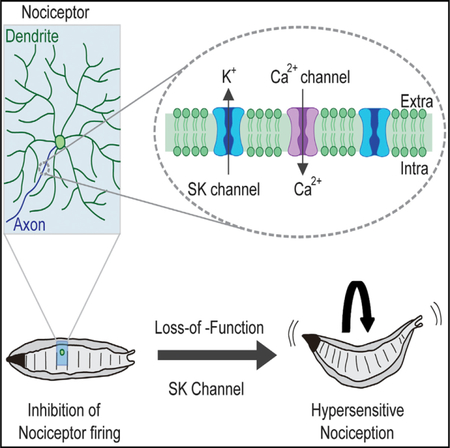
SUMMARY
Inhibition of nociceptor activity is important for the prevention of spontaneous pain and hyperalgesia. To identify the critical K+ channels that regulate nociceptor excitability, we performed a forward genetic screen using a Drosophila larval nociception paradigm. Knockdown of three K+ channel loci, the small conductance calcium-activated potassium channel (SK), seizure, and tiwaz, causes marked hypersensitive nociception behaviors. In more detailed studies of SK, we found that hypersensitive phenotypes can be recapitulated with a genetically null allele. Optical recordings from nociceptive neurons showed a significant increase in mechanically activated Ca2+ signals in SK mutant nociceptors. SK is expressed in peripheral neurons, including nociceptive neurons. Interestingly, SK proteins localize to axons of these neurons but are not detected in dendrites. Our findings suggest a major role for SK channels in the regulation of nociceptor excitation and are inconsistent with the hypothesis that the important site of action is within dendrites.
Graphical Abstract
In Brief
Walcott et al. performed a forward genetic screen and identify three potassium channel subunits that negatively regulate nociception in Drosophila larvae. In a more detailed investigation of the SK channel, null mutants, rescue experiments, optical recordings, and protein localization studies indicate a functional role for SK in nociceptor excitability.
INTRODUCTION
The sensation of pain is important for avoiding exposure to noxious environmental stimuli that have the potential to cause tissue damage. These stimuli are detected by nociceptors, which are the primary sensory neurons that detect noxious mechanical, noxious chemical, and/or noxious temperatures. Transduction of noxious thermal, mechanical, and chemical stimuli is initiated by sensory receptor ion channels, which depolarize the sensory neuron plasma membrane and trigger action potentials (Dubin and Patapoutian, 2010). In the absence of such stimuli, healthy nociceptors remain relatively silent, with little spontaneous activity (Ritter and Mendell, 1992; Xiang et al., 2010) due to the action of potassium (K+) channels and chloride (Cl—) channels, which oppose depolarizing sodium (Na+) and calcium (Ca2+) currents. Despite their importance in keeping nociceptive neurons silent, the identity of the K+ channels that play the most critical roles in negatively regulating nociceptor excitability remains largely undetermined.
To identify these critical channels, we conducted a forward genetic screen using a modified Drosophila larval nociception paradigm that was optimized for detecting hypersensitive nociception phenotypes (Honjo et al., 2016).
RESULTS AND DISCUSSION
Tissue-Specific Knockdown of K+ Channel Genes in Drosophila Uncovers Hypersensitive Thermal Nociception Phenotypes
A collection of transgenic RNAi strains from the Vienna Drosophila Resource Center (VDRC) and the Transgenic RNAi Project (TRiP) allow for in vivo tissue-specific gene silencing under control of the Gal4/UAS system (Dietzl et al., 2007; Brand and Perrimon, 1993; Ni et al., 2009). We identified 53 UAS-inverted repeat (UAS-IR) RNAi lines in these collections that targeted 34 K+ channels with few predicted off-target effects (Table S1). All known Drosophila K+ channels are represented in our assembled collection.
We first investigated the effects of knocking down the K+ channels under control of the GAL4109(2)80;UAS-Dicer2 (md-Gal4;UAS-Dicer2) driver strain. This strain drives UAS transgene expression in the class I, II, III, and IV md neurons (Gao et al., 1999). Evidence suggests that the major nociceptive function is mediated by the class IV md neurons, but class II and class III neurons are also involved (Hwang et al., 2007; Hu et al., 2017). The use of UAS-Dicer2 in the driver strain results in more efficient gene silencing (Dietzl et al., 2007). To perform the screen, the md-Gal4;UAS-Dicer2 driver strain was crossed to each of the 53 UAS-RNAi strains targeting the K+ channels and the nocifensive escape locomotion (NEL) response latency of the larval progeny stimulated with a 42°C heat probe were measured. The crossed progeny from UAS-RNAi lines targeting three distinct K+ channel subunits showed a significantly more rapid response relative to the genetic background control strain: the small conductance calcium-activated potassium channel (SK) (Abou Tayoun et al., 2011), the seizure channel (in the ether-a-gogo family) (Jackson et al., 1984), and the tiwaz gene (encodes a protein with homology to the potassium channel tetramerization domain) (Williams et al., 2014) (Figures 1A and 1C) (Table S2 reports all transgenic flies used in this study). Although phenotypes were not observed for other tested K+ channels, this method for RNAi is prone to false negatives, so our screen cannot rule out potential involvement for other channels. To test whether the effects of the RNAi were specific to the nociceptive class IV sensory neurons, we next tested animals expressing UAS-RNAi targeting these three candidates under control of ppk-Gal4;UAS-Dicer2 (Ainsley et al., 2003). The hypersensitive responses persisted in SK-RNAi and seizure-RNAi animals (Figures 1B and 1C).
Figure 1. Thermal Nociception Responses of K+ Channel Knockdown Larvae.
