Abstract
Background
Geraniol is an acyclic monoterpene alcohol, which is extracted from the ethereal oils of aromatic plants. A systematic analysis of its mechanism of action has not yet been carried out.
Methods
In this study, the druggability of geraniol was assessed via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP), and the potential targets of geraniol were identified using the Comparative Toxicogenomics Database (CTD). Additionally, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using WebGestalt. Drug-target-pathway networks were constructed using Cytoscape to give a visual view.
Results
Our findings showed that geraniol has superb druggability with 38 putative identified target genes. GO, KEGG, and network analyses revealed that these targets were associated with cancer, inflammatory immunoreactions, and other physiological processes.
Conclusion
Geraniol is predicted to target multiple proteins and pathways that shape a network which can exert systematic pharmacological effects.
Keywords: geraniol, druggability, target prediction, enrichment analysis, network pharmacology
Introduction
Natural products and traditional Chinese medicine (TCM) are the most abundant resources of active compounds for drug discovery. Monoterpenes, for example, are dietary compounds extracted from the ethereal oils of many vegetables, fruits, and especially TCM. Geraniol (Figure 1A) is an acyclic monoterpene alcohol, which is found in the ethereal oils of aromatic plants.1 Geraniol has been shown to exert a wide spectrum of pharmacological activities, for example anti-inflammatory, antimicrobial, antitumor, and so on.2–5 Close attention has been paid to geraniol due to its potential role in the treatment of a variety of diseases, such as chronic or allergic rhinitis, lung cancer, etc.6–8 These results suggest that geraniol can be utilized as a valuable chemical probe or a chemical moiety for the dissection of complex biological processes, discovery of hidden molecular relationships, and identification of therapeutic target molecules and pathways. Accordingly, the molecular mechanisms which geraniol induces and the resulting changes in cellular phenotypes are rarely studied. Meanwhile, employment of computational methodologies for the identification of drug targets and the underlying mechanisms is becoming mainstream in order to save money, time, and effort.9,10 In particular, computational target identification and following the molecular mechanisms can accelerate the drug discovery and drug design processes.
Figure 1.
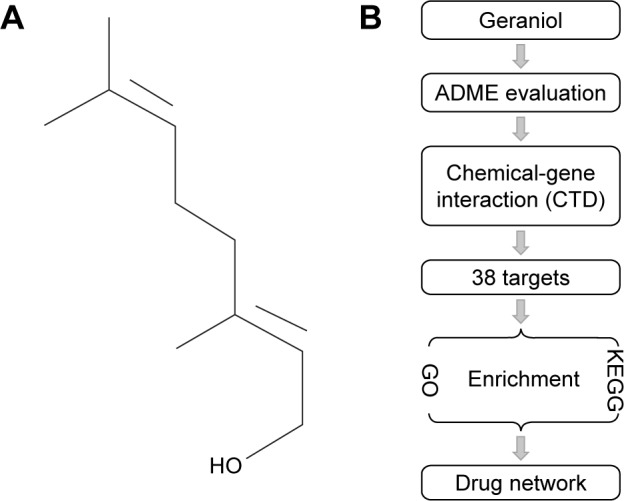
(A) Chemical structure of geraniol downloaded from the PubChem database (CID: 637566); (B) Workflow for the identification of potential geraniol target genes that integrates ADME evaluation, chemical-gene interaction, GO and KEGG pathway analyses and network construction.
Abbreviations: ADME, absorption, distribution, metabolism, and excretion; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CTD, Comparative Toxicogenomics Database.
Therefore, we elucidated the pharmacological actions of geraniol systematically using computational methodologies. First, the druggability of geraniol was assessed using the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) server.11 Next the potential candidate target genes were predicted by chemical-gene interaction analysis.12 Furthermore, gene ontology and pathway analyses were investigated using the identified target genes. Finally, drug-target network was constructed to provide a systematic overview of the potential target genes and the mechanism of action for geraniol. A schematic diagram of the analysis procedures for geraniol target gene prediction is shown in Figure 1B.
