Abstract
Background
Human and animal studies have raised concerns that supplemental selenium can increase the risk of metabolic disorders, but underlying mechanisms are unclear.
Objective
We used an integrated transcriptome and metabolome analysis of liver to test for functional pathway and network responses to supplemental selenium in mice.
Methods
Male mice (8-wk-old, C57BL/6J) fed a standard diet (0.41 ppm Se) were given selenium (Na2SeO4, 20 μmol/L) or vehicle (drinking water) for 16 wk. Livers were analyzed for selenium concentration, activity of selenoproteins, reduced glutathione (GSH) redox state, gene expression, and high-resolution metabolomics. Transcriptomic and nontargeted metabolomic data were analyzed with biostatistics, bioinformatics, pathway enrichment analysis, and combined transcriptome–metabolome-wide association study (TMWAS).
Results
Mice supplemented with selenium had greater body mass gain from baseline to 16 wk (55% ± 5%) compared with controls (40% ± 3%) (P < 0.05); however, no difference was observed in liver selenium content, selenoenzyme transcripts, or enzyme activity. Selenium was higher in the heart, kidney, and urine of mice supplemented with selenium. Gene enrichment analysis showed that supplemental selenium altered pathways of lipid and energy metabolism. Integrated transcriptome and metabolome network analysis showed 2 major gene-metabolite clusters, 1 centered on the transcript for the bidirectional glucose transporter 2 (Glut2) and the other centered on the transcripts for carnitine-palmitoyl transferase 2 (Cpt2) and acetyl-CoA acyltransferase (Acaa1). Pathway analysis showed that highly associated metabolites (P < 0.05) were enriched in fatty acid metabolism and bile acid biosynthesis, including acylcarnitines, triglycerides and glycerophospholipids, long-chain acyl–coenzyme As, phosphatidylcholines, and sterols. TMWAS of body weight gain confirmed changes in the same pathways.
Conclusions
Supplemental selenium in mice alters hepatic fatty acid and energy metabolism and causes increases in body mass. A lack of effect on hepatic selenium content suggests that signaling involves an extrahepatic mechanism.
Keywords: high-resolution metabolomics, metal nutrition, nutritional toxicology, pathway enrichment analysis
Introduction
Selenium is an essential nutrient for humans and animals. Twenty-five human selenoproteins have selenocysteine (Sec) at their active sites (1) and function in a wide range of oxidation-reduction reactions, redox signaling, antioxidant defense, thyroid hormone metabolism, and immune responses (1). Well-characterized categories of selenoproteins include glutathione peroxidases (GPxs), thioredoxin reductases (TrxRs), and iodothyronine deiodinases (DIOs), all of which catalyze metabolic redox reactions (1). The physiologic and pathologic effects of selenium deficiency are linked to decreased abundance and activity of selenoproteins (2–4), and lowered GPx activity resulted in decreasing metabolic syndromes associated with type 2 diabetes (T2D) (4). Low selenium concentrations in serum have been associated with poor immune function, cognitive decline, and increased risk of mortality (5).
Knowledge of oxidative mechanisms in disease and antioxidant properties of selenium has provided a basis to test selenium supplementation for the prevention of cancers, T2D, and cardiovascular disease (5). Results showed a U-shaped relation in which both low and high selenium intakes or status have adverse health effects (5). Of concern, participants receiving selenium supplementation (200 μg/d) in the Nutritional Prevention of Cancer trials had an apparent increased risk of T2D (6). This and other human and animal studies (7–10) raise concerns that selenium supplementation by individuals with adequate selenium intake could lead to altered energy regulation and cause liver, cardiovascular, and related organ system dysfunction.
High doses of selenium cause increased body weight and hyperinsulinemia, hyperglycemia, insulin resistance, glucose intolerance, and altered lipid metabolism in mice, rats, and pigs (10–12). Possible mechanisms involve a more reduced state and elevated activity of GPx, with overscavenging of hydrogen peroxide, a second messenger for insulin signaling (1, 11, 13). Research also shows, however, that plasma and tissue selenoprotein concentrations and activities reach a plateau after slightly exceeding adequate intake (AI) (3, 14, 15), and selenium ≤20 times the required intake amount does not change the transcript levels of 22 detected selenoproteins (3). Similarly, a liver transcriptomics study in rats by Raines and Sunde (16) showed that toxic amounts of selenium (5 μg Se/g diet) largely affected liver transcriptome, whereas selenium deficiency or high but nontoxic selenium intake (0.08, 0.24, 0.8, and 2.0 μg Se/g diet) elicited relatively few changes. This study also showed that selenium intake did not have a significant effect on activity of Gpx1 and Gpx4 in liver and plasma Gpx3 (16), which is similar to results of the current study. Together, the present study shows that the mechanisms by which supplemental selenium disrupts energy metabolism would appear to involve mechanisms other than those controlling the selenoproteins.
High-resolution metabolomics (HRM) measures metabolites in most metabolic pathways (17–20). We developed integrated omics methods (21–23), which allow the use of HRM data with other omics data to define central hubs in complex systems (24, 25). In particular, transcriptome–metabolome-wide association study (TMWAS) (21) provides an approach to address complexity in nutrition and health, because this can show how specific nutrients interact with genes, proteins, and metabolites (26). Both transcriptomics and HRM are global in nature, with transcriptomics measuring essentially every transcript and HRM measuring metabolites from most metabolic pathways (18). In principle, comparison of nutrient intakes near the Tolerable Upper Intake Level (UL) with those at AI or RDA levels could reveal central organizational structure of biological responses to selenium over an otherwise adequate range of intake (18, 21).
In the present research, we used TMWAS of mouse liver to examine biological responses to supplemental selenium. Mice were fed standard mouse diet with water or water supplemented with 20 µmol sodium selenate/L. The estimated selenium intake in the supplemented mice was ∼4-fold that found in standard mouse diet, creating a range similar to the difference between the human adult UL for selenium (400 μg; 7-fold of the RDA) and the RDA, thereby mimicking selenium supplementation in individuals with AI. We specifically studied the liver because of its central role in energy metabolism.
Methods
Animals
Animal protocols were approved by the Institutional Animal Care and Use Committee at Emory University. Eight-week-old male C57BL/6J mice were purchased from Jackson Laboratory and housed under conventional conditions of 21–24°C and a 12-h light-dark cycle with commercial standard mouse diet (Laboratory Rodent Diet 5001; LabDiet) containing 0.41 ppm Se (27), with water provided ad libitum. Twenty-five mice were randomly assigned to vehicle control [purified water by reverse osmosis system, no detectable range of selenium as measured by inductively coupled plasma MS (<0.00025 ppm Se) with 0 μM Na2SeO4; n = 15] and selenium treatment [1.6 ppm Se (20 μM Na2SeO4) in purified water; n = 10] groups and maintained for 16 wk; subsets were used for sample collections for different assays as described below. The 5 extra mice in the control group were used for body weight assessment to quantify metals in different tissues to establish reference levels in untreated normal mice. For all other examinations, we used equal numbers of mice. For assays not conducted on the day of tissue collection, samples were immediately frozen and maintained at –80 °C until assay. Mice were group-housed and individual food and water consumption was not determined. Urine samples were collected from the bladder at the time of tissue collection.
