Abstract
Background.
Streptococcus agalactiae can cause urinary tract infection (UTI). The role of the S. agalactiae global virulence regulator, CovR, in UTI pathogenesis is unknown.
Methods.
We used murine and human bladder uroepithelial cell models of UTI and S. agalactiae mutants in covR and related factors, including β-hemolysin/cytolysin (β-h/c), surface-anchored adhesin HvgA, and capsule to study the role of CovR in UTI.
Results.
We found that covR-deficient serotype III S. agalactiae 874391 was significantly attenuated for colonization in mice and adhesion to uroepithelial cells. Mice infected with covR-deficient S. agalactiae produced less proinflammatory cytokines than those infected with wild-type 874391. Acute cytotoxicity in uroepithelial cells triggered by covR-deficient but not wild-type 874391 was associated with significant caspase 3 activation. Mechanistically, covR mutation significantly altered the expression of several genes in S. agalactiae 874391 that encode key virulence factors, including β-h/c and HvgA, but not capsule. Subsequent mutational analyses revealed that HvgA and capsule, but not the β-h/c, exerted significant effects on colonization of the murine urinary tract in vivo.
Conclusions.
S. agalactiae CovR promotes bladder infection and inflammation, as well as adhesion to and viability of uroepithelial cells. The pathogenesis of S. agalactiae UTI is complex, multifactorial, and influenced by virulence effects of CovR, HvgA, and capsule.
Keywords: Urinary tract infection, Streptococcus agalactiae, covR, bladder, uroepithelium, virulence
Streptococcus agalactiae can cause urinary tract infection (UTI), including cystitis, pyelonephritis, and asymptomatic bacteriuria [1]. Immunocompromised adults and elderly individuals are particularly prone to UTI with S. agalactiae. Among the 10 serotypes of S. agalactiae, serotype III has been more frequently associated with acute disease than the other serotypes [1]. The mechanisms underlying the pathogenesis of S. agalactiae UTI are not well understood. Prior studies have shown that S. agalactiae UTI involves binding of bacteria to the uroepithelium followed by activation of immune responses, including the production of interleukin 1α, 1β, 9, 10 and macrophage inflammatory protein 1α and 1β within 24 hours after infection [2–4]. The bacterial virulence factor β-hemolysin/cytolysin (β-h/c) promotes inflammation in the bladder [5] and colonization for some strains of S. agalactiae [6] but not others [4]. Capsular sialic acids also enhance S. agalactiae colonization in the bladder [4], and augment the ability of uropathogenic Escherichia coli to survive in the bladder in coinfection assays [7]. Similar to findings in uropathogenic E. coli UTI [8–11], these studies suggest that distinct mechanisms of pathogenesis underpin S. agalactiae UTI.
CovR is the DNA-binding regulatory element of the major global regulatory system of S. agalactiae, CovR/CovS (alternate designation: CsrR/CsrS) [12]. This 2-component system confers an ability to sense environmental signals via a sensor histidine kinase (CovS), which relays signals through the response regulator (CovR). CovR and/or CovS contribute to S. agalactiae pathogenesis by regulating the expression of many genes, including those that encode virulence factors, such as β-h/c [13, 14], pili, capsule and surface proteins, such as HvgA [15] and BsaB [16]. HvgA is of particular interest in pathogenesis studies because it enables persistent colonization by group B Streptococcus strains of the so-called “hypervirulent” clonal complex 17 lineage [17]. CovR positively regulates its own expression and the expression of >150 other genes that may influence the ability of S. agalactiae to transition between commensalism and virulence [18]. In the current work, we studied the role of CovR in experimental UTI in mice and in human uroepithelial cells to determine whether the S. agalactiae global virulence regulator affects the pathogenesis of UTI.
METHODS
Murine Model
Female C57BL/6 mice (Animal Resources Centre) were infected with 5 × 108 colony-forming units (CFUs) of wild-type (WT) S. agalactiae 874391 [19], or its covR-deficient, cylE-deficient, hvgA-deficient, or cpsE-deficient mutant, using transurethral challenge [2, 20]. Urine samples were collected 24 hours after challenge; subsequently, mice were euthanized, and bladders, and kidneys were collected and homogenized in sterile PBS. Urine and tissue homogenates were used for colony counts or analysis using the Bio-Plex Pro Mouse Cytokine 23-Plex Immunoassay (Bio-Rad). The Griffith University Animal Ethics Committee approved this research (MSC/03/12/AEC) in accordance with the guidelines of the National Health and Medical Research Council (NHMRC).
Bacteria
The S. agalactiae strains used in this study are listed in Supplementary Table 1. Separate mutations in covR (strain GU2400), hvgA (strain GU2488), and cpsE (strain GU2522) were made in S. agalactiae 874391 (clonal complex 17/ST-17 strain [21]) using allelic replacement by homologous recombination with pHY304-aad9 and a chloramphenicol resistance cassette (from pLZ12) with primers listed in Supplementary Table 1, essentially as described elsewhere [19, 22]. Isogenic mutants were verified by polymerase chain reaction (PCR), and the mutated regions were sequenced for confirmation. Complementation of the covR− strain was performed by marker rescue. A 1742–base pair region containing intact covR was amplified by PCR using genomic DNA from S. agalactiae 874391 and primers (per Supplementary Table 1) and cloned into pHY304-aad9, forming pGU2571. This was electroporated into covR− strain GU2400, and allelic replacement was carried out as described above, without antibiotic selection. Colonies were screened for the loss of resistance to chloramphenicol and spectinomycin, and confirmed by PCR and sequencing.