Materials and methods
Assessment of pharmacokinetics properties using TCMSP database (http://lsp.nwu.edu.cn/tcmsp.php) is a resource of systems pharmacology for TCMs or related compounds.11 It can provide information on the absorption, distribution, metabolism, and excretion (ADME) properties of a drug with potential biological effects at a systematic level, for example, oral bioavailability (OB), drug likeness (DL), Caco-2 permeability (Caco-2), blood–brain barrier (BBB), and so on.13,14
Of all pharmacokinetics properties, OB is the foremost feature of orally administered drugs, since it acts as a vital role in evaluating the efficacy of the drug distribution to the systemic circulation. In TCMSP database, OB was calculated on OBioavail1.1 based on an in-house model.11,13 For orally administered drugs, the movement across the intestinal epithelial barrier is one of the biggest obstacles of human absorption and its bioavailability.11,13 In present study, the chemical name “geraniol” was entered to the search box and its pharmacokinetic properties were investigated at the molecular level.
Target identification by the Comparative Toxicogenomics Database
The Comparative Toxicogenomics Database (CTD, http://ctdbase.org/), is a robust, publicly available database for toxicogenomic information. It provides manually curated key information about chemical-gene/protein interactions, chemical-disease and gene-disease relationships, from peer-reviewed scientific literature. Currently, CTD includes more than 30.5 million toxicogenomic relationships related to chemicals, proteins, and so on.12 Given a compound, CTD can provide the corresponding target genes sorted by the interactions between them by descending order. Candidate targets of geraniol were predicted using CTD with default parameters.
Analysis by GeneMANIA
GeneMANIA (http://www.genemania.org), is an user-friendly and flexible web server, for generating hypotheses in regard to gene function, analyzing gene lists, and prioritizing genes for functional assays.15
Given a query list, GeneMANIA can list the genes that have shared properties, or function similarly with the original query. It also shows a functional relationship network, expounding the association among the list as well as curated genomics and proteomics data. The potential candidate target genes were entered into the search bar after selecting Homo sapiens from the organism option, and the results were further collated.
Gene function and pathway enrichment analysis
Web-based gene set analysis toolkit (WebGestalt, http://www.webgestalt.org/option.php) can be utilized to thoroughly understand the functional and pathway enrichment information on the gene of interest.16 Potential candidate targets were input to the WebGestalt server using over-representation enrichment analysis method with the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
GO analysis is a commonly used approach for annotating genes and gene products with functions including molecular function, biological pathways, and cellular components.17 KEGG is a useful resource for systematic analysis of gene functions and related high-level genome functional information.18,19
Network construction
In order to understand the complex relationships among compound, targets, and diseases, we used Cytoscape (v 3.6.1; https://www.nigms.nih.gov/) to construct and analyze the three-layer networks.
Results
Pharmacokinetics properties of geraniol
ADME describes the disposition of a pharmaceutical compound and TCMCP provides information on 12 very important characteristics on ADME-related properties like Caco-2, human OB, BBB, and Lipinski’s rule of five for drug screening and evaluation.11 AMDE-related properties of geraniol were thoroughly investigated using TCMSP. Notably, the OB of geraniol was calculated to be 23.93%.
Targets identification of geraniol
Potential targets of geraniol were predicted using the CTD as described in the “Materials and methods” section.12 In total, 41 candidate target genes were identified by CTD. Afterwards, we filtered these genes by using the threshold chemical-gene interaction ≥1 and removed the non-human genes. Finally, 38 unique target genes for geraniol remained (Table 1). These 38 identified interacting genes were used for further investigation.
Table 1.
