Abstract
Autism spectrum disorder (ASD) is a highly heritable and heterogeneous group of neurodevelopmental phenotypes diagnosed in more than 1% of children. Common genetic variants contribute substantially to ASD susceptibility, but to date no individual variants have been robustly associated with ASD. With a marked sample size increase from a unique Danish population resource, we report a genome-wide association meta-analysis of 18,381 ASD cases and 27,969 controls that identifies five genome-wide significant loci. Leveraging GWAS results from three phenotypes with significantly overlapping genetic architectures (schizophrenia, major depression, and educational attainment), seven additional loci shared with other traits are identified at equally strict significance levels. Dissecting the polygenic architecture, we find both quantitative and qualitative polygenic heterogeneity across ASD subtypes. These results highlight biological insights, particularly relating to neuronal function and corticogenesis and establish that GWAS performed at scale will be much more productive in the near term in ASD.
Editorial Summary
A genome-wide association meta-analysis of 18,381 austim spectrum disorder (ASD) cases and 27,969 controls identifies 5 risk loci. The authors find quantitative and qualitative polygenic heterogeneity across ASD subtypes.
ASD is the term for a group of pervasive neurodevelopmental disorders characterized by impaired social and communication skills along with repetitive and restrictive behavior. The clinical presentation is very heterogeneous, including individuals with severe impairment and intellectual disability as well as individuals with above average IQ and high levels of academic and occupational functioning. ASD affects 1–1.5% of individuals and is highly heritable, with both common and rare variants contributing to its etiology1–4. Common variants have been estimated to account for a major part of ASD liability2 as has been observed for other common neuropsychiatric disorders. By contrast, de novo mutations, mostly copy number variants (CNVs) and gene disrupting point mutations, have larger individual effects, but collectively explain < 5% of the overall liability1–3 and far less of the heritability. While a number of genes have been convincingly implicated via excess statistical aggregation of de novo mutations, the largest GWAS to date (n = 7,387 cases scanned) – while providing compelling evidence for the bulk contribution of common variants – did not conclusively identify single variants at genome-wide significance5–7. This underscored that common variants, as in other complex diseases such as schizophrenia, individually have low impact and that a substantial scale-up in sample numbers would be needed.
Here we report the first common risk variants robustly associated with ASD by more than doubling the discovery sample size compared to previous GWAS5–8. We describe strong genetic correlations between ASD and other complex disorders and traits, confirming shared etiology, and we show results indicating differences in the polygenic architecture across clinical sub-types of ASD. Leveraging these relationships and recently introduced computational techniques9, we identify additional novel ASD-associated variants that are shared with other phenotypes. Furthermore, by integrating with complementary data from Hi-C chromatin interaction analysis of fetal brains and brain transcriptome data, we explore the functional implications of our top-ranking GWAS results.
Results
GWAS
As part of the iPSYCH project10, we collected and genotyped a Danish nation-wide population-based case-cohort sample including nearly all individuals born in Denmark between 1981 and 2005 and diagnosed with ASD (according to ICD-10) before 2014. We randomly selected controls from the same birth cohorts (Supplementary Table 1). We have previously validated registry-based ASD diagnoses11,12 and demonstrated the accuracy of genotyping DNA extracted and amplified from bloodspots collected shortly after birth13,14. Genotypes were processed using Ricopili15, performing stringent quality control of data, removal of related individuals, exclusion of ancestry outliers based on principal component analysis, and imputation using the 1000 Genomes Project phase 3 reference panel. After this processing, genotypes from 13,076 cases and 22,664 controls from the iPSYCH sample were included in the analysis. As is now standard in human complex trait genomics, our primary analysis was a meta-analysis of the iPSYCH ASD results with five family-based trio samples of European ancestry from the Psychiatric Genomics Consortium (PGC; 5,305 cases and 5,305 pseudo controls)16. All PGC samples had been processed using the same Ricopili pipeline for QC, imputation and analysis as employed here.
Supporting the consistency between the study designs, the iPSYCH population-based and PGC family-based analyses showed a high degree of genetic correlation with rG = 0.779 (SE = 0.106; P = 1.75 × 10−13), similar to the genetic correlations observed between datasets in other mental disorders17. Likewise, polygenicity as assessed by polygenic risk scores (PRS) showed consistency across the samples supporting homogeneity of effects across samples and study designs (see the results below regarding PRS on a five-way split of the sample). The SNP heritability was estimated to be 0.118 (SE = 0.010), for a population prevalence of 0.01218.
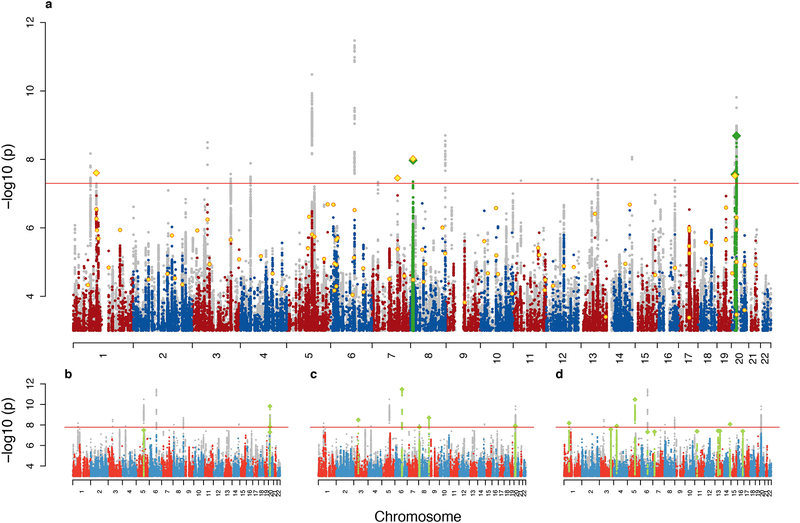
The main GWAS meta-analysis totaled 18,381 ASD cases and 27,969 controls, and applied an inverse variance-weighted fixed effects model. To ensure that the analysis was well-powered and robust, we examined markers with minor allele frequency (MAF) ≥ 0.01, imputation INFO score ≥ 0.7, and supported by an effective sample size in > 70% of the total. This final meta-analysis included results for 9,112,387 autosomal markers and yielded 93 genome-wide significant markers in three separate loci (Figure 1; Table 1a; Supplementary Figures 1–44). Each locus was strongly supported by both the Danish case-control and the PGC family-based data. While modest inflation was observed (lambda = 1.12, lambda1000 = 1.006), LD score regression analysis19 indicates this is arising from polygenicity (> 96%, see Methods) rather than confounding. The strongest signal among 294,911 markers analyzed on chromosome X was P = 7.8 × 10−5.
Figure 1. Manhattans plots:
with the x axis showing genomic position (chromosomes 1–22) and the y axis showing statistical significance as −log10 (P) of z statistics. a: The main ASD scan (18,381 cases and 27,969 controls) with the results of the combined analysis with the follow-up sample (2,119 cases and 142,379 controls) in yellow in the foreground. Genome-wide significant clumps are painted green with index SNPs as diamonds. b-d: Manhattan plots for three MTAG scans of ASD together with, respectively, schizophrenia15 (34,129 cases and 45,512 controls), educational attainment20 (N = 328,917) and major depression21 (111,902 case and 312,113 controls). See Supplementary Figures 45–48 for full size plots. In all panels the results of the composite of the five analyses (consisting for each marker of the minimal p-value of the five) is shown in grey in the background.
Table 1. Genome-wide significant loci from ASD scans and MTAG analyses.
Panel a shows the loci reaching genome-wide significance in analysis of the ASD phenotype alone. The column “Analysis” indicates the minimum p-value arising from the original scan (ASD) and the combined analysis with the follow-up sample (Comb ASD). The columns “Support from other scans” list the other analyses (including MTAG) that further support the locus at genome-wide significance. For the ASD scan results, this shows the genome-wide significant results in the locus from the other scans, and for Comb ASD it displays the result from ASD. Panel b presents additional genome-wide significant loci identified in the three MTAG analyses. The three analyses are ASD with schizophrenia (SCZ)15, educational attainment (Edu)20 and major depression (MD)21. Here the “Analysis” column points to which MTAG analysis gave the results (ASD-Edu or ASD-MD), and the columns “Support from other scans” provide the corresponding scan results in ASD alone. In both panels, independent loci are defined to have r2 < 0.1 and distance > 400 kb and the index variant is displayed in the column “Index var”. Other columns are: chromosome (CHR), chromosomal position (BP), alleles (A1/A2), allele frequency of A1 (FRQ), estimate of effect (β) with respect to A1, standard error of β (SE), and the association p-value of the index variant (P). The column “Nearest genes” lists nearest genes from within 50 kb of the region spanned by all SNPs with r2 ≥ 0.6 to the index variant.
*Indicate different lead SNP than the index variant
| Index var | CHR | BP | Analysis | P | β | SE | A1/A2 | FRQ | Support from other scans | Nearest genes | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scan | P | β | |||||||||||
| a | rs910805 | 20 | 21248116 | ASD | 2.04 × 10−9 | −0.096 | 0.016 | A/G | 0.76 | ASD-SCZ ASD-Edu* |
1.5 × 10−10 2.0 × 10−8 |
−0.069 −0.061 |
KIZ, XRN2, NKX2–2, NKX2–4 |
| rs10099100 | 8 | 10576775 | ASD | 1.07 × 10−8 | 0.084 | 0.015 | C/G | 0.331 | Comb ASD ASD-Edu |
9.6 × 10−9 1.6 × 10−8 |
0.078 0.056 |
C8orf74, SOX7, PINX1 | |
| rs201910565 | 1 | 96561801 | Comb ASD | 2.48 × 10−8 | −0.077 | 0.014 | A/AT | 0.689 | ASD | 3.4 × 10−7 | −0.033 | LOC102723661, PTBP2 | |
| rs71190156 | 20 | 14836243 | ASD | 2.75 × 10−8 | −0.078 | 0.014 | GTTTT TTT/G |
0.481 | Comb ASD ASD-Edu |
3.0 × 10−8 1.2 × 10−8 |
−0.072 0.053 |
MACROD2 | |
| rs111931861 | 7 | 104744219 | Comb ASD | 3.53 × 10−8 | −0.216 | 0.039 | A/G | 0.966 | ASD | 1.1 × 10−7 | −0.094 | KMT2E, SRPK2 | |
| b | rs2388334 | 6 | 98591622 | ASD-Edu | 3.34 × 10−12 | −0.065 | 0.009 | A/G | 0.517 | ASD | 1.0 × 10−6 | −0.068 | MMS22L, POU3F2 |
| rs325506 | 5 | 104012303 | ASD-MD | 3.26 × 10−11 | 0.057 | 0.009 | C/G | 0.423 | ASD | 3.5 × 10−7 | 0.071 | NUD12 | |
| rs11787216 | 8 | 142615222 | ASD-Edu | 1.99 × 10−9 | −0.058 | 0.010 | T/C | 0.364 | ASD | 2.6 × 10−6 | −0.030 | MROH5 | |
| rs1452075 | 3 | 62481063 | ASD-Edu | 3.17 × 10−9 | 0.061 | 0.010 | T/C | 0.721 | ASD | 2.1 × 10−7 | 0.035 | CADPS | |
| rs1620977 | 1 | 72729142 | ASD-MD | 6.66 × 10−9 | 0.056 | 0.010 | A/G | 0.26 | ASD | 1.2 × 10−4 | 0.062 | NEGR1 | |
| rs10149470 | 14 | 104017953 | ASD-MD | 8.52 × 10−9 | −0.049 | 0.008 | A/G | 0.487 | ASD | 8.5 × 10−5 | −0.056 | MARK3, CKB, TRMT61A, BAG5, APOPT1, KLC1, XRCC3 | |
| rs16854048 | 4 | 42123728 | ASD-MD | 1.29 × 10−8 | 0.069 | 0.012 | A/C | 0.858 | ASD | 5.9 × 10−5 | 0.082 | SLC30A9, BEND4, TMEM33, DCAF4L1 | |
We next obtained replication data for the top 88 loci with p-values < 1 × 10−5 in five cohorts of European ancestry totaling 2,119 additional cases and 142,379 controls (Supplementary Table 2 and 3). An overall replication of direction of effects was observed (53 of 88 (60%) of P < 1 × 10−5; 16 of 23 (70%) at P < 1 × 10−6; sign tests P = 0.035 and P = 0.047, respectively) and two additional loci achieved genome-wide significance in the combined analysis (Table 1a). More details on the identified loci can be found in Supplementary Table 4 and selected candidates are described in Box1.
