Abstract
Chlamydia is an obligate intracellular bacterium that relies on host cells for essential nutrients and adenosine triphosphate (ATP) for a productive infection. Although the unfolded protein response (UPR) plays a major role in certain microbial infectivity, its role in chlamydial pathogenesis is unknown. We hypothesized that Chlamydia induces UPR and exploits it to upregulate host cell uptake and metabolism of glucose, production of ATP, phospholipids, and other molecules required for its replicative development and host survival. Using a combination of biochemical and pathway inhibition assays, we showed that the 3 UPR pathway transducers—protein kinase RNA-activated (PKR)-like ER kinase (PERK), inositol-requiring enzyme-1α (IRE1α), and activating transcription factor-6α (ATF6α)—were activated during Chlamydia infection. The kinase activity of PERK and ribonuclease (RNase) of IRE1α mediated the upregulation of hexokinase II and production of ATP via substrate-level phosphorylation. In addition, the activation of PERK and IRE1α promoted autophagy formation and apoptosis resistance for host survival. Moreover, the activation of IRE1α resulted in the generation of spliced X-box binding protein 1 (sXBP1) and upregulation of lipid production. The vital role of UPR pathways in Chlamydia development and pathogenesis could lead to the identification of potential molecular targets for therapeutics against Chlamydia.
Keywords: Chlamydia, pathogenesis, unfolded protein response
Genital infection of women by the obligate intracellular bacteria of the genus Chlamydia causes complications such as pelvic inflammatory disease, ectopic pregnancy, and tubal-factor infertility [1]. The 1.4 million reported cases in the United States in 2014 was a 2.8% increase over 2013, despite awareness programs. The rising infections with attendant cost constitute a major public health concern, requiring a better understanding of Chlamydia pathogenesis, and the design of an efficacious, safe human vaccine. Chlamydial pathogenesis requires the productive infection of the host cells and host’s pathologic inflammatory responses. As an energy and nutrient parasite, lacking key enzymes of glucose metabolic pathways, including hexokinase, and essential phospholipids and envelope lipid components (ie, phosphatidylcholine [2], cholesterol, and sphingomyelin [2–5]), Chlamydia relies on the host cell for a productive infection. Chlamydial inclusion membrane proteins include transporters of ATP, lipids, and nucleotides that mediate a unidirectional nutrient flow into the inclusion. Interestingly, chlamydial-infected cells upregulate expression of glucose transporters, ATP production via substrate-level phosphorylation with higher pyruvate and its derivatives [6], and phospholipids synthesis [7]. Thus, Chlamydia has evolved to alter the host’s metabolic pathways to favor its energy and nutrient needs, although the biochemical processes and mechanisms were unknown.
Unfolded protein response (UPR) is a cellular response to pathological and physiological perturbation of the protein folding machinery of the endoplasmic reticulum (ER) resulting in a condition called ER stress [8]. The hallmark of UPR is the activation of 3 major signaling pathways enzymes—protein kinase RNA-activated (PKR)-like ER kinase (PERK), inositol-requiring enzyme-1α (IRE1α), and activating transcription factor-6α (ATF6α)—aimed at restoration of cellular homeostasis or activation of apoptosis if homeostasis cannot be restored [9]. Under normal physiological conditions, the 3 transducers of UPR (PERK, IRE1α, and ATF6α) are kept inactive by binding to glucose-regulated protein (GRP78), also called immunoglobulin heavy-chain binding protein (BiP), on their luminal domains [10]. However, during ER stress, the excess unfolded/misfolded proteins in the ER lumen are bound to BiP, thus dissociating and titrating it away from PERK, IRE1α, and ATF6α, hence their activation [11]. Activated PERK phosphorylates and inactivates the eukaryotic initiation factor 2-α (eIF2α) [12], causing a general cellular translation shutdown that lowers the amount of nascent peptides and proteins accumulating in the ER [13]. However, select open-reading frames that are preferentially translated under this condition include activating transcription factor 4 (ATF4) [13] and C/EBP homologous protein (CHOP) [14], transcription factors that activate genes involved in restoration of homeostasis or apoptosis [15]. PERK is activated in some neurodegenerative