Quantification of selenium
Selenium was measured in mouse organs, including liver, heart, kidney, and lung, and in urine with the use of inductively coupled plasma MS (iCap Q; ThermoFisher Scientific) following procedures to measure trace metals that conformed to accuracy (100% ± 10%) and precision standards (Relative standard deviation <12%). Briefly, 50-mg tissues were homogenized in 500 μL purified water and digested in 2% nitric acid to a final volume of 10 mL. Selenium standard (1000 mg/L, 2% HNO3) was purchased from Ricca Chemical (Arlington, Texas) and diluted in series for standard curve. For urine analysis, samples were injected along with internal standards, nitric acid, and hydrogen peroxide without digestion. Selenium concentration in urine was normalized to creatinine level (Creatinine Urinary Colorimetric Assay; Cayman Chemical). Protein concentration was measured in 5 μL tissue homogenates with the use of a Bio-Rad DC kit.
GPx and TrxR activity
Selenium-dependent GPx activity was measured by the decrease in NAD(P)H absorbance (340 nm) in liver tissue homogenates (n = 10) with a recycling assay and hydrogen peroxide as substrate (28). NAD(P)H-dependent TrxR activity was assayed in liver homogenates by using the catalytic reduction of DTNB [5,5-dithio-bis-(2-nitrobenzoic acid)] to 5-thio-2-nitrobenzoate measured at 412 nm (29).
Transcriptomics
Total RNA was isolated from mouse liver (n = 5/group) with the mirVana RNA isolation kit (Life Technologies) and stabilized in RNAlater (Qiagen). The preserved RNA was hybridized on Affymetrix Mouse Gene ST 2.0 exon chips after NuGEN Ovation RNA Amplification. Robust multiarray average (RMA) was used to pre-process and summarize the chip data as an Affymetrix Expression set, resulting in normalized expression levels of 33,793 transcripts. Gene Set Enrichment Analysis (GSEA) was used to determine enriched gene sets and genes that differed between the biological states (30). Gene set enrichment statistics were obtained with the GSEA applet (v. 2.2; Broad Institute) with a false discovery rate (FDR)–adjusted q < 0.05 for significance. Genes that contributed to the enrichment of significant pathways were used for TMWAS as described below.
HRM
Ultra-high-resolution MS with LC was used for metabolic profiling (24) of liver extracts from control (n = 8) and selenium-treated (n = 8) mice as described previously (24). Samples were extracted at 0°C by homogenization in acetonitrile:water (2:1) containing a mixture of stable isotope–labeled internal standards (31). The acetonitrile:water mixture was added to a weighed tissue sample (∼20 mg tissue) at a ratio of 15 μL/mg tissue. After homogenization and incubation on ice for 30 min, extracts were centrifuged to remove protein (24), randomized, and 10-μL aliquots were analyzed with 3 technical replicates with the use of C18 LC and MS (50–2000 m/z) with positive electrospray ionization on an LTQ-Velos Orbitrap mass spectrometer (Thermo Fisher) (17, 32). MS data were extracted by using apLCMS (33) and xMSanalyzer (34). Data were prefiltered to retain only features with nonzero values in >80% in all samples, retaining 12,461 features.
TMWAS
Transcriptome and metabolome data from the same set of samples (n = 5/group) were integrated by using xMWAS (22). We used partial least-squares regression (35–37), a variable selection and dimensionality reduction method, to conduct pairwise correlation analysis between transcriptome (5 samples × 80 significant transcripts selected by GSEA as described above) and metabolome (5 samples × 12,421 metabolic features selected by relative SD). Selected metabolic features were mapped to metabolic pathways by using mummichog (38) and annotated with xMSannotator (19) with the use of HMDB (Human Metabolome Database (http://www.hmdb.ca/)) (39). These approaches provide level 5 identification by the criteria of Schymanski et al. (40); confidence scores for annotation by xMSannotator are derived from a multistage clustering algorithm. Confirmed identities were obtained by co-elution and ion dissociation MS (MS/MS) relative to authentic standards [level 1 identification by criteria of Schymanski et al. (40)] or by MS/MS relative to the METLIN (https://metlin.scripps.edu) spectral database (41) [level 2 identification by criteria of Schymanski et al. (40)]. Metabolic features correlated with body mass increase (grams per week) were selected by using linear correlation analysis at P < 0.05 (32) followed by module and pathway enrichment analyses with the use of mummichog (38). This approach protects against type 2 statistical error by including all features at P < 0.05 and protects against type 1 statistical error by permutation testing in pathway enrichment analysis (42).
Quantification of acetyl-CoA
Concentrations of acetyl-CoA in liver samples (n = 5/group) were assayed with a fluorometric assay kit (Sigma-Aldrich) following procedures provided by manufacturer.
Statistical analysis
Data are presented as means ± SEMs. Student's t test (2-tailed with Welch's correction for unequal variance) was used to test statistical significance. For weight-gain comparison, a linear mixed model with repeated measures was used (IBM SPSS Statistics 24). The significance level was P < 0.05 for all tests; the Benjamini and Hochberg FDR was used for multiple comparisons (43). Pearson's correlation coefficient and t test were used to assess the significance of correlations between genes and metabolites.
Results
Selenium supplementation caused body mass increase with no apparent change in liver selenium content
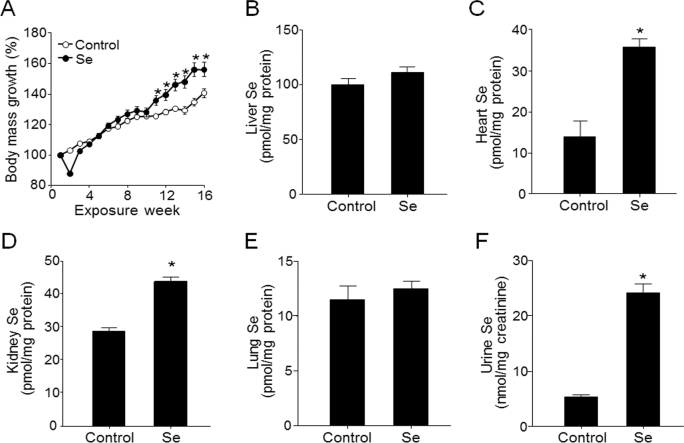
Selenium-supplemented mice had a 55% ± 5% increase in body mass at 16 wk compared with a 40% ± 3% increase in controls, with the difference becoming apparent after 10 wk (P < 0.05; Figure 1A). Selenium content in liver measured at the final time point of the experiment did not differ between selenium and control groups (Figure 1B) and was higher (control: 102 ±5 pmol/mg protein; selenium: 113 ± 5 pmol/mg protein) than in other organs [10–40 pmol/mg protein; heart (Figure 1C), kidney (Figure 1D), lung (Figure 1E)]. Selenium values in liver [9.7–11.8 nmol (0.8–0.9 μg)/g tissue] were similar to those (10 nmol/g wet tissue, 0.2–2 μg/g tissue) shown in the previous studies in mice and humans (3, 44, 45). Selenium content did not differ between groups for lung (Figure 1E) but was higher in the selenium group for heart (Figure 1C), kidney (Figure 1D), and urine (Figure 1F). Thus, the selenium content and storage in the liver appeared to have achieved plateau levels with the selenium provided by the mouse diet (3), even though supplemental selenium caused increased urinary selenium and an increase in some nonhepatic tissues.
