PCR targeting the internal transcribed spacer (ITS) region of rRNA, followed by sequence analysis to determine the genotype(s) of E. bieneusi.
Results
The overall prevalence of E. bieneusi was 45.2% (71/157), with rates of 53.6% (52/97) in Korean water deer, 35.4% (17/48) in raccoon dogs and 16.7% (2/12) in other taxa. We detected seven ITS genotypes, including one known (genotype D) and six new genotypes (Korea-WL1–Korea-WL6). Phylogenetically, all detected genotypes clustered with counterparts belonging to group 1, which includes isolates from different animal hosts and humans, suggesting their zoonotic potential.
Conclusions
Our survey results indicate that E. bieneusi circulates widely in wild animals in Korea. These findings address the role of wildlife as a potential source of microsporidiosis in domestic animals and humans.
Keywords: Enterocytozoon bieneusi, Prevalence, Wildlife, Korean water deer, Raccoon dog, Genotyping, South Korea
Background
Enterocytozoon bieneusi is a cosmopolitan microsporidian that infects a wide range of vertebrate and invertebrate hosts, including humans, domestic animals and wild game [1–6]. Enterocytozoon bieneusi infection is associated with enteropathy, resulting in diarrhea, malabsorption and occasionally growth impairment, especially in pediatric and immunocompromised individuals [7–12]. In addition, extra-intestinal microsporidian infections due to E. bieneusi are frequently reported [13–15]. Fecal-oral transmission is the most common route of infection through spore-contaminated food and/or water [9, 16–18]. Airborne transmission in cases of respiratory infections is still largely controversial [13]. Treatment options for E. bieneusi infections are limited, and few drugs have shown anti-microsporidian activity [9, 19].
Microscopic investigations of E. bieneusi spores in stool samples are inadequate to discriminate between genotypes owing to a lack of morphological differences. Molecular biology analyses are widely used for the sensitive detection and genotyping of microsporidian species [20, 21]. The genetic structure of the internal transcribed spacer (ITS) region of rRNA shows high diversity among isolates [22], providing the ability to differentiate between host-adapted and zoonotic genotypes [1, 2, 23] as well as the characterization of the geographical distribution of certain genotypes [24].
Enterocytozoon bieneusi is commonly detected in different wildlife, either in captive or free-living populations [5, 25–31], with ample range of genotypes of both host-adapted and those lacking host specificity [1, 32]. Thus, wildlife animals are recognized as environmental reservoir for many human and animal infections [32, 33]. In Korea, a few studies have reported the molecular detection of E. bieneusi in livestock [34, 35] and in bats [36]; however, the infection burden of this microsporidian fungal pathogen in wildlife is largely unknown. Therefore, in the present study, the prevalence and genotypes of this parasite in wild animal populations in Korea were determined. Fecal samples were collected from wild animals at wildlife centers. Samples were screened for parasite occurrence by nested-PCR targeting the ITS region of rRNA. Genotypes were determined by sequence analysis of the positive samples. The relatedness of the obtained sequences was determined by alignment with reference sequences in GenBank and phylogenetic analysis.
Methods
Collection of samples
This study was performed from April to September, 2018, using samples collected from wild animals at five Korean wildlife centers (Fig. 1). A total of 157 fecal samples were collected from different animal taxa (Table 1), including 97 samples from Korean water deer (Hydropotes inermis argyropus), 48 from raccoon dogs (Nyctereutes procyonoides), 2 from roe deer (Capreolus pygargus), 3 from leopard cats (Prionailurus bengalensis), 3 from Eurasian otters (Lutra lutra), 3 from the Siberian weasel (Mustela sibirica) and 1 from a Eurasian badger (Meles meles). Fresh samples were placed in labeled plastic cups, mixed with 10 volumes of 70% ethanol, and stored at 4 °C. The samples were transported in an ice box to the laboratory of Veterinary Laboratory Medicine at the Veterinary Medical Center, CNBU, Cheongju, Korea.
Fig. 1.