diseases, diabetes, tumors, and during interferon-γ (IFN-γ)–induced persistent chlamydial infection [16], and plays a role in energy and antioxidant production [6, 17]. Activation of IRE1α results in its acquisition of kinase and endonuclease activities [18]. The endonuclease activity specifically splices the messenger RNA (mRNA) of XBP1, excising a 26-base pair (bp) fragment to generate a potent transcription factor sXBP1 [19] that downregulates secretory proteins [20] but upregulates enzymes in the lipid biosynthesis pathway [21]. Also, the IRE1α/TRAF2 complex formed between activated IRE1α and tumor necrosis factor (TNF)–α receptor-associated factor 2 (TRAF2) [22] activates c-Jun to promote cell survival through upregulation of the microtubule-associated protein 1 light chain 3 (MAP-LC3) [23] and downregulation of tumor suppressors such as the phosphatase and tensin homolog (PTEN) [24]. Activation of ATF6α, a 90-kDa transmembrane basic leucine-zipper (bZIP) transcription factor, unmasks its Golgi localization signal [25, 26] and its translocation to the Golgi apparatus where it is cleaved by proteases to the active form [26] that upregulates chaperone proteins, such as BiP and 58-kDa inhibitor protein kinase (p58IPK), which increase the ER folding capacity to restore homeostasis [27]. Activated ATF6α also inhibits the PERK through its upregulation of p58IPK that reduces the phosphorylation of eIF2α and the expression of CHOP [28, 29]. The blocking of CHOP, a proapoptotic molecule, ensures the survival of ER-stressed cells [30]. Mutation in the ATF6 gene, causing a loss of the inhibitory function on the PERK, is associated with photoreceptor degeneration disease [31] and achromatopsia [32].
In addition to chemicals and nutrient-deprivation conditions, certain microbial agents, including viruses and bacteria, induce UPR [33], and it may be required for Brucella replication [34]. A potential role for UPR in chlamydial pathogenesis is suggested by its ability to enhance ATP via substrate-level phosphorylation and provide other metabolites in rapidly dividing cancer cells [17]. Also, UPR may provide the conditions for resistance of the host cell to apoptosis [35]. We tested the hypothesis that Chlamydia activates the major UPR signaling pathways the sources of the extra host energy, metabolites, and essential lipids parasitized for its development and initiation of disease [2]. Our results show that the 3 transducers of UPR (PERK, IRE1α, and ATF6α) are activated during Chlamydia infection, and they are required for replication and complications. Thus, targeting host UPR machinery could be an effective means of treating/preventing Chlamydia infection and its complications.
MATERIAL AND METHODS
Chlamydia Stocks, Mammalian Cells, and Mice
Chlamydia muridarum (the agent of mouse pneumonitis, MoPn) used in this study was propagated in McCoy cells and purified elementary bodies (EBs) were tittered as infectious-forming units per millimeter (IFU/mL) using standard procedures described previously [36]. Mammalian cells were mouse oviduct epithelial cells (C57epi.1) kindly provided by Dr Raymond Johnson (Indiana Univ., Indianapolis, IN). For in vitro studies, cells were maintained in epithelial cell medium (1:1 Dulbecco’s modified Eagle medium-F12K; Sigma, St. Louis, MO), supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT) and 2 mM L-glutamine (Glutamax I; Gibco/Invitrogen), and infection was in sucrose phosphate glutamate medium as previously described [37]. Female wild-type C57BL/6Ntac mice 5–8 weeks old were obtained from Taconic Farms, Inc. (Hudson, NY). Mice were housed at the Centers for Disease Control and Prevention (CDC) animal facility and fed with food and water ad libitum. A 12-hour light and a 12-hour dark cycle was maintained throughout the duration of the experiment. Five days prior to infection with Chlamydia, mice received subcutaneous injection of 2.5 μg/mouse Depo Provera (medroxy-progesterone acetate; Pfizer Inc, New York, NY). Mice were infected with 20 μl Chlamydia intravaginally at 105 IFU/mouse in phosphate-buffered saline while under the long-acting anesthetic sodium pentobarbital (30 μg/body weight) (Sigma-Aldrich, St Louis, MO). Productive infection was monitored by isolating and counting Chlamydia (IFU/mL) from genital swabs obtained from infected mice. Cell culture experiments were repeated at least 3 times, while animal experiments were repeated twice. All animal protocols were approved by the CDC Institutional Animal Care and Use Committee (IACUC).