FIGURE 1.
Relative body mass (A) and liver (B), heart (C), kidney (D), lung (E), and urine (F) selenium concentrations in C57BL/6 male mice that received water with or without supplemental selenium for 16 wk. Values are means ± SEMs, n = 15 (control) or 10 (Se). Selenium's effect on body mass was tested by linear mixed model (P < 0.05). *Significant body mass differences between Se and control mice by using Student's t test at specific time points, P < 0.05.
Supplemental selenium did not alter liver selenoprotein gene expression or activity of GPx or TrxR
Consistent with previous research on selenium supplementation (3, 15) and the lack of effect on hepatic selenium as shown above, transcript abundances for the 21 selenoproteins in the Affymetrix Mouse Gene ST 2.0 exon chips did not differ between selenium and control groups (Supplemental Table 1). Similarly, the enzymatic activity of 2 major selenoproteins, GPx and TrxR, which are used as molecular biomarkers for selenium status, showed no differences in activity in liver between the selenium and control groups (Supplemental Figure 1). Thus, we conclude that the dietary selenium was adequate to support known selenium requirements and that the selenium provided in drinking water represented a supplemental level above the required intake of selenium.
Expression levels of 80 genes for cholesterol homeostasis, pancreatic β cell signaling, and FA metabolism were elevated by supplemental selenium
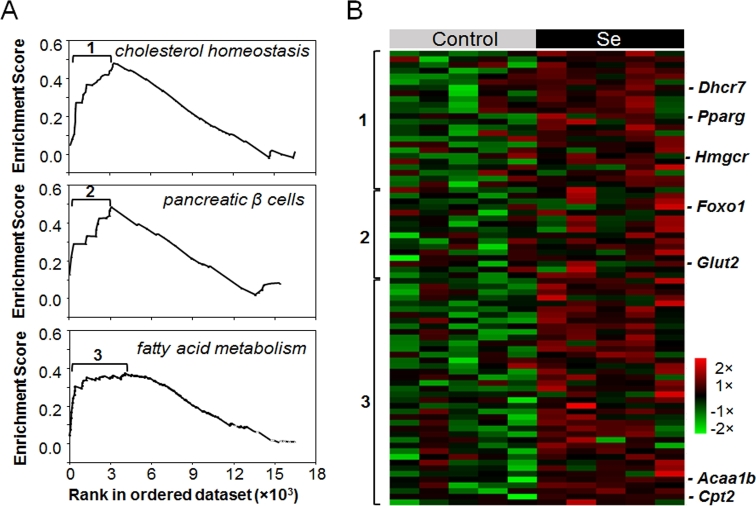
To determine whether supplemental selenium affected hepatic transcription, we performed GSEA on liver transcriptome data. The results showed that 3 pathways in the liver transcriptome of selenium-treated mice were enriched compared with controls. These included genes for cholesterol homeostasis, pancreatic β cell signaling, and FA metabolism (Figure 2A; FDR: q < 0.05) (Table 1), consistent with previous research showing that selenium regulates homeostasis of glucose, lipids, and amino acids (17, 46, 47). RMA-normalized expression levels of the 80 genes that contributed to the gene set enrichment split into 3 clusters corresponding to 3 pathways (Figure 2B, Supplemental Table 2). Twenty-four genes were in a cluster associated with cholesterol homeostasis (cluster 1; Figure 2A, top) and included genes for 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) and 11 other cholesterol biosynthesis enzymes, as well as regulators of cholesterol metabolism such as peroxisome proliferator-activated receptor (Pparg). Cluster 2 included 15 genes in pancreatic β cell signaling (Figure 2A, middle), including glucose transporter 2 (Glut2) and mediators for glucose metabolism. Cluster 3 consisted of 41 genes for proteins in FA metabolism (Figure 2A, bottom), including acetyl-CoA acyltransferase (Acaa1) and carnitine palmitoyltransferase (Cpt) 2 (Cpt2) (Figure 2B). The RMA expression levels in the heat map showed that supplemental selenium tended to increase (P < 0.10) 31 transcripts for genes in pathways regulating glucose and FA homeostasis (Figure 2B; P values shown in Supplemental Table 2).
FIGURE 2.
GSEA of supplemental selenium effect on mouse hepatic transcriptome. Enrichment plots of 3 significant pathways (FDR q < 0.05) from GSEA of supplemental selenium effect on mouse liver transcriptome (n = 5) (A) and heat map showing RMA normalized expression levels of genes that contributed to the enrichment (B) (marked as 1, 2 and 3 in panel A). Acaa1b, Acetyl-CoA acyltransferase B (also called peroxisomal 3-oxoacyl-CoA thiolase B); Cpt2, carnitine palmitoyltransferase 2; Dhcr7, 7-dehydrocholesterol reductase; FDR, false discovery rate; Foxo1, forkhead box protein class O; Glut2, glucose transporter 2; GSEA, Gene Set Enrichment Analysis; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; RMA, robust multiarray average.
TABLE 1.
Gene set enrichment results for the liver transcriptome of selenium-treated and control mice1
| Upregulated in group and pathway name | Gene set size | Genes in enrichment, n | Different genes, n (P < 0.05) | Enrichment score | P | FDR q value2 |
|---|---|---|---|---|---|---|
| Selenium | ||||||
| Cholesterol homeostasis | 72 | 25 | 8 | 1.8 | <0.001 | 0.006 |
| Pancreas β cell signaling | 37 | 14 | 0 | 1.62 | 0.013 | 0.027 |
| Fatty acid metabolism | 147 | 41 | 2 | 1.57 | 0.002 | 0.032 |
| IFN-α response | 89 | 25 | 13 | 1.4 | 0.028 | 0.149 |
| Epithelial mesenchymal transition | 191 | 54 | 5 | 1.28 | 0.03 | 0.335 |
| Control | ||||||
| Mitotic spindle | 194 | 69 | 13 | −1.47 | 0.002 | 0.1 |
| UV response | 143 | 34 | 10 | −1.47 | 0.005 | 0.142 |
| Myc targets | 52 | 35 | 2 | −1.57 | 0.014 | 0.123 |
| Unfolded protein response | 109 | 38 | 7 | −1.39 | 0.025 | 0.157 |
| Androgen response | 91 | 48 | 10 | −1.38 | 0.033 | 0.132 |
| IL6/JAK/STAT3 signaling | 87 | 26 | 5 | −1.34 | 0.041 | 0.157 |
1Top pathways (ranked by P values) for each group and number of genes that contribute to core enrichment as well as genes that differ between the groups are shown (P < 0.05). The Hallmark gene set database was used for the analysis. FDR, false discovery rate; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3.
2FDR q < 0.05 pathways were considered significant.