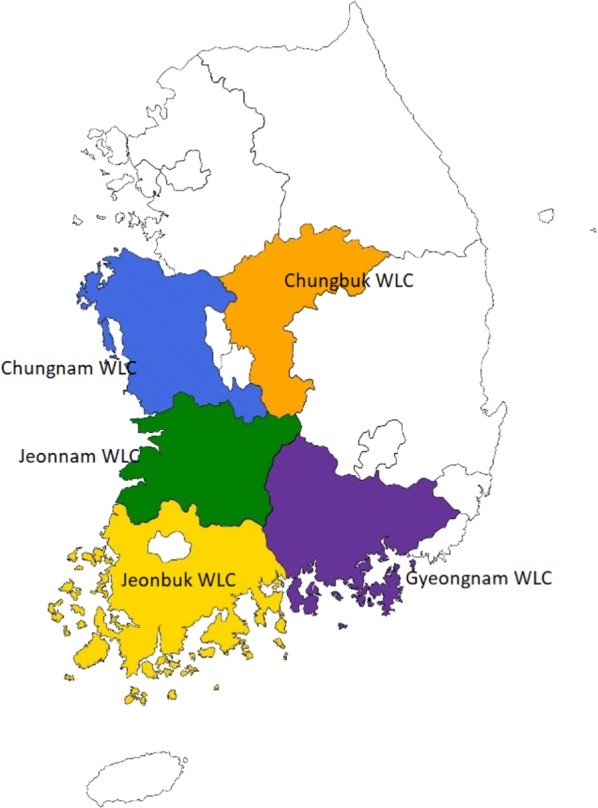
Site map of wildlife centers which contributed to the collection of samples analyzed in the present study
Table 1.
Distribution of samples and prevalence of E. bieneusi for each wildlife center and animal species
| Location | Korean water deer (n = 97) | Raccoon dog (n = 48) | Other taxa (n = 12) | No. infected | Total number | Infection rate (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Infected | Uninfected | Infected | Uninfected | Infected | Uninfected | ||||
| Chungbuk | 7 | 11 | 4 | 11 | 0 | 4 | 11 | 37 | 29.7 |
| Jeonbuk | 16 | 12 | 13 | 7 | 1 | 2 | 30 | 51 | 58.8 |
| ChungNam | 3 | 6 | 0 | 9 | 1 | 1 | 4 | 20 | 20.0 |
| JeonNam | 3 | 7 | 0 | 4 | 0 | 3 | 3 | 17 | 17.7 |
| GyungNam | 23 | 9 | 0 | 0 | 0 | 0 | 23 | 32 | 71.9 |
| Total | 52 | 45 | 17 | 31 | 2 | 10 | 71 | 157 | 45.2a |
| Infection rate (%) | 53.6 | 35.4 | 16.7 | ||||||
aOverall rate
DNA extraction and PCR amplification
Ethanol was washed off the fecal samples with Milli Q water by centrifugation. Genomic DNA was extracted from 0.2 ml of fecal slurry using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according the manufacturer’s instructions. DNA preparations were screened for E. bieneusi by nested PCR as previously described [5], with reagent grade water as a negative control. PCR was performed using 50-µl reaction mixtures consisting of 1× PCR buffer, 200 µM dNTPs, 3 mM MgCl2, 260 nM primers, and 1.5 units of Taq DNA polymerase (Takara, Tokyo, Japan). Amplified fragments were electrophoresed on a 1.5% gel stained with EcoDye™ and visualized using UV light.
DNA sequence analysis
Secondary PCR products of positive samples were sequenced in both directions using Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3130 Genetic Analyzer (Applied Biosystems). Generated sequences were assembled using ChromasPro (https://technelysium.com.au/wp/chromaspro/ v.2.1.8) and aligned with reference sequences in GenBank using ClustalX (http://www.clustal.org/) to determine the occurrence and genotype of E. bieneusi. Phylogenetic analysis was performed using maximum likelihood (ML) as implemented in MEGA v.7.0 (https://www.megasoftware.net/) with the Kimura 2-parameter model.