UPR Pathways Inhibition Assays
In vitro studies to analyze the effect of UPR pathways inhibition on Chlamydia replication was performed by incorporating 20 μM PERK inhibitor (GSK2606414, Tocris Bioscience, Bristol, United Kingdom), or 120 μM IRE1α inhibitor (4 μ8C, Axon Medchem LLC, Reston, VA), or equal amount of dimethylsulphoxide (DMSO) solution (0.08% v/v) that was used in preparing each inhibitor in the infection medium. Briefly, 1.0 × 106 or 1.4 × 105 cells/well were seeded into 6- or 24-well plates, respectively, and infected with MoPn at a multiplicity of infection (MOI) of 1 or 5 for 24, 48, and 72 hours. Inhibitors for PERK kinase and IRE1α-RNase activities were simultaneously added during infection.
To determine the effect of UPR pathways inhibition on Chlamydia replication in vivo, 24 hours after infection with Chlamydia, the UPR inhibition experimental group was treated intravaginally with 20 μM PERK kinase inhibitor or 120 μM IRE1α RNase inhibitor, or an equal amount of DMSO solution that was used in preparing each inhibitor.
Western Blot Analysis
Total protein was prepared from cells by lysing in situ with M-PER Mammalian Protein Extraction Reagent (Thermo Scientific, Rockford, IL) supplemented with ethylenediaminetetraacetic acid-free protease inhibitor cocktail according to manufacturer’s instruction. Briefly, 1 mL of protein extraction reagent supplemented with 1× protease inhibitor was added to 107 cells and mixed gently for 5 minutes. Lysed cells were aspirated into a 15 mL tube and vortexed for 30 seconds. Protein concentration was determined using the Bradford method. Equal amounts of protein were loaded onto 4–20% PROTEAN TGX protein gel (Bio-Rad, Hercules, CA) and electrophoresed for 1 hour, and the separated bands were transferred onto nitrocellulose membrane (Bio-Rad). Membranes were washed, blocked with 5% casein or 5% nonfat milk or 5% bovine serum albumin for 1 hour, and incubated with primary antibody overnight at 4°C. The primary antibodies used recognized epitopes in the following proteins: phosphorylated-IRE1α and ATF6α (Novus-Bio, Littleton, CO), phosphorylated-PERK, phosphorylated-eIF2α, sXBP1, phosphorylated-c-Jun, ATF4, hexokinase II and glucose transporter 1 (Cell Signaling Technology, Beverly, MA); MAP-LC3β, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Inc, Dallas, TX). Washed membranes were incubated with horseradish peroxidase-conjugated goat antimouse or antirabbit immunoglobulin G secondary antibody for 1 hour, then analyzed using a ChemiDoc XRS+ system (Bio-Rad). Band intensity analysis was performed using Image Lab software.
Reverse Transcription–Polymerase Chain Reaction and Normal Polymerase Chain Reaction Analysis
Total RNA was isolated from cell cultures using RNeasy Plus Mini Kit (Qiagen, Germantown, MD) according to manufacturer’s instruction. All isolated RNAs were stored at −80°C until used. According to manufacturer’s instructions, reverse transcription–polymerase chain reaction (RT-PCR) was performed using RT2 First Strand Kit to reverse transcribe the isolated RNA into complementary DNA (cDNA). Primers for the detection of spliced and unspliced XBP1 and GAPDH mRNA were designed using National Center for Biotechnology Information Primer-Blast software. The primer pair selected for spliced and unspliced XBP1 mRNA were the same but their PCR product were 179-bp and 205-bp, respectively. The PCR product for GAPDH primer pair was 450-bp. The XBP1 primer pair used was as follows: Forward (TM 56.58°C; % GC 47.62): 5’- GAACCAGGAGTTAAGAACACG -3’; Reverse (59.61; % GC 55.00): 5’ – AGGCAACAGTGTCAGAGTCC -3’. For synthesis of cDNA, the concentration of RNA used was 1 μg per reaction mix of 50 μl while a 50 μl reaction mix for PCR contained 0.5 μg of cDNA. Genomic DNA was eliminated from RNA preparations using the genomic DNA elimination reagent of the RNeasy Plus Mini Kit. The analysis included the identification of a spliced RNA that is the conversion of a 205-bp to 179-bp band.