Integration of transcriptome and metabolome data identified 2 major hubs
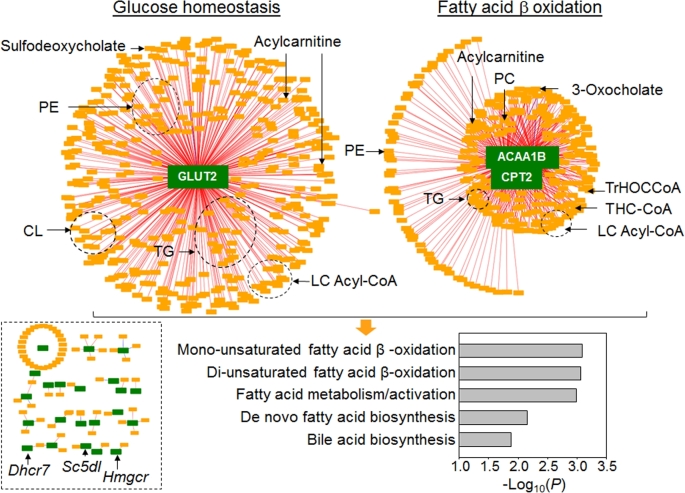
To examine metabolic responses associated with selenium-dependent transcriptional changes, we used xMWAS (22) to test for correlations of the 12,421 metabolic features detected by HRM with the 80 genes from the gene sets that were associated with supplemental selenium (Supplemental Table 2). The community detection tool within this approach provided an association network visualizing the top interactions among transcripts and metabolites (Figure 3). With lower stringency criteria (e.g., |ρ| ≥ 0.5), the network structures were complex (data not shown), but by increasing the correlation threshold to |ρ| ≥ 0.9, top interaction network substructures representing central hubs of strongly associated transcripts and metabolites were visualized (Figure 3). Two major hubs were apparent, each with hundreds of metabolites correlating with 1 or 2 functional transcripts. Hub 1 was centered around Glut2, a bidirectional plasma membrane glucose transporter that is essential for glucose uptake into hepatocytes and that mediates glucose release during gluconeogenesis (47). In this hub, 421 metabolites were correlated with Glut2, with 420 having a positive association and only 1 having a negative association. Hub 2 was centered on genes for 2 enzymes, Cpt2 (long-chain FA transporter to mitochondrial inner membrane for β-oxidation) (48) and Acaa1b (thiolase in peroxisomal β-oxidation of bile acid intermediates) (49) (Figure 3). In this hub, 287 of 288 metabolites were positively correlated with Acaa1b and Cpt2, and only 1 metabolite was negatively correlated. These 2 major hubs reflected 2 significant clusters of genes in the GSEA (cluster 2: pancreatic β cell signaling; cluster 3: FA metabolism; Figure 2B). Cluster 1 for cholesterol homeostasis in the GSEA was not represented in this integrative network analysis because tight gene-metabolite hubs were not present. For instance, the cluster 1 genes, 7-dehydrocholesterol reductase (Dhcr7), sterol-C5-desaturase (Sc5dl), Hmgcr, etc., had very few metabolite associations and thus were present with scattered connections (Figure 3, bottom left). Overall, these network substructures depict peroxisomal and mitochondrial FA oxidation, and glucose homeostasis as critical processes altered by supplemental selenium.
FIGURE 3.
Metabolome-wide association study of transcripts in 3 gene sets of mouse liver (from Figure 2) altered by supplemental selenium. Associations between 80 significant genes and the metabolome (12,421 features) from mouse liver (n = 5) are visualized at |ρ| ≥ 0.9, showing 2 central gene hubs and scattered connections (bottom left) and pathway associations of metabolites (bottom right). ACAA1B, peroxisomal 3-oxoacyl-CoA thiolase B; CL, cardiolipin; CPT2, carnitine palmitoyltransferase 2; Dhcr7, 7-dehydrocholesterol reductase; GLUT2, glucose transporter 2; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; LC, long-chain; PC, phosphatidylcholine; PE, phosphatidylethanolamine; Sc5dl, sterol-C5-desaturase; THC-CoA, 3α,7α,12α-trihydroxy-5β-cholest-24-enoyl-CoA; TrHOCCoA, 3α,7α,12α -trihydroxy-5β-24-oxocholestanoyl-CoA.
Annotation of metabolites in the 2 major transcriptome-metabolome hubs
Metabolic pathway enrichment analysis was performed on the 709 (421 + 288) metabolic features associated with the 2 major transcriptome-metabolome hubs (Glut2 and Acaa1b/Cpt2) (38). The results showed enrichment in 5 pathways, including mono- and di-unsaturated FA β-oxidation, FA metabolism/activation, de novo FA biosynthesis, and bile acid biosynthesis (Figure 3, bottom right). These results suggest that altered FA metabolism is part of the central organizational structure of biological responses to supplemental selenium (Supplemental Figure 2).
Metabolic features in the top 2 transcriptome-metabolome hubs were annotated with the use of xMSannotator (19). Seventy-six metabolites in the Glut2 hub were annotated as follows: 57 are different lipid species including TGs, phosphatidylethanolamines (PEs), and cardiolipins (CLs), 2 sterols, and 17 are lipid degradation intermediates and other lipid metabolites such as long-chain acyl-CoAs and acylcarnitines (Supplemental Table 3, upper list). In addition, 36 metabolic features in the Cpt2/Acaa1 hub were annotated, and they are 24 lipid species, including TGs, PEs, and CLs and 12 lipid degradation intermediates and products (Supplemental Table 3, lower list). All of the lipid species in the 2 hubs were positively associated with central genes (Glut2, Cpt2, and Acaa1b) and their abundances were increased in selenium-treated mice.
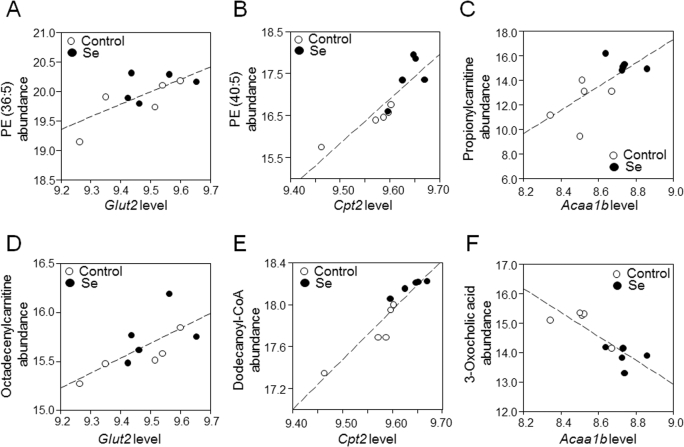
Correlations of central genes with selected metabolites on the basis of high confidence in their annotation by xMSannotator (19) (confidence levels of ≥2) and representative of related functional pathway are shown in Figure 4. These included lipid species, such as PE, propionylcarnitine, and octadecenylcarnitine, each positively correlated with hub genes (Acaa1b and Glut2; Figure 4A–D). We extended analysis of other correlated metabolites in the categories of acylcarnitine, bile acids, and long-chain FAs with |ρ| < 0.9 and found that, consistent with the propionylcarnitine and octadecenylcarnitine, 6 of all 22 metabolic features annotated as acylcarnitines showed a trend of increased abundance (P < 0.1) in selenium-treated mice (Supplemental Table 4). Dodecanoyl-CoA was positively correlated with Cpt2 (Figure 4E; P < 0.05), and other long-chain acyl-CoA intermediates (Supplemental Table 4) were also higher in selenium-supplemented mice. By contrast, the abundance of bile acids (represented by 3-oxocholic acid) were negatively correlated with the transcript for Acaa1b (Figure 4F), and decreased levels of metabolic features annotated as primary and secondary bile acids were detected across the whole metabolome (Supplemental Table 4).