Results
Occurrence of Enterocytozoon bieneusi
Based on PCR and sequencing, we detected E. bieneusi in 71 out of 157 investigated samples, with an overall infection rate of 45.2%. The prevalence was 53.6% (52/97) and 35.4% (17/48) in Korean water deer and raccoon dogs, respectively (Table 1). In addition, one positive sample was detected in each of the roe deer (1/2) and leopard cat (1/3). Samples from other taxa, including three otters, three weasels and one Eurasian badger, were negative. Infection was distributed in animals from all wildlife centers involved in the present study, with the highest infection rate detected at Gyungnam wildlife center (71.9%), followed by Jeonbuk (58.8%), Chungbuk (29.7%), Chungnam (20.0%) and Jeonnam (17.7%).
Enterocytozoon bieneusi genotypes
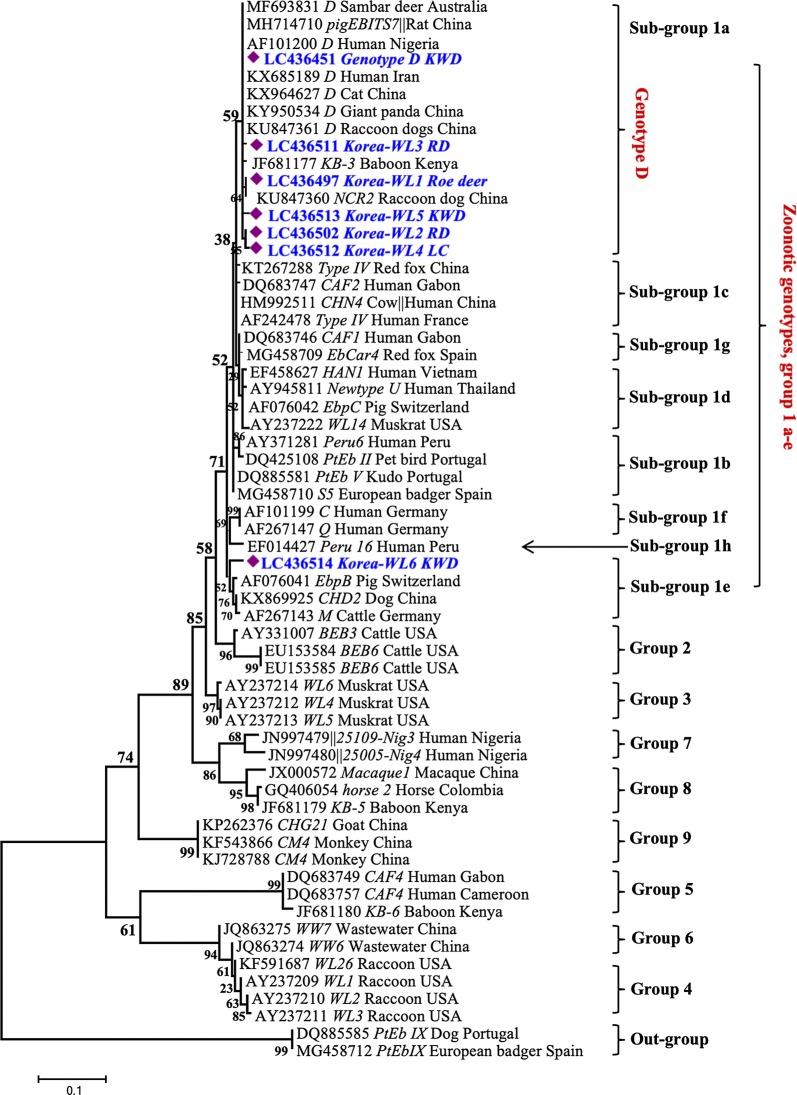
Based on sequence analysis of the ITS gene marker, we detected seven genotypes, including one known (genotype D) and six novel genotypes (Korea-WL1-WL6). The known genotype D (detected in 35 samples isolated from 29 Korean water deer and 6 raccoon dogs) was identical to KX685189 from a human in Iran [11], KY950543 from a giant panda in China [28] and MF693831 from a sambar deer in Australia [5]. The novel genotypes Korea-WL1 (detected in 21 samples isolated from 12 Korean water deer, 8 raccoon dogs and 1 roe deer), Korea-WL2 genotype (detected in 11 samples isolated from 6 raccoon dogs and 5 Korean water deer) and Korea-WL3 (detected in one sequence isolated from a raccoon dog) showed one nucleotide substitution each at positions 174 (A/G), 277 (T/G) and 232 (A/G), respectively, compared to our genotype D sequences and the reference sequence (MF693831). We detected Korea-WL4 in one sequence (derived from sample of a leopard cat) with two substitutions (A/G and T/G) at positions 152 and 277, respectively. Korea-WL5, also detected in one sequence derived from a sample of Korean water deer, showed a 2-bp deletion at positions 152 and 153 as well as substitutions of T/G and G/A at positions 259 and 298, respectively, compared to genotype D (Table 2). Moreover, Korea-WL6 (generated from sample of a Korean water deer) showed a sequence identity of 98% with KR902354 detected in wastewater in China [37] and KP262358 detected from a goat in China [38], with seven nucleotide substitutions. Phylogenetically, all detected genotypes clustered with counterparts belonging to zoonotic group 1. Genotype D and novel genotypes Korea-WL1-WL5 clustered with other sequences of group 1a, whereas Korea-WL6 clustered with sequences in group 1e (Fig. 2).