ATP and Lipid Assays
The ATP content of noninfected and Chlamydia-infected cells were determined using the ATP Colorimetric/Fluorometric Assay Kit (Bio Vision, Milpitas, CA) according manufacturer’s instruction. Briefly, cells were lysed in appropriate buffer and deproteinized. Lysed cells were centrifuged at 12 × g for 20 min and the supernatant was mixed with glycerol containing reagents to generate a product that was quantified using SPECTRAmax PLUS384 (Sunnyvale, CA).
The lipid content of cell cultures were measured using Phospholipid Assay Kit (Sigma-Aldrich Co. LLC, St. Louis, MO) according to manufacturer’s instructions.
Statistical Analysis
Statistical analyses were performed with SigmaPlot software. The data derived from different experiments were analyzed and compared by performing a 1- or 2-tailed t test, and the relationship between different experimental groupings was assessed by analysis of variance. Statistical significance was judged at P < .05.
RESULTS
Induction of the Three Major Signaling Pathways of UPR During Chlamydial Infection
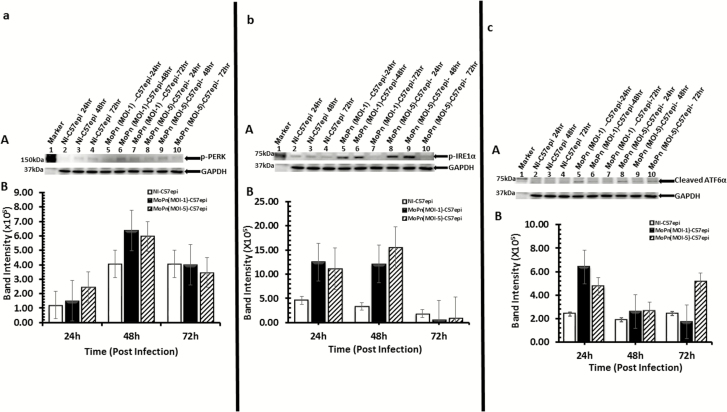
In our initial investigation of the possibility that Chlamydia may exploit UPR for its energy and nutrient requirements, we undertook rigorous experimental validations to establish that Chlamydia induces UPR. First, we measured the levels of activated IRE1α, PERK, and ATF6α representing the 3 major pathways of UPR following infection of the murine oviduct epithelial cells (C57epi) with C. muridarum (MoPn). Monolayers of the C57epi cells were infected with MoPn at different MOIs, and cultures were harvested at 24, 48, and 72 hours. Results presented in Figure 1A–C show the activation of PERK, IRE1α, and ATF6α at different times postinfection. The activation of UPR in infected cells occurred within 24 and 48 hours, the period of active replication and inclusion development.
Figure 1.
C57epi cells infected with C. muridarum induced the activation of the UPR transducers [PERK (1A), IRE1α (1B) and ATF6α (1C)]. A, Ten μg total protein from cells infected with C. muridarum (MoPn) for 24, 48, and 72 hours were prepared for Western blot analysis. Blot was probed with primary antibody against phosphorylated PERK or phosphorylated IRE1α or ATF6α. Lane 1: protein molecular weight marker. Lanes 2–4: samples from noninfected C57epi cells at 24, 48, and 72 hours. Lanes 5–7: samples from C57epi cells infected with C. muridarum (MOI 1) at 24, 48, and 72 hours. Lanes 8–10: samples from C57epi cells infected with C. muridarum (MOI 5) at 24, 48, and 72 hours. B, Bar chart of phosphorylated PERK (A), phosphorylated IRE1α (B), and ATF6α (C) intensity, showing the level of activation at the different MOIs and times postinfection (white bars: noninfected; black bars: MoPn [MOI 1] infected; hatched bars: MoPn [MOI 5] infected). Abbreviations: ATF6α, activating transcription factor-6α; C57epi cells, mouse oviduct epithelial cells; IRE1α, inositol-requiring enzyme-1α; MOI, multiplicity of infection; MoPn, mouse pneumonitis; NI, noninfected; PERK, protein kinase RNA-activated–like ER kinase; UPR, unfolded protein response.