Putative targets of geraniol
| Num. | Gene ID | Gene symbol | Gene name |
|---|---|---|---|
|
| |||
| 1 | 1260 | CNGA2 | Cyclic nucleotide-gated channel alpha 2 |
| 2 | 1350 | COX7C | Cytochrome c oxidase subunit 7C |
| 3 | 1537 | CYC1 | Cytochrome c1 |
| 4 | 1555 | CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 |
| 5 | 2099 | ESR1 | Estrogen receptor 1 |
| 6 | 2100 | ESR2 | Estrogen receptor 2 |
| 7 | 2554 | GABRA1 | Gamma-aminobutyric acid type A receptor alpha1 subunit |
| 8 | 2560 | GABRB1 | Gamma-aminobutyric acid type A receptor beta1 subunit |
| 9 | 2641 | GCG | Glucagon |
| 10 | 26762 | HAVCR1 | Hepatitis A virus cellular receptor 1 |
| 11 | 2769 | GNA15 | G protein subunit alpha 15 |
| 12 | 2774 | GNAL | G protein subunit alpha L |
| 13 | 2936 | GSR | Glutathione-disulfide reductase |
| 14 | 3162 | HMOX1 | Heme oxygenase 1 |
| 15 | 3265 | HRAS | HRas proto-oncogene, GTPase |
| 16 | 3575 | IL7R | Interleukin 7 receptor |
| 17 | 3576 | CXCL8 | C-X-C motif chemokine ligand 8 |
| 18 | 3949 | LDLR | Low density lipoprotein receptor |
| 19 | 43 | ACHE | Acetylcholinesterase |
| 20 | 4790 | NFKB1 | Nuclear factor kappa B subunit 1 |
| 21 | 4953 | ODC1 | Ornithine decarboxylase 1 |
| 22 | 5111 | PCNA | Proliferating cell nuclear antigen |
| 23 | 5594 | MAPK1 | Mitogen-activated protein kinase 1 |
| 24 | 5595 | MAPK2 | Mitogen-activated protein kinase 2 |
| 25 | 5743 | PTGS2 | Prostaglandin-endoperoxide synthase 2 |
| 26 | 581 | BAX | BCL2 associated X, apoptosis regulator |
| 27 | 5894 | RAF1 | Raf-1 proto-oncogene, serine/threonine kinase |
| 28 | 596 | BCL2 | BCL2, apoptosis regulator |
| 29 | 7083 | TK1 | Thymidine kinase 1 |
| 30 | 7157 | TP53 | Tumor protein p53 |
| 31 | 7298 | TYMS | Thymidylate synthetase |
| 32 | 7442 | TRPV1 | Transient receptor potential cation channel subfamily V member1 |
| 33 | 836 | CASP3 | Caspase 3 |
| 34 | 8383 | OR1A1 | Olfactory receptor family 1 subfamily A member 1 |
| 35 | 8390 | OR1G1 | Olfactory receptor family 1 subfamily G member 1 |
| 36 | 841 | CASP8 | Caspase 8 |
| 37 | 842 | CASP9 | Caspase 9 |
| 38 | 847 | CAT | Catalase |
GeneMANIA analysis
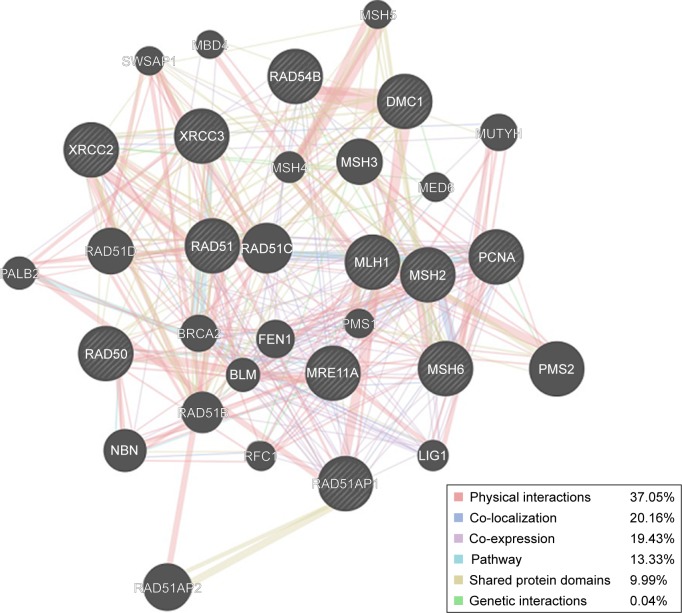
Among the 38 targets and their interacting proteins, it was found that 37.05% had physical interactions, 20.16% exerted co-localization, and 19.43% displayed similar co-expression characteristics. Other results, including pathway, shared protein domains and genetic interactions, are shown in Figure 2.
Figure 2.
Protein network of geraniol. Black nodes represent target proteins, and connecting colors indicate different correlations. Functional associations between targets were investigated using GeneMANIA. Genes in black circles were query terms while these in gray circle indicate genes associated with query genes.