Table 2.
Positions of nucleotide changes within E. bieneusi genotype D-related sequences
| Genotype/sequence position | 152 | 159 | 160 | 174 | 232 | 259 | 277 | 298 | Host animal | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| Korea genotype D (35 sequences)/MF693831 | G | T | G | G | G | G | G | A | KWD, RD | LC436451-LC436482 |
| Korea-WL1 (21 sequences) | . | . | . | A | . | . | . | . | KWD, RD, roe deer | LC436483- LC436501 |
| Korea-WL2 (11 sequences) | . | . | . | . | . | . | T | . | KWD, RD | LC436502-LC436510 |
| Korea-WL3 (1 sequence) | . | . | . | . | A | . | . | . | RD | LC436511 |
| Korea-WL4 (1 sequence) | A | . | . | . | . | . | T | . | LC | LC436512 |
| Korea-WL5 (1 sequence) | . | – | – | . | . | T | . | G | KWD | LC436513 |
Abbreviations: KWD, Korean water deer; RD, raccoon dog; LC, leopard cat; –, deletion
Fig. 2.
Phylogenetic relationships among Enterocytozoon bieneusi from wildlife in Korea and isolates with reference sequences in GenBank. Evolutionary relationships were inferred based on internal transcribed spacer (ITS) sequences using the Maximum Likelihood (ML) method implemented in MEGA7. Branch support on the ML tree was calculated based on 1000 bootstrap replicates. Sequences obtained in this study are marked with diamonds on the tree. In all cases, the branch label includes accession number, followed by genotype name, host animal and country of origin. Abbreviations: KWD, Korean water deer; RD, raccoon dog; LC, leopard cat
Discussion
Our results provide the first description of the occurrence and genotypes of the microsporidian E. bieneusi in wild animal populations in Korea. We observed an overall prevalence of 45.2%, with the highest infection rate (53.6%) detected in samples from Korean water deer, followed by raccoon dogs (35.4%) and other taxa (16.7%). Little is known about the occurrence of E. bieneusi in Korean water deer and free-range raccoon dogs. However, a low infection rate of 4.1% was recently reported in a survey of wild deer in water catchments in Melbourne, Australia [5], with all infections restricted to the sambar deer. The average E. bieneusi infection rates are 16.8% in wild reindeers [39], 34.0% in wild Pere David’s deer in China [40] and 32.5% in wild white-tailed deer in the USA [41]. In farmed deer, the prevalence is 7–35% in sika deer, 20–37% in red deer [42–45] and ~17% in musk deer [31] in China. Compared with the 35.4% E. bieneusi infection rate in raccoon dogs in our study, a lower prevalence of 4–22% has been reported in farmed raccoon dogs (N. procyonoides) in China [29, 46, 47]. Moreover, rates of E. bieneusi occurrence differ according to the animal species and geographical region. Danišová et al. [48] reported an infection rate of 1.07% in wild mice in Slovakia, far lower than estimates of 38.9% in different wild rodents in Poland [25]. Additionally, a prevalence of 16.7% was observed in red-bellied tree squirrels in China [27]. A comparable prevalence of 41.2% was detected in Eurasian wild boars in China [28]. Similar rates of infection (~11%) have been observed in the herbivorous giant panda in China [26] and in wild carnivores (~13%) in Spain [33]. Humidity and temperature due to precipitation rate and vegetation types in forests in Korea are favorable environmental factors that maintain viability of E. bieneusi spores for long time and the social behavior of the Korean water deer and raccoon dogs that facilitates contact among families and groups of animals may be responsible for the high infection rate detected in the present study.