Box1. Selected loci and candidates (ordered by chromosome).
| Gene | Locus* and supporting evidence | Gene function |
|---|---|---|
| NEGR1 | Chr1:72,729,142 Shared ASD-MDD locus Locus also significant in depression21,22, educational attainment20, intelligence23, obesity and BMI24–28 and in an ASD-schizophrenia meta-analysis5. NEGR1 is the only protein-coding gene in the locus NEGR1 is supported by brain Hi-C and eQTL analyses21 |
NEGR1 (neuronal growth regulator 1) is an adhesion molecule modulating synapse formation in hippocampal neurons29,30 and neurite outgrowth31,32. It is a member of the IgLON protein family implicated in synaptic plasticity and axon extension33–35. Predominantly expressed (and developmentally upregulated) in hippocampus and cortex36 and also hypothalamus37. |
| PTBP2 | Chr1:96,561,801 ASD locus Locus also significant in BMI24,25,27 weight25 and educational attainment20. In schizophrenia, the locus shows a p-value of 6.5 × 10−6 15 PTBP2 is the nearest protein-coding gene, approx. 625 kb from index SNP. De novo and rare variants in PTBP2 have been reported in ASD cases1,3,38. PTBP2 is supported by Hi-C results in this study (Fig. 5d) |
PTBP2 is also known as nPTB (neuronal PTB) or brPTB (brain PTB) and is a splicing regulator. PTBP1 and its paralog PTBP2 bind to intronic polypyrimidine tracts in pre-mRNAs and target large sets of exons to coordinate alternative splicing programs during development39. Several switches in the expression of PTBP1 and PTBP2 regulate alternative splicing during neurogenesis and neuronal differentiation40–43. |
| CADPS | Chr3:62,481,063 Shared ASD-Educational attainment locus Locus also significant in study of cognitive decline rate44 CADPS is supported by Hi-C results in this study (Fig. 5a). |
CADPS encodes a calcium-binding protein involved in exocytosis of neurotransmitters and neuropeptides. In line with CAPDS mRNA being mainly expressed in brain and pituitary (GTEx portal- see URLs), immunoreactive CAPS-1 is localized in neural and various endocrine tissues45. In hippocampal synapses, CADPS regulates the pool of readily releasable vesicles at pre-synaptic terminals46,47 |
| KCNN2 | Chr5: 113,801,423 ASD locus (gene-wise analysis) Locus also significant in educational attainment20,48. KCNN2 synaptic levels are regulated by the E3 ubiquitin ligase UBE3A49, of which overexpression has been linked to ASD risk49,50. |
KCNN2 is a voltage-independent Ca2+-activated K+ channel that responds to changes in intracellular calcium concentration and couples calcium metabolism to potassium flux and membrane excitability. In CNS neurons, activation of KCNN2 modulates neuronal excitability by causing membrane hyperpolarization51. Hippocampal KCNN2 has roles in the formation of new memory52, encoding and consolidation of contextual fear53, and in drug-induced plasticity54. |
| KMT2E | Chr7:104,744,219 ASD locus Locus also significant in schizophrenia15,55 and in ASD-schizophrenia meta-analysis5. KMT2E de novo mutations are associated with ASD at FDR < 0.156 A KMT2E credible SNP is a loss-of-function variant (Supplementary Table 16) |
KMT2E encodes Histone-lysine N-methyltransferase 2E and forms a family together with SETD557,58. Evidence suggest that recognition of the histone H3K4me3 mark by the KMT2E PHD finger can facilitate the recruitment of KMT2E to transcription-active chromatin regions59,60. KMT2E has been implicated in chromatin regulation, control of cell cycle progression, and maintaining genomic stability61. |
| MACROD2 | Chr20: 14836243 ASD locus Locus found significant in previous ASD GWAS62 but not supported in larger study63 MACROD2 is the only protein-coding gene in the locus |
MACROD2 is a nuclear enzyme that binds to mono-ADP-ribosylated (MARylated) proteins and functions as an eraser of mono-ADP-ribosylation64. Intracellular MARylated histones and GSK3β are substrates of MACROD2, and the removal of MAR from GSK3β is responsible for reactivating of its kinase activity64. This gene is expressed in lung and multiple regions of the brain. Low or no expression across most other tissue (GTEx portal- see URLs). |
position of index SNP is listed.
Correlation with other traits and multi-trait GWAS
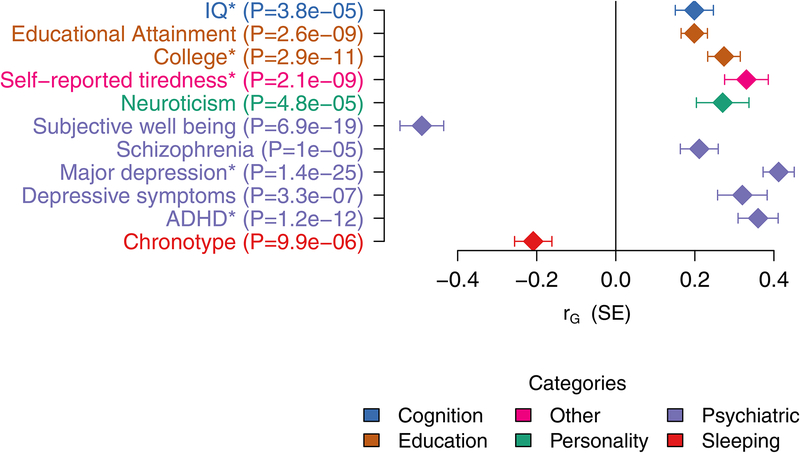
To investigate the extent of genetic overlap between ASD and other phenotypes we estimated the genetic correlations with a broad set of psychiatric and other medical diseases, disorders, and traits available at LD Hub65 using bivariate LD score regression (Figure 2, Supplementary Table 5). Significant correlations were found for several traits including schizophrenia15 (rG = 0.211, P = 1.03 × 10−5) and measures of cognitive ability, especially educational attainment20 (rG = 0.199, P = 2.56 × 10−9), indicating a substantial genetic overlap with these phenotypes and corroborating previous reports5,66–68. In contrast to previous reports16, we found a strong and highly significant correlation with major depression21 (rG = 0.412, P = 1.40 × 10−25), and we report a prominent overlap with ADHD69 (rG = 0.360, P = 1.24 × 10−12). Moreover, we confirm the genetic correlation with social communication difficulties at age 8 in a non-ASD population sample reported previously based on a subset of the ASD sample70 (rG = 0.375, P = 0.0028).
Figure 2. Genetic correlation with other traits.
Significant genetic correlations between ASD (N = 46,350) and other traits after Bonferroni correction for testing a total of 234 traits available at LDhub with the addition of a handful of new phenotypes. Estimates and tests by LDSC19. The results here correspond to the following GWAS analyses: IQ23 (N = 78,308), educational attainment20 (N = 328,917), college71 (N = 111,114), self-reported tiredness72 (N = 108,976), neuroticism67 (N = 170,911), subjective well-being67 (N = 298,420), schizophrenia15 (N = 82,315), major depression21 (N = 480,359), depressive symptoms67(N = 161,460), attention deficit/hyperactivity disorder (ADHD)69 (N = 53,293), and chronotype73 (N = 128,266). See Supplementary Table 5 for the full output of this analysis.
* Indicates that the values are from in-house analyses of new summary statistics not yet included in LD Hub.
In order to leverage these observations for the discovery of loci that may be shared between ASD and these other traits, we selected three particularly well-powered and genetically correlated phenotypes. These were schizophrenia (N = 79,641)15, major depression (N = 424,015)21 and educational attainment (N = 328,917)20. We utilized the recently introduced MTAG method9 which, briefly, generalizes the standard inverse-variance weighted meta-analysis for multiple phenotypes. In this case, MTAG takes advantage of the fact that, given an overall genetic correlation between ASD and a second trait, the effect size estimate and evidence for association to ASD can be improved by appropriate use of the association information from the second trait. The results of these three ASD-anchored MTAG scans are correlated to the primary ASD scan (and to each other) but given the exploration of three scans, we utilized a more conservative threshold of 1.67 × 10−8 for declaring significance across these secondary scans giving an estimated maximum false discovery rate (maxFDR) of 0.021. In addition to stronger evidence for several of the ASD hits defined above, variants in seven additional regions achieved genome-wide significance, including three loci shared with educational attainment and four shared with major depression (Table 1b, Box 1, Supplementary Table 6, Supplementary Figures 49–55). We note that in these seven instances, the effect size estimate is stronger in ASD than the secondary trait, that the result is not characteristic of the strongest signals in these other scans (Supplementary Table 7–9) (and in fact 3 of these 7 were not significant in the secondary trait and constitute potentially novel findings). Moreover, we benchmarked against MTAG running two very large and heritable traits (height74, N = 252,288, and body mass index (BMI)24, N = 322,154) with no expected links to ASD and no significant loci were added to the list of ASD-only significant associations.
Gene and gene-set analysis
Next, we performed gene-based association analysis on our primary ASD meta-analysis using MAGMA75, testing for the joint association of all markers within a locus (across all protein-coding genes in the genome). This analysis identified 15 genes surpassing the significance threshold (Supplementary Table 10). As expected, the majority of these genes were located within the genome-wide significant loci identified in the GWAS, but seven genes are located in four additional loci including KCNN2, MMP12, NTM and a cluster of genes on chromosome 17 (KANSLl, WNT3, MAPT and CRHRl) (Supplementary Figures 57–71). In particular, KCNN2 was strongly associated (P = 1.02 × 10−9), far beyond even single-variant statistical thresholds and is included in the descriptions in Box 1.