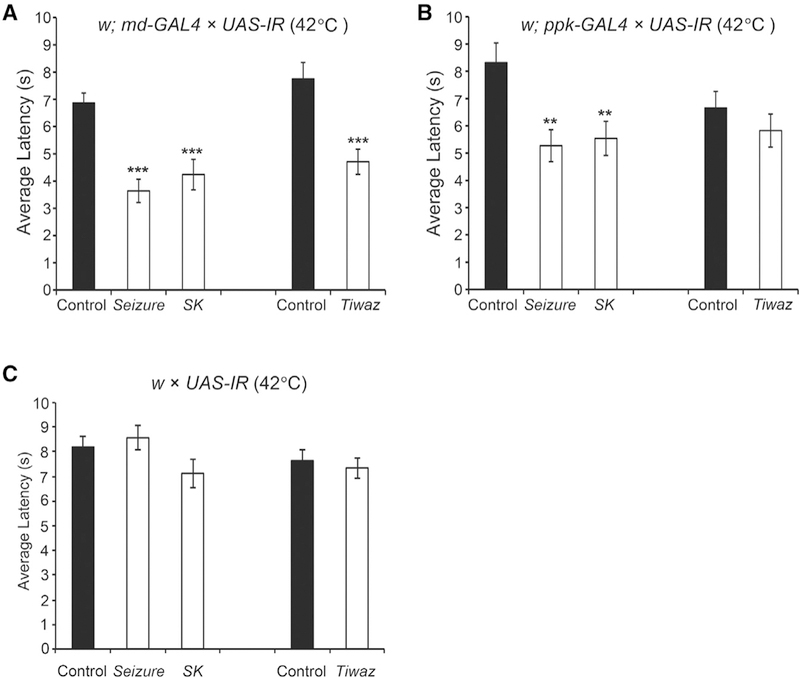
(A) In response to a noxious temperature (42°C), larval crossed progeny from w; md-Gal4; UAS-Dicer2 and UAS-IR lines targeting seizure (P{KK105733}v104698) or SK (P{KK107699} v103985) show reduced latency compared to control animals. One-way ANOVA with Dunnett’s test, ***p < 0.001. Larval crossed progeny from w;md-Gal4; UAS-Dicer2 and a UAS-IR line targeting tiwaz (P{TRiP.JF01867}) show hypersensitivity to 42°C noxious heat compared to control animals. Student’s t test, ***p < 0.001.
(B) The average NEL latency of larval progeny from w; ppk-Gal4; UAS-Dicer2 crossed to UAS-IR lines targeting SK, seizure, and tiwaz when stimulated with a 42°C probe. SK-RNAi and seizure-RNAi animals show reduced latency to perform nociception behaviors compared to control animals. One-way ANOVA with Dunnett’s test, **p < 0.01. tiwaz-RNAi animals do not show significant hypersensitivity to noxious heat compared to control animals. Student’s t test, p = 0.29. Data are presented as mean ± SEM.
(C) UAS-IR RNAi transgenes alone do not show thermal nociception defects. Control strains from the VDRC-KK and TRiP collections and RNAi transgenes targeting SK, seizure, and tiwaz were each crossed to the w1118 strain. No difference was detected relative to the control animals. Oneway ANOVA with Dunnett’s test. Similarly, animals carrying the UAS-IR transgene targeting tiwaz in the absence of the driver are not different from the control strain. Data are presented as mean ± SEM, see Table S2 for sample sizes.
SK Negatively Regulates Thermal Nociception Behavior
A prior pharmacological study on mammalian sensory neurons suggested an SK-mediated pathway for nociceptor excitability (Pagadala et al., 2013); however, the cellular role of this ion channel specifically in nociception remains largely unexplored and has not been verified with genetic mutants. The mammalian genome contains three genes that encode SK channel subunits (Köhler et al., 1996), while the Drosophila genome encodes only a single SK locus on the X chromosome (Abou Tayoun et al., 2011; Adelman et al., 2012; Köhler et al., 1996) that is 60 kb in length and is predicted to encode at least 14 distinct transcripts (Gramates et al., 2017) (Figure 2A [for simplicity, only two transcripts are shown]). The Drosophila SK locus has been found to mediate, a slow Ca2+-activated K+ current in photoreceptor neurons and muscle as well as playing a role in learning and memory (Abou Tayoun et al., 2011, 2012; Gertner et al., 2014). To further investigate the function of SK, we generated a DNA null mutant by deleting the gene with Flippase (FLP) and FLP recombination target (FRT)-containing transposons (Thibault et al., 2004; Parks et al., 2004) (Figures 2A, S1A, and S1B). Consistent with the hypothesis that SK is an important negative regulator of nociception, SK null mutants (∆SK) showed a pronounced hypersensitive response at 42°C. These animals showed an average response latency to a 42°C stimulus of 3.2 s, which was significantly faster than the control background strain (Exelixis isogenic white) response of 6.2 s (Figure 2B).
Figure 2. Expression of SK Rescues the Nociception Phenotypes.
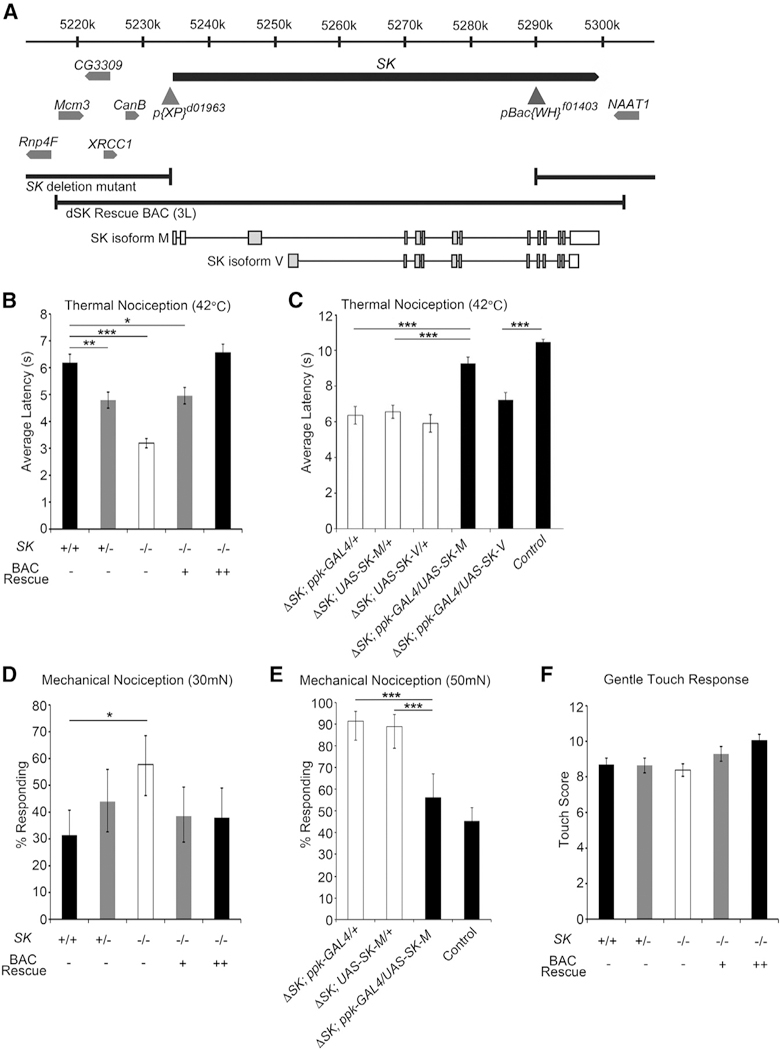
(A) Schematic representation of the genetic interval surrounding SK locus. The locations of pBac and P transposable elements containing FRT sites used to generate SK null mutants are shown as gray triangles, and the breakpoints of the ΔSK allele are denoted. The SK BAC rescue and SK isoforms, SK-M and SK-V, are schematically shown.