GO and pathway analysis
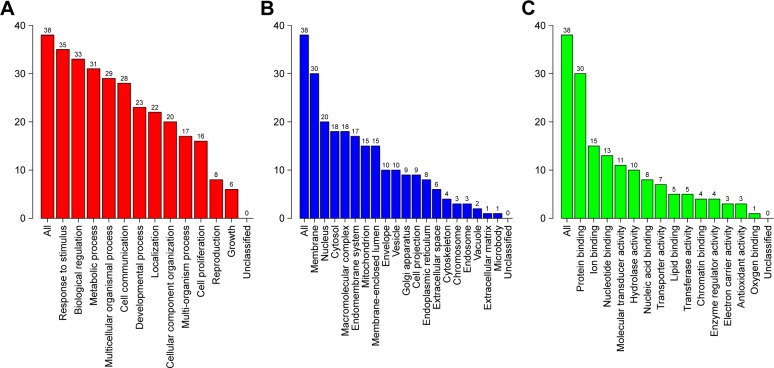
In order to study further the 38 identified target genes, GO and KEGG enrichment analyses were carried out using WebGestalt. As shown in Figure 3, the top seven functions were used as response to stimulus (35/38), biological regulation (33/38), metabolic process (31/38), membrane (30/38), protein binding (30/38), multicellular organismal process (29/38), and cell communication (28/38). These functional terms are highly relevant to anti-inflammatory activities, especially for chronic or allergic rhinitis.
Figure 3.
GO map of putative target genes. (A) Biological process categories. (B) Cellular component categories. (C) Molecular function categories.
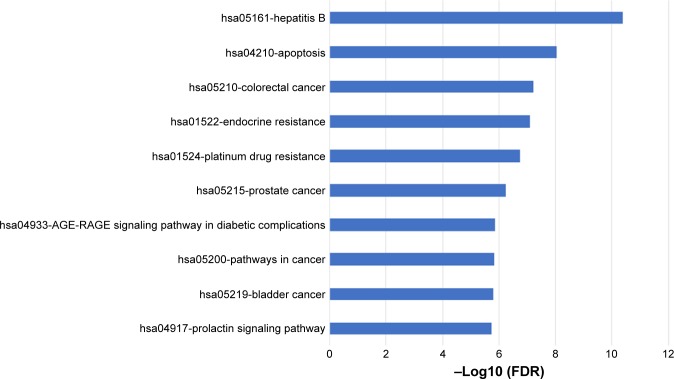
As for pathway analysis, the 38 targets participate in 10 KEGG pathways with significant false discovery rate (FDR)-adjusted P-value including apoptosis, pathways in cancer, and so on, which were shown in Figure 4.
Figure 4.
KEGG pathway analysis of putative target genes.
Network analysis
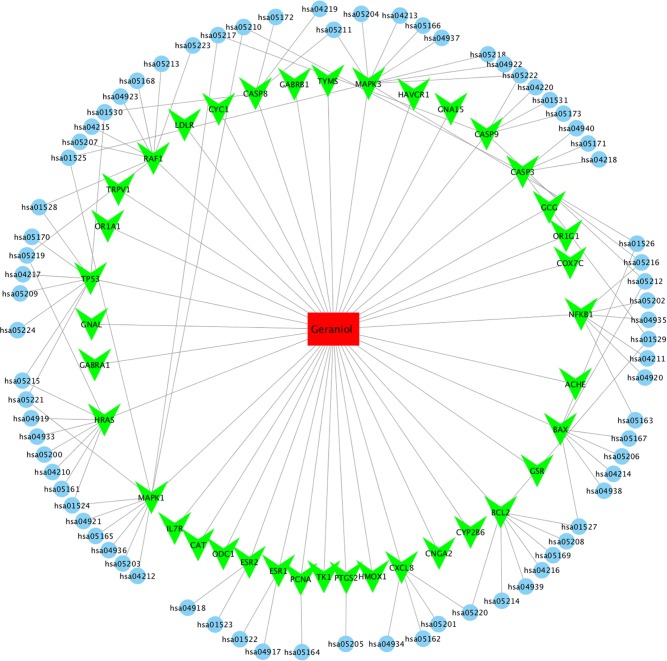
Based on target and pathway analyses, an entire compound, targets and diseases network was constructed using Cytoscape (v 3.6.1). As shown in Figure 5, this compound, targets, and diseases interaction network has 80 nodes and 129 edges. The red oblong, green inverted triangles, and blue circles correspond to geraniol, target genes, and pathways, respectively.