In Vitro Infections
Human 5637 uroepithelial cells were used for adhesion-invasion assays [3]. The antibiotic concentrations used to kill extracellular bacteria were 250 U/mL penicillin, 250 U/mL streptomycin, and 50 µg/mL gentamicin (used combined). A total of 3 × 105 5637 cells/mL in 24-well tissue culture plates (Nunc) were grown for 24 hours at 37°C in 5% carbon dioxide. The multiplicity of infection (MOI) was 150–250 bacteria per cell. Culture supernatants were analyzed for lactate dehydrogenase (LDH) using the TOX7 kit (Sigma-Aldrich), or using the Bio-Plex Pro Human Cytokine 27-Plex Immunoassay (Bio-Rad). Cells were stained with trypan blue for viability measures. Cells grown on coverslip inserts were stained for active caspase 3 (G7481, 1/250; Promega), with anti-rabbit immunoglobulin G secondary antibody Alexa Fluor 488 conjugate (A-11008, 1/125; Life Technologies) and Hoechst 33258 (20 μg/mL phosphate-buffered saline [PBS]; Sigma-Aldrich). Cells were mounted in 0.2% n-propyl gallate, and viewed using a AxioImager.M2 microscope (Carl Zeiss MicroImaging) with Zen 2012 SP2 imaging software.
Hemolysis Assays
Hemolysis assays were performed as described elsewhere [23], with modifications. Briefly, 10-mL overnight THB cultures were washed twice in PBS and resuspended in 10-mL of PBS plus 0.2% glucose. Then, 100-μL aliquots (approximately 107 CFUs) in a 96-well plate were mixed with an equal volume of 1% (vol/vol) horse erythrocytes (Thermo Fisher Scientific), suspended in PBS-glucose, and incubated at 37°C for 2 hours. As positive and negative controls, 100 μL of 2% Triton X-100 or PBS-glucose were used, respectively. At each time point, clarified supernatants were diluted 1:5 in PBS, and hemoglobin release was measured at an optical density of 420 nm (OD420). Data are shown as erythrocyte lysis, as a percentage of the positive control value.
RNA Extractions and Quantitative PCR
Ten-milliliter Todd-Hewitt broth (THB) cultures, grown to OD600 0.4–0.5 (109 CFUs), were added to 4 mL of ice-cold 95% ethanol/5% phenol (vol/vol) solution and incubated on ice for 30 minutes. Cell pellets (stored at –80°C) were resuspended in 1 mL of TE buffer containing 100 U of mutanolysin and 30 mg/mL lysozyme (Sigma-Aldrich) and incubated for 1 hour at 37°C before RNA isolation performed using an SV Total RNA isolation kit (Promega). Trace DNA contamination was removed using Turbo DNA-free DNase (Life Technologies) and confirmed by means of PCR. RNA was quantified, and integrity confirmed using an Experion Automated Electrophoresis platform (Bio-Rad). Total RNA (1 µg) was reverse-transcribed using Superscript III reverse transcriptase (Life Technologies) and random hexamers. Complementary DNA was diluted 1:5 with water for quantitative PCR. Primers (Supplementary Table 1) were designed using Primer3 Plus software [24], to amplify 100–150–base pair products, with a melting temperature (Tm) of approximately 60°C and used at a final concentration of 0.4 µmol/L. Transcripts were quantified using a SensiFAST SYBR No ROX kit (Bioline) and a Lightcycler 480 II system (Roche). All assays conformed to MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines [25]. RNA from 3 independent biological replicates was analyzed, and amplification efficiencies were determined using 10-fold serially diluted genomic DNA. Relative fold-change values were calculated as described elsewhere [26].
Statistical Analysis
Mann–Whitney U tests, unpaired t tests, Kruskal-Wallis analysis of variance with Dunn’s multiple comparisons, and area under the curve analysis and analysis of variance were used where appropriate to analyze the data derived from murine, in vitro, gene expression, and cytokine assays, as indicated in the figure legends. The analyses were performed using GraphPad Prism software (version 6).
RESULTS
Effect of S. agalactiae CovR on Bladder Colonization and Proinflammatory Cytokine Synthesis
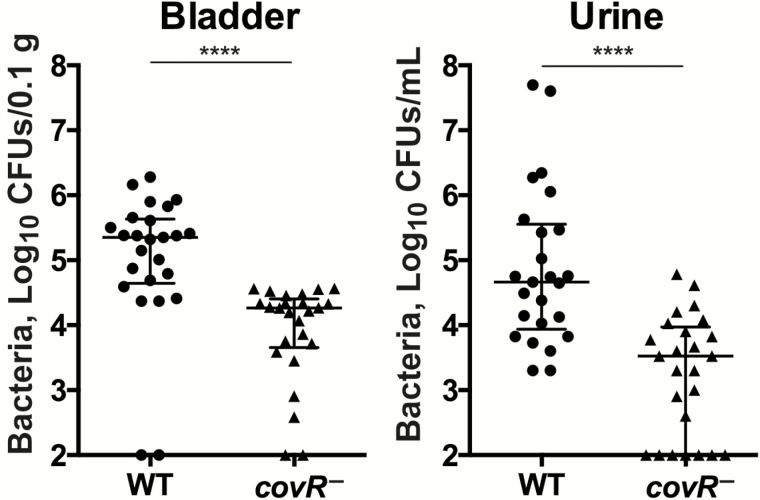
Infection assays using S. agalactiae 874391 and its covR-deficient mutant revealed significant attenuation in the ability of the mutant to colonize the bladder and urine in mice. The median bacterial loads and interquartile ranges (IQRs) of the WT and mutant in the bladder 24 hours after infection were 5.4 log10 CFUs/0.1 g (IQR, 4.6–5.6), and 4.3 log10 CFUs/0.1 g (3.7–4.4), respectively (Figure 1). The median bacterial loads in urine 24 hours after infection were also significantly different: 4.7 log10 CFUs/mL (IQR, 3.9–5.5) for WT versus 3.5 log10 CFUs/mL (2.0–4.0) for mutant (Figure 1). Neither the WT nor the mutant consistently colonized the kidneys (colonization in 2 of 25 for WT vs 4 of 25 for mutant; data not shown).
Figure 1.
Effect of Streptococcus agalactiae covR on colonization of the bladder and urine in mice during experimental urinary tract infection. Bacterial colonization is shown for the bladder and urine in C57BL/6 mice after transurethral infection. Data are pooled from 3 independent experiments, each containing ≥8 mice per group (medians shown for n = 25) and compared using a Mann–Whitney U test; ****P < .001. Bars represent medians with interquartile ranges. Abbreviations: CFUs, colony-forming units; WT, wild type.