Enrichment analyses using gene co-expression modules from human neocortex transcriptomic data (M13, M16 and M17 from Parikshak et al. 201376) and loss-of-function intolerant genes (pLI > 0.9)77,78, which previously have shown evidence of enrichment in neurodevelopmental disorders69,76,79, yielded only nominal significance for the latter (P = 0.014) and M16 (P = 0.050) (Supplementary Table 11). Genes implicated in ASD by studies or rare variants in Sanders et al.56 was just shy of showing nominally significant enrichment (P = 0.063) while enrichment in the curated gene list from the SPARK consortium80 was significant (P = 0.0034). Likewise, analysis of Gene Ontology sets81,82 for molecular function from MsigDB83 showed no significant sets after Bonferroni correction for multiple testing (Supplementary Table 12).
Dissection of the polygenic architecture
As ASD is a highly heterogeneous disorder, we explored how partitioned across phenotypic sub-categories in the iPSYCH sample and estimated the genetic correlations between these groups using GCTA84. We examined cases with and without intellectual disability (ID, N = 1,873) and the ICD-10 diagnostic sub-categories: childhood autism (F84.0, N = 3,310), atypical autism (F84.1, N = 1,607), Asperger’s syndrome (F84.5, N = 4,622), and other/unspecified pervasive developmental disorders (PDD, F84.8–9, N = 5,795), reducing to non-overlapping groups when doing pairwise comparisons (see Supplementary Table 13). While the pairwise genetic correlations were consistently high between all subgroups (95% CIs including 1 in all comparisons), the of Asperger’s syndrome (, SE = 0.001 was found to be twice the of both childhood autism (, SE = 0.009, P = 0.001) and the group of other/unspecified PDD (, SE = 0.008, P = 0.001) (Supplementary Tables 14 and 15, Supplementary Figures 82 and 83). Similarly, the of ASD without ID (, SE = 0.005) was found to be three times higher than for cases with ID (, SE = 0.013, P = 0.015).
To further examine the apparent polygenic heterogeneity across subtypes, we investigated how PRS trained on different phenotypes were distributed across distinct ASD subgroups. We focused on phenotypes showing strong genetic correlation with ASD (e.g., educational attainment), but included also traits with little or no correlation to ASD (e.g., BMI) as negative controls. In this analysis, we regressed the normalized scores on ASD subgroups while including covariates for batches and principal components in a multivariate regression. Of the eight phenotypes we evaluated, only the cognitive phenotypes showed strong heterogeneity (educational attainment20 P = 1.8 × 10−8, IQ23 P = 3.7 × 10−9) (Supplementary Figure 84). Interestingly, all case-control groups with or without intellectual disability showed significantly different loading for the two cognitive phenotypes: controls with intellectual disability have the lowest score followed by ASD cases with intellectual disability, and ASD cases without intellectual disability have significantly higher scores again than any other group (P = 2.6 × 10−12 for educational attainment, P = 8.2 × 10−12 for IQ).
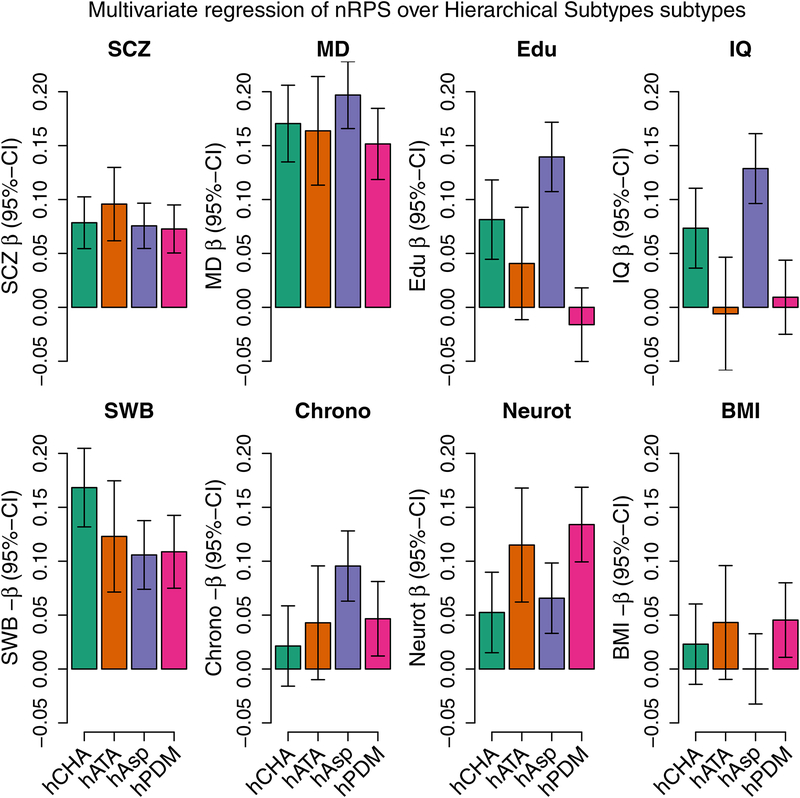
With respect to the diagnostic sub-categories constructed hierarchically from ASD subtypes (Supplementary Table 13), it was again the cognitive phenotypes that showed the strongest heterogeneity across the diagnostic classes (educational attainment P = 2.6 × 10−11, IQ P = 3.4 × 10−8), while neuroticism67 (P = 0.0015), chronotype73 (P = 0.011) and subjective well-being67 (P = 0.029) showed weaker but nominally significant degree of heterogeneity, and schizophrenia (SCZ), major depressive disorder (MD) and BMI24 were non-significant across the groups (P > 0.19) (Figure 3). This pattern weakened only slightly when excluding subjects with intellectual disability (Supplementary Figure 85). For neuroticism, there was a clear split with atypical and other/unspecified PDD cases having significantly higher PRS than childhood autism and Asperger’s, P = 0.00013. Considering the genetic overlap of each subcategory with each phenotype, the hypothesis of homogeneity across sub-phenotypes was strongly rejected (P = 1.6 × 10−11), thereby establishing that these sub-categories indeed have differences in their genetic architectures.
Figure 3. Profiling PRS load across distinct ASD sub-groups for 8 different phenotypes.
(schizophrenia (SCZ)15, major depression (MD)21, educational attainment (Edu)20, human intelligence (IQ)23, subjective well-being (SWB)67, chronotype73, neuroticism67 and body mass index (BMI)24. The bars show coefficients from multivariate multivariable regression of the 8 normalized scores on the distinct ASD sub-types of 13,076 cases and 22,664 controls, adjusting for batches and principal compenents. The subtypes are the hierarchically defined subtypes for childhood autism (hCHA, N = 3,310), atypical autism (hATA, N = 1,494), Asperger’s (hAsp, N = 4,417), and the lumped pervasive disorders developmental group (hPDM, N = 3,855). Please note that the orientation of the scores for subjective well-being, chronotype and BMI have been switched to improve graphical presentation. The corresponding plot where subjects with intellectual disability have been excluded can be seen in Supplementary Figure 85, and with intellectual disability as a subtype in Supplementary Figure 84. Applying the same procedure to the internally trained ASD score did not display systematic heterogeneity (P = 0.068) except as expected for the ID groups (P = 0.00027) (Supplementary Figure 88). Linear hypotheses tested using the Pillai test.
Focusing on educational attainment, significant enrichment of PRS was found for Asperger’s syndrome (P = 2.0 × 10−17) in particular, and for childhood autism (P = 1.5 × 10−5), but not for the group of other/unspecified PDD (P = 0.36) or for atypical autism (P = 0.13) (Figure 3). Excluding individuals with intellectual disability only changes this marginally, with atypical autism becoming nominally significant (P = 0.020) (Supplementary Figure 85). These results show that the genetic architecture underlying educational attainment is indeed shared with ASD but to a variable degree across the disorder spectrum. We find that the observed excess in ASD subjects of alleles positively associated with education attainment85,86 is confined to Asperger’s and childhood autism, and it is not seen here in atypical autism nor in other/unspecified PDD.
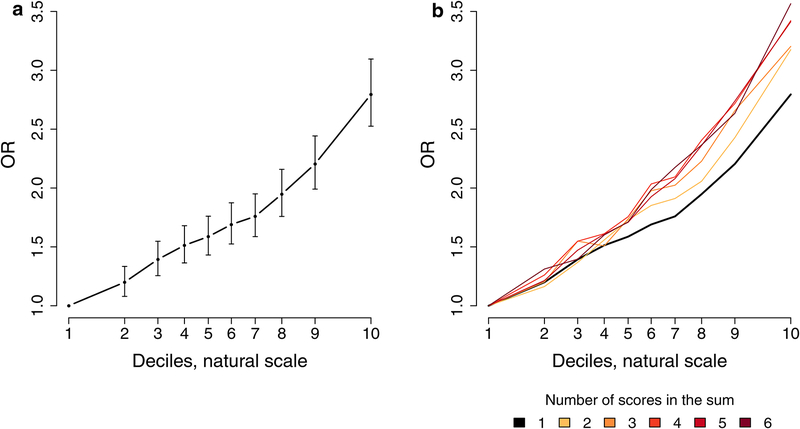
Finally, we evaluated the predictive ability of ASD PRS using five different sets of target and training samples within the combined iPSYCH-PGC sample. The observed mean variance explained by PRS (Nagelkerke’s R2) was 2.45% (P = 5.58 × 10−140) with a pooled PRS-based case-control odds ratio OR = 1.33 (CI.95% 1.30–1.36) (Supplementary Figures 89 and 91). Dividing the target samples into PRS decile groups revealed an increase in OR with increasing PRS. The OR for subjects with the highest PRS increased to OR = 2.80 (CI.95% 2.53–3.10) relative to the lowest decile (Figure 4a and Supplementary Figure 92). Leveraging correlated phenotypes in an attempt to improve prediction of ASD, we generated a multi-phenotype PRS as a weighted sum of phenotype specific PRS (see Methods). As expected, Nagelkerkes’s R2 increased for each PRS included attaining its maximum at the full model at 3.77% (P = 2.03 × 10−215) for the pooled analysis with an OR = 3.57 (CI.95% 3.22–3.96) for the highest decile (Figure 4b and Supplementary Figure 93 and 94). These results demonstrate that an individual’s ASD risk depends on the level of polygenic burden of thousands of common variants in a dose-dependent way, which can be reinforced by adding SNP-weights from ASD correlated traits.
Figure 4. Decile plots.