(B) SK null mutant animals are hypersensitive to noxious heat (42°C). ΔSK larvae (white bar) show hypersensitivity to noxious heat compared to control animals (black bar). Replacing SK in the genome with two copies of the BAC transgenes fully rescue the nociception defect (black bar). The SK mutant hypersensitivity phenotype is semi-dominant as animals heterozygous for the deletion (gray bars) show quicker responses compared to control animals. One-way ANOVA with Dunnett’s test, ***p < 0.001, **p < 0.01, *p < 0.05. Data are presented as mean ± SEM.
(C) Latency to respond to a 42°C probe in SK mutant larvae expressing SK-M in the class IV neurons (black bar) is significantly different from ΔSK larvae (white bars) but not control animals (back bar). Latency to respond to a 42°C probe in SK mutant larvae expressing SK-V in the class IV neurons (black bar) is significantly different from control larvae (black bar) but not ΔSK mutant larvae (white bars). Data are presented as mean ± SEM. One-way ANOVA followed by Tukey’s test. ***p < 0.001.
(D) Mechanical nociception assay using a 30-mN von Frey fiber. ΔSK mutants (white bar) show hypersensitivity to noxious mechanical stimulation compared to control animals (black bar). Replacing SK in the genome with two copies of the BAC transgenes rescue the nociception defect (black bar). The SK mutant hypersensitivity phenotype is semi-dominant as animals heterozygous for the deletion (gray bars) show hypersensitivity compared to control animals. Fisher’s exact test with Holm-Bonferroni correction. Data are presented as percentages ± 95% confidence intervals. *p < 0.05.
(E) Expression of SK isoform M in class IV neurons rescues the mechanical nociception phenotype of SK mutant. Fisher’s exact test with Holm-Bonferroni correction. Data are presented as percentages ± 95% confidence intervals. ***p < 0.001.
(F) ΔSK mutant animals show normal gentle touch responses. One-way ANOVA with Dunnett’s test, p > 0.05. Data are presented as mean ± SEM, see Table S2 for sample sizes.
To confirm that loss of SK was responsible for the nociception defect, we performed a genetic rescue experiment through transgenic insertion of an ~80-kb bacterial artificial chromosome (BAC) that covered the entire SK locus (Venken et al. 2006, f mutant background to create rescue animals containing either one or two copies of the BAC transgene covering the SK genomic region (Figure 2A). The nociception hypersensitivity phenotype was fully reverted in rescue animals containing two copies of the BAC transgene (Figure 2B). Interestingly, animals heterozygous for the SK mutation exhibit hypersensitivity to noxious heat but to a lesser degree than homozygotes (4.8 s). Note that testing of heterozygotes can only be performed in female larvae as SK is located on the X chromosome (thus, female larvae were used in all experiments to allow for consistent comparisons). One copy of the BAC rescue transgene provides only partial rescue of the hypersensitivity phenotype (5.0 s) but two copies fully rescued (Figure 2B). These data combined support a dosage-sensitive, semi-dominant thermal nociception defect for SK mutants that requires two copies of the BAC transgene for phenotypic full rescue.
To test for a nociceptor-specific requirement for SK, we expressed the SK-M transcript under control of the ppk-GAL4 driver in the SK mutant background (Figure 2A). This manipulation fully rescued the hypersensitive nociception phenotype of the SK mutant animals (Figure 2C), confirming the site of action for SK in the nociceptor neurons. SK-M is one of eight long protein isoforms that are annotated on Flybase, and there are an additional six predicted short isoforms (Gramates et al., 2017). As well, we have cloned a cDNA for a transcript encoding a seventh short SK isoform (SK-V, Figure 2A). Unlike our experiments with SK-M, expression of the SK-V transcript in nociceptors did not result in a rescue of the hypersensitive mutant phenotype (Figure 2C). These experiments suggest that long SK isoforms may be more important than short isoforms for suppressing the thermal sensitivity of nociceptors.
SK Negatively Regulates Mechanical Nociception Behavior
The elaborately branched class IV neurons function as polymodal nociceptors, playing a role in both thermal (≥39°C) and mechanical nociception (≥30 mN). Channels expressed in class IV neurons such as such Painless and dTRPA1 are required for both thermal and mechanical nociception, while Pickpocket, Balboa/ PPK26, and Piezo have more specific roles in mechanical nociception (Tracey et al., 2003; Zhong et al., 2010, 2012; Neely et al., 2011; Kim et al., 2012; Mauthner et al., 2014; Gorczyca et al., 2014; Guo et al., 2014). Interestingly, SK mutant larvae showed enhanced nocifensive responses to a 30-mN mechanical stimulus compared to parental strain animals (Figure 2D). With this stimulus, ∆SK mutant animals rolled in response to the noxious force stimuli in 58% of trials, while control animals responded in only 31%. As with thermal nociception, replacing SK in the genome by BAC transgene restored the mechanical nociception response to wild-type levels with 38% of BAC rescue animals responding to the 30-mN stimulus (Figure 2D). However, the mechanical nociception phenotype was less sensitive to dosage. Animals heterozygous for the ΔSK mutation as well as animals containing one copy of the BAC rescue transgene respond similarly to wild-type animals (44% and 39%, respectively). As with thermal nociception, UAS-SK-M expressed under control of the ppk-GAL4 driver fully rescued the SK mutant mechanical nociception phenotype (Figure 2E).
To determine whether SK disruption affects mechanosensation in general, we tested SK mutant larvae in an established gentle touch assay (Kernan et al., 1994). Gentle touch responses in SK mutants appeared normal (Figure 2F). Thus, the somatosensory effects of SK were more specific to the nociception pathway, regulating nociceptor activity both in response to noxious thermal and mechanical stimuli.
SK Negatively Regulates Nociceptor Excitability
Next, we performed optical recordings from control (Exelixis isogenic white) and SK mutant larvae expressing the genetically encoded Ca2+ indicator, GCaMP3.0 (Tian et al., 2009), under the control of the nociceptor-specific driver, ppk-Gal4. In this filleted larval preparation, the md neurons expressing GCaMP3.0 were imaged through the transparent cuticle using high-speed, time-lapse confocal microscopy while stimulated with a 50-mN probe. ppk-Gal4-expressing neurons imaged in this preparation showed rapidly increasing GCaMP3.0 signals during the initial application of force and this signal rapidly declined (Figure 3A). In SK mutant animals, the peak calcium response (measured at the cell soma) was significantly increased relative to wild-type, and the signal remained elevated above the baseline for several seconds following the mechanical stimulus (Figure 3B). Restoration of SK-M to the mutant background rescued the elevated peak response but did not fully suppress the prolonged signal seen in the mutant (Figure 3B). Thus, although the SK-M isoform can rescue behavioral phenotypes and peak calcium responses in class IV neurons, it is possible that one or more of the 13 other isoforms is required for complete restoration of wild-type responses in this Ca2+ imaging assay.