Analysis of innate cytokine responses induced by infection in the bladder showed that >25% of significant responses develop in a covR-dependent manner. The most striking response was for proinflammatory cytokines, including interleukin 6 and 17A and interleukin 12(p70) (IL-12[p70]), and chemokines, including granulocyte-macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein (MCP) 1 and keratinocyte chemoattractant (Supplementary Figure 1); WT infection induced significantly higher levels of all these factors compared to infection with the covR-deficient mutant (total number of factors, 6 of 23 [26.1%]). In contrast, IL-12(p40), macrophage inflammatory protein 1α and 1β, granulocyte colony-stimulating factor, and RANTES (regulated on activation of normal T cells expressed and secreted) were significantly induced by both WT infection and the covR-deficient mutant compared with noninfected controls, and the responses did not differ significantly between the infected groups (ie, independent of covR).
We observed significant suppression of eotaxin by S. agalactiae independent of covR (Supplementary Figure 1). The levels of other factors tested did not significantly differ between the groups. Thus, a significant proportion of innate immune mediators are induced in the bladder in a CovR-dependent manner during UTI (6 of 23; P = .01 [Fisher exact test]); a significant proportion are also induced via CovR-independent mechanisms (5 of 23; P = .049 [Fisher exact test]; ie, both the WT and mutant deviated significantly from the PBS control, to a similar extent and in the same direction).
Effect of S. agalactiae CovR on Adhesion to and Invasion of Human Bladder Uroepithelial Cells and Role in Cytotoxicity
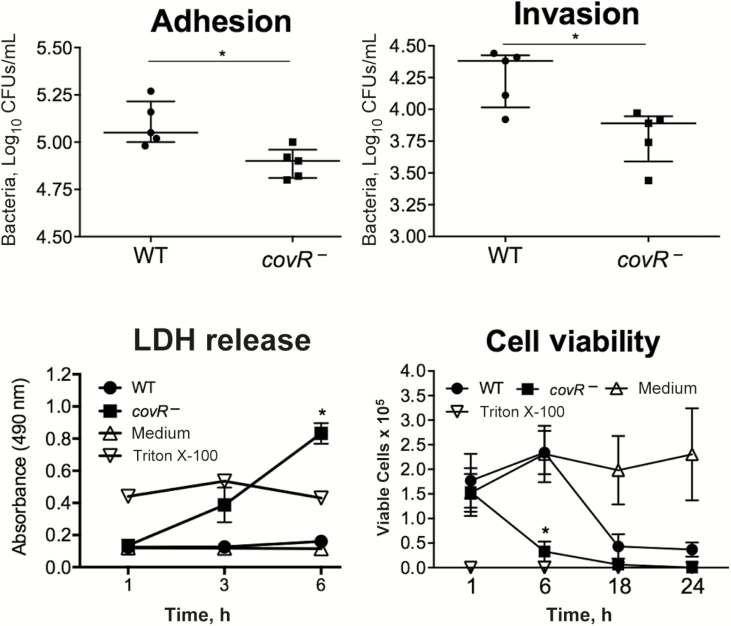
To test whether the attenuated phenotype of the covR− mutant in vivo might parallel differential adhesion of the WT and mutant in human uroepithelial cells, we performed in vitro assays using 5637 bladder cells. The covR− mutant exhibited significantly less adhesion than the WT at 1 hour after infection, with 38% fewer mutant bacteria recovered (P = .01; Figure 2). Invasion assays at 3 hours after infection showed 79.3% fewer covR− bacteria recovered compared with the WT (P = .001). Analysis of invasion levels at 3 hours adjusted for the differences in adhesion between WT and mutant (at 1 hour) showed that 65% fewer covR− bacteria invaded the host cells compared with WT (P = .01; Figure 2).
Figure 2.
Role of Streptococcus agalactiae covR in adhesion, invasion, and cytotoxicity in human uroepithelial cells in vitro. The data for bacterial adhesion to and invasion of 5637 cells are from 1 experiment (n = 5), representative of 4 independent experiments, and were compared using Student t test; *P < .05. The level of cytotoxicity is shown according to lactate dehydrogenase (LDH) release and cell viability measures based on live/dead cell counts using trypan blue exclusion. The positive and negative controls were 0.1% Triton X-100 and medium, respectively. Data are pooled from ≥3 independent experiments, each containing quadruplicate samples, and were compared using Mann–Whitney U test; *P < .05. Bars represent medians with interquartile ranges. P values for the LDH release and cell viability plots are for the comparisons of wild-type (WT) S. agalactiae and its covR− mutant.