Figure 5.
Geraniol-target-pathway network.
Discussion
Poor pharmacokinetics and toxicity are the most important causes of costly delays in drug discovery and development. There is, therefore, growing belief that certain features in the drug discovery process should be prioritized.20 In silico analysis can improve predictions and pharmacokinetic modeling, as well as metabolic and toxicity endpoints; all of which accelerate and streamline the drug discovery process.9,10
Lipinski’s rule of five can identify some very important drug properties, which should be taken into account for compounds developed with the aim of oral delivery.21 The rule of five takes into consideration molecular weights (MWs) <500 Da, a LogP <5, as well as numbers of hydrogen-bond donors and acceptors less than 5 and 10, respectively. Today, the rule of five is generally referred to as a guideline for drug optimization.22 As shown in Table 2, the pharmacokinetic properties of geraniol meet these requirements, meaning geraniol is a superior candidate for drug development.
Table 2.
Pharmacological and molecular properties of geraniol
| Name | MW | AlogP | Hdom | Hacc | OB (%) | Caco-2 | BBB | DL | FSAF | TPSA | RBN |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Geraniol | 154.28 | 2.93 | 1 | 1 | 23.93 | 1.19 | 1.14 | 0.02 | 0.27 | 20.23 | 4 |
Abbreviations: Caco-2, Caco-2 permeability; OB, oral bioavailability; DL, drug likeness; BBB, blood–brain barrier.
In drug discovery, target gene identification is the first step. More and more active compounds or drugs are being shown to interact with multiple genes or proteins.23–26 A variety of in silico target identification approaches have been developed and are broadly applied toward this aim. As listed in Table 1, 38 potential targets of geraniol were identified using computational methods. The results of GeneMANIA provided information on physical interactions, co-localization, co-expression as well as shared protein domains, and implied that the targets and their interacting proteins may have identical or similar functions.
We identified an inflammatory role for geraniol in allergic rhinitis. Similarly, Madankumar et al reported geraniol exerts antimicrobial, antioxidant, antitumor, and anti-inflammatory activities via activation of apoptotic pathways.6,27 These results closely coincide with our findings from GO and KEGG analyses.
The drug-target network shown in Figure 5 also revealed that geraniol has multiple targets and further indicated that it possesses multiple pharmacological activities. Cho et al have also revealed that geraniol exerts systematic pharmacological effects by targeting multiple proteins and pathways.28,29 Multiple target therapeutic medicaments are more effective for the treatment of complex diseases, for instance allergic rhinitis, cancers, and are less vulnerable to adaptive resistance. Hence, geraniol could be a promising resource that may be utilized as chemical moiety, lead compound, or an active ingredient for future drug discovery.
In summary, we would like to underline that geraniol is an active ingredient or a promising compound for the development of a safe and effective multi-targeted anticancer medicament. This study provides novel insight into the perspectives and challenges for geraniol research and its application in future clinical investigation.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lapczynski A, Bhatia SP, Foxenberg RJ, Letizia CS, Api AM. Fragrance material review on geraniol. Food Chem Toxicol. 2008;46(Suppl 11):S160–S170. doi: 10.1016/j.fct.2008.06.048. [DOI] [PubMed] [Google Scholar]
- 2.Solórzano-Santos F, Miranda-Novales MG. Essential oils from aromatic herbs as antimicrobial agents. Curr Opin Biotechnol. 2012;23(2):136–141. doi: 10.1016/j.copbio.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Tiwari M, Kakkar P. Plant derived antioxidants-Geraniol and camphene protect rat alveolar macrophages against t-BHP induced oxidative stress. Toxicol In vitro. 2009;23(2):295–301. doi: 10.1016/j.tiv.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 4.de Carvalho KI, Bonamin F, Dos Santos RC, et al. Geraniol-a flavoring agent with multifunctional effects in protecting the gastric and duodenal mucosa. Naunyn Schmiedebergs Arch Pharmacol. 2013;387(4):355–365. doi: 10.1007/s00210-013-0947-z. [DOI] [PubMed] [Google Scholar]