Figure 3. Optical Recordings Show SK Mutant Enhances Force-Triggered Ca2+ Response of Class IV Neurons.
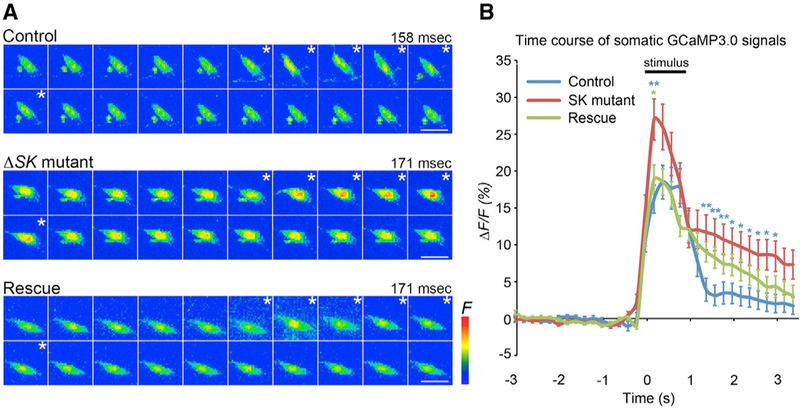
(A) Maximum-intensity projections from Z-stack time series. (Control: upper) Images of representative class IV ddaC neuron expressing GCaMP3.0, displaying increased GCaMP3.0 fluorescence during stimulation with a 50 mN probe (asterisks) (1 frame = 158 ms). (SK mutant: middle) Images of representative SK mutant ddaC neuron shows higher increase in GCaMP3.0 intensity relative to controls during stimulation (asterisks) (1 frame = 171 ms). (Rescue: bottom) Images of representative ddaC neuron repressing UAS-SK isoform M in SK mutant background (1 frame = 171 ms). Scale bars: 10 μm.
(B) Traces showing average peak ΔF/F0% of ddaC neurons in each genotype before, during, and after mechanical stimulation. The bar above the trace shows the average length of the mechanical stimulus (0.93 ± 0.02 s). Statistical analysis was performed using a one-way ANOVA with Tukey’s HSD post hoc for pairwise comparisons. Data are presented as mean ± SEM. Blue asterisks show control versus SK mutant. The green asterisk shows SK mutant versus rescue. **p < 0.01 and *p < 0.05 in each time point.
The SK Gene Is Expressed in Class IV md Sensory Neurons
To evaluate the expression of SK, we examined a transgenic Drosophila strain from the Minos-mediated integration cassette (MiMIC) collection (Venken et al., 2011). A MiMIC element inserted in the proper orientation into the 5’ non-coding intron of SK long isoform transcripts should express EGFP in the native pattern of SK (i.e., at endogenous levels in appropriate tissues). We observed EGFP expression in the larval peripheral nervous system in a subset of type I and type II sensory neurons that included the class IV md neurons (Figure 4A). These results reveal that transcripts encoding long-isoform SK proteins are endogenously expressed in the nociceptors and provide additional validation of our tissue-specific UAS-SK-M rescue experiments that restored normal nociception responses to SK mutant larvae (Figures 2C and 2E).
Figure 4. SK Is Expressed in Class IV md Neurons and SK::V5 Proteins Localize to the Proximal Axon.
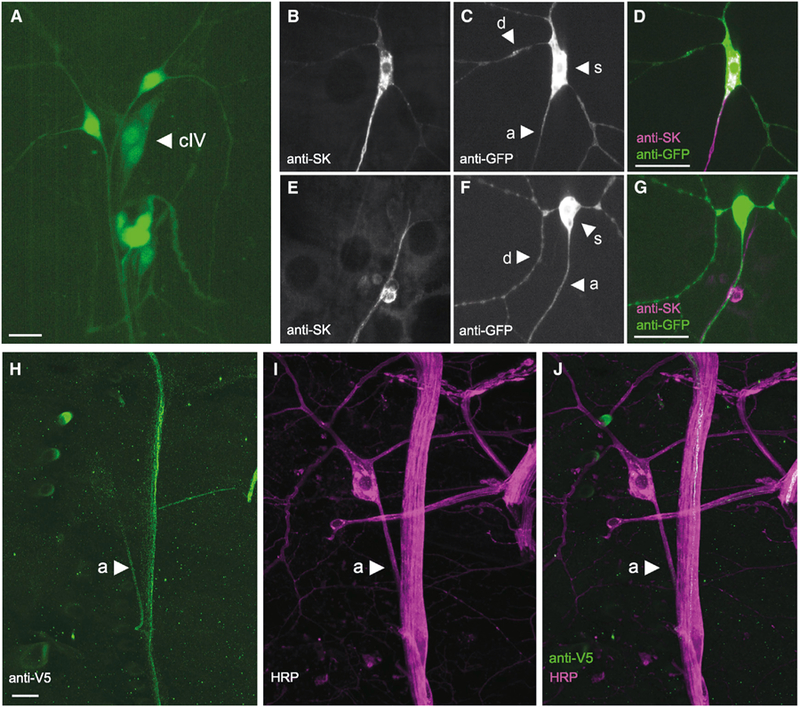
Maximum-intensity projection from z-stack images of the dorsal md neuron cluster (segment A4–A6).
(A) A MiMIC element inserted in the 5’ UTR of SK expresses GFP in md neurons (arrowhead indicates the class IV neuron ddaC) and es-neurons.
(B–D) Anti-SK antibodies label SK-M proteins expressed in class IV neurons in the ΔSK mutant background. SK protein expression (B and magenta in D) and class IV neuron (C and green in D).
(E–G) Endogenous SK protein is detected in axons of sensory neurons in control larvae. SK protein expression (E and magenta in G) and class IV neuron (F and green in G).
(H–J) Anti-V5 antibodies label class IV axons in SK::V5 larval filet preparation (green; H) that was costained with anti-HRP (magenta; I) and merge of (H) and (I) (J). d, dendrite; s, soma; a, axon. Scale bars: 10 μm.
SK Proteins Are Localized to a Proximal Compartment of Sensory Axons
The potassium currents mediated by SK channels in mammalian neurons make an important contribution to the afterhyperpolarization (AHP) of action potentials (Faber and Sah, 2002; Pedarzani et al., 2005). However, the precise subcellular function of SK channels is thought to vary depending on the subcellular compartment where it resides. SK proteins have been found in somatic or dendritic compartments in some cell types (reviewed by Rudolph and Thanawala, 2015), and in axons or presynaptic compartments in others (Abou Tayoun et al., 2011). Thus, we wished to identify the subcellular compartment containing SK channels in the nociceptive md neurons.
