Ever since the sequence of the enzymatic assembly line that is responsible for forging the carbon backbone of the polyketide antibiotic erythromycin was determined 27 years ago, collections of domains that are repeated throughout such polyketide synthases (PKSs) have been termed “modules”, in analogy with the order of domains in the mammalian fatty acid synthase.[1] Very recently, Abe and co-workers reported that the groups of domains that genetically migrate together during the evolution of PKS assembly lines do not match this definition.[2]
Students often learn about modular PKSs in an introductory biochemistry course when taught the biosynthesis of palmitate by the mammalian fatty acid synthase.[3] They learn that enzymatic domains cooperate with acyl carrier protein (ACP) domains to elongate a priming acyl group with carbon extender units and generate a longer acyl chain whose chemistry is set by the enzymes encountered during its synthesis. An acyltransferase (AT) selects the monomer to be condensed with the growing chain by a ketosynthase (KS), and processing enzymes perform reactions around the β-keto group generated by the condensation. The common processing enzymes of polyketide assembly lines are equivalent to those of the mammalian fatty acid synthase, in which a ketoreductase (KR) stereoselectively reduces the β-keto group, a dehydratase (DH) dehydrates the resulting β-hydroxy group, and an enoylreductase (ER) reduces the β-carbon to a methylene group. A thioesterase (TE) at the C-terminus of these synthases typically offloads the final product through hydrolysis or cyclization. As the domain order of the mammalian fatty acid synthase is KS+AT+DH+ER+KR+ACP+TE, the modules of the polyketide assembly line had been defined equivalently to begin with KS and end with ACP, with optional processing enzymes in the middle.
Abe and co-workers have now reported that processing enzymes migrate during assembly line evolution with the KS that is downstream, not upstream, of them (Figure 1). They observed this by comparing four of the largest known PKSs (each 25–30 modules long and ca. 5 times larger than the bacterial ribosome), which produce the aminopolyols mediomycin, neomediomycin, ECO-02301, and neotetrafibricin. Bioinformatic analysis showed the evolutionary relationships between equivalent domains from the giant assembly lines and enabled the authors to determine which domains move together and which domains move independently during evolution. Instead of the domain order KS+AT+DH+ER+KR+ACP, a module containing all of the common processing enzymes was found to possess the domain order AT+DH+ER+KR+ACP+KS.
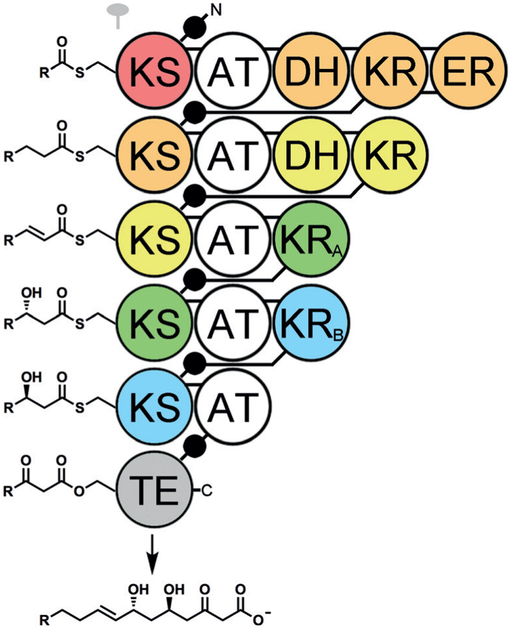
Figure 1.
Evolutionary comigration of PKS domains. Analysis of four related polyketide assembly lines of 25–30 modules revealed the enzymatic domains that move together during the evolution of the assembly lines (colored equivalently in this hypothetical synthase). The definition of a module needs to be revised such that processing enzymes, such as the ketoreductase (KR), dehydratase (DH), and enoylreductase (ER), are grouped with the ketosynthase (KS) that is downstream, not upstream, of them. The analysis also shows that acyltransferase (AT) domains, which select the carbon monomers, move independently between different types of modules during assembly line evolution. Polyketide intermediates are shown bound to KSs instead of the acyl carrier protein (ACP) domains (black circles) as this study and other investigations have indicated that KSs select between intermediates presented to them. Abe and co-workers described recombination hotspots upstream and downstream of the AT domain that may be utilized in evolution-guided engineering to produce new polyketides. The symbol for a twofold axis indicates that only half of the homodimeric assembly line is shown. A- and B-type KRs produce l- and d-oriented hydroxy groups, respectively.