(Odds Ratio (OR) by PRS within each decile for 13,076 cases and 22,664 controls): a. Decile plot with 95%-CI for the internally trained ASD score (P-value threshold is 0.1). b. Decile plots on a weighted sums of PRSs starting with the ASD score of panel a and successively adding the scores for major depression21, subjective well-being67, schizophrenia15, educational attainment20, and chronotype73. In all instances the P-value threshold for the score used is the one with the highest Nagelkerke’s R2. Supplementary Figures 92 and 94 show the stability across leave-one out groups that was used to create these combined results.
Functional annotation
In order to obtain information on possible biological underpinnings of our GWAS results we conducted several analyses. First, we examined how the ASD partitioned on functional genomic categories as well as on cell type-specific regulatory elements using stratified LD score regression87. This analysis identified significant enrichment of heritability in conserved DNA regions and H3K4me1 histone marks88, as well as in genes expressed in central nervous system (CNS) cell types as a group (Supplementary Figures 95 and 96), which is in line with observations in schizophrenia15, major depression21, and bipolar disorder66. Analyzing the enhancer associated mark H3K4me1 in individual cell/tissue88, we found significant enrichment in brain and neuronal cell lines (Supplementary Figure 97). The highest enrichment was observed in the developing brain, germinal matrix, cortex-derived neurospheres, and embryonic stem cell (ESC)-derived neurons, consistent with ASD as a neurodevelopmental disorder with largely prenatal origins, as supported by data from analysis of rare de novo variants76.
Common variation in ASD is located in regions that are highly enriched with regulatory elements predicted to be active in human corticogenesis (Supplementary Figures 95–97). As most gene regulatory events occur at a distance via chromosome looping, we leveraged Hi-C data from germinal zone (GZ) and post-mitotic zones cortical plate (CP) in the developing fetal brain to identify potential target genes for these variants89. We performed fine mapping of 28 loci to identify the set of credible variants containing likely causal genetic risk90 (see Methods). Credible SNPs were significantly enriched with enhancer marks in the fetal brain (Supplementary Figure 98), again confirming the likely regulatory role of these SNPs during brain development.
Based on location or evidence of physical contact from Hi-C, the 380 credible SNPs (28 loci) could be assigned to 95 genes (40 protein-coding), including 39 SNPs within promoters that were assigned to 9 genes, and 16 SNPs within the protein coding sequence of 8 genes (Supplementary Table 16, Supplementary Figure 98). Hi-C identified 86 genes, which interacted with credible SNPs in either the CP or GZ during brain development. Among these genes, 34 are interacting with credible SNPs in both CP and GZ, which represent a high-confidence gene list. Notable examples are illustrated in Figure 5 and highlighted in Box 1. By analyzing their mean expression trajectory, we observed that the identified ASD candidate genes (Supplementary Table 16) show highest expression during fetal corticogenesis, which is in line with the enrichment of heritability in the regulatory elements in developing brain (Figure 5e–g). Interestingly, both common and rare variation in ASD preferentially affects genes expressed during corticogenesis76, highlighting a potential spatiotemporal convergence of genetic risk on this specific developmental epoch, despite the disorder’s profound genetic heterogeneity.
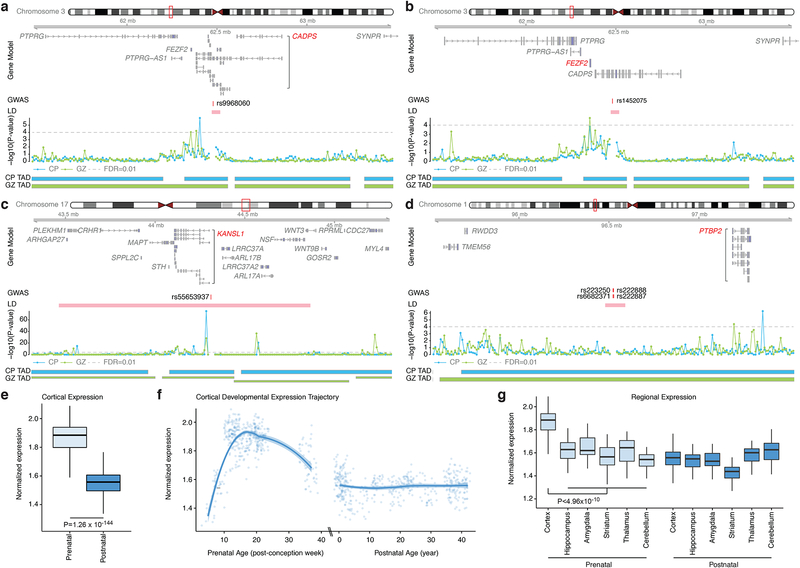
Figure 5. Chromatin interactions identify putative target genes of ASD loci.
a-d. Chromatin interaction maps of credible SNPs to the 1 Mb flanking region, providing putative candidate genes that physically interact with credible SNPs. Gene Model is based on Gencode v19 and putative target genes are marked in red; Genomic coordinate for a credible SNP is labeled as GWAS; −log10(P-value), significance of the interaction between a SNP and each 10-kb bin, grey dotted line for FDR = 0.01 (one-sided significance test calculated as the probability of observing a higher contact frequency under the fitted Weibull distribution matched by chromosome and distance); Topologically associated domain (TAD) borders in cortical plate (CP) and germinal zone (GZ). e-f. Developmental expression trajectories of ASD candidate genes show highest expression in prenatal periods. Significance by t-test (N = 410 and 453 for prenatal and postnatal samples, respectively). Box-plots showing median, interquartile range (IQR) with whiskers adding IQR to the 1st and 3rd quartile (e and g). LOESS smooth curve plotted with actual data points (f) g. ASD candidate genes are highly expressed in the developing cortex as compared to other brain regions. One-way ANOVA and posthoc Tukey test, FDR-corrected. (N = 410/453, 39/36, 33/37, 48/34, 37/36, 32/39 for prenatal/postnatal cortex, hippocampus, amygdala, striatum, thalamus, and cerebellum, respectively).
Discussion
The high heritability of ASD has been recognized for decades and remains among the highest for any complex disease despite many clinical diagnostic changes over the past 30–40 years resulting in a broader phenotype that characterizes more than 1% of the population. While early GWAS permitted estimates that common polygenic variation should explain a substantial fraction of the heritability of ASD, individually significant loci remained elusive. This was suspected to be due to limited sample size since studies of schizophrenia – with similar prevalence, heritability and reduced fitness – and major depression achieved striking results only when sample sizes five to ten times larger than available in ASD were employed. This study has finally borne out that expectation with definitively demonstrated significant “hits”.
Here we report the first common risk variants robustly associated with ASD by using unique Danish resources in conjunction with results of the earlier PGC data – more than tripling the previous largest discovery sample. Of these, five loci were defined in ASD alone, and seven additional suggested at a stricter threshold utilizing GWAS results from three correlated phenotypes (schizophrenia, depression and educational attainment) and a recently introduced analytic approach, MTAG. Both genome-wide LD score regression analysis and the fact that even among the loci defined in ASD alone there was additional evidence in these other trait scans indicate that the polygenic architecture of ASD is significantly shared with risk to adult psychiatric illness and higher educational attainment and intelligence. It should be noted that the MTAG analyses were carried out as three pairwise analyses. This way we avoid the complex interactions that could arise if we ran three or four correlated phenotypes at a time9. Indeed, what we get, despite the secondary summary statistics coming from large, high-powered studies, are relatively modest weights to the contributions from these statistics, because the genetic correlations are modest. The largest weight was 0.27 for schizophrenia, followed by 0.24 for major depression, and 0.11 for educational attainment. Moreover, the estimated worst case FDR was just 0.021 up just 0.001 from that of the ASD GWAS alone. Thus all loci identified by MTAG were found with an acceptable degree of certainty and have substantial contributions from ASD alone (Table 1a, b and Supplementary Table 6). Our expectation is that most or all such loci will likely be identified in future ASD-only GWAS as sample sizes are increased substantially – however, given how new these methods are, the precise phenotypic consequences of these particular variants awaits expansion of all these trait GWAS.
In most GWAS studies there has been little evidence of heterogeneity of association across phenotypic subgroups. In this study, however, we see strong heterogeneity of genetic overlap with other traits when our ASD samples are broken into distinct subsets. In particular, the excess of alleles associated with higher intelligence and educational attainment was only observed in the higher functioning categories (particularly Asperger’s syndrome and individuals without comorbid intellectual disability) – and not in the other/unspecified PDD and intellectual disability categories. This is reminiscent, and logically inverted, from the much greater role of spontaneous mutations in these latter categories, particularly in genes known to have an even larger impact in cohorts ascertained for intellectual disability/developmental delay91. Interestingly, other/unspecified PDD and atypical autism also have a significantly higher PRS for neuroticism than childhood autism and Asperger’s. These different enrichment profiles observed provide evidence for a heterogeneous and qualitatively different genetic architecture between sub-types of ASD, which should inform future studies aiming at identifying etiologies and disease mechanisms in ASD.
The strong differences in estimated SNP heritability between ASD cases with and without intellectual disability, and highest in Asperger’s provide genetic evidence of longstanding observations. In particular, this aligns well with the observation that de novo variants are more frequently observed in ASD cases with intellectual disability compared to cases without comorbid intellectual disability, that IQ correlates positively with family history of psychiatric disorders92 and that severe intellectual disability (encompassing many syndromes that confer high risk to ASD) show far less heritability than is observed for mild intellectual disability93, intelligence in general94 and ASDs. Thus it is perhaps unsurprising that our data suggests that the contribution of common variants may be more prominent in high-functioning ASD cases such as Asperger’s syndrome.
We further explored the functional implications of these results with complementary functional genomics data including Hi-C analyses of fetal brains and brain transcriptome data. Analyses at genome-wide scale (partitioned (Supplementary Figures 95–97) and brain transcriptome enrichment (Figure 5e–g)) as well as at single loci (Figure 5a–d, Box 1) highlighted the involvement of processes relating to brain development and neuronal function. Notably, a number of genes located in the identified loci have previously been linked to ASD risk in studies of de novo and rare variants (Box 1, Supplementary Table 4), including PTBP2, CADPS, and KMT2E, which were found to interact with credible SNPs in the Hi-C analysis (PTBP2, CADPS) or contain a loss-of-function credible SNP (KMT2E). Interestingly, aberrant splicing of CADPS’ sister gene CADPS2, which has almost identical function, has been found in autism cases and Cadps2 knockout mice display behavioral anomalies with translational relevance to autism95. PTBP2 encodes a neuronal splicing factor and alterations in alternative splicing have been identified in brains from individuals diagnosed with ASD96.
In summary, we have established a first robust set of common variant associations in ASD and have begun laying the groundwork through which the biology of ASD and related phenotypes will inevitably be better articulated.