Using a previously generated anti-SK antibody (Abou Tayoun et al., 2011), we first observed subcellular localization of the SK-M protein used in our nociceptor-specific rescue experiment (Figures 4B–4D). Surprisingly, we found that the rescuing SK-M protein was clearly detectable in the class IV axons and soma (Figures 4B and 4D) but only weakly detectable in proximal dendrites (Figures 4C and 4D). This was consistent with staining in wild-type animals (Figures 4E–4G), where we were able to detect SK protein in axons (Figures 4E and 4G) of sensory neurons but we were unable to detect any expression in the dendrites or soma of class IV neurons (Figures 4F and 4G). Note that, because the SK antibody detects sensory neuron axons of multiple types in the dorsal sensory neuron cluster, it was often impossible to unambiguously assign the axonal staining observed in wild-type animals to the ddaC class IV axon. However, the anti-SK staining was completely eliminated in null mutant animals confirming specificity of the antibody (Figures S1C–S1E).
These results raised the possibility that SK proteins are required in axons and not in dendrites for nociception. The anti-SK antibody was raised against a purified SK fragment fusion protein largely composed of the N-terminal domain but the exact epitope it detects is not known (Abou Tayoun et al., 2011). Thus, it remained possible that isoforms of SK not detected by this antibody might localize to dendrites. Therefore, in order to detect as many SK protein isoforms as possible, and at their endogenous expression levels, we used clustered regularly interspaced short palindromic repeats (CRISPR)-mediated homologous repair (Gratz et al., 2013; Jinek et al., 2012; Cong et al., 2013) to introduce a V5 epitope tag (Hampel et al., 2011) at the SK locus (Figures S2A–S2C). The inserted V5 epitope tag is encoded by an exon present in 13 of the 14 known SK transcripts (with the exception of the SK-J) located immediately downstream of the sequence encoding the SK calmodulin-binding domain (Figures S2A and S2A’). Interestingly, anti-V5 directed immunofluorescence in animals with our genomic modification was present in the axons of peripheral sensory neurons. Specifically, we observed strong labeling of axons of a subset of type I (external sensory [es] and chordotonal; data not shown) neurons and type II md neurons, including the class IV (Figures 4H–4J). The latter could be unambiguously identified in the v’ada class IV cell because its axon does not bundle with other axons prior to entering the nerve. Interestingly SK::V5 proteins concentrate within a proximal compartment of axons in class IV sensory neurons (Figures 4H and 4J) but were not detected in nociceptor axon terminals in the CNS (Figures S3A–S3C’) or in sensory dendrites of the nociceptive neuron arbor (not shown). The subcellular distribution of SK::V5 proteins along the axon of sensory neurons corroborated labeling of similar neuronal structures with anti-SK antibodies targeting long isoforms of SK (Abou Tayoun et al., 2011) (Figures 4E–4G). Note that the SK-J protein contains the N-terminal domain that was used for raising the anti-SK antibody, making it unlikely that it localizes to dendrites. Thus, our evidence combined suggests SK proteins in axons, and not in dendrites, are important for nociception. As with all antibody staining approaches, we cannot exclude the possibility that SK channels present in dendrites exist but are beneath the limits of detection by this approach.
Our finding on the axonal localization of SK channel proteins is interesting in several respects. First, it suggests that the enhanced Ca2+ signals that we observed in nociceptor soma are potentially caused by backpropagation of action potentials rather than a hyperexcitable soma or dendrites. Second, the proximal axonal localization is consistent with recently described evidence that other GFP-tagged Drosophila K+ channels (Shal and Elk) localize to the axon initial segment in the class I md neurons (Jegla et al., 2016). It is possible that SK regulates the AHP of action potentials as in other systems, and this, in turn, regulates firing frequency. It has been proposed that bursts and pauses in firing of the Drosophila nociceptor neurons may be necessary for robust nociception responses (Terada et al., 2016). Additionally, an “unconventional spike” that is triggered by a large dendritic calcium transient has been proposed to be important (Terada et al., 2016). Indeed, a recent study also provided evidence that SK channels could regulate firing, because RNAi against SK was found to cause increases in the firing frequency of nociceptive neurons (Onodera et al., 2017). That study proposed that dendritically localized SK channels might respond to a dendritic Ca2+ transient. Although our investigation of the localization of the SK channel proteins makes them well positioned to regulate firing of nociceptive neurons, our finding that SK is localized to axons is inconsistent with the hypothesis that SK is directly regulated by dendritic Ca2+. Nevertheless, our comprehensive analysis, including the generation of null mutant alleles, and genomic and tissue-specific rescue experiments, demonstrates a genuine involvement for SK channels in Drosophila nociception. Our studies of protein localization by a CRISPR engineered tagged SK channel, and anti-SK staining of an untagged rescuing cDNA suggest that the likely site of action for this important channel resides in the proximal axon segment and not in dendrites. An interesting question for the future will be to investigate whether Seizure and Tiwaz show a similar axonal localization.