Our PCR and sequence analysis of the ITS region revealed a substantial degree of genetic diversity in the form of nucleotide substitutions and deletions. We identified seven distinct genotypes, including one known (genotype D) and six novel genotypes (Korea-WL1-WL6). Comparable results for genetic diversity and new genotypes have been obtained for isolates from different species of wild and farmed deer in Australia and China [5, 39, 40, 44]. Contrasting our findings, more new genotypes (up to 25) were detected in wild white-tailed deer in the USA [41] and in farmed sika deer in China [45], and only one or two new genotypes were detected in musk deer, red deer and Siberian roe deer in China [31, 43]. Genotype D of E. bieneusi detected in raccoon dogs in our study is a common genotype in farmed raccoon dogs in China [46], along with various (3–7) additional known and new genotypes [29, 46, 47]. Results of this study expand the geographical and host range of these genotypes.
Phylogenetic analysis (Fig. 2) revealed that E. bieneusi genotypes cluster in roughly eight groups. Genotypes of group 1 have no host specificity and were retrieved from different wildlife, domestic animals and humans, and those belonging to groups 2–8 are almost all host-specific [22, 49, 50]. In this study, all detected genotypes were assigned to E. bieneusi group 1; 70 sequences belonged to 1a and one sequence belonged to 1e. Furthermore, isolates of genotype D, Korea-WL1 and Korea-WL2 reported in the present study were obtained from different hosts including Korean water deer, raccoon dogs and roe deer, indicating a lack of host specificity. However, the situation of host specificity for Korea-WL3 to Korea-WL6 genotypes is difficult due to isolation from only one sample. Similarly, most previously reported genotypes detected in wild and farmed deer and raccoon dogs belonged to group 1 [5, 29, 39–41, 45–47]. The transmission of group 1 genotypes between animals and humans is likely, indicating that they have zoonotic potential [32, 51]. The exclusive identification of genotypes belonging to group 1 in this study emphasizes the role of wild populations as a source of environmental contamination; these genotypes are potentially hazardous for domestic animals and human health in Korea.
Conclusions
Our results provide the first evidence for the presence of E. bieneusi in wildlife in Korea. All infected animals carried potentially zoonotic pathogenic genotypes of this microsporidian. This study was limited by the uneven collection of samples from different wildlife centers and animal species, which my lead to biases in the results. Therefore, a larger study covering more geographical regions of Korea with even sample collection is underway to ascertain the role of wildlife as a reservoir for zoonotic gastrointestinal pathogen. The obtained results expand the geographical and host range of the detected genotypes and improve our understanding of the epidemiology of E. bieneusi infection in humans and animals in Korea.
Acknowledgements
The staff at contributing wildlife centers and College of Veterinary Medicine, Chungbuk National University are greatly appreciated.
Funding
This work was supported by the Brain Pool Programme funded by the Ministry of Science and ICT through the National Research Foundation of Korea (2018H1D3A2002236).
Availability of data and materials
Nucleotide sequences generated in this study are available in the GenBank database under accession numbers LC436451-LC436514.
Authors’ contributions
SA and KJN conceived of the study. SA, SK and JIH collected the samples and conducted the experiments. SA, JIH and KJN analyzed the data. SA and KJN prepared the report for publication. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- KWD
Korean water deer (Hydropotes inermis argyropus)
- ITS
internal transcribed spacer of nuclear ribosomal DNA
Contributor Information
Said Amer, Email: mssamer5@yahoo.com.
Sungryong Kim, Email: vet08dannyk@gmail.com.
Jae-Ik Han, Email: jihan@jbnu.ac.kr.
Ki-Jeong Na, Email: sigol@cbnu.ac.kr.