The evolutionary comigration of processing enzymes and the KS domain downstream of them in assembly lines that possess embedded AT domains (cis-AT PKSs, such as the mediomycin synthase) is not completely surprising. Assembly lines without embedded AT domains (trans-AT PKSs, such as the bryostatin synthase) show strong correlations between the chemical transformations performed by the processing enzymes and the type of KS that is downstream of those processing enzymes,[1d,e, 4] as well as genetic comigration of these domains.[5] The substrate selectivities of such KSs have also been demonstrated.[6] KSs from cis-AT PKSs have also been observed to act as gatekeepers: the second KS of the erythromycin synthase heavily favors the stereochemistry set by the preceding KR, the second KS of the nanchangmycin synthase selects between two epimers interconverted by a preceding epimerase, and the second KS of the aureothin synthase selects a chain generated by two rounds of elongation over a chain that has been elongated once.[7] Engineered PKSs were found to be more active when the KS downstream to a set of processing enzymes is the KS that is downstream to those domains in the native PKS.[8] Recently, a single point mutation to the third KS of the erythromycin synthase was observed to greatly relax its substrate specificity.[9] This mutation is in a loop that, as Abe and co-workers describe, might play the role of a filter that enables KSs to specify a single intermediate generated by the preceding processing enzymes. The authors also determined that ATs evolutionarily migrate between module types independent of other domains, noting that the ATs of one assembly line are more closely related than equivalent ATs from different assembly lines.
Why has it taken so long to properly define the module? The most studied PKSs (e.g., the erythromycin and pikromycin synthases) are relatively small, and the evolutionary relationships between their domains are not very strong. In order to be convinced that they were indeed observing the comigration of domains, Abe and co-workers had to find larger assembly lines that are close to one another on the evolutionary timescale and did so with the help of the new technology of genome mining. Furthermore, the polypeptide boundaries of the hundreds of sequenced cis-AT PKSs seemed to agree with the traditional definition of a module.[10] This is especially deceptive in the stigmatellin PKS, which appears to house one module within each of its nine polypeptides.[11] In fact, short docking domains at the termini of each polypeptide enable them to noncovalently self-assemble such that modules are formed at each polypeptide junction.[12] Redefining the PKS module raises the question whether module boundaries should also be adjusted for another class of assembly line, the nonribosomal peptide synthetases (NRPSs), such that the condensation domain, which often serves as a gatekeeper, is placed at the downstream end.[13]
While the change in nomenclature will take a while to catch on, opportunities may immediately be afforded by employing the updated PKS module boundaries (Figure 2). The domains of the fifth module of the pikromycin synthase, as traditionally defined, have been visualized by electron microscopy.[14] Constructs that include the downstream KS may yield complementary structures by electron microscopy or even X-ray crystallography. Many resources have been spent over the years attempting to engineer PKSs using the traditional definition of a module. The updated module blueprints provide evolution-validated guidelines for PKS engineers tweaking an assembly line to produce derivatives of known pharmaceuticals or promising drug leads. Indeed, the updated boundaries were recently utilized to successfully excise a module from neoaureothin synthase and generate the anticipated product, homoaureothin.[15] With the paradigm shift proposed by Abe and co-workers comes renewed hope for engineering PKSs to produce designer polyketides.
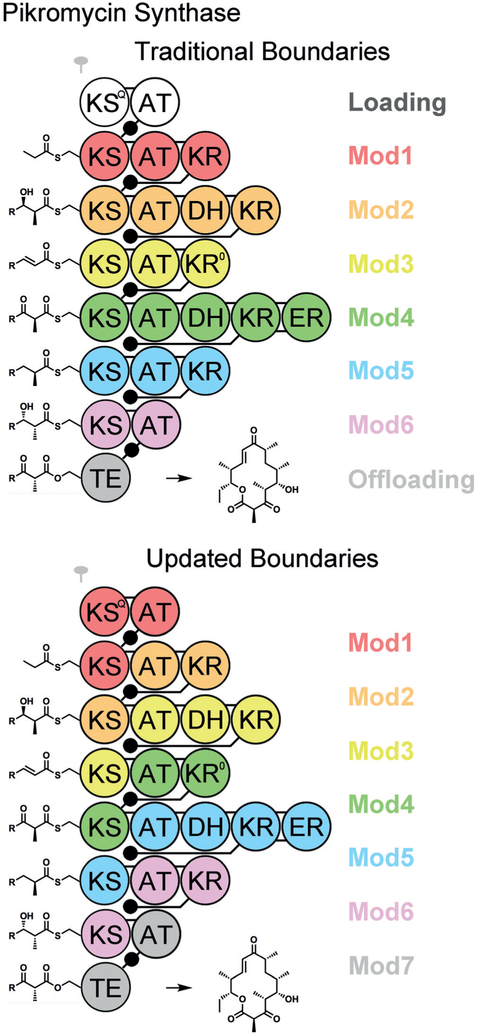
Figure 2.