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, W. Dan Tracey (dtracey@indiana.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Strains
All fly strains used in this study are detailed in Key Resources Table and Table S1. We identified K+ channel genes using the controlled vocabulary function at FlyBase (http://flybase.org/) for all genes with K+ channel activity (http://flybase.org/cgi-bin/cvreport.html?id=GO:0005267). We used these fly strains: w; md-Gal4;UAS-Dicer2 (primary screen), w; ppk-Gal4;UAS-Dicer2 (secondary screen), and w; ppk-Gal4 UAS-GCaMP3.0. Fly stocks with UAS lines and TRiP RNAi lines were provided by the Bloomington Stock Center. RNAi lines for screening were provided by the Vienna Drosophila RNAi Center (Dietzl et al., 2007). Drosophila stocks were raised on standard cornmeal molasses fly food medium at 25°C. Where possible, we used balancers containing the Tb marker so that the inheritance of the UAS insertion(s) could be followed. Exelixis lines, P{XP}d01963 and pBAC{WH}SKf01403, were provided by the Exelixis collection at Harvard Medical School (Thibault et al., 2004) and used to generate FLP-FRT mediated SK mutant flies (crossing strategy outlined in (Parks et al., 2004)). SK genomic rescue strains were generated by injection of BAC CH321–90C05 (Venken et al., 2009) performed by Rainbow Transgenic Flies, Inc. for PhiC31-mediated chromosome integration into the attP2 docking site. Transgenic flies containing the BAC rescue construct were crossed into SK deletion background to generate the genomic rescue strain. BestGene, Inc. performed microinjections of pUAST-SK-M and pUAST-SK-V vectors into w1118 and transgenic flies were generated via standard P-element mediated transformation.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-dSK (polyclonal antibody targeting SK’s N-terminal domain) | Patrick Dolph (Abou Tayoun et al., 2011) | N/A |
| Mouse anti-V5 | Thermo Fisher Scientific | Cat# R960–25;RRID:AB_2556564 |
| Rabbit anti-GFP | Thermo Fisher Scientific | Cat# A-11122;RRID:AB_221569 |
| Rabbit anti-HRP | Jackson ImmunoResearch Laboratories | Cat# 323–005-021;RRID:AB_2314648 |
| Alexa Fluor. 488 goat anti-mouse | Thermo Fisher Scientific | Cat# A-11029;RRID:AB_2534088 |
| Alexa Fluor. 488 goat anti-rabbit | Thermo Fisher Scientific | Cat# A-11034;RRID:AB_2576217 |
| Cat# A-11008;RRID:AB_143165 | ||
| Alexa Fluor. 546 goat anti-mouse | Thermo Fisher Scientific | Cat# A-11030;RRID:AB_2534089 |
| Alexa Fluor. 546 goat anti-rabbit | Thermo Fisher Scientific | Cat# A-11035;RRID:AB_2534093 |
| Deposited Data | ||
| SK-V isoform sequence | GenBank | MH001552 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: RNAi strains targeting K+ channels (See Table S1) | Vienna Drosophila Resource Center; Bloomington Drosophila Stock Center | See Table S1 |
| D. melanogaster: y1 w*; md-GAL4 (aka GAL4109(2)80) | Bloomington Drosophila Stock Center | BDSC: 8769; FlyBase: FBti0015554 |
| D. melanogaster: y1 w*; md-GAL4 (aka GAL4109(2)80) | Bloomington Drosophila Stock Center | BDSC: 8769; FlyBase: FBti0015554 |
| D. melanogaster: w*; ppk-GAL4 (aka ppk1.9-GAL4) | Ainsley et al., 2003 | FlyBase: FBal0152201 |
| D. melanogaster: w1118; UAS-Dicer2 | Vienna Drosophila Resource Center | 60009 |
| D. melanogaster: w;; UAS-GCamP3.0attp2 | Bloomington Drosophila Stock Center | BDSC: 32236 |
| D. melanogaster: y1 w67c23;; attp2 | Bloomington Drosophila Stock Center | BDSC: 8622; FlyBase: FBti0040535 |
| D. melanogaster: w* P{XP}d01963 | The Exelixis Collection at the Harvard Medical School | d01963; FlyBase: FBti0054688 |
| D. melanogaster: w* pBac{WH}SKf01403 | The Exelixis Collection at the Harvard Medical School | f01403; FlyBase: FBst1016953 |
| D. melanogaster: w1118 ΔSK | Generated in this study | N/A |
| D. melanogaster: w1118 SK::V5 | Generated in this study | N/A |
| D. melanogaster: w1118 ; UAS-SK-M | Generated in this study | N/A |
| D. melanogaster: w1118 ; UAS-SK-V | Generated in this study | N/A |
| Oligonucleotides | ||
| See Table S3 | N/A | N/A |
| Recombinant DNA | ||
| BAC: CH321–90C05 (aka CHORI-321–90C05) | BACPAC Resources Center (BPRC) | CH321–90C05 |
| TOPO-XL | Thermo Fisher Scientific | Cat# K8050 |
| pENTR (Gateway Entry Vector) | Thermo Fisher Scientific | Cat# K240020 |
| pTW (Drosophila Gateway Vector Collection) | Drosophila Genomics Resource Center | Gateway Collection |
METHOD DETAILS
Molecular Biology
To preliminarily screen for deletion of SK, polymerase chain reaction (PCR) was performed on genomic DNA extracted from wandering third-instar larvae with the putative FLP/FRT-mediated SK deletion. The QIAGEN DNeasy® Blood and Tissue kit was used for DNA extraction and the forward primer: 5ʹ-ATGTCAATTCAGAAGCTTAACGACAC-3ʹ and the reverse primer: 5ʹ-TGCAGTTGCTGCT GATGCAGAT-3ʹ were used to test for absence of SK gene product by PCR amplification (primer pair 2, Figures S1A and S1B). Three additional primer pairs were used to confirm the SK deletion. From the upstream gene, CanB, into the p{XP} element: forward primer 5ʹ-GCAGTCGTGTATTTGCTGTCG-3ʹ and reverse primer 5ʹ-TACTATTCCTTTCACTCGCACTTATTG-3ʹ (primer pair 1, Figures S1A and S1B). From within the SK locus: forward primer 5ʹ-TTATTCATGATAGATAACTGCGCTGACG-302B9 and reverse primer 5ʹ-CAC CATGCCGCAGGTGAGGGTGATA-3ʹ (primer pair 3, Figures S1A and S1B). From the pBac{WH} into C-terminal end of SK: forward primer 5ʹ-CCTCGATATACAGACCGATAAAAC-3ʹ and reverse primer 5ʹ-ATCCTGGTGCCCTACGATCTATGT-3ʹ (primer pair 4, Figures S1A and S1B). Lastly, control primers targeting Caa1D, forward primer: 5ʹ-CAACCGGATGTGAAGTGCG-3ʹ and the reverse primer: 5ʹ-CTTGGCACTT CGCCTGAAGG-3ʹ, were used to confirm integrity of DNA preps (control primer pair, Figure S1B).
For the cloning of SK-M and SK-V rescue constructs, RT-PCR was performed on total RNA extracts (TRIzol, Life Technologies) from Canton-S third instar larvae. The SuperScript III reverse transcriptase enzyme (Life Technologies) and Oligo(dT)12–18 primer (Life Technologies) were used to direct the synthesis of first strand cDNA. For SK-M and SK-V cDNA amplification, PCR was performed using the reverse primer 5ʹ-TCAGCTAGAATGTGGAAACAGCAT-3ʹ, and either the forward primer 5ʹ-ATGTCAATTCA GAAGCTTAACGAC-3ʹ to target the SK-M isoform or the forward primer 5ʹ-ATGTCGCCGGCCTTCTGC-3ʹ to target the SK-V isoform. The SK-M and SK-V PCR products were subsequently cloned into the TOPO-XL vector (Life Technologies) and fully sequenced. The complete sequence for the newly cloned SK-V isoform was deposited in GenBank (accession number MH001552). The following primers were used to target the desired SK-M and SK-V amplicons for subcloning into the pENTR/DTOPO vector (Thermo Fisher Scientific): 5ʹ-CACCATGTCAATTCAGAAGCTTAACGAC-3ʹ and 5ʹ-TCAGCTAGAATGTGGAAACAG CATG-3ʹ for SK-M, or 5ʹ-CACCATGTCGCCGGCCTTCT-3ʹ and 5ʹ-TCAGCTAGAATGTGGAAACAGCATGGGC-3ʹ for SK-V. Gateway recombination technology was used to further subclone the SK-M and SK-V cDNAs from pENTR into the final pTW destination vector (i.e., pUASt-SK-M and pUASt-SK-V) via the LR Clonase enzyme (Life Technologies).