FIGURE 4.
Correlations of selected metabolites with central genes (Glut2, Cpt2, and Acaa1b) in the major transcriptome-metabolome hubs (from Figure 3). (A) PE (36:5): m/z 738.5082, retention time 579 s (ρ = 0.72, P = 0.02); (B) PE (40:5): m/z 794.5710, retention time 582 s, confirmed by MS/MS (ρ = 0.88, P < 0.01); (C) propionylcarnitine: m/z 218.1274, retention time 45 s, confirmed by MS/MS (ρ = 0.71, P = 0.02); (D) octadecenylcarnitine: m/z 426.3561, time 563 s, confirmed by MS/MS (ρ = 0.67, P = 0.03); (E) dodecanoyl-CoA: m/z: 950.2873, retention time 38 s (ρ = 0.94, P < 0.01); (F) 3-oxocholic acid, m/z 407.2790, retention time 436 s (ρ = -0.84, P < 0.01). Values for the transcripts represent RMA average expression levels, and values for the metabolites represent log2 transformed, quantile-normalized mass spectral intensities; fold-changes for nontransformed values are provided in Supplemental Table 4. Acaa1b, peroxisomal 3-oxoacyl-CoA thiolase B; Cpt2, carnitine palmitoyltransferase 2; Glut2, glucose transporter 2; PE, phosphatidylethanolamine; RMA, robust multiarray average.
The changes in metabolites in these 2 major hubs suggested that supplemental selenium intake altered mitochondrial FA oxidation and peroxisomal lipid degradation, causing accumulation of acylcarnitines and reduced biosynthesis of bile acids, respectively. Acylcarnitines are important mitochondrial fuels, and to more specifically test for effect on mitochondrial bioenergetics, we quantified acetyl-CoA, an end product of β-oxidation that is used for ATP synthesis. Results showed that selenium supplementation substantially decreased acetyl-CoA concentration in the liver by >50% (control: 13.2 ± 2.2 nmol/g; selenium: 6.5 ± 0.8 nmol/g tissue; n = 10; P < 0.05). Consistent with this, the citric acid cycle intermediate, succinate, and the related amino acid, glutamine, which were measured by HRM, were decreased in selenium-treated mice (Supplemental Table 3), further supporting the evidence for altered mitochondrial energy metabolism.
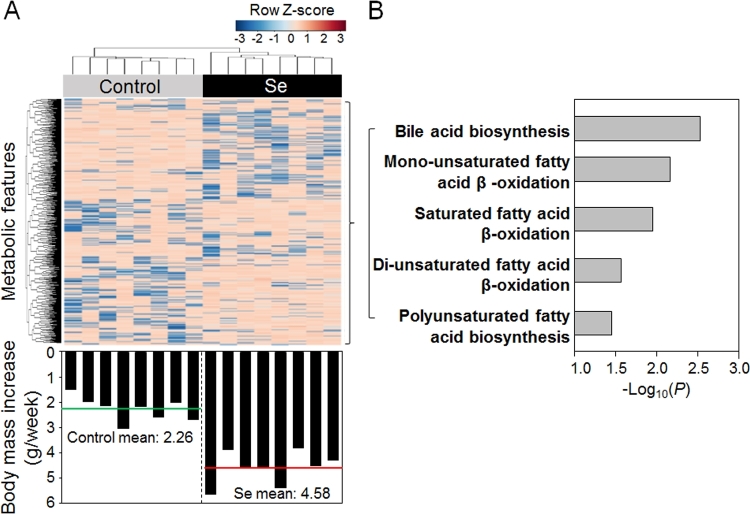
Metabolic pathways for lipid, cholesterol, and sugar metabolism were associated with excess body weight gain induced by selenium
The analyses described above show that supplemental selenium is associated with weight gain and also with changes in transcription that were associated with changes in mitochondrial and peroxisomal lipid metabolism. To examine associations between metabolic changes and weight gain, we performed a metabolome-wide association study of the nontargeted HRM data from liver (32) with body mass increase (grams per week) of individual mice over 16 wk of the experimental period without regard to selenium supplementation status (Supplemental Table 5). An unsupervised hierarchical cluster analysis of the 239 mass spectral features that correlated (raw P < 0.05) with the body mass increase showed clustering of selenium-treated mice from controls (Figure 5A). Pathway enrichment analysis with mummichog showed that the enriched pathways were similar to those from the TMWAS described above (Figure 3, bottom). Specifically, 3 of the pathways were the same, including bile acid biosynthesis and mono- and di-unsaturated FA β-oxidation (Figure 5B compared with Figure 3, bottom). Other pathways associated with body mass increase, SFA β-oxidation and PUFA biosynthesis (Figure 5B), were similar to FA metabolism and activation and de novo FA biosynthesis pathways found in the TMWAS analysis (Figure 3, bottom). Twenty-three of the 239 metabolites were annotated by xMSannotator including long- and medium-chain fatty acyl-CoA (substrates and intermediates of mitochondrial FA β-oxidation), bile acid intermediates (side chain of cholesterol, peroxisomal β-oxidation intermediates), and related metabolites such as carnitine and FAD (Supplemental Table 6). Thus, the analyses directly link changes in lipid metabolism to weight gain, reinforcing the data showing that supplemental selenium affects transcription and metabolism of FA and energy metabolism and this is directly related to body mass increase.
FIGURE 5.
Metabolome-wide association study of body mass increase. (A) Two-way unsupervised hierarchical clustering analysis of 239 metabolic features correlated with body mass increase (Supplemental Table 5) in individual mice showing 2 clustering groups according to selenium supplementation. The corresponding body mass increase for each mouse is provided on the bottom of the panel. (B) Mummichog pathway enrichment analysis of 239 metabolic features.
Discussion
Selenium supplementation has divergent effects on populations with low and high selenium status (5), and consideration of overall nutritional status, health status, and genetic variations is necessary to balance benefits and risks of selenium supplementation (50). The RDA for selenium is 55 µg/d, based in part on the amount needed to maximize plasma GPx activity (14). The UL is based on chronic selenosis and set at only 7.3-fold higher (400 µg/d) (14). Although supplementation by individuals with intakes near the UL is not advisable, the usefulness of supplementation by other individuals within the AI range is less clear. To examine the effect of supplementation under conditions of AI without exceeding UL in mice, we compared a control group [standard diet (0.41 ppm Se) and water ad libitum (<0.25 ppb)] and a selenium-supplement group fed the same diet (0.41 ppm Se) and water containing 1.6 ppm Se (20 µM Na2SeO4). Estimated daily selenium intake from the diet was 0.07 mg/kg for each group, on the basis of the average daily diet consumption (4 g) by C57BL/6J mice (25 g body weight) (51). Estimated intake from drinking water was 0.00 mg/kg for the control group and 0.25 mg/kg body weight for the selenium-supplemented groups, on the basis of the average daily water consumption (4 mL) by C57BL/6J mice (25 g body weight) (51). Combined, food and water provided ∼0.07 mg/kg for the control group, which meets the minimum intake value for mice (52). The combined food plus water intake was ∼0.32 mg ⋅ kg body weight−1 ⋅ d−1 for the selenium group, which is under the highest reported mouse no-observed-adverse-effect level for mice given the more toxic selenite (53), and represents a relatively high selenium supplement content in animal nutritional research (3, 11). With recognition that mice are imperfect models for human nutrition, the conditions used provide a mammalian model comparing 2 oral intakes within a low-adequate to high-adequate intake range.