References
- 1.Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, 3rd, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003;69:4495–4501. doi: 10.1128/AEM.69.8.4495-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, Xiao L. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol Res. 2004;92:328–334. doi: 10.1007/s00436-003-1049-5. [DOI] [PubMed] [Google Scholar]
- 3.Didier ES, Weiss LM. Microsporidiosis: current status. Curr Opin Infect Dis. 2006;19:485–492. doi: 10.1097/01.qco.0000244055.46382.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen D, Wang SS, Zou Y, Li Z, Xie SC, Shi LQ, et al. Prevalence and multi-locus genotypes of Enterocytozoon bieneusi in black-boned sheep and goats in Yunnan Province, southwestern China. Infect Genet Evol. 2018;65:385–391. doi: 10.1016/j.meegid.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Koehler AV, Wang T, Haydon SR, Gasser RB. First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourneʼs water catchments in Australia. Parasit Vectors. 2018;11:2. doi: 10.1186/s13071-017-2577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou Y, Hou JL, Li FC, Zou FC, Lin RQ, Ma JG, et al. Prevalence and genotypes of Enterocytozoon bieneusi in pigs in southern China. Infect Genet Evol. 2018;66:52–56. doi: 10.1016/j.meegid.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Kotler DP, Orenstein JM. Clinical syndromes associated with microsporidiosis. Adv Parasitol. 1998;40:321–349. doi: 10.1016/S0065-308X(08)60126-8. [DOI] [PubMed] [Google Scholar]
- 8.Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, et al. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–925. doi: 10.4269/ajtmh.2005.73.921. [DOI] [PubMed] [Google Scholar]
- 9.Anane S, Attouchi H. Microsporidiosis: epidemiology, clinical data and therapy. Gastroenterol Clin Biol. 2010;34:450–464. doi: 10.1016/j.gcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Ghoshal U, Dey A, Ranjan P, Khanduja S, Agarwal V, Ghoshal UC. Identification of opportunistic enteric parasites among immunocompetent patients with diarrhoea from Northern India and genetic characterisation of Cryptosporidium and Microsporidia. Indian J Med Microbiol. 2016;34:60–66. doi: 10.4103/0255-0857.174114. [DOI] [PubMed] [Google Scholar]
- 11.Ghoyounchi R, Ahmadpour E, Spotin A, Mahami-Oskouei M, Rezamand A, Aminisani N, et al. Microsporidiosis in Iran: a systematic review and meta-analysis. Asian Pac J Trop Med. 2017;10:341–350. doi: 10.1016/j.apjtm.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Hassan NA, Lim YAL, Mahmud R, Mohd-Shaharuddin N, Wan Sulaiman WY, Ngui R. Molecular diagnosis of microsporidia among immunocompromised patients in Kuala Lumpur, Malaysia. Am J Trop Med Hyg. 2018;99:1562–1566. doi: 10.4269/ajtmh.17-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Aguila C, Lopez-Velez R, Fenoy S, Turrientes C, Cobo J, Navajas R, et al. Identification of Enterocytozoon bieneusi spores in respiratory samples from an AIDS patient with a 2-year history of intestinal microsporidiosis. J Clin Microbiol. 1997;35:1862–1866. doi: 10.1128/jcm.35.7.1862-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botterel F, Minozzi C, Vittecoq D, Bourée P. Pulmonary localization of Enterocytozoon bieneusi in an AIDS patient: case report and review. J Clin Microbiol. 2002;40:4800–4801. doi: 10.1128/JCM.40.12.4800-4801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kicia M, Sędzimirska M, Sak B, Kváč M, Wesołowska M, Hendrich AB, et al. Respiratory microsporidiosis caused by Enterocytozoon bieneusi in an HIV-negative hematopoietic stem cell transplant recipient. Int J Infect Dis. 2018;77:26–28. doi: 10.1016/j.ijid.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Jedrzejewski S, Graczyk TK, Slodkowicz-Kowalska A, Tamang L, Majewska AC. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl Environ Microbiol. 2007;73:4071–4073. doi: 10.1128/AEM.00477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decraene V, Lebbad M, Botero-Kleiven S, Gustavsson AM, Löfdahl M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol Infect. 