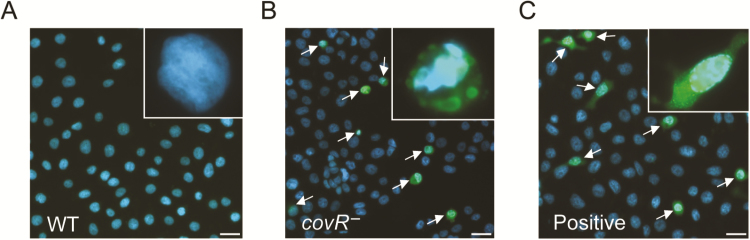
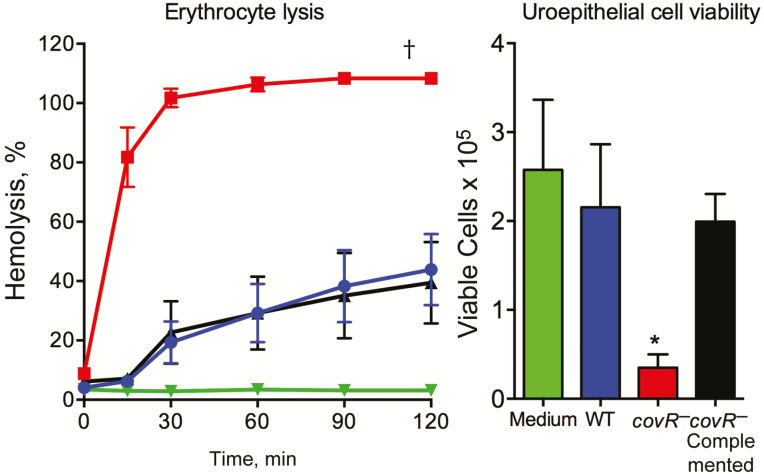
We observed acute uroepithelial cell cytotoxicity in response to the mutant compared with WT at 6 hours after infection, according to LDH release measures and counts of live/dead uroepithelial cells (Figure 2). Caspase 3 was activated by the covR-deficient mutant in contrast to infection with WT (Figure 3). A higher level of host cell cytotoxicity induced by the covR-deficient mutant was correlated with higher activity of β-h/c, according to the erythrocyte lysis assay (Figure 4). Interestingly, we also noted that WT strain 874391 is significantly less hemolytic than a previously characterized strain 807 [6] (Supplementary Figure 2). Cytotoxic effects on 5637 cells and erythrocytes were negated in comparative assays using a covR-deficient mutant derivative in which the mutation was repaired by chromosomal complementation using full-length covR from WT 874391 (Figure 4). The adhesion, invasion, LDH release, and caspase 3 activation phenotypes observed in 5637 cells in vitro after infection with the complemented strain were also similar to WT levels (data not shown).
Figure 3.
Contrasting levels of capase 3 activation in human 5637 uroepithelial cells after in vitro infection with wild-type (WT) Streptococcus agalactiae and its covR-mutant. Comparative infections with WT S. agalactiae (A), its covR− mutant (B), and the positive control (40 μmol/L staurosporine) (C) are shown. Arrows indicate cells with a high degree of caspase 3 activation. All images were captured 6 hours after infection and are representative of 3 independent assays. Scales bars represent approximately 20 μm.
Figure 4.
Complementation of covR mutation in Streptococcus agalactiae. Left, Hemolytic activity was assessed using horse erythrocytes incubated with wild-type (WT) (blue), covR− (red), or covR−-complemented (black) bacteria or phosphate-buffered saline (PBS) (negative control; green). Erythrocyte lysis data were compared by area under the curve analysis followed by analysis of variance (ANOVA), which showed signifiantly higher hemolytic activity of covR−S. agalactiae compared with WT or covR-complemented strains; †P < .001. Right, Uroepithelial cell cytotoxicity was assessed by live/dead cell counts at 6 hours after infection with WT (blue), covR− (red), or covR− complemented (black) bacteria or medium (negative control; green). Cytotoxicity data were compared using Kruskal Wallis ANOVA, which showed significantly higher cytotoxicity of covRS. agalactiae compared with WT or covR-complemented strains; *P < .01. Bars show means and standard errors of the mean from ≥2 independent experiments.
Effect of S. agalactiae CovR on Human Uroepithelial Cell Cytokine Production
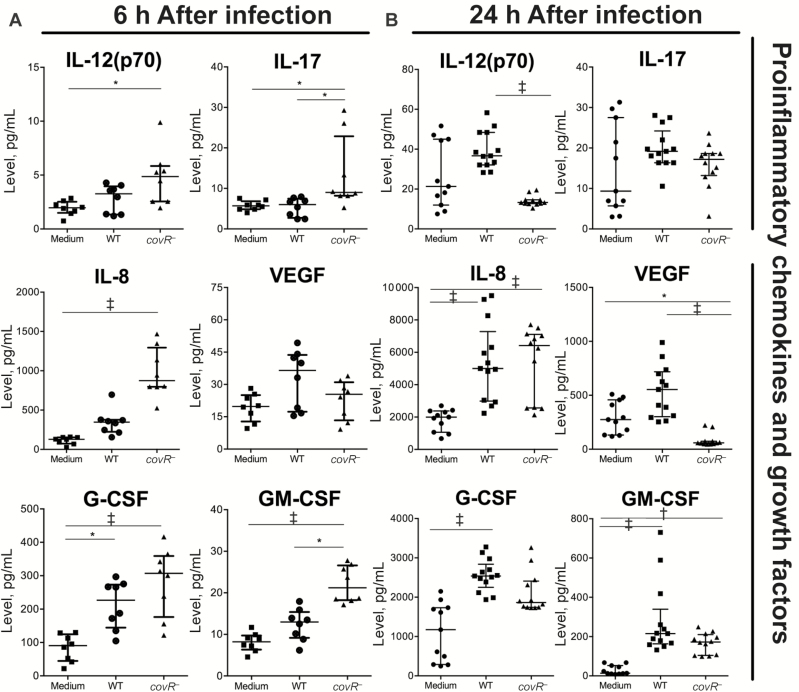
Multiplex assays of 5637 uroepithelial cells were used to test whether innate immune cytokine responses in human bladder cells develop in a covR-dependent manner and whether covR deficiency leads to attenuated innate cytokine responses in vitro. Surprisingly, the covR-deficient mutant induced >50% of cytokines (14 of 27), including IL-12(p70), interleukin 17 and 8, and GM-CSF, at significantly higher levels than noninfected controls at 6 hours after infection (Figure 5 and Supplementary Figure 3); only 1 cytokine, granulocyte colony-stimulating factor, was significantly induced by WT compared with noninfected controls at or before 6 hours (Figure 5 and Supplementary Figure 3). Some of the responses (eg, interleukin 8 and 1β and GM-CSF) began as early as 3 hours after infection (Supplementary Figure 3).
Figure 5.
Influence of Streptococcus agalactiae covR on cytokine responses in human 5637 uroepithelial cells. A, At 6 hours after infection, covR− mutation was associated with higher levels of cytokines, including interleukin 17 (IL-17) and interleukin 8 (IL-8). B, At 24 hours after infection, covR− mutation was associated with lower levels of cytokines, including interleukin 12(p70) (IL-12[p70]) and vascular endothelial growth factor (VEGF). Data are pooled from ≥3 independent experiments, each including quadruplicate samples. Data were compared using Kruskal-Wallis analysis of variance (ANOVA); *P < .05; †P < .01; ‡P < .001. Bars represent medians with interquartile ranges. Abbreviations: G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; WT, wild type.