Methods
Subjects
iPSYCH sample
The iPSYCH ASD sample is a part of a population based case-cohort sample extracted from a baseline cohort10 consisting of all children born in Denmark between May 1st 1981 and December 31st 2005. Eligible were singletons born to a known mother and resident in Denmark on their one-year birthday. Cases were identified from the Danish Psychiatric Central Research Register (DPCRR)12, which includes data on all individuals treated in Denmark at psychiatric hospitals (from 1969 onwards) as well as at outpatient psychiatric clinics (from 1995 onwards). Cases were diagnosed with ASD in 2013 or earlier by a psychiatrist according to ICD10, including diagnoses of childhood autism (ICD10 code F84.0), atypical autism (F84.1), Asperger’s syndrome (F84.5), other pervasive developmental disorders (F84.8), and pervasive developmental disorder, unspecified (F84.9). As controls we selected a random sample from the set of eligible children excluding those with an ASD diagnosis by 2013.
The samples were linked using the unique personal identification number to the Danish Newborn Screening Biobank (DNSB) at Statens Serum Institute (SSI), where DNA was extracted from Guthrie cards and whole genome amplified in triplicates as described previously13,97. Genotyping was performed at the Broad Institute of Harvard and MIT (Cambridge, MA, USA) using the PsychChip array from Illumina (CA, San Diego, USA) according to the instructions of the manufacturer. Genotype calling of markers with minor allele frequency (MAF) > 0.01 was performed by merging callsets from GenCall98 and Birdseed99 while less frequent variants were called with zCall100. Genotyping and data processing was carried out in 23 waves.
All analyses of the iPSYCH sample and joint analyses with the PGC samples were performed at the secured national GenomeDK high performance-computing cluster in Denmark.
The study was approved by the Regional Scientific Ethics Committee in Denmark and the Danish Data Protection Agency.
Psychiatric Genomic Consortium (PGC) samples
In brief, five cohorts provided genotypes to the sample (N denoting the number of trios for which genotypes were available): The Geschwind Autism Center of Excellence (ACE; N = 391), the Autism Genome Project62 (AGP; N = 2,272), the Autism Genetic Resource Exchange101,102 (AGRE; N = 974), the NIMH Repository, the Montreal103/Boston Collection (MONBOS; N = 1,396, and the Simons Simplex Collection104,105(SSC; N = 2,231). The trios were analyzed as cases and pseudo controls. A detailed description of the sample is available on the PGC web site and even more details are provided in Anney et al5. Analyses of the PGC genotypes were conducted on the computer cluster LISA at the Dutch HPC center SURFsara.
Follow-up samples
As follow-up for the loci with p-values less than 10−6 we asked for look up in five samples of Nordic and Eastern European origin with altogether 2,119 cases and 142,379 controls: BUPGEN (Norway: 164 cases and 656 controls), PAGES (Sweden: 926 cases and 3,841 controls not part of the PGC sample above), the Finnish autism case-control study (Finland: 159 cases and 526 controls), deCODE (Iceland 574 cases and 136,968 controls; Eastern Europe: 296 cases and 388 controls). See Supplementary Note for details.
Statistical analyses
All statistical tests are two-sided unless otherwise stated. Software versions etc. can be found in Life Sciences Reporting Summary.
GWAS analysis
Ricopili15, the pipeline developed by the Psychiatric Genomics Consortium (PGC) Statistical Analysis Group was used for quality control, imputation, principle component analysis (PCA) and primary association analysis. For details see the Supplementary Note. The data were processed separately in the 23 genotyping batches in the case of iPSYCH and separately for each study in the PGC sample. Phasing was achieved using SHAPEIT106 and imputation done by IMPUTE2107,108 with haplotypes from the 1000 Genomes Project, phase 3109 (1kGP3) as reference.
After excluding regions of high linkage disequilibrium (LD)110, the genotypes were pruned down to a set of roughly 30k markers. See supplementary Note for details. Using PLINK’s111 identity by state analysis, pairs of subjects were identified with and one subject of each such pair was excluded at random (with a preference for keeping cases). PCA was carried out using smartPCA112,113. In iPSYCH, a subsample of European ancestry was selected as an ellipsoid in the space of PC1–3 centred and scaled using the mean and eight standard deviation of the subsample whose parents and grandparents were all known to have been born in Denmark (N = 31,500). In the PGC sample the European (CEU) subset was chosen using a Euclidian distance measure weighted by the variance explained by each of the first 3 principal components. Individuals more distant than 10 standard deviations from the combined CEU and Toscani in Italy (TSI) HapMap reference populations were excluded. We conducted a secondary PCA on the remaining 13,076 cases and 22,664 controls to provide covariates for the association analyses. Numbers of subjects in the data generation flow for the iPSYCH sample are found in Supplementary Table 1.
Association analyses were done by applying PLINK 1.9 to the imputed dosage data (the sum of imputation probabilities P(A1A2) + 2P(A1A1)). In iPSYCH we included the first four principal components (PCs) as covariates as well as any PC beyond that, which were significantly associated with ASD in the sample, while the case-pseudo-controls from the PGC trios required no PC covariates. Combined results for iPSYCH and for iPSYCH with the PGC was achieved by meta-analysing batch-wise and study-wise results using METAL114 (July 2010 version) employing an inverse variance weighted fixed effect model115. On chromosome X males and females were analyzed separately and then meta-analyzed together. Subsequently we applied a quality filter allowing only markers with an imputation info score ≥ 0.7, MAF ≥ 0.01 and an effective sample size (see Supplementary Note) of at least 70% of the study maximum. The degree to which the deviation in the test statistics can be ascribed to cryptic relatedness and population stratification rather than to polygenicity was measured from the intercept in LD score regression19 (LDSC) as the ratio of (intercept-1) and (mean(χ2)-1).
MTAG9 was applied with standard settings. The iPSYCH-PGC meta-analysis summary statistics were paired up with the summary statistics for each of major depression21 (excluding the Danish sampled but including summary statistics from 23andMe22; 111,902 cases, 312,113 controls, and mean χ2 = 1.477), schizophrenia15 (also excluding the Danish samples; 34,129 cases, 45,512 controls, and mean χ2 = 1.804) and educational attainment20 (328,917 samples and mean χ2 = 1.648). These are studies that have considerably more statistical power than the ASD scan, but the genetic correlations are modest in the context of MTAG, so the weights ascribed to the secondary phenotypes in the MTAG analyses remain relatively low (no higher than 0.27). The maximum FDR was estimated as recommended in the MTAG paper9. See Supplementary Note for details. The results were clumped and we highlighted loci of interest by selecting those that were significant at 5 × 10−8 in the iPSYCH-PGC meta-analysis or the meta-analysis with the follow-up sample or were significant at 1.67 × l0−8 in any of the three MTAG analyses. The composite GWAS consisting of the minimal p-values at each marker over these five analyses was used as a background when creating Manhattan plots for the different analyses showing both what is maximally achieved and what the individual analysis contributes to that.
Gene-based association and gene-set analyses.
MAGMA 1.0675 was applied to the ASD GWAS summary statistics to test for gene-based association. Using NCBI 37.3 gene definitions and restricting the analysis to SNPs located within the transcribed region, mean SNP association was tested with the sum of −log(SNP p-value) as test statistic. The resulting gene-based p-values were further used in competitive gene-set enrichment analyses in MAGMA. One analysis explored the candidate sets M13, M16 and M17 from Parikshak et al. 201376, constrained, loss-of-function intolerant genes (pLI > 0.9)77,78 derived from data of the Exome Aggregation Consortium (see Supplementary Note for details), as well as gene sets found in studies of rare variants in autism by Sanders et al.56 and the curated gene list from the SPARK consortium80. Another was an agnostic analysis of the Gene Ontology sets81,82 for molecular function from MsigDB 6.083. We analyzed only genes outside the broad MHC region (hg19:chr6:25–35M) and included only gene sets with 10–1,000 genes. The gene set from Sanders et al. and SPARK include only one gene in MHC and was exempt from the MHC exclusion to be as true to the set as possible. All gene sets with significant enrichment were inspected to ensure that the signal was not driven by one or a few associated loci with multiple genes in close LD.
SNP heritability
SNP heritability, , was estimated using LDSC19 for the full ASD GWAS sample and GCTA84,116,117 for subsamples too small for LDSC. For LDSC we used precomputed LD scores based on the European ancestry samples of the 1000 Genomes Project118 restricted to HapMap3119 SNPs. The summary stats with standard LDSC filtering were regressed onto these scores. For liability scale estimates, we used a population prevalence for Denmark of 1.22%18. Lacking proper prevalence estimates for subtypes, we scaled the full spectrum prevalence based on the composition of the case sample.
For subsamples too small for LDSC, the GREML approach of GCTA84,116,117 was used. On best guess genotypes (genotype probability > 0.8, missing rate < 0.01 and MAF > 0.05) with INDELs removed, a genetic relatedness matrix (GRM) was fitted for the association sample (i.e. the subjects of European ancestry with ) providing a relatedness estimate for all pairwise combinations of individuals. Estimation of the phenotypic variance explained by the SNPs (REML) was performed including PC 1–4 as continuous covariates together with any other PC that was nominally significantly associated to the phenotype as well as batches as categorical indicator covariates. Testing equal heritability for non-overlapping groups was done by permutation test (with 1000 permutations) keeping the controls and randomly assigning the different case labels.
Following Finucane et al.87, we conducted an enrichment analysis of the heritability for SNPs for functional annotation and for SNPs located in cell-type-specific regulatory elements. Using first the same 24 overlapping functional annotations (stripped down from 53) as in Finucane et al. we regressed the χ2 from the ASD GWAS summary statistics on to the cell-type specific LD scores download from the site mentioned above with baseline scores, regression weights and allele frequencies based on European ancestry 1000 Genome Project data. The enrichment of a category was defined as the proportion of SNP heritability in the category divided by the proportion of SNPs in that category. Still following Finucane et al. we did a similar analysis using 220 cell type–specific annotations divided into 10 overlapping groups. In addition to this, we conducted an analysis based on annotation derived from data on H3K4Me1 imputed gapped peaks data from the Roadmap Epigenomics Mapping Consortium120; more specifically information excluding the broad MHC-region (chr6:25–35MB).
Genetic correlation
For the main ASD samples, SNP correlations, rG were estimated using LDSC19 and for the analysis of ASD subtypes and subgroups where the sample sizes were generally small, we used GCTA84. In both cases, we followed the same procedures as explained above. For all but a few phenotypes, LDSC estimates of correlation were achieved by upload to LD hub65 for comparison to all together 234 phenotypes.
Polygenic risk scores
For the polygenic risk scores (PRS) we clumped the summary stats applying standard Ricopili parameters (see Supplementary Note for details). To avoid potential strand conflicts, we excluded all ambiguous markers for summary statistics not generated by Ricopili using the same imputation reference. PRS were generated at the default p-value thresholds (5e-8, 1e-6, 1e-4, 0.001, 0.01, 0.05, 0.1, 0.2, 0.5 and 1) as a weighted sum of the allele dosages in the ASD GWAS sample. Summing over the markers abiding by the p-value threshold in the training set and weighing by the additive scale effect measure of the marker (log(OR) or β) as estimated in the training set. Scores were normalized prior to analysis.