- 5.Rekha KR, Selvakumar GP, Sethupathy S. Geraniol ameliorates the motor behavior and neurotrophic factors inadequacy in MPTP-induced mice model of Parkinson’s disease. J Mol Neurosci. 2013;51(3):851–862. doi: 10.1007/s12031-013-0074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Yang XL, Ni YH, Xu ZM. Geraniol suppresses proinflammatory mediators in phorbol 12-myristate 13-acetate with A23187-induced HMC-1 cells. Drug Des Dev Ther. 2018;12:2897–2903. doi: 10.2147/DDDT.S145702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong TP, Heidor R, de Conti A, Dagli MLZ, Moreno FS. Farnesol and geraniol chemopreventive activities during the initial phases of hepatocarcinogenesis involve similar actions on cell proliferation and DNA damage, but distinct actions on apoptosis, plasma cholesterol and HMGCoA reductase. Carcinogenesis. 2006;27(6):1194–1203. doi: 10.1093/carcin/bgi291. [DOI] [PubMed] [Google Scholar]
- 8.Galle M, Crespo R, Kladniew BR, Villegas SM, Polo M, de Bravo MG. Suppression by geraniol of the growth of A549 human lung adenocarcinoma cells and inhibition of the mevalonate pathway in culture and in vivo: potential use in cancer chemotherapy. Nutr Cancer. 2014;66(5):888–895. doi: 10.1080/01635581.2014.916320. [DOI] [PubMed] [Google Scholar]
- 9.Bi YH, Zhang LH, Chen SJ, Page GL. Antitumor mechanisms of curcumae rhizoma based on network pharmacology. Evid Based Compl Altern Med. 2018;2018:9. doi: 10.1155/2018/9567061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YC, Chang CW, Wu CR. Analysis of the action mechanism of Fang Ji Huang Qi decoction in treating rheumatoid arthritis by network pharmacology. Modernization Traditional Med. 2018;3(6):286–294. [Google Scholar]
- 11.Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017;45(D1):D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei T, Zheng C, Huang C, et al. Systematic understanding the mechanisms of vitiligo pathogenesis and its treatment by Qubaibabuqi formula. J Ethnopharmacol. 2016;190:272–287. doi: 10.1016/j.jep.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Tao Q, Guo Z, et al. Systems pharmacology dissection of the integrated treatment for cardiovascular and gastrointestinal disorders by traditional Chinese medicine. Sci Rep. 2016;6:32400. doi: 10.1038/srep32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology Consortium Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov. 2003;2(3):192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 21.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 22.Leeson PD. Molecular inflation, attrition and the rule of five. Adv Drug Deliv Rev. 2016;101:22–33. doi: 10.1016/j.addr.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Schenone M, Dančík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Xie XQ. Computational target fishing: what should chemo-genomics researchers expect for the future of in silico drug design and discovery? Future Med Chem. 2014;6(3):247–249. doi: 10.4155/fmc.14.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Gao L, Lee YM, et al. Target identification of natural and traditional medicines with quantitative chemical proteomics approaches. Pharmacol Ther. 2016;162:10–22. doi: 10.1016/j.pharmthera.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Madankumar A, Tamilarasi S, Premkumar T, Gopikrishnan M, Nagabhishek N, Devaki T. Geraniol attenuates 4NQO-induced tongue carcinogenesis through downregulating the activation of NF-kappaB in rats. Mol Cell Biochem. 2017;434(1–2):7–15. doi: 10.1007/s11010-017-3030-0. [DOI] [PubMed] [Google Scholar]
- 28.Cho M, So I, Chun JN, Jeon JH. The antitumor effects of geraniol: modulation of cancer hallmark pathways (Review) Int J Oncol. 2016;48(5):1772–1782. doi: 10.3892/ijo.2016.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crespo R, Montero Villegas S, Abba MC, de Bravo MG, Polo MP. Transcriptional and posttranscriptional inhibition of HMGCR and PC biosynthesis by geraniol in 2 Hep-G2 cell proliferation linked pathways. Biochem Cell Biol. 2013;91(3):131–139. doi: 10.1139/bcb-2012-0076. [DOI] [PubMed] [Google Scholar]