Cytokine responses at 24 hours after infection comprised WT bacteria inducing stronger responses for 4 of 27 cytokines compared with the mutant (IL-12[p70], vascular endothelial growth factor [VEGF], MCP-1, and interleukin 5). The mutant induced significantly higher levels of 3 of 27 cytokines (interleukin 1β, fibroblast growth factor basic, and tumor necrosis factor α) compared with WT at 24 hours (Supplementary Figure 3). The other cytokines were equivalent between groups or not detected (Supplementary Figure 3). The levels of several cytokines (eg, fibroblast growth factor basic, interleukin 9, and RANTES) decreased over time, coincident with host cell death. Together, these data show that covR deficiency does not attenuate innate cytokine responses in human uroepithelial cells in vitro; covR mutation actually enhances the early production of many cytokines in human cells in vitro.
Regulatory Effects of covR on β-h/c, HvgA, and Capsule Gene Expression in S. agalactiae 874391 and Influence on Colonization During UTI
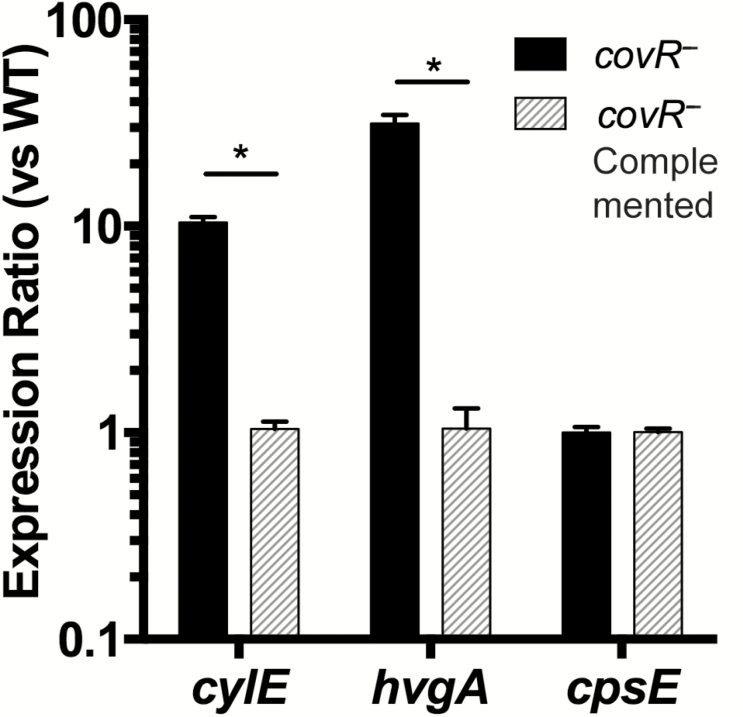
Given the effects of covR on virulence, we tested whether covR mutation affected the expression of genes that encode the virulence factors β-h/c (cylE), HvgA (hvgA), and capsule (cpsE). CovR deficiency in strain 874391 caused significantly higher expression of cylE and hvgA, but the expression of cpsE was similar to that with WT (Figure 6). The complemented strain had equivalent expression of these genes compared with WT, demonstrating a functional restoration of CovR (Figure 6).
Figure 6.
Effect of Streptococcus agalactiae covR mutation on virulence gene expression for cylE (β-hemolysin/cytolysin), hvgA (HvgA) and cpsE (capsule). Transcripts were quantified from cultures of S. agalactiae 874391::covR− (black bars) or the complemented strain 874391::covR−::covR (gray bars) and compared with wild-type (WT) S. agalactiae using quantitative reverse-transcription polymerase chain reaction. Bars show means and standard errors of the mean for 3 independent biological replicates. Differences in expression ratios were compared using unpaired t tests; *P < .001.
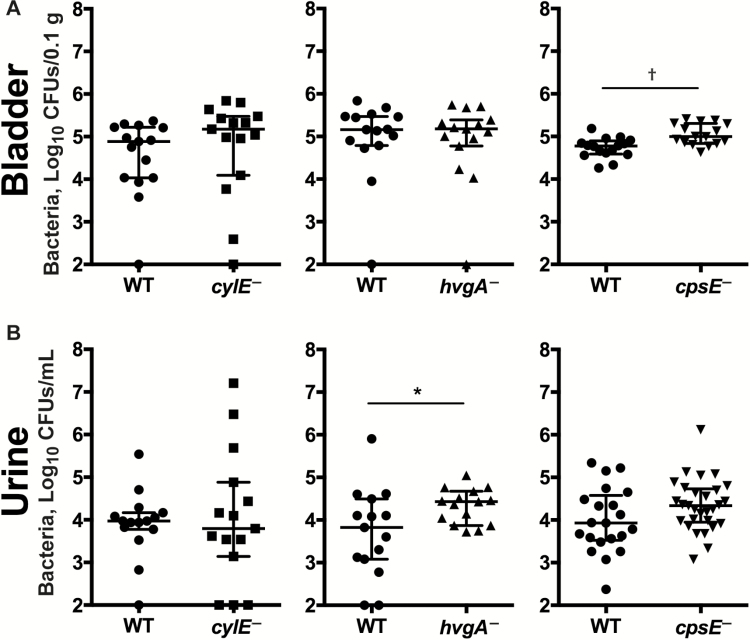
Owing to the contrasting expression levels of cylE and hvgA in the covR− background, we tested their role in colonization in mice, independent of CovR. We also examined cpsE despite the observed lack of regulation by CovR that contrasts with a prior report in a different S. agalactiae strain [27]. Compared with WT bacteria, the following findings were noted: (1) cylE-deficient S. agalactiae had similar median bacterial loads in the bladder (WT vs cylE−, 4.9 vs 5.2 log10 CFUs/0.1 g; IQR, 4.0–5.2 vs 4.1–5.5), and urine (WT vs cylE−, 4.0 vs 3.8 log10 CFUs/mL; IQR, 3.8–4.2 vs 3.1–4.9); (2) hvgA-deficient S. agalactiae had significantly higher loads in the urine (WT vs hvgA−, 3.8 vs 4.4 log10 CFUs/mL; IQR, 3.1–4.5 vs 3.9–4.7); and (3) cpsE-deficient S. agalactiae had significantly higher loads in the bladder (WT vs cpsE−, 4.8 vs 5.0 log10 CFUs/0.1 g; IQR, 4.6–4.9 vs 4.8–5.3) (Figure 7). Together, these data indicate that in S. agalactiae strain 874391, HvgA and capsule significantly impair colonization in the murine UTI model, in contrast to β-h/c, which has no significant effect.