Our research confirms previous studies that in selenium-adequate mice, supplemental selenium does not increase the activities of 2 major selenoenzymes (GPx and TrxR) or change the transcript abundance of any selenoprotein measured (Supplemental Figure 1, Supplemental Table 1). There was no apparent phenotypic sign of obesity in the selenium-supplemented group, and the present research did not examine adipose tissue to support fat accumulation metabolism. Despite this, supplemental selenium caused increased body mass, consistent with evidence for potential health risks from high selenium status due to altered energy metabolism and disruption in lipid biosynthesis and metabolism (44, 54). Previous studies showed that plasma selenium concentrations were significantly associated with nonalcoholic fatty liver disease and increased visceral fat in patients with T2D (4, 54). High selenium intake in humans was associated with decreased serum triiodothyronine (T3) and increased thyrotropin, suggesting a subclinical hypothyroid response that may contribute to increased body weight (55). In a well-characterized pig study, a high-selenium diet induced metabolic alterations (fasting insulin and cholesterol concentrations) that did not reach a significant level compared with a selenium-adequate diet; the pattern suggested, however, a potential predisposition to T2D (56). Molecular changes, including upregulation of forkhead box protein class O (Foxo1), the key controller of energy metabolism, were found in the skeletal muscle but not in the liver (56).
Although we did not examine selenium amounts in all organs in the present study, the results showed that supplemental selenium had different effects on selenium content in organs of mice, causing no change in selenium content in the liver or lung and a significant increase in the kidney and heart and an increase in urine (Figure 1). The liver plays a critical role in selenium metabolism and homeostasis of selenium by receiving selenium directly from small intestine, removing selenium from the dietary form (selenomethionine), and producing excretory metabolites of selenium (57, 58). Given this central role of the liver in selenium disposition, no detectable increase in liver selenium in the selenium-supplemented group could be the result of an elevated release of selenium in a form that could be taken up by other organs (e.g., heart and kidney) or by production of excretory metabolites in the supplemental selenium group. In addition to hepatic selenium, plasma selenium is also important in interorgan selenium balance. Two major selenoproteins contribute to plasma selenium, including selenoprotein-P and GPx-3. We did not measure plasma selenium concentrations in the present study, so possible effects would be speculative. Liver GPx was not affected by selenium supplementation, suggesting that there could be no effect. On the other hand, selenoprotein-P is a major selenium transport form and is mostly produced by the liver (59); consequently, liver production of selenoprotein-P could be increased even without a detectable increase in hepatic selenium content. To improve our understanding of selenium nutritional biology, future studies on metabolic responses in plasma to the varied selenium concentrations regulated by selenoprotein-P or GPx-3 are warranted. In the pig study (56), biosynthesis of selenoproteins was saturated by adequate selenium so that the effects of high selenium intake in that study may have derived from mechanisms other than the organ-specific activities of selenoenzymes.
To explore the complex effects of selenium in the liver, we used newly developed integrative omics methods providing unprecedented, large-scale views of the molecular interactions (60). The results showed that, even though supplemental selenium had no effect on liver selenium content, 31 transcripts of lipid and glucose metabolism genes tended to increase along with concordant changes in the lipid metabolome. We interpret these alterations to be molecular signatures of bioenergetic changes associated with the phenotypic increase in body mass in selenium-supplemented mice. Visualization of transcriptome and metabolome interaction networks with community detection (Figure 3) connected a central hepatic glucose transporter, mitochondrial FA β-oxidation, and peroxisomal lipid degradation as central hubs responding to supplemental selenium. The associated accumulation of FA metabolites (e.g., acylcarnitines, long-chain acyl-CoAs) along with decreased metabolites of bile acid biosynthesis suggest disrupted lipid metabolism. Decreased acetyl-CoA and TCA cycle intermediates with accumulation of FA intermediates (Supplemental Table 4) suggests a metabolic imbalance found in many disease models, in particular in skeletal muscle insulin resistance (61–63). Increases in genes for FA synthase (Fasn), acyl-CoA synthetase (Acss2, Acss3), and type I acyl-CoA thioesterase (Acot1 and Acot2) with increased fatty acyl-CoA suggests that selenium supplementation may elevate lipid synthesis. On the other hand, decreased Cpt1a could also lead to a build-up of long-chain acylcarnitine. Thus, the evidence suggests that supplemental selenium contributes to increased FA synthesis as well as decreased capability for mitochondrial β-oxidation.
Consistent with this, we found that selenium supplementation affected major lipid metabolism systems that may be critical for glucose homeostasis. The selenium-impaired peroxisomal oxidation and biosynthesis of bile acids may lead to disruption of normal physiologic signaling and glucose homeostasis, because bile acids regulate glucose metabolism and increase insulin sensitivity through farnesoid X receptor or TGR5 (a G protein–coupled receptor) (64–67). On the other hand, amino acid–derived acylcarnitines (e.g., propionylcarnitine and malonylcarnitine) and medium- and long-chain acylcarnitines from incomplete oxidation (Figure 3, Supplemental Table 4) can activate proinflammatory signaling, causing insulin resistance (68, 69), and thus strongly associate with diabetes (70).
Collectively, our data show that supplemental selenium in this mouse model induced metabolic alterations serving as a nexus for lipid metabolism and glucose homeostasis (Supplemental Figure 2), which may be central to selenium's effects on human disorders, including T2D, obesity, and hyperlipidemia. However, the correlation analyses in the current study do not provide data to determine the sequence of molecular responses to supplemental selenium. Accordingly, either response could occur by supplemental selenium: 1) selenium → metabolic disruption → transcriptional and signaling disruption or 2) selenium → transcriptional and signaling disruption → metabolic disruption (Supplemental Figure 2). Future studies with cell experiments are needed to improve our understanding of cellular and molecular responses to supplemental selenium but may be complicated by possible extrahepatic effects. Given the lack of increase in selenium in the liver, the unexplained reduction in hepatic GSH redox state, and the complexity of metabolic and transcriptional responses, mechanistic interpretations of hepatic effects are largely speculative. These include possible modification of protein thiols by low-molecular-weight selenium compounds or metabolites (71), modulation of redox-regulated transcription factors that are critical for energy metabolism such as PPAR-γ (72), and effects of reactive selenium intermediates on thioredoxin systems (73). In addition, chemical reactivity with thiols varies significantly among inorganic and organic forms of dietary selenium (71, 73, 74), as well as the incorporation of selenium into proteins (e.g., by selenomethionine) (75). We are also aware that urinary selenium excretion differs by sex (76), and our data are limited to males only. These factors may affect protein function even without changes in total hepatic selenium.