We evaluated the predictive power using Nagelkerke’s R2 and plots of odds ratios and confidence intervals over score deciles. Both R2 and odds ratios were estimated in regression analyses including the relevant PCs and indicator variables for genotyping waves.
Lacking a large ASD sample outside of iPSYCH and PGC, we trained a set of PRS for ASD internally in the following way. We divided the sample in five subsamples of roughly equal size respecting the division into batches. We then ran five GWAS leaving out each group in turn from the training set and meta-analyzed these with the PGC results. This produced a set of PRS for each of the five subsamples trained on their complement. Prior to analyses, each score was normalized on the group where it was defined. We evaluated the predictive power in each group and on the whole sample combined.
To exploit the genetic overlap with other phenotypes to improve prediction, we created a series of new PRS by adding to the internally trained ASD score the PRS of other highly correlated phenotypes in a weighted sum. See supplementary info for details.
To analyze ASD subtypes in relation to PRS we defined a hierarchical set of phenotypes in the following way: First hierarchical subtypes was childhood autism, hierarchical atypical autism was defined as everybody with atypical autism and no childhood autism diagnosis, hierarchical Asperger’s as everybody with an Asperger’s diagnosis and neither childhood autism nor atypical autism. Finally, we lumped other pervasive developmental disorders and pervasive developmental disorder, unspecified into pervasive disorders developmental mixed, and the hierarchical version of that consists of everybody with such a diagnosis and none of the preceding ones (Supplementary Table 13). We examined the distribution over the distinct ASD subtypes of PRS for a number of phenotypes showing high rG with ASD (as well as a few with low rG as negative controls), by doing multivariate regression of the scores on the subtypes while adjusting for relevant PCs and wave indicator variables in a linear regression. See Supplementary Note for details.
Hi-C analysis
The Hi-C data were generated from two major cortical laminae: the germinal zone (GZ), containing primarily mitotically active neural progenitors, and the cortical and subcortical plate (CP), consisting primarily of post-mitotic neurons89. We first derived a set of credible SNPs (putative causal SNPs) from the identified top ranking loci in the ASD GWAS using CAVIAR90. The 30 loci showing the strongest association was intersected with the Hi-C reference data resulting in 28 loci for analysis. To test whether credible SNPs are enriched in active marks in the fetal brain120, we employed GREAT as previously described89,121. Credible SNPs were sub-grouped into those without known function (unannotated) and functionally annotated SNPs (SNPs in the gene promoters and SNPs that cause nonsynonymous variants) (Supplementary Figure 98). Then we integrated unannotated credible SNPs with chromatin contact profiles during fetal corticogenesis89, defining genes physically interacting with intergenic or intronic SNPs (Supplementary Figure 98).
The spatiotemporal transcriptomic atlas of human brain was obtained from Kang et al122. We used transcriptomic profiles of multiple brain regions with developmental epochs that span prenatal (6–37 post-conception week, PCW) and postnatal (4 months-42 years) periods. Expression values were log-transformed and centered to the mean expression level for each sample using a scale(center=T, scale=F)+1 function in R. ASD candidate genes identified by Hi-C analyses (Supplementary Figure 98) were selected for each sample and their average centered expression values were calculated and plotted.
Availability of summary statistics
The summary statistics are available for download the iPSYCH and at the PGC download sites (see the URL section).
Availability of genotype data
For access to genotypes from the PGC samples and the iPSYCH sample, researchers should contact the lead PIs Mark J. Daly and Anders D. Børglum for PGC-ASD and iPSYCH-ASD respectively.
URLs
The GenomeDK high performance-computing cluster in Denmark, https://genome.au.dk; the iPSYCH project, http://ipsych.au.dk, the iPSYCH download page, http://ipsych.au.dk/downloads/; the NIMH Repository, https://www.nimhgenetics.org/available_data/autism/; the PGC download site, https://www.med.unc.edu/pgc/results-and-downloads; the LISA cluster at SURFsara, https://userinfo.surfsara.nl/systems/lisa; plink 1.9, www.cog-genomics.org/plink/1.9/; LDSC and associated files, https://github.com/bulik/ldsc; LD hub, http://ldsc.broadinstitute.org/ldhub/; GTExportal, https://gtexportal.org/home/
Supplementary Material
Acknowledgements
The iPSYCH project is funded by the Lundbeck Foundation (grant numbers R102-A9118 and R155-2014-1724) and the universities and university hospitals of Aarhus and Copenhagen. Genotyping of iPSYCH and PGC samples was supported by grants from the Lundbeck Foundation, the Stanley Foundation, the Simons Foundation (SFARI 311789 to MJD), and NIMH (5U01MH094432–02 to MJD). The Danish National Biobank resource was supported by the Novo Nordisk Foundation. Data handling and analysis on the GenomeDK HPC facility was supported by NIMH (1U01MH109514–01 to M O’Donovan and ADB). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to ADB). Drs. S De Rubeis and JD Buxbaum were supported by NIH grants MH097849 (to JDB) and MH111661 (to JDB), and by the Seaver Foundation (to SDR and JDB). Dr J Martin was supported by the Wellcome Trust (grant no: 106047). O. Andreassen received funding from Research Council of Norway (#213694, #223273, #248980, #248778), Stiftelsen KG Jebsen and South-East Norway Health Authority. We thank the research participants and employees of 23andMe for making this work possible.
Consortia Members
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium
Naomi R. Wray55,56, Maciej Trzaskowski55, Enda M. Byrne55, Abdel Abdellaoui57, Mark J. Adams58, Tracy M. Air59, Till F.M. Andlauer60,61, Silviu-Alin Bacanu62, Aartjan T.F. Beekman63, Tim B. Bigdeli62,64, Elisabeth B. Binder60,65, Douglas H.R. Blackwood58, Julien Bryois23, Henriette N. Buttenschøn1,2,66, Na Cai67,68, Enrique Castelao69, Toni-Kim Clarke58, Jonathan R.I. Coleman70, Lucía Colodro-Conde71, Baptiste Couvy-Duchesne72,73, Nick Craddock74, Gregory E. Crawford75,76, Gail Davies77, Ian J. Deary77, Franziska Degenhardt78,79, Eske M. Derks71, Nese Direk80,81, Conor V. Dolan57, Erin C. Dunn6,82,83, Thalia C. Eley70, Valentina Escott-Price84, Farnush Farhadi Hassan Kiadeh85, Hilary K. Finucane36,86, Andreas J. Forstner78,79,87,88, Josef Frank89, Héléna A. Gaspar70, Michael Gill90, Fernando S. Goes91, Scott D. Gordon71, Lynsey S. Hall58,92, Thomas F. Hansen93,94,95, Stefan Herms78,79,88, Ian B. Hickie96, Per Hoffmann78,79,88, Georg Homuth97, Carsten Horn98, Jouke-Jan Hottenga57, Marcus Ising99, Rick Jansen63,63, Eric Jorgenson100, James A. Knowles101, Isaac S. Kohane102,103,104, Julia Kraft105, Warren W. Kretzschmar106, Jesper Krogh107, Zoltán Kutalik108,109, Yihan Li106, Penelope A. Lind71, Donald J. MacIntyre110,111, Dean F. MacKinnon91, Robert M. Maier56, Wolfgang Maier112, Jonathan Marchini113, Hamdi Mbarek57, Patrick McGrath114, Peter McGuffin70, Sarah E. Medland71, Divya Mehta56,115, Christel M. Middeldorp57,116,117, Evelin Mihailov118, Yuri Milaneschi63,63, Lili Milani118, Francis M. Mondimore91, Grant W. Montgomery55, Sara Mostafavi119,120, Niamh Mullins70, Matthias Nauck121,122, Bernard Ng120, Michel G. Nivard57, Dale R. Nyholt123, Paul F. O’Reilly70, Hogni Oskarsson124, Michael J. Owen16, Jodie N. Painter71, Roseann E. Peterson62,125, Erik Pettersson23, Wouter J. Peyrot63, Giorgio Pistis69, Danielle Posthuma126,127, Jorge A. Quiroz128, John P. Rice129, Brien P. Riley62, Margarita Rivera70,130, Saira Saeed Mirza80, Robert Schoevers131, Eva C. Schulte132,133, Ling Shen100, Jianxin Shi134, Stanley I. Shyn135, Engilbert Sigurdsson136, Grant C.B. Sinnamon137, Johannes H. Smit63, Daniel J. Smith138, Fabian Streit89, Jana Strohmaier89, Katherine E. Tansey139, Henning Teismann140, Alexander Teumer141, Wesley Thompson1,14,15,94,142, Pippa A. Thomson143, Thorgeir E. Thorgeirsson144, Matthew Traylor145, Jens Treutlein89, Vassily Trubetskoy105, André G. Uitterlinden146, Daniel Umbricht147, Sandra Van der Auwera148, Albert M. van Hemert149, Alexander Viktorin23, Peter M. Visscher55,56, Yunpeng Wang1,14,15,94, Bradley T. Webb125, Shantel Marie Weinsheimer1,94, Jürgen Wellmann140, Gonneke Willemsen57, Stephanie H. Witt89, Yang Wu55, Hualin S. Xi150, Jian Yang56,151, Futao Zhang55, Volker Arolt152, Bernhard T. Baune59, Klaus Berger140, Dorret I. Boomsma57, Sven Cichon78,88,153,154, Udo Dannlowski152, EJC de Geus57,155, J Raymond DePaulo91, Enrico Domenici156, Katharina Domschke157, Tõnu Esko22,118, Hans J. Grabe148, Steven P. Hamilton158, Caroline Hayward159, Andrew C. Heath129, Kenneth S. Kendler62, Stefan Kloiber99,160,161, Glyn Lewis162, Qingqin S. Li163, Susanne Lucae99, Pamela A.F. Madden129, Patrik K. Magnusson23, Nicholas G. Martin71, Andrew M. McIntosh58,77, Andres Metspalu118,164, Bertram Müller-Myhsok60,61,165, Markus M. Nöthen78,79, Michael C. O’Donovan16, Sara A. Paciga166, Nancy L. Pedersen23, Brenda WJH Penninx63, Roy H. Perlis82,167, David J. Porteous143, James B. Potash168, Martin Preisig69, Marcella Rietschel89, Catherine Schaefer100, Thomas G. Schulze89,91,133,169,170, Jordan W. Smoller6,82,83, Henning Tiemeier80,171,172, Rudolf Uher173, Henry Völzke141, Myrna M. Weissman114,174, Cathryn M. Lewis70,175, Douglas F. Levinson176, and Gerome Breen70,177
23andMe Research Team
Michelle Agee178, Babak Alipanahi178, Adam Auton178, Robert K Bell178, Katarzyna Bryc178, Sarah L Elson178, Pierre Fontanillas178, Nicholas A Furlotte178, Bethann S Hromatka178, Karen E Huber178, Aaron Kleinman178, Nadia K Litterman178, Matthew H McIntyre178, Joanna L Mountain178, Elizabeth S Noblin178, Carrie AM Northover178, Steven J Pitts178, J Fah Sathirapongsasuti178, Olga V Sazonova178, Janie F Shelton178, Suyash Shringarpure178, Joyce Y Tung178, Vladimir Vacic178, and Catherine H Wilson178
Affiliations unique to the consortia:
55. Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
56. Queensland Brain Institute, The University of Queensland, Brisbane, QLD, Australia
57. Departmentof Biological Psychology & EMGO+ Institute for Health and Care Research, Vrije Universiteit, Amsterdam, Amsterdam, Netherlands
58. Division of Psychiatry, University of Edinburgh, Edinburgh, UK
59. Discipline of Psychiatry, University of Adelaide, Adelaide, SA, Australia