CRISPR Gene Tagging
A V5 epitope tag was introduced at the SK locus (Figures S2A, S2Aʹ, and S2B) using a single-stranded oligodeoxynucloetide (ssODN) homology directed repair CRISPR-Cas9 approach (Gratz et al., 2013). The guide RNA sequence 5ʹ-CCATTCAAGCGC CAACGCCC-3ʹ was cloned into the BbsI site of pU6-BbsI-chiRNA plasmid (Gratz et al., 2013) and co-injected at a concentration of 250 ng/mL with the ssODN donor repair template 5ʹ-GAGCGGATCGAGCAGCGGCGGAAC TTTTTACATCCTGACACAGCTG CAGTTGCCCCCATTGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTAGATTCTACGCAAGCGCCAACGCCCCAATCGATGTTCA ATGCAGCGCCCATGCTGTTTCCACATTCTAGG-3ʹ (Integrated DNA Technologies) at a concentration of 100ng/uL into w1118;; PBac{vas-Cas9}VK00027 embryos. Underlined sequence in the ssODN repair template indicates the introduced in-frame V5 epitope tag with a diagnostic XbaI restriction site (bold text). F0 flies were crossed to w1118 flies and single F1 founders were mated with an FM7c balancer strain to establish independent lines. To screen for V5 integrations events, PCR and Xba1 RFLP analysis were conducted on genomic DNA extracted from individual F1 candidate flies. PCR was performed with the following primer pair: 5ʹ-GAGCGTTTAACCAACCTAGAG-3ʹ and 5ʹ-GCAGTTAGTGTTCGTCCAAAG-3ʹ. PCR amplified products showing the desired XbaI cleavage were selected for further sequence verification of proper in-frame SK::V5 integration (Figure S2B). Candidate strains were sequenced both in genomic DNA and cDNA (Figure S2C).
Thermal Nociception Assay
Thermal nociception assays were as described previously (Honjo et al., 2014, 2016). In a primary screen, approximately 10–20 lines were tested per day and each day included a control cross for the driver line to the relevant isogenic background (md-Gal4/+;UAS-Dicer 2/+). We assigned alternative numeric labels to the VDRC lines so that the identity of the genes was blind to the tester throughout the entire screening process. The gene candidates were decoded following completion of the screen. Five to fifteen larvae were tested in the initial screen and an average latency was calculated. UAS-IR lines were selected for retesting if they displayed a latency that was one or more standard deviation from the control strain mean (w; md-Gal4/+;UAS-Dicer2/+, w; ppk-Gal4/+;UAS-Dicer2/+). We re-tested approximately 20–40 larvae for each line kept after the initial screen. For SK genetic mutant testing and rescue experiments, only female wandering, third-instar larvae were tested to measure dosage sensitivity.
Mechanical Nociception Assay
Crosses were prepared as with the thermal nociception assay. Wandering, third-instar larvae were collected between days five and seven, and stimulated with calibrated (~30–50 mN) von Frey fibers as previously described (Hwang et al., 2007; Zhong et al., 2010; Mauthner et al., 2014). Noxious mechanical stimuli were delivered by the rapid depression and release of the fiber so that the fiber would begin to bend on the dorsal surface of the larva. The stimulus was delivered between abdominal segments four, five, and six. The percent response following the stimulus was calculated for each genotype. For SK genetic mutant testing and rescue experiments, only female wandering, third-instar larvae were tested to measure dosage sensitivity.
Gentle Touch Assay
The gentle touch behavioral assay was performed as previously described (Kernan et al., 1994; Tsubouchi et al., 2012). For SK genetic mutant testing and rescue experiments, only female wandering, third-instar larvae were tested to measure dosage sensitivity.
Calcium Imaging
Calcium imaging experiments were performed as described previously (Tsubouchi et al., 2012) but with a 50 mN probe.
Immunostaining
The anti-SK was kindly provided by the Dolph lab (Abou Tayoun et al., 2011). The anti-SK antibody was used at a concentration of 1:2000, mouse anti-V5 antibody (Thermo Fisher Scientific) was used at a concentration of 1:200, anti-GFP antibody was used at a concentration of 1:250 (Thermo Fisher Scientific), and an anti-HRP antibody was used at concentration of 1:100 (Jackson ImmunoResearch Lab). All secondary antibodies were used at a concentration of 1:600 or 1:1000 (Alexa Flours from Thermo Fisher Scientific). Detailed staining protocols are available upon request.
Confocal Imaging
For live GFP imaging, larvae were anesthetized and mounted as previously described in (Mauthner et al., 2014). Confocal Z stacks of the dorsal cluster from the SKMI1037 MiMIC line were taken on a Zeiss LSM 5 LIVE. For immunostained tissue, anti-SK images were taken on a LSM 5 LIVE confocal microscope using a 40X objective and anti-V5 images were taken on an LSM 880 63x objective.
QUANTIFICATION AND STATISTICAL ANALYSIS
The following statistical analyses were used in this study: Fisher’s Exact test with Holm-Bonferroni correction for nonparametric data analysis, Student’s t test for comparing two means, one-way ANOVA with post hoc Tukey’s HSD test for multiple comparisons of parametric data, or one-way ANOVA with Dunnett’s test for comparisons of parametric data. All statistical analyses were performed using Graphpad, Prism, or R and are specified in the figure legends (including p value significance thresholds). The larval sample sizes for the behavioral assays are noted in Table S2 or in STAR Methods. Error bars denoting standard error of the mean or confidence intervals are also indicated in relevant figure legend
Supplementary Material
Highlights.
Specific potassium channels regulate nociceptor excitability
SK channels have a critical function in nociception
SK channels specifically localize to sensory axons
SK channels are not detectable in sensory dendrites
ACKNOWLEDGMENTS
We thank the Vienna Drosophila Resource Center, Transgenic RNAi Project, and the Exelixis Collection at Harvard Medical School (NIH/NIGMS R01-GM084947). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and vectors obtained from the Drosophila Genomics Resource Center (NIH 2P40OD010949) were used in this study. We also thank Sarah Sweeney-Howard, Katherine H. Fisher, and Luuli Tran for technical assistance and members of the Tracey laboratory for helpful discussion.
This work was supported by grants from the NIH (5R21DC010222, 5R01GM086458, and 5R01NS054899) (to W.D.T.). K.C.E.W. and J.R. were supported by pre-doctoral fellowships from the National Science Foundation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The accession number for the SK-V isoform sequence reported in this paper is GenBank: MH001552.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and three tables can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.070.