Figure 7.
Effect of Streptococcus agalactiae cylE, hvgA, and cpsE on colonization of the bladder and urine in mice during experimental urinary tract infection. Bacterial colonization is shown for the bladder (A) and urine (B) in C57BL/6 mice at 24 hours after infection. Data are pooled from 2 independent experiments (medians shown for n = 15) and compared using Mann–Whitney U test; *P < .05; †P < .01. Bars represent medians with interquartile ranges. Abbreviations: CFUs, colony-forming units; WT, wild type.
DISCUSSION
This study establishes a major role for S. agalactiae CovR in UTI pathogenesis. The central findings are as follows: (1) CovR-regulated factors exacerbate bladder infection and influence local cytokine production in mice; (2) CovR-regulated factors affect human uroepithelial cell death; (3) covR deficiency increases the expression of β-h/c and HvgA in S. agalactiae 874391; (4) despite regulation of β-h/c by covR in S. agalactiae 874391, β-h/c does not significantly affect colonization of the urinary tract by this strain, in contrast to S. agalactiae strain 807; and (5) hvgA deficiency exacerbates S. agalactiae bacteriuria. Together, these findings support a complex role for CovR in UTI, probably due to its effects on many genes in S. agalactiae. Finally, the finding that capsule expression in S. agalactiae 874391 is unaffected by covR but impairs bladder colonization is consistent with a reported role of capsule in UTI pathogenesis [4].
The idea that S. agalactiae CovR affects the pathogenesis of UTI via complex mechanisms that involve multiple genes is supported by reports that CovR regulates a multitude of virulence factors, including adhesins (eg, HvgA, FbsA, and Rib/αC protein), cytotoxins (eg, β-h/c), immune subversion factors (eg, C5a peptidase) [12, 27], ABC transporters (Fhu/iron), and transcriptional regulators [14, 18]. CovR/CovS controls acid responses [28] and regulates strain-specific targets [13], which the current study confirms are important in S. agalactiae UTI based on a 2016 report of a role for β-h/c in UTI alongside strain-specific effects [6]. Significantly higher hemolytic activity in S. agalactiae strain 807 compared with strain 874391 (as shown in this study) might explain strain-specific effects related to the role of β-h/c in UTI. Another study showed that CovRS supports survival of S. agalactiae in macrophages [29], and a loss of regulation by CovR heightened chemokine responses during genital tract infection [30]. The attenuated phenotype for the covR− mutant in vivo is likely to reflect a complex interplay between multiple virulence factors in addition to β-h/c and HvgA; given that CovR regulates >150 genes [18, 31], future work to define the relationship between CovR and factors not examined in this study will be of interest.
In murine sepsis, CovR deficiency causes higher brain bacterial titers, perhaps owing to increased invasiveness [18], which is a pathogenesis strategy [32]. Other observations suggest that CovR deficiency reduces invasiveness for host cells, regardless of cell type: CovR-deficient S. agalactiae exhibited reduced invasion of brain endothelial and lung epithelial cells and altered inflammatory responses [18] and were less invasive (but more adherent) in human vaginal epithelial cells than WT [30]. The current findings, showing reduced invasiveness of CovR-deficient S. agalactiae on bladder cells are consistent with these prior reports and support a model of CovR deficiency exerting distinct effects in different host environments.
How could S. agalactiae use CovR deficiency as a pathogenesis strategy given that it is naturally present in S. agalactiae? Altering levels of expression of covR in vivo to alter virulence factor expression during disease could be one explanation. As suggested elsewhere, the CovR/CovS system is likely to be tightly regulated in space and time to adapt bacterial virulence capacity to the infected compartments of the host [33], and fine-tune gene expression for adaptation [34]. Reports of throat infection due to S. agalactiae with a natural CovR mutation [35], and covR mutations in strains transmitted vertically [36], highlight the clinical relevance of understanding the complex role of covR in S. agalactiae–mediated diseases.
S. agalactiae that was covR deficient induced multiple cytokines in human bladder uroepithelial cells as early as 3 hours after infection at significantly higher levels than induced by WT; the levels of several cytokines decreased over time, coincident with host cell death induced by covR-deficient S. agalactiae. This finding is consistent with the results of a model of genital tract infection, in which a loss of regulation by CovR heightened cytokine responses in vitro at 5 hours after infection [30]. Thus, CovR plays a crucial role in modulating epithelial barrier function and innate immune activation early after infection.
The finding that covR-deficient S. agalactiae activates caspase 3 in uroepithelial cells is consistent with findings in macrophages [37, 38] that undergo apoptosis via multiple mechanisms [19, 39–41]. Cell death may also reflect caspase 1–mediated pyroptosis [42–45]. However, rapid cellular LDH release after infection with the covR− mutant might also imply cell rupture by nonapoptotic mechanisms. Consistent with prior reports, our covR− mutant was hyperhemolytic, which amplified cytotoxicity. The amplified early cytokine response in uroepithelial cells induced by the covR− mutant may reflect increased β-h/c expression, because this toxin enhances cytokine production in bladder cells [5]. The β-h/c activates the nucleotide oligomerization domain (Nod)-like receptor (NLR) family, pyrin domain-containing 3 (NLRP3) inflammasome [46], and hyperhemolytic S. agalactiae∆covR activates NF-κB [47]. Importantly, complementation of the covR mutation restored phenotypes related to adhesion, invasion, cytotoxicity, hemolysis, and virulence gene expression, confirming the specificity of the effects related to CovR deficiency.