60. Department of Translational Research in Psychiatry, Max Planck Institute of Psychiatry, Munich, Germany
61. Munich Cluster for Systems Neurology (SyNergy), Munich, Germany
62. Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
63. Department of Psychiatry, Vrije Universiteit Medical Center and GGZ inGeest, Amsterdam, Netherlands
64. Virginia Institute for Psychiatric and Behavior Genetics, Richmond, VA, USA
65. Department of Psychiatry and Behavioral Sciences, Emory University School of Medicine, Atlanta, GA, USA
66. Department of Clinical Medicine, Translational Neuropsychiatry Unit, Aarhus University, Aarhus, Denmark
67. Human Genetics, Wellcome Trust Sanger Institute, Cambridge, UK
68. Statistical genomics and systems genetics, European Bioinformatics Institute (EMBL-EBI), Cambridge, UK
69. Department of Psychiatry, University Hospital of Lausanne, Prilly, Vaud, Switzerland
70. MRC Social Genetic and Developmental Psychiatry Centre, King’s College London, London, UK
71. Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
72. Centre for Advanced Imaging, The University of Queensland, Saint Lucia, QLD, Australia
73. Queensland Brain Institute, The University of Queensland, Saint Lucia, QLD, Australia
74. Psychological Medicine, Cardiff University, Cardiff, UK
75. Center for Genomic and Computational Biology, Duke University, Durham, NC, USA
76. Department of Pediatrics, Division of Medical Genetics, Duke University, Durham, NC, USA
77. Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh, UK
78. Institute of Human Genetics, University of Bonn, Bonn, Germany
79. Life&Brain Center, Department of Genomics, University of Bonn, Bonn, Germany
80. Epidemiology, Erasmus MC, Rotterdam, Zuid-Holland, Netherlands
81. Psychiatry, Dokuz Eylul University School Of Medicine, Izmir, Turkey
82. Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA
83. Psychiatric and Neurodevelopmental Genetics Unit (PNGU), Massachusetts General Hospital, Boston, MA, USA
84. Neuroscience and Mental Health, Cardiff University, Cardiff, UK
85. Bioinformatics, University of British Columbia, Vancouver, BC, Canada
86. Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA, USA
87. Department of Psychiatry (UPK), University of Basel, Basel, Switzerland
88. Human Genomics Research Group, Department of Biomedicine, University of Basel, Basel, Switzerland
89. Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty, Mannheim, Heidelberg University, Mannheim, Baden-Württemberg, Germany
90. Department of Psychiatry, Trinity College Dublin, Dublin, Ireland
91. Department of Psychiatry and Behavioral Sciences, Johns Hopkins University, Baltimore, MD, USA
92. Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
93. Danish Headache Centre, Department of Neurology, Rigshospitalet, Glostrup, Denmark
94. Institute of Biological Psychiatry, Mental Health Center Sct. Hans, Mental Health Services Capital Region of Denmark, Copenhagen, Denmark
95. iPSYCH, The Lundbeck Foundation Initiative for Psychiatric Research, Copenhagen, Denmark
96. Brain and Mind Centre, University of Sydney, Sydney, NSW, Australia
97. Interfaculty Institute for Genetics and Functional Genomics, Department of Functional Genomics, University Medicine and Ernst Moritz Arndt University Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany
98. Roche Pharmaceutical Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd, Basel, Switzerland
99. Max Planck Institute of Psychiatry, Munich, Germany
100. Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA
101. Psychiatry & The Behavioral Sciences, University of Southern California, Los Angeles, CA, USA
102. Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA
103. Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA
104. Informatics Program, Boston Children’s Hospital, Boston, MA, USA
105. Department of Psychiatry and Psychotherapy, Universitätsmedizin Berlin Campus Charité Mitte, Berlin, Germany
106. Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK
107. Department of Endocrinology at Herlev University Hospital, University of Copenhagen, Copenhagen, Denmark
108. Institute of Social and Preventive Medicine (IUMSP), University Hospital of Lausanne, Lausanne, VD, Switzerland
109. Swiss Institute of Bioinformatics, Lausanne, VD, Switzerland
110. Division of Psychiatry, Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK
111. Mental Health, NHS 24, Glasgow, UK
112. Department of Psychiatry and Psychotherapy, University of Bonn, Bonn, Germany
113. Statistics, University of Oxford, Oxford, UK
114. Psychiatry, Columbia University College of Physicians and Surgeons, New York, NY, USA
115. School of Psychology and Counseling, Queensland University of Technology, Brisbane, QLD, Australia
116. Child and Youth Mental Health Service, Children’s Health Queensland Hospital and Health Service, South Brisbane, QLD, Australia
117. Child Health Research Centre, University of Queensland, Brisbane, QLD, Australia
118. Estonian Genome Center, University of Tartu, Tartu, Estonia
119. Medical Genetics, University of British Columbia, Vancouver, BC, Canada
120. Statistics, University of British Columbia, Vancouver, BC, Canada
121. DZHK (German Centre for Cardiovascular Research), Partner Site Greifswald, University Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany
122. Institute of Clinical Chemistry and Laboratory Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany
123. Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia
124. Humus, Reykjavik, Iceland
125. Virginia Institute for Psychiatric & Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, USA
126. Clinical Genetics, Vrije Universiteit Medical Center, Amsterdam, Netherlands
127. Complex Trait Genetics, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
128. Solid Biosciences, Boston, MA, USA
129. Department of Psychiatry, Washington University in Saint Louis School of Medicine, Saint Louis, MO, USA
130. Department of Biochemistry and Molecular Biology II, Institute of Neurosciences, Center for Biomedical Research, University of Granada, Granada, Spain
131. Department of Psychiatry, University of Groningen, University Medical Center Groningen, Groningen, Netherlands
132. Department of Psychiatry and Psychotherapy, Medical Center of the University of Munich, Campus Innenstadt, Munich, Germany
133. Institute of Psychiatric Phenomics and Genomics (IPPG), Medical Center of the University of Munich, Campus Innenstadt, Munich, Germany
134. Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA
135. Behavioral Health Services, Kaiser Permanente Washington, Seattle, WA, USA
136. Faculty of Medicine, Department of Psychiatry, University of Iceland, Reykjavik, Iceland
137. School of Medicine and Dentistry, James Cook University, Townsville, QLD, Australia
138. Institute of Health and Wellbeing, University of Glasgow, Glasgow, UK
139. College of Biomedical and Life Sciences, Cardiff University, Cardiff, UK
140. Institute of Epidemiology and Social Medicine, University of Münster, Münster, Nordrhein-Westfalen, Germany
141. Institute for Community Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany
142. Department of Psychiatry, University of California, San Diego, San Diego, CA, USA
143. Medical Genetics Section, CGEM, IGMM, University of Edinburgh, Edinburgh, UK
144. deCODE Genetics / Amgen, Reykjavik, Iceland
145. Clinical Neurosciences, University of Cambridge, Cambridge, UK
146. Internal Medicine, Erasmus MC, Rotterdam, Zuid-Holland, Netherlands
147. Roche Pharmaceutical Research and Early Development, Neuroscience, Ophthalmology and Rare Diseases Discovery & Translational Medicine Area, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd, Basel, Switzerland
148. Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany
149. Department of Psychiatry, Leiden University Medical Center, Leiden, Netherlands
150. Computational Sciences Center of Emphasis, Pfizer Global Research and Development, Cambridge, MA, USA
151. Institute for Molecular Bioscience, Queensland Brain Institute, The University of Queensland, Brisbane, QLD, Australia
152. Department of Psychiatry, University of Münster, Münster, Nordrhein-Westfalen, Germany
153. Institute of Medical Genetics and Pathology, University Hospital Basel, University of Basel, Basel, Switzerland
154. Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, Juelich, Germany
155. Amsterdam Public Health Institute, Vrije Universiteit Medical Center, Amsterdam, Netherlands
156. Centre for Integrative Biology, Università degli Studi di Trento, Trento, Trentino-Alto Adige, Italy
157. Department of Psychiatry and Psychotherapy, Medical Center, University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
158. Psychiatry, Kaiser Permanente Northern California, San Francisco, CA, USA
159. Medical Research Council Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK
160. Department of Psychiatry, University of Toronto, Toronto, ON, Canada
161. Centre for Addiction and Mental Health, Toronto, ON, Canada
162. Division of Psychiatry, University College London, London, UK
163. Neuroscience Therapeutic Area, Janssen Research and Development, LLC, Titusville, NJ, USA
164. Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia
165. University of Liverpool, Liverpool, UK
166. Human Genetics and Computational Biomedicine, Pfizer Global Research and Development, Groton, CT, USA
167. Psychiatry, Harvard Medical School, Boston, MA, USA
168. Psychiatry, University of Iowa, Iowa City, IA, USA
169. Department of Psychiatry and Psychotherapy, University Medical Center Göttingen, Goettingen, Niedersachsen, Germany
170. Human Genetics Branch, NIMH Division of Intramural Research Programs, Bethesda, MD, USA
171. Child and Adolescent Psychiatry, Erasmus MC, Rotterdam, Zuid-Holland, Netherlands
172. Psychiatry, Erasmus MC, Rotterdam, Zuid-Holland, Netherlands
173. Psychiatry, Dalhousie University, Halifax, NS, Canada
174. Division of Epidemiology, New York State Psychiatric Institute, New York, NY, USA
175. Department of Medical & Molecular Genetics, King’s College London, London, UK
176. Psychiatry & Behavioral Sciences, Stanford University, Stanford, CA, USA
177. NIHR BRC for Mental Health, King’s College London, London, UK
178. 23andMe, Inc., Mountain View, CA, USA
Footnotes
Competing Interests Statement
Hreinn Stefansson, Kari Stefansson, Stacy Steinberg, and G. Bragi Walters are employees of deCODE genetics/Amgen. The 23andMe Research Team are employed by 23andMe. Daniel H Geschwind is a scientific advisor for Ovid Therapeutic, Falcon Computing and Axial Biotherapeutics. Thomas Werge has acted as scientific advisor and lecturer for H. Lundbeck A/S.