REFERENCES
- Abou Tayoun AN, Li X, Chu B, Hardie RC, Juusola M, and Dolph PJ (2011). The Drosophila SK channel (dSK) contributes to photoreceptor performance by mediating sensitivity control at the first visual network. J. Neurosci 31, 13897–13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Tayoun AN, Pikielny C, and Dolph PJ (2012). Roles of the Drosophila SK channel (dSK) in courtship memory. PLoS One 7, e34665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman JP, Maylie J, and Sah P (2012). Small-conductance Ca2+-activated K+ channels: form and function. Annu. Rev. Physiol 74, 245–269. [DOI] [PubMed] [Google Scholar]
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, and Johnson WA (2003). Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol 13, 1557–1563. [DOI] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- Dubin AE, and Patapoutian A (2010). Nociceptors: the sensors of the pain pathway. J. Clin. Invest 120, 3760–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber ES, and Sah P (2002). Physiological role of calcium-activated potassium currents in the rat lateral amygdala. J. Neurosci 22, 1618–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, and Jan YN (1999). Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev 13, 2549–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertner DM, Desai S, and Lnenicka GA (2014). Synaptic excitation is regulated by the postsynaptic dSK channel at the Drosophila larval NMJ. J. Neurophysiol 111, 2533–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca DA, Younger S, Meltzer S, Kim SE, Cheng L, Song W, Lee HY, Jan LY, and Jan YN (2014). Identification of Ppk26, a DEG/ENaC channel functioning with Ppk1 in a mutually dependent manner to guide locomotion behavior in Drosophila. Cell Rep 9, 1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, et al. ; the FlyBase Consortium (2017). FlyBase at 25: looking to the future. Nucleic Acids Res 45, D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O’Connor-Giles KM (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Wang Q, and Wang Z (2014). The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep 9, 1183–1190. [DOI] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, and Simpson JH (2011). Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat. Methods 8, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo K, Robertson J, and Tracey WD (2014). Nociception. In Behavioral Genetics of the Fly (Drosophila melanogaster), Dubnau J, ed. (Cambridge University Press; ), pp. 66–76. [Google Scholar]
- Honjo K, Mauthner SE, Wang Y, Skene JHP, and Tracey WD Jr. (2016). Nociceptor-enriched genes required for normal thermal nociception. Cell Rep 16, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Petersen M, Hoyer N, Spitzweck B, Tenedini F, Wang D, Gruschka A, Burchardt LS, Szpotowicz E, Schweizer M, et al. (2017). Sensory integration and neuromodulatory feedback facilitate Drosophila mechanonociceptive behavior. Nat. Neurosci 20, 1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, and Tracey WD (2007). Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol 17, 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Wilson SD, Strichartz GR, and Hall LM (1984). Two types of mutants affecting voltage-sensitive sodium channels in Drosophila melanogaster. Nature 308, 189–191. [DOI] [PubMed] [Google Scholar]
- Jegla T, Nguyen MM, Feng C, Goetschius DJ, Luna E, van Rossum DB, Kamel B, Pisupati A, Milner ES, and Rolls MM (2016). Bilaterian giant ankyrins have a common evolutionary origin and play a conserved role in patterning the axon initial segment. PLoS Genet 12, e1006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernan M, Cowan D, and Zuker C (1994). Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron 12, 1195–1206. [DOI] [PubMed] [Google Scholar]
- Kim SE, Coste B, Chadha A, Cook B, and Patapoutian A (2012). The role of Drosophila Piezo in mechanical nociception. Nature 483, 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, and Adelman JP (1996). Small-conductance, calcium-activated potassium channels from mammalian brain. Science 273, 1709–1714. [DOI] [PubMed] [Google Scholar]
- Mauthner SE, Hwang RY, Lewis AH, Xiao Q, Tsubouchi A, Wang Y, Honjo K, Skene JH, Grandl J, and Tracey WD Jr. (2014). Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr. Biol 24, 2920–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely GG, Keene AC, Duchek P, Chang EC, Wang QP, Aksoy YA, Rosenzweig M, Costigan M, Woolf CJ, Garrity PA, and Penninger JM (2011). TrpA1 regulates thermal nociception in Drosophila. PLoS One 6, e24343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. (2009). A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera K, Baba S, Murakami A, Uemura T, and Usui T (2017). Small conductance Ca2+-activated K+ channels induce the firing pause periods during the activation of Drosophila nociceptive neurons. eLife 6, e29754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagadala P, Park CK, Bang S, Xu ZZ, Xie RG, Liu T, Han BX, Tracey WD Jr., Wang F, and Ji RR (2013). Loss of NR1 subunit of NMDARs in primary sensory neurons leads to hyperexcitability and pain hypersensitivity: involvement of Ca2+-activated small conductance potassium channels. J. Neurosci 33, 13425–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. (2004). Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet 36, 288–292. [DOI] [PubMed] [Google Scholar]
- Pedarzani P, McCutcheon JE, Rogge G, Jensen BS, Christophersen P, Hougaard C, Strøbaek D, and Stocker M (2005). Specific enhancement of SK channel activity selectively potentiates the afterhyperpolarizing current I(AHP) and modulates the firing properties of hippocampal pyramidal neurons. J. Biol. Chem 280, 41404–41411. [DOI] [PubMed] [Google Scholar]
- Ritter AM, and Mendell LM (1992). Somal membrane properties of physiologically identified sensory neurons in the rat: effects of nerve growth factor. J. Neurophysiol 68, 2033–2041. [DOI] [PubMed] [Google Scholar]
- Rudolph S, and Thanawala MS (2015). Location matters: somatic and dendritic SK channels answer to distinct calcium signals. J. Neurophysiol 114, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S, Matsubara D, Onodera K, Matsuzaki M, Uemura T, and Usui T (2016). Neuronal processing of noxious thermal stimuli mediated by dendritic Ca2+ influx in Drosophila somatosensory neurons. eLife 5, e12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet 36, 283–287. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD Jr., Wilson RI, Laurent G, and Benzer S (2003). painless,a Drosophila gene essential for nociception. Cell 113, 261–273. [DOI] [PubMed] [Google Scholar]
- Tsubouchi A, Caldwell JC, and Tracey WD (2012). Dendritic filopodia, Ripped Pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr. Biol 22, 2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, and Bellen HJ (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster . Science 314, 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, and Bellen HJ (2011). MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ, Goergen P, Rajendran J, Klockars A, Kasagiannis A, Fredriksson R, and Schiöth HB (2014). Regulation of aggression by obesitylinked genes TfAP-2 and Twz through octopamine signaling in Drosophila. Genetics 196, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, and Jan YN (2010). Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, and Tracey WD (2010). Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr. Biol 20, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Bellemer A, Yan H, Ken H, Jessica R, Hwang RY, Pitt GS, and Tracey WD (2012). Thermosensory and nonthermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat-sensor domains of a thermoTRP channel. Cell Rep 1, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