In summary, the present study of selenium supplementation in mice shows changes in hepatic gene expression and associated metabolites in FA metabolism and sterol and glucose homeostasis. Integrated analysis identified 2 central transcript-metabolite hubs at the network level, showing that increased expression of genes of glucose transport and FA β-oxidation were associated with the accumulation of acylcarnitines and other lipid metabolites and decreased bile acid metabolites. These characteristics suggest that selenium supplementation alters FA oxidation and creates an imbalance with downstream energy metabolism dependent on acetyl-CoA. Such a metabolic disruption could contribute to an increased body mass observed with supplemental selenium. Importantly, the hepatic effects occur without detectable changes in liver selenium, indicating that responses are part of interorgan systems that control energy balance.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—Y-MG and DPJ: designed the research and had primary responsibility for the final content; XH, JDC, MLO, LH, and KL: performed experiments and data analysis; KU: provided support on statistical and bioinformatics analysis; and all authors: read and approved the final manuscript.
Notes
Y-MG and DPJ contributed equally to this work as corresponding and senior authors.
Supported by National Institute of Environmental Health Sciences grants R01 ES023485 (DPJ and YMG), R21 ES025632 (DPJ and YMG), and P30 ES019776 (DPJ); NIH grant S10 OD018006 (DPJ); and National Heart, Lung, and Blood Institute grant F32 HL132493 (JDC).
Author disclosures: XH, JDC, MLO, LH, KL, KU, Y-MG, and DPJ, no conflicts of interest.
Supplemental Tables 1–6 and Supplemental Figures 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: Acaa, acetyl-CoA acyltransferase; AI, Adequate Intake; Cpt, carnitine palmitoyltransferase; FDR, false discovery rate; Glut2, glucose transporter 2; GPx, glutathione peroxidase; GSEA, Gene Set Enrichment Analysis; HRM, high-resolution metabolomics; RMA, robust multiarray average; T2D, type 2 diabetes; TMWAS, transcriptome–metabolome-wide association study; TrxR, thioredoxin reductase; UL, Tolerable Upper Intake Level.
References
- 1. Lu J, Holmgren A. Selenoproteins. J Biol Chem 2009;284:723–7. [DOI] [PubMed] [Google Scholar]
- 2. Brigelius-Flohé R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med 1999;27:951–65. [DOI] [PubMed] [Google Scholar]
- 3. Sunde RA, Raines AM. Selenium regulation of the selenoprotein and nonselenoprotein transcriptomes in rodents. Adv Nutr 2011;2:138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan X, Pepper MP, Vatamaniuk MZ, Roneker CA, Li L, Lei XG. Dietary selenium deficiency partially rescues type 2 diabetes–like phenotypes of glutathione peroxidase-1–overexpressing male mice. J Nutr 2012;142:1975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rayman MP. Selenium and human health. Lancet 2012;379:1256–68. [DOI] [PubMed] [Google Scholar]
- 6. Stranges S, Marshall JR, Natarajan R, Donahue RP, Trevisan M, Combs GF, Cappuccio FP, Ceriello A, Reid ME. Effects of long-term selenium supplementation on the incidence of type 2 diabetes: A randomized trial. Ann Intern Med 2007;147:217–23. [DOI] [PubMed] [Google Scholar]
- 7. Jablonska E, Reszka E, Gromadzinska J, Wieczorek E, Krol MB, Raimondi S, Socha K, Borawska MH, Wasowicz W. The effect of selenium supplementation on glucose homeostasis and the expression of genes related to glucose metabolism. Nutrients 2016;8(12):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahmoodianfard S, Vafa M, Golgiri F, Khoshniat M, Gohari M, Solati Z, Djalali M. Effects of zinc and selenium supplementation on thyroid function in overweight and obese hypothyroid female patients: a randomized double-blind controlled trial. J Am Coll Nutr 2015;34:391–9. [DOI] [PubMed] [Google Scholar]
- 9. Calamari L, Abeni F, Bertin G. Metabolic and hematological profiles in mature horses supplemented with different selenium sources and doses. J Anim Sci 2010;88:650–9. [DOI] [PubMed] [Google Scholar]
- 10. Zeng M-S, Li X, Liu Y, Zhao H, Zhou J-C, Li K, Huang J-Q, Sun L-H, Tang J-Y, Xia X-J et al. . A high-selenium diet induces insulin resistance in gestating rats and their offspring. Free Radic Biol Med 2012;52:1335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J, Huang K, Lei XG. Selenium and diabetes—evidence from animal studies. Free Radic Biol Med 2013;65:1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Z, Barcus M, Kim J, Lum KL, Mills C, Lei XG. High dietary selenium intake alters lipid metabolism and protein synthesis in liver and muscle of pigs. J Nutr 2016;146:1625–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Labunskyy VM, Lee BC, Handy DE, Loscalzo J, Hatfield DL, Gladyshev VN. Both maximal expression of selenoproteins and selenoprotein deficiency can promote development of type 2 diabetes-like phenotype in mice. Antioxid Redox Signal 2011;14:2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bendich A; Institute of Medicine Dietary Reference Intakes for vitamin C, vitamin E, selenium, and carotenoids. Washington (DC): National Academies Press; 2001. [PubMed] [Google Scholar]
- 15. Barnes KM, Evenson JK, Raines AM, Sunde RA. Transcript analysis of the selenoproteome indicates that dietary selenium requirements of rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J Nutr 2009;139:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raines AM, Sunde RA. Selenium toxicity but not deficient or super-nutritional selenium status vastly alters the transcriptome in rodents. BMC Genomics 2011;12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Go Y-M, Sutliff RL, Chandler JD, Khalidur R, Kang B-Y, Anania FA, Orr M, Hao L, Fowler BA, Jones DP. Low-dose cadmium causes metabolic and genetic dysregulation associated with fatty liver disease in mice. Toxicol Sci 2015;147:524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr 2012;32:183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uppal K, Walker DI, Jones DP. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal Chem 2017;89:1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walker DI, Go Y-M, Liu K, Pennell KD, Jones DP. Population screening for biological and environmental properties of the human metabolic phenotype: implications for personalized medicine: Elsevier; 2016, E. Holmes, J.K. Nicholson, A.W. Darzi & J.C. Lindon (Eds), London, UK. [Google Scholar]
- 21. Roede JR, Uppal K, Park Y, Tran V, Jones DP. Transcriptome–metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicol Rep 2014;1:435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uppal K, Ma C, Go Y-M, Jones DP. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 2018;34:701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, Jones DP. MetabNet: an R package for metabolic association analysis of high-resolution metabolomics data. Front Bioeng Biotechnol 2015;3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chandler JD, Hu X, Ko E-J, Park S, Lee Y-T, Orr M, Fernandes J, Uppal K, Kang S-M, Jones DP et al. . Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am J Physiol Regul Integr Comp Physiol 2016;311:R906–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L, Fitch A, Greenblatt RM, Kingsley L, Guidot DM et al. . Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 2016;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohlhorst SD, Russell R, Bier D, Klurfeld DM, Li Z, Mein JR, Milner J, Ross AC, Stover P, Konopka E. Nutrition research to affect food and a healthy life span. J Nutr 2013;143:1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. LabDiet Standard diets. Available from: http://www.labdiet.com/Products/StandardDiets/. (accessed on January 3rd, 2018). [Google Scholar]