References
- 1.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaugler T et al. Most genetic risk for autism resides with common variation. Nat Genet 46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumm N et al. Excess of rare, inherited truncating mutations in autism. Nat Genet 47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anney RJL et al. Meta-analysis of gwas of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Molecular Autism 8, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma D et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Annals of human genetics 73, 263–273 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devlin B, Melhem N & Roeder K Do common variants play a role in risk for autism? evidence and theoretical musings. Brain research 1380, 78–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anney R et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet 21, 4781–4792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turley P et al. Multi-trait analysis of genome-wide association summary statistics using mtag. Nature Genetics 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen CB et al. The ipsych2012 case-cohort sample: new directions for unravelling the genetic and environmental architecture of severe mental disorders. Molecular Psychiatry 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauritsen MB et al. Validity of childhood autism in the danish psychiatric central register: Findings from a cohort sample born 1990–1999. J Autism Dev Disord 40, 139–148 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Mors O, Perto GP & Mortensen PB The danish psychiatric central research register. Scandinavian journal of public health 39, 54–57 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Hollegaard M et al. Robustness of genome-wide scanning using archived dried blood spot samples as a dna source. BMC Genet 12, 58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollegaard MV et al. Genome-wide scans using archived neonatal dried blood spot samples. BMC Genomics 10, 297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross-Disorder Group of the Psychiatric Genomics Consortium et al. Genetic relationship between five psychiatric disorders estimated from genome-wide snps. Nature genetics 45, 984–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gratten J, Wray NR, Keller MC & Visscher PM Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature neuroscience 17, 782–790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen SN, Overgaard M, Andersen PK & Parner ET Estimating a population cumulative incidence under calendar time trends. BMC medical research methodology 17, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK et al. Ld score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depressive disorder. Nature Genetics 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyde CL et al. Identification of 15 genetic loci associated with risk of major depression in individuals of european descent. Nature genetics 48, 1031–1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sniekers S et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nature genetics 49, 1546–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locke AE et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorleifsson G et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature genetics 41, 18–24 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Willer CJ et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics 41, 25–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speliotes EK et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics 42, 937–948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berndt SI et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nature genetics 45, 501–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto T, Yamada M, Maekawa S, Nakashima T & Miyata S Iglon cell adhesion molecule kilon is a crucial modulator for synapse number in hippocampal neurons. Brain research 1224, 1–11 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto T, Maekawa S & Miyata S Iglon cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell biochemistry and function 27, 496–498 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Pischedda F et al. A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Molecular & cellular proteomics 13, 733–748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pischedda F & Piccoli G The iglon family member negr1 promotes neuronal arborization acting as soluble factor via fgfr2. Frontiers in molecular neuroscience 8, 89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marg A et al. Neurotractin, a novel neurite outgrowth-promoting ig-like protein that interacts with cepu-1 and lamp. The Journal of cell biology 145, 865–876 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funatsu N et al. Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (kilon), a member of the iglon cell adhesion molecule family. The Journal of biological chemistry 274, 8224–8230 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Sanz R, Ferraro GB & Fournier AE Iglon cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. The Journal of biological chemistry 290, 4330–4342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schäfer M, Bräuer AU, Savaskan NE, Rathjen FG & Brümmendorf T Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Molecular and cellular neurosciences 29, 580–590 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Lee AWS et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PloS one 7, e41537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doan RN et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341–354.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuong JK et al. Ptbp1 and ptbp2 serve both specific and redundant functions in neuronal pre-mrna splicing. Cell reports 17, 2766–2775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boutz PL et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & development 21, 1636–1652 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makeyev EV, Zhang J, Carrasco MA & Maniatis T The microrna mir-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mrna splicing. Molecular cell 27, 435–448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spellman R, Llorian M & Smith CWJ Crossregulation and functional redundancy between the splicing regulator ptb and its paralogs nptb and rod1. Molecular cell 27, 420–434 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng S et al. Psd-95 is post-transcriptionally repressed during early neural development by ptbp1 and ptbp2. Nature neuroscience 15, 381–8, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li QS, Parrado AR, Samtani MN, Narayan VA & Initiative ADN Variations in the fra10ac1 fragile site and 15q21 are associated with cerebrospinal fluid aβ1–42 level. PloS one 10, e0134000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wassenberg JJ & Martin TFJ Role of caps in dense-core vesicle exocytosis. Annals of the New York Academy of Sciences 971, 201–209 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Shinoda Y et al. Caps1 stabilizes the state of readily releasable synaptic vesicles to fusion competence at ca3-ca1 synapses in adult hippocampus. Scientific reports 6, 31540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farina M et al. Caps-1 promotes fusion competence of stationary dense-core vesicles in presynaptic terminals of mammalian neurons. eLife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rietveld CA et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. PNAS 111, 13790–13794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J et al. Ube3a regulates synaptic plasticity and learning and memory by controlling sk2 channel endocytosis. Cell reports 12, 449–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook EH et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. American journal of human genetics 60, 928–934 (1997). [PMC free article] [PubMed] [Google Scholar]
- 51.Lin MT, Luján R, Watanabe M, Adelman JP & Maylie J Sk2 channel plasticity contributes to ltp at schaffer collateral-ca1 synapses. Nature neuroscience 11, 170–177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammond RS et al. Small-conductance ca2+-activated k+ channel type 2 (sk2) modulates hippocampal learning, memory, and synaptic plasticity. The Journal of neuroscience 26, 1844–1853 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murthy SRK et al. Small-conductance ca2+-activated potassium type 2 channels regulate the formation of contextual fear memory. PloS one 10, e0127264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fakira AK, Portugal GS, Carusillo B, Melyan Z & Morón JA Increased small conductance calcium-activated potassium type 2 channel-mediated negative feedback on n-methyl-d-aspartate receptors impairs synaptic plasticity following context-dependent sensitization to morphine. Biological psychiatry 75, 105–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goes FS et al. Genome-wide association study of schizophrenia in ashkenazi jews. American journal of medical genetics. Part B, Neuropsychiatric genetics 168, 649–659 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Sanders S et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mas-Y-Mas S et al. The human mixed lineage leukemia 5 (mll5), a sequentially and structurally divergent set domain-containing protein with no intrinsic catalytic activity. PloS one 11, e0165139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun X-J et al. Genome-wide survey and developmental expression mapping of zebrafish set domain-containing genes. PloS one 3, e1499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ali M et al. Molecular basis for chromatin binding and regulation of mll5. PNAS 110, 11296–11301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemak A et al. Solution nmr structure and histone binding of the phd domain of human mll5. PloS one 8, e77020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X, Novera W, Zhang Y & Deng L-W Mll5 (kmt2e): structure, function, and clinical relevance. Cellular and molecular life sciences 74, 2333–2344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anney R et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 19, 4072–4082 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torrico B et al. Lack of replication of previous autism spectrum disorder gwas hits in european populations. Autism research 10, 202–211 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Feijs KLH, Forst AH, Verheugd P & Lüscher B Macrodomain-containing proteins: regulating new intracellular functions of mono(adp-ribosyl)ation. Nature reviews. Molecular cell biology 14, 443–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng J et al. Ld hub: a centralized database and web interface to perform ld score regression that maximizes the potential of summary level gwas data for snp heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nature Genetics 47, 1061–4036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okbay A et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature genetics 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarke T-K et al. Common polygenic risk for autism spectrum disorder (asd) is associated with cognitive ability in the general population. Mol Psychiatry 21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demontis D et al. Discovery of the first genome-wide significant risk loci for adhd. Nature Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pourcain BS et al. Asd and schizophrenia show distinct developmental profiles in common genetic overlap with population-based social communication difficulties, Molecular Psychiatry 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies G et al. Genome-wide association study of cognitive functions and educational attainment in uk biobank (n=112,151). Molecular psychiatry 21, 758–767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deary V et al. Genetic contributions to self-reported tiredness. Molecular psychiatry 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones SE et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS genetics 12, e1006125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood AR et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Leeuw CA, Mooij JM, Heskes T & Posthuma D Magma: generalized gene-set analysis of gwas data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parikshak NN et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samocha KE et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 46, 944–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pardiñas A et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nature Genetics 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.SPARK Consortium. Spark: A us cohort of 50,000 families to accelerate autism research. Neuron 97, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashburner M et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nature genetics 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Consortium, G. O. Gene ontology consortium: going forward. Nucleic acids research 43, D1049–D1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Lee SH, Goddard ME & Visscher PM Gcta: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson EB et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nature Genetics 48, 552–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weiner D et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders, Nature Genetic 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature Genetics 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shlyueva D, Stampfel G & Stark A Transcriptional enhancers: from properties to genome-wide predictions. Nature Reviews Genetics 15, 272–286 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Won H et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B & Eskin E Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kosmicki JA et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nature genetics 49, 504–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson EB et al. Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. PNAS 111, 15161–15165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reichenberg A et al. Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. PNAS 113, 1098–1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Polderman TJC et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature genetics 47, 702–709 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Sadakata T et al. Autistic-like phenotypes in cadps2-knockout mice and aberrant cadps2 splicing in autistic patients. The Journal of clinical investigation 117, 931–943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parikshak NN et al. Genome-wide changes in lncrna, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references
- 97.Børglum AD et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection identifies new schizophrenia loci. Molecular Psychiatry 19, 325–333 (2014). Published online 29 January 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Illumina gencall data analysis software Tech. Rep, Illumina, Inc. (2005). [Google Scholar]
- 99.Korn JM et al. Integrated genotype calling and association analysis of snps, common copy number polymorphisms and rare cnvs. Nature genetics 40, 1253–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goldstein JI et al. zcall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics (Oxford, England) 28, 2543–2545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lajonchere CM & Consortium, A. Changing the landscape of autism research: the autism genetic resource exchange. Neuron 68, 187–191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Geschwind DH et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. American journal of human genetics 69, 463–466 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gauthier J et al. Autism spectrum disorders associated with x chromosome markers in french-canadian males. Molecular psychiatry 11, 206–213 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Fischbach GD & Lord C The simons simplex collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010). [DOI] [PubMed] [Google Scholar]
- 105.Chaste P et al. A genome-wide association study of autism using the simons simplex collection: Does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry 77, 775–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Delaneau O, Marchini J & Zagury J-F A linear complexity phasing method for thousands of genomes. Nature methods 9, 179–181 (2011). [DOI] [PubMed] [Google Scholar]
- 107.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44, 955–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Howie BN, Donnelly P & Marchini J A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Consortium, . G. P. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price AL et al. Long-range ld can confound genome scans in admixed populations. Am J Hum Genet 83, 132–5; author reply 135–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang CC et al. Second-generation plink: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). Original DateCompleted: 20150227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Willer CJ, Li Y & Abecasis GR Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Begum F, Ghosh D, Tseng GC & Feingold E Comprehensive literature review and statistical considerations for gwas meta-analysis. Nucleic acids research 40, 3777–3784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee SH, Wray NR, Goddard ME & Visscher PM Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang J et al. Common snps explain a large proportion of the heritability for human height. Nat Genet 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Consortium, I. H.. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Consortium, R. E. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McLean CY et al. Great improves functional interpretation of cis-regulatory regions. Nature biotechnology 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kang HJ et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.