Comparing immune responses in mice with human uroepithelial cells revealed conserved responses for only a few cytokines, including IL-12(p70), GM-CSF, and MCP-1. Higher production of some factors in response to WT versus covR−S. agalactiae in mice differed from the in vitro response patterns of interleukin 6, and tumor necrosis factor α, which were induced at higher levels by the mutant. Our analysis of multiple time points in vitro enabled identification of responses that were induced in a CovR-dependent mechanism over time; this was reflected in cytokine production patterns at 24 hours after infection not observed at 6 hours. The more rapid effect of the covR− mutant on immune responses may be attributed to higher cytotoxicity and might explain the lower recovery of bacteria from mice at 24 hours after infection. Overall, these findings support the concept of immune activation and suppression in S. agalactiae UTI [3].
O-acetylation of S. agalactiae capsular sialic acid has been linked to UTI pathogenesis [4]. Interestingly, covR-deficient S. agalactiae exhibit an enlarged, fibrous extracellular matrix [27] that may include surface-anchored molecules, such as capsule. The first description of a CovRS homologous 2-component system in Streptococcus was that of CsrRS (capsule synthesis regulator) [48], which represses capsule synthesis in Streptococcus pyogenes. In covR−S. agalactiae, capsule is derepressed in a strain-dependent manner [12, 18, 27]. In this study, we analyzed cpsE expression and found no difference between covR− and WT S. agalactiae, consistent with previous studies using S. agalactiae strains 2603V/R and 515, but in contrast to strains A909 and NEM316 [12, 18]. Other covR-activated virulence factors that would be of interest for future UTI research include hyaluronate lyase, streptococcal histidine triad protein, and CAMP factor [18].
Our study has several limitations. First, the distinct cytokine response patterns observed in vitro and in vivo at 24 hours after infection probably reflect inherent differences between these models; for example, in species, in cell culture versus tissue complexity, and in host-pathogen interactions that can differ in vitro and in vivo [49]. We used uroepithelial cell monocultures at a single MOI; cocultures can better reflect host-pathogen interactions in some circumstances [49]; the relationship between the MOI used in vitro and the challenge dose used in vivo is unclear. We limited in vivo assays to 24 hours after infection, and it would be useful to study earlier time points that can offer added insight into the murine UTI model [20]. Differences in cell numbers in vivo and in vitro could be important and might reflect the death of uroepithelial cells in vitro, leading to bacteria being washed away during the assay, whereas bacteria in vivo are voided by micturition and attacked by innate immune responses that do not occur in a closed in vitro monoculture [3]. The benefits and limitations of in vitro and in vivo models are reviewed elsewhere [20, 49]. The current study should thus be considered alongside these limitations to avoid simplistic comparisons of data sets derived from 2 very distinct models.
We conclude that CovR enhances the ability of S. agalactiae to cause UTI by promoting bladder colonization, and by affecting local inflammation and adhesion to and viability of uroepithelial cells. CovR repression of hvgA in S. agalactiae partly explains the apparent complex role of CovR in promoting UTI. Additional work is now needed to explore the regulation of CovR during UTI.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank Harry Sakelaris for critical review of the manuscript.
Financial support. This work was supported by the NHMRC (project grants (APP1005315 and APP1084889 to G. C. U. and the Peter Doherty fellowship [APP1052464] to A. J. C.), the Griffith Health Institute, the Conselho Nacional de Desenvolvimento Científico e Tecnológico–Brazil (S. Y. L.), and the Australian Research Council (Future Fellowship FT110101048 to G. C. U.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ulett KB, Benjamin WH, Jr, Zhuo F, et al. Diversity of group B Streptococcus serotypes causing urinary tract infection in adults. J Clin Microbiol 2009; 47:2055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ulett GC, Webb RI, Ulett KB, et al. Group B Streptococcus (GBS) urinary tract infection involves binding of GBS to bladder uroepithelium and potent but GBS-specific induction of interleukin 1α. J Infect Dis 2010; 201:866–70. [DOI] [PubMed] [Google Scholar]
- 3. Tan CK, Carey AJ, Cui X, et al. Genome-wide mapping of cystitis due to Streptococcus agalactiae and Escherichia coli in mice identifies a unique bladder transcriptome that signifies pathogen-specific antimicrobial defense against urinary tract infection. Infect Immun 2012; 80:3145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kline KA, Schwartz DJ, Lewis WG, Hultgren SJ, Lewis AL. Immune activation and suppression by group B Streptococcus in a murine model of urinary tract infection. Infect Immun 2011; 79:3588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kulkarni R, Randis TM, Antala S, Wang A, Amaral FE, Ratner AJ. β-hemolysin/cytolysin of group B Streptococcus enhances host inflammation but is dispensable for establishment of urinary tract infection. PLoS One 2013; 8:e59091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leclercq SY, Sullivan MJ, Ipe DS, Smith JP, Cripps AW, Ulett GC. Pathogenesis of Streptococcus urinary tract infection depends on bacterial strain and β-hemolysin/cytolysin that mediates cytotoxicity, cytokine synthesis, inflammation and virulence. Sci Rep 2016; 6:29000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kline KA, Schwartz DJ, Gilbert NM, Hultgren SJ, Lewis AL. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection by uropathogenic Escherichia coli. Infect Immun 2012; 80:4186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hunstad DA, Justice SS. Intracellular lifestyles and immune evasion strategies of uropathogenic Escherichia coli. Annu Rev Microbiol 2010; 64:203–21. [DOI] [PubMed] [Google Scholar]
- 9. Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, Schembri MA. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol 2013; 16:100–107. [DOI] [PubMed] [Google Scholar]
- 10. Sivick KE, Mobley HL. Waging war against uropathogenic Escherichia coli: winning back the urinary tract. Infect Immun 2010; 78:568–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ragnarsdottir B, Lutay N, Gronberg-Hernandez J, Koves B, Svanborg C. Genetics of innate immunity and UTI susceptibility. Nat Rev Urol 2011; 8:449–68. [DOI] [PubMed] [Google Scholar]