2012;140:519–527. doi: 10.1017/S095026881100077X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galván AL, Magnet A, Izquierdo F, Fenoy S, Rueda C, Fernández Vadillo C, et al. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: a year-long longitudinal study. Appl Environ Microbiol. 2013;79:449–459. doi: 10.1128/AEM.02737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han B, Weiss LM. Therapeutic targets for the treatment of microsporidiosis in humans. Expert Opin Ther Targets. 2018;22:903–915. doi: 10.1080/14728222.2018.1538360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. 2012;2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyworth MF. Molecular diagnosis of human microsporidian infections. Trans R Soc Trop Med Hyg. 2017;111:382–383. doi: 10.1093/trstmh/trx070. [DOI] [PubMed] [Google Scholar]
- 22.Santín M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56:34–38. doi: 10.1111/j.1550-7408.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- 23.Widmer G, Akiyoshi DE. Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi. Infect Genet Evol. 2010;10:122–128. doi: 10.1016/j.meegid.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henriques-Gil N, Haro M, Izquierdo F, Fenoy S, del Aguila C. Phylogenetic approach to the variability of the microsporidian Enterocytozoon bieneusi and its implications for inter- and intrahost transmission. Appl Environ Microbiol. 2010;76:3333–3342. doi: 10.1128/AEM.03026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perec-Matysiak A, Buńkowska-Gawlik K, Kváč M, Sak B, Hildebrand J, Leśniańska K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet Parasitol. 2015;214:242–246. doi: 10.1016/j.vetpar.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Tian GR, Zhao GH, Du SZ, Hu XF, Wang HB, Zhang LX, et al. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect Genet Evol. 2015;34:32–35. doi: 10.1016/j.meegid.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Deng L, Li W, Yu X, Gong C, Liu X, Zhong Z, et al. First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS One. 2016;11:e0163605. doi: 10.1371/journal.pone.0163605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Deng L, Wu K, Huang X, Song Y, Su H, et al. Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasit Vectors. 2017;10:10. doi: 10.1186/s13071-016-1942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, Ma X, Zhang H, Zhang XX, Zhao JP, Ba HX, et al. Prevalence, risk factors and molecular characterization of Enterocytozoon bieneusi in raccoon dogs (Nyctereutes procyonoides) in five provinces of northern China. Acta Trop. 2016;161:68–72. doi: 10.1016/j.actatropica.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Cong W, Qin SY, Meng QF. Molecular characterization and new genotypes of Enterocytozoon bieneusi in minks (Neovison vison) in China. Parasite. 2018;25:34. doi: 10.1051/parasite/2018038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y, Li W, Liu H, Zhong Z, Luo Y, Wei Y, et al. First report of Giardia duodenalis and Enterocytozoon bieneusi in forest musk deer (Moschus berezovskii) in China. Parasit Vectors. 2018;11:204. doi: 10.1186/s13071-018-2681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leśniańska K, Perec-Matysiak A. Wildlife as an environmental reservoir of Enterocytozoon bieneusi (Microsporidia) - analyses of data based on molecular methods. Ann Parasitol. 2017;63:265–281. doi: 10.17420/ap6304.113. [DOI] [PubMed] [Google Scholar]
- 33.Santín M, Calero-Bernal R, Carmena D, Mateo M, Balseiro A, Barral M, et al. Molecular characterization of Enterocytozoon bieneusi in wild carnivores in Spain. J Eukaryot Microbiol. 2018;65:468–474. doi: 10.1111/jeu.12492. [DOI] [PubMed] [Google Scholar]
- 34.Jeong DK, Won GY, Park BK, Hur J, You JY, Kang SJ, et al. Occurrence and genotypic characteristics of Enterocytozoon bieneusi in pigs with diarrhea. Parasitol Res. 2007;102:123–128. doi: 10.1007/s00436-007-0740-3. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH. Prevalence and molecular characteristics of Enterocytozoon bieneusi in cattle in Korea. Parasitol Res. 2007;101:391–396. doi: 10.1007/s00436-007-0468-0. [DOI] [PubMed] [Google Scholar]