The pikromycin synthase, color-coded with the traditional and updated module boundaries. The numbering of KS domains is not altered but the numbering of all other domains is increased by one. Employing the updated module boundaries, an ACP visits its own AT to acquire an extender unit, the KS of the previous module to collect the growing acyl chain through carbon–carbon bond formation, and its own processing enzymes before delivering the properly processed intermediate to its own KS. KSQ generates the priming propionyl group for the first KS through the decarboxylation of a methylmalonyl group, and belongs to the first module. The KS from module 4 selects between two intermediates interconverted by the epimerase KR0.
Acknowledgements
A.T.K. is supported by the NIH (GM106112) and the Welch Foundation (F-1712).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1] a).Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF, Nature 1990, 348, 176–178 [DOI] [PubMed] [Google Scholar]; b) Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L, Science 1991, 252, 675–679 [DOI] [PubMed] [Google Scholar]; c) Keatinge-Clay AT, Nat. Prod. Rep. 2012, 29, 1050–1073; [DOI] [PubMed] [Google Scholar]; d) Piel J, Nat. Prod. Rep. 2010, 27, 996–1047; [DOI] [PubMed] [Google Scholar]; e) Helfrich EJ, Piel J, Nat. Prod. Rep. 2016, 33, 231–316. [DOI] [PubMed] [Google Scholar]
- [2].Zhang L, Hashimoto T, Qin B, Hashimoto J, Kozone I, Kawahara T, Okada M, Awakawa T, Ito T, Asakawa Y, Ueki M, Takahashi S, Osada H, Wakimoto T, Ikeda H, Shin-ya K, Abe I, Angew. Chem. Int. Ed. 2017, 56, 1740–1745; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 1766–1771. [Google Scholar]
- [3].Maier T, Leibundgut M, Ban N, Science 2008, 321, 1315–1322. [DOI] [PubMed] [Google Scholar]
- [4].Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J, Nat. Biotechnol. 2008, 26, 225–233. [DOI] [PubMed] [Google Scholar]
- [5] a).Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J, Nat. Chem. Biol. 2009, 5, 494–501; [DOI] [PubMed] [Google Scholar]; b) Ueoka R, Uria AR, Reiter S, Mori T, Karbaum P, Peters EE, Helfrich EJN, Morinaka BI, Gugger M, Takeyama H, Matsunaga S, Piel J, Nat. Chem. Biol. 2015, 11, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jenner M, Frank S, Kampa A, Kohlhass C, Pöplau P, Briggs GS, Piel J, Oldham NJ, Angew. Chem. Int. Ed. 2013, 52, 1143–1147; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 1181–1185. [Google Scholar]
- [7] a).Wu J, Kinoshita K, Khosla C, Cane DE, Biochemistry 2004, 43, 16301–16310; [DOI] [PubMed] [Google Scholar]; b) Guo X, Liu T, Deng Z, Cane DE, Biochemistry 2012, 51, 879–887; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Busch B, Ueberschaar N, Behnken S, Sugimoto Y, Werneburg M, Traitcheva N, He J, Hertweck C, Angew. Chem. Int. Ed. 2013, 52, 5285–5289; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 5393–5397. [Google Scholar]
- [8] a).Ranganathan A, Timoney M, Bycroft M, Cortés J, Thomas IP, Wilkinson B, Kellenberger L, Hanefeld U, Galloway IS, Staunton J, Leadlay PF, Chem. Biol. 1999, 6, 731–741; [DOI] [PubMed] [Google Scholar]; b) Chandran SS, Menzella HG, Carney JR, Santi DV, Chem. Biol. 2006, 13, 469–474. [DOI] [PubMed] [Google Scholar]
- [9].Murphy AC, Hong H, Vance S, Broadhurst RW, Leadlay PF, Chem. Commun. 2016, 52, 8373–8376. [DOI] [PubMed] [Google Scholar]
- [10].Medema M et al. , Nat. Chem. Biol. 2015, 11, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gaitatzis N, Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, Muller R, J. Biol. Chem. 2002, 277, 13082–13090. [DOI] [PubMed] [Google Scholar]
- [12] a).Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ, Chem. Biol. 2003, 10, 723–731; [DOI] [PubMed] [Google Scholar]; b) Whicher JR, Smaga SS, Hansen DA, Brown WC, Gerwick WH, Sherman DH, Smith JL, Chem. Biol. 2013, 20, 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Winn M, Fyans JK, Zhuo Y, Micklefield J, Nat. Prod. Rep. 2016, 33, 317–347. [DOI] [PubMed] [Google Scholar]
- [14] a).Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan AR, Håkansson K, Sherman DH, Smith JL, Skiniotis G, Nature 2014, 510, 512–517; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Whicher JR, Dutta S, Hansen DA, Hale WA, Chemler JA, Dosey AM, Narayan AR, Håkansson K, Sherman DH, Smith JL, Skiniotis G, Nature 2014, 510, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sugimoto Y, Ishida K, Traitcheva N, Busch B, Dahse HM, Hertweck C, Chem. Biol. 2015, 22, 229–240. [DOI] [PubMed] [Google Scholar]