- 12. Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in group B Streptococcus. J Bacteriol 2005; 187:1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang SM, Ishmael N, Dunning Hotopp J, et al. Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J Bacteriol 2008; 190:1956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rajagopal L, Vo A, Silvestroni A, Rubens CE. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol Microbiol 2006; 62:941–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tazi A, Disson O, Bellais S, et al. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med 2010; 207:2313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landwehr-Kenzel S, Henneke P. Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front Immunol 2014; 5:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teatero S, McGeer A, Low DE, et al. Characterization of invasive group B Streptococcus strains from the greater Toronto area, Canada. J Clin Microbiol 2014; 52:1441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lembo A, Gurney MA, Burnside K, et al. Regulation of CovR expression in group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol 2010; 77:431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ulett GC, Bohnsack JF, Armstrong J, Adderson EE. β-Hemolysin-independent induction of apoptosis of macrophages infected with serotype III group B Streptococcus. J Infect Dis 2003; 188:1049–53. [DOI] [PubMed] [Google Scholar]
- 20. Carey AJ, Tan CK, Ipe DS, et al. Urinary tract infection of mice to model human disease: practicalities, implications and limitations. Crit Rev Microbiol 2016; 42:780–99. [DOI] [PubMed] [Google Scholar]
- 21. Chattopadhyay D, Carey AJ, Caliot E, et al. Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of group B Streptococcus. Microbes Infect 2011; 13:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ipe DS, Ben Zakour NL, Sullivan MJ, et al. Discovery and characterization of human-urine utilization by asymptomatic-bacteriuria-causing Streptococcus agalactiae. Infect Immun 2015; 84:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nizet V, Gibson RL, Chi EY, Framson PE, Hulse M, Rubens CE. Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect Immun 1996; 64:3818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611–22. [DOI] [PubMed] [Google Scholar]
- 26. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lamy MC, Zouine M, Fert J, et al. CovS/CovR of group B Streptococcus: a two-component global regulatory system involved in virulence. Mol Microbiol 2004; 54:1250–68. [DOI] [PubMed] [Google Scholar]
- 28. Santi I, Grifantini R, Jiang SM, et al. CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J Bacteriol 2009; 191:5387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cumley NJ, Smith LM, Anthony M, May RC. The CovS/CovR acid response regulator is required for intracellular survival of group B Streptococcus in macrophages. Infect Immun 2012; 80:1650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patras KA, Wang NY, Fletcher EM, et al. Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol 2013; 15:1154–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Di Palo B, Rippa V, Santi I, et al. Adaptive response of group B Streptococcus to high glucose conditions: new insights on the CovRS regulation network. PLoS One 2013; 8:e61294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dando SJ, Mackay-Sim A, Norton R, et al. Pathogens penetrating the central nervous system: infection pathways and the cellular and molecular mechanisms of invasion. Clin Microbiol Rev 2014; 27:691–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Firon A, Tazi A, Da Cunha V, et al. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog 2013; 9:e1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin WJ, Walthers D, Connelly JE, et al. Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Mol Microbiol 2009; 71:1477–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Whidbey C, Burnside K, Martinez RM, et al. A hyperhemolytic/hyperpigmented group B Streptococcus strain with a CovR mutation isolated from an adolescent patient with sore throat. Clin Res Infect Dis 2015; 2:1018. [PMC free article] [PubMed] [Google Scholar]
- 36. Almeida A, Villain A, Joubrel C, et al. Whole-genome comparison uncovers genomic mutations between group B streptococci sampled from infected newborns and their mothers. J Bacteriol 2015; 197:3354–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ulett GC, Adderson EE. Regulation of apoptosis by gram-positive bacteria: mechanistic diversity and consequences for immunity. Curr Immunol Rev 2006; 2:119–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulett GC, Maclean KH, Nekkalapu S, Cleveland JL, Adderson EE. Mechanisms of group B streptococcal-induced apoptosis of murine macrophages. J Immunol 2005; 175:2555–62. [DOI] [PubMed] [Google Scholar]
- 39. Ulett GC, Adderson EE. Nitric oxide is a key determinant of group B Streptococcus-induced murine macrophage apoptosis. J Infect Dis 2005; 191:1761–70. [DOI] [PubMed] [Google Scholar]
- 40. Liu GY, Doran KS, Lawrence T, et al. Sword and shield: linked group B streptococcal β-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc Natl Acad Sci U S A 2004; 101:14491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buratta S, Fettucciari K, Mambrini R, Fetriconi I, Marconi P, Mozzi R. Group B Streptococcus (GBS) modifies macrophage phosphatidylserine metabolism during induction of apoptosis. FEBS Lett 2002; 520:68–72. [DOI] [PubMed] [Google Scholar]
- 42. Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev 2011; 243:206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miao EA, Leaf IA, Treuting PM, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 2010; 11:1136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009; 7:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Croker BA, O’Donnell JA, Gerlic M. Pyroptotic death storms and cytopenia. Curr Opin Immunol 2014; 26:128–37. [DOI] [PubMed] [Google Scholar]
- 46. Costa A, Gupta R, Signorino G, et al. Activation of the NLRP3 inflammasome by group B streptococci. J Immunol 2012; 188:1953–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whidbey C, Harrell MI, Burnside K, et al. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 2013; 210:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol 1998; 30:209–19. [DOI] [PubMed] [Google Scholar]
- 49. Duell BL, Cripps AW, Schembri MA, Ulett GC. Epithelial cell coculture models for studying infectious diseases: benefits and limitations. J Biomed Biotechnol 2011; 2011:852419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zundler S, Neurath MF. Interleukin-12: functional activities and implications for disease. Cytokine Growth Factor Rev 2015; 26:559–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.