- 36.Lee SH, Oem JK, Lee SM, Son K, Jo SD, Kwak D. Molecular detection of Enterocytozoon bieneusi from bats in South Korea. Med Mycol. 2018;56:1033–1037. doi: 10.1093/mmy/myx125. [DOI] [PubMed] [Google Scholar]
- 37.Ma J, Feng Y, Hu Y, Villegas EN, Xiao L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J Water Health. 2016;14:411–423. doi: 10.2166/wh.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi K, Li M, Wang X, Li J, Karim MR, Wang R, et al. Molecular survey of Enterocytozoon bieneusi in sheep and goats in China. Parasit Vectors. 2016;9:23. doi: 10.1186/s13071-016-1304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Nie C, Zhang L, Wang R, Liu A, Zhao W, Li H. First detection and genotyping of Enterocytozoon bieneusi in reindeers (Rangifer tarandus): a zoonotic potential of ITS genotypes. Parasit Vectors. 2015;8:526. doi: 10.1186/s13071-015-1155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Huang J, Karim MR, Zhao J, Dong H, Ai W, et al. Zoonotic Enterocytozoon bieneusi genotypes in Pere Davidʼs deer (Elaphurus davidianus) in Henan, China. Exp Parasitol. 2015;155:46–48. doi: 10.1016/j.exppara.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Santín M, Fayer R. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol. 2015;62:34–43. doi: 10.1111/jeu.12155. [DOI] [PubMed] [Google Scholar]
- 42.Zhao W, Zhang W, Wang R, Liu W, Liu A, Yang D, et al. Enterocytozoon bieneusi in sika deer (Cervus nippon) and red deer (Cervus elaphus): deer specificity and zoonotic potential of ITS genotypes. Parasitol Res. 2014;113:4243–4250. doi: 10.1007/s00436-014-4100-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhao W, Wang J, Yang Z, Liu A. Dominance of the Enterocytozoon bieneusi genotype BEB6 in red deer (Cervus elaphus) and Siberian roe deer (Capreolus pygargus) in China and a brief literature review. Parasite. 2017;24:54. doi: 10.1051/parasite/2017056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang XX, Cong W, Liu GH, Ni XT, Ma JG, Zheng WB, et al. Prevalence and genotypes of Enterocytozoon bieneusi in sika deer in Jilin Province, northeastern China. Acta Parasitol. 2016;61:382–388. doi: 10.1515/ap-2016-0050. [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Zhang Z, Yang Y, Wang R, Zhao J, Jian F, et al. New genotypes of Enterocytozoon bieneusi isolated from sika deer and red deer in China. Front Microbiol. 2017;8:879. doi: 10.3389/fmicb.2017.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Lin Y, Li Q, Zhang S, Tao W, Wan Q, et al. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotype D in farmed foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in China: first identification and zoonotic concern. Parasitol Res. 2015;114:4341–4348. doi: 10.1007/s00436-015-4714-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhao W, Zhang W, Yang Z, Liu A, Zhang L, Yang F, et al. Genotyping of Enterocytozoon bieneusi in farmed blue foxes (Alopex lagopus) and raccoon dogs (Nyctereutes procyonoides) in China. PLoS One. 2015;10:e0142611. doi: 10.1371/journal.pone.0142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danišová O, Valenčáková A, Stanko M, Luptáková L, Hasajová A. First report of Enterocytozoon bieneusi and Encephalitozoon intestinalis infection of wild mice in Slovakia. Ann Agric Environ Med. 2015;22:251–252. doi: 10.5604/12321966.1152075. [DOI] [PubMed] [Google Scholar]
- 49.Thellier M, Breton J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite. 2008;15:349–358. doi: 10.1051/parasite/2008153349. [DOI] [PubMed] [Google Scholar]
- 50.Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, Li J, Wang R, Li S, Li X, Rume FI, Ning C. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One. 2015;10:e0117991. doi: 10.1371/journal.pone.0117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kruse H, Kirkemo AM, Handeland K. Wildlife as source of zoonotic infections. Emerg Infect Dis. 2004;10:2067–2072. doi: 10.3201/eid1012.040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Nucleotide sequences generated in this study are available in the GenBank database under accession numbers LC436451-LC436514.