- 28. Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc 2010;5:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandler JD, Nichols DP, Nick JA, Hondal RJ, Day BJ. Selective metabolism of hypothiocyanous acid by mammalian thioredoxin reductase promotes lung innate immunity and antioxidant defense. J Biol Chem 2013;288:18421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Go YM, Kim CW, Walker DI, Kang DW, Kumar S, Orr M, Uppal K, Quyyumi AA, Jo H, Jones DP. Disturbed flow induces systemic changes in metabolites in mouse plasma: a metabolomics study using ApoE(-)/(-) mice with partial carotid ligation. Am J Physiol Regul Integr Comp Physiol 2015;308:R62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Go Y-M, Walker DI, Soltow QA, Uppal K, Wachtman LM, Strobel FH, Pennell K, Promislow DEL, Jones DP. Metabolome-wide association study of phenylalanine in plasma of common marmosets. Amino Acids 2015;47:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu T, Park Y, Johnson JM, Jones DP. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25:1930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, Jones DP. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lê Cao K-A, González I, Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics 2009;25:2855–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liquet B, Lê Cao K-A, Hocini H, Thiébaut R. A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinformatics 2012;13:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. González I, Lê Cao K-A, Davis MJ, Déjean S. Visualising associations between paired ‘omics’ data sets. BioData Mining 2012;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E et al. . HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res 2013;41:D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol 2014;48:2097–8. [DOI] [PubMed] [Google Scholar]
- 41. Available from: http://metlin.scripps.edu/index.php. [Google Scholar]
- 42. Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol 2016;29:1956–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Method 1995;57:289–300. [Google Scholar]
- 44. Hu L, Wang C, Zhang Q, Yan H, Li Y, Pan J, Tang Z. Mitochondrial protein profile in mice with low or excessive selenium diets. Int J Mol Sci 2016;17:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oster O, Schmiedel G, Prellwitz W. The organ distribution of selenium in German adults. Biol Trace Elem Res 1988;15:23–45. [DOI] [PubMed] [Google Scholar]
- 46. Lee S, Mardinoglu A, Zhang C, Lee D, Nielsen J. Dysregulated signaling hubs of liver lipid metabolism reveal hepatocellular carcinoma pathogenesis. Nucleic Acids Res 2016;44:5529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rui L. Energy metabolism in the liver. Compr Physiol 2014;4(1):177–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rinaldo P, Matern D, Bennett MJ. Fatty acid oxidation disorders. Annu Rev Physiol 2002;64:477–502. [DOI] [PubMed] [Google Scholar]
- 49. Van Veldhoven PP. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J Lipid Res 2010;51:2863–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rayman MP. Food-chain selenium and human health: emphasis on intake. Br J Nutr 2008;100:254–68. [DOI] [PubMed] [Google Scholar]
- 51. Johns Hopkins University Animal Care and Use Committee. The mouse. Available from: http://web.jhu.edu/animalcare/procedures/mouse.html, (accessed on January 3rd, 2018). [Google Scholar]
- 52. National Research Council Nutrient requirements of laboratory animals. 4th revised ed. Washington (DC): National Academies Press; 1995. [PubMed] [Google Scholar]
- 53. Risher J. Toxicological profile for selenium. Agency for Toxic Substances and Disease Registry; 2003, Atlanta, GA, USA. [PubMed] [Google Scholar]
- 54. Yang Z, Yan C, Liu G, Niu Y, Zhang W, Lu S, Li X, Zhang H, Ning G, Fan J et al. . Plasma selenium levels and nonalcoholic fatty liver disease in Chinese adults: a cross-sectional analysis. Sci Rep 2016;6:37288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hawkes WC, Keim NL. Dietary selenium intake modulates thyroid hormone and energy metabolism in men. J Nutr 2003;133:3443–8. [DOI] [PubMed] [Google Scholar]
- 56. Pinto A, Juniper DT, Sanil M, Morgan L, Clark L, Sies H, Rayman MP, Steinbrenner H. Supranutritional selenium induces alterations in molecular targets related to energy metabolism in skeletal muscle and visceral adipose tissue of pigs. J Inorg Biochem 2012;114:47–54. [DOI] [PubMed] [Google Scholar]
- 57. Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem 2003;278:13640–6. [DOI] [PubMed] [Google Scholar]
- 58. Esaki N, Nakamura T, Tanaka H, Suzuki T, Morino Y, Soda K. Enzymatic synthesis of selenocysteine in rat liver. Biochemistry 1981;20:4492–6. [DOI] [PubMed] [Google Scholar]
- 59. Kato T, Read R, Rozga J, Burk RF. Evidence for intestinal release of absorbed selenium in a form with high hepatic extraction. Am J Physiol 1992;262:G854–8. [DOI] [PubMed] [Google Scholar]
- 60. Joyce AR, Palsson BØ. The model organism as a system: integrating 'omics' data sets. Nat Rev Mol Cell Biol 2006;7:198–210. [DOI] [PubMed] [Google Scholar]
- 61. Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 2008;7:45–56. [DOI] [PubMed] [Google Scholar]
- 62. Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab 2012;15:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 2005;280:33588–98. [DOI] [PubMed] [Google Scholar]
- 64. Renga B, Mencarelli A, Vavassori P, Brancaleone V, Fiorucci S. The bile acid sensor FXR regulates insulin transcription and secretion. Biochim Biophys Acta 2010;1802:363–72. [DOI] [PubMed] [Google Scholar]
- 65. Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M et al. . TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 2009;10:167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 2009;89:147–91. [DOI] [PubMed] [Google Scholar]
- 67. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov 2008;7:678–93. [DOI] [PubMed] [Google Scholar]
- 68. Rutkowsky JM, Knotts TA, Ono-Moore KD, McCoin CS, Huang S, Schneider D, Singh S, Adams SH, Hwang DH. Acylcarnitines activate proinflammatory signaling pathways. Am J Physiol Endocrinol Metab 2014;306:E1378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aguer C, McCoin CS, Knotts TA, Thrush AB, Ono-Moore K, McPherson R, Dent R, Hwang DH, Adams SH, Harper M-E. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J 2015;29:336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines. Diabetes 2013;62:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gopalakrishna R, Gundimeda U. Antioxidant regulation of protein kinase C in cancer prevention. J Nutr 2002;132(Suppl):3819S–23S. [DOI] [PubMed] [Google Scholar]
- 72. Gandhi UH, Kaushal N, Ravindra KC, Hegde S, Nelson SM, Narayan V, Vunta H, Paulson RF, Prabhu KS. Selenoprotein-dependent up-regulation of hematopoietic prostaglandin D2 synthase in macrophages is mediated through the activation of peroxisome proliferator-activated receptor (PPAR) γ. J Biol Chem 2011;286:27471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis 1999;20:1657–66. [DOI] [PubMed] [Google Scholar]
- 74. Beilstein M, Whanger P. Chemical forms of selenium in rat tissues after administration of selenite or selenomethionine. J Nutr 1986;116:1711–9. [DOI] [PubMed] [Google Scholar]
- 75. Schrauzer GN. Selenomethionine: a review of its nutritional significance, metabolism and toxicity. J Nutr 2000;130:1653–6. [DOI] [PubMed] [Google Scholar]
- 76. Combs GF., Jr Biomarkers of selenium status. Nutrients 2015;7:2209–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.