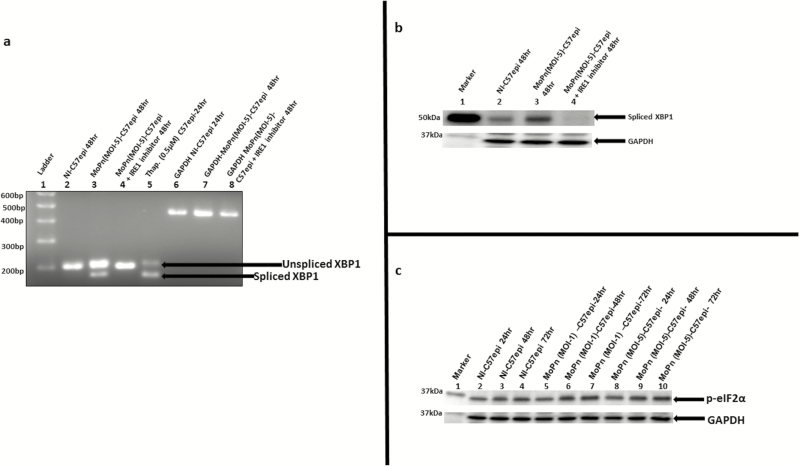
A key confirmation of UPR is the acquisition of the endonuclease activity by IRE1α, resulting in the splicing of XBP1 mRNA and upregulation of spliced XBP1 protein. The splicing of XBP1 mRNA was investigated using a combination of reverse transcription and conventional PCR approaches. The PCR primers used in the amplification of XBP1 gene was adopted from [38]. The primers were designed such that the unspliced mRNA of XBP1 will give a PCR product of 205-bp, while the spliced mRNA will give a PCR product of 179-bp. As shown in Figure 2A, the infection of C57epi cells with MoPn resulted in the splicing of XBP1 mRNA, and the inhibition of the RNase activity of activated IRE1α abrogated the splicing of XBP1. Also, at the protein level, Figure 2B shows that MoPn-infected C57epi cells showed the upregulation of XBP1 protein, and the inhibition of IRE1α RNase activity resulted in downregulation of XBP1. In addition, a confirmatory enzymatic activity that is the hallmark of UPR is the phosphorylation of eIF2α by PERK along a second signaling pathway of UPR [13]. Figure 2C shows that chlamydial infection resulted in the upregulation of phosphorylated eIF2α. We conclude that Chlamydia induces UPR, which may be exploited for energy and nutrients requirements during a productive infection.
Figure 2.
A, The mRNA of XBP1 is spliced by the endonuclease activity of IRE1α. Total RNA was extracted from C57epi cells cultured in various experimental conditions. Primer pair (forward: 5’-GAACCAGGAGTTAAGAACACG -3’; reverse: 5’AGGCAACAGTGTCAGAGTCC -3’) was used in PCR amplification. The DNA template was the complementary DNA reverse transcribed from the RNA extracted from each culture condition. The PCR product from each culture condition was ran on 2% agarose gel and stained with ethidium bromide. Lane 1: molecular weight marker. Lane 2: noninfected C57epi cells cultured for 48 hours showing the presence of unspliced 205-bp band used as negative control. Lane 3: C57epi cells infected with C. muridarum (MoPn) at MOI 5 for 48 hours showing the presence of unspliced 205-bp band as well as spliced 179-bp band. Lane 4: C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of IRE1α RNase activity for 48 hours showing the presence of unspliced 205-bp band. Lane 5: Noninfected C57epi cells treated with thapsigargin (chemical inducer of UPR) cultured for 48 hours showing the presence of unspliced 205-bp band as well as spliced 179-bp band used as positive control. Lanes 6–8: PCR amplification of GAPDH gene products used as loading control for noninfected C57epi cells infected with MoPn at MOI 5 and C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of IRE1α RNase activity for 48 hours showing the presence of unspliced 205-bp band. B, C57epi cells infected with C. muridarum induced the upregulation of XBP1 protein and inhibition of IRE1α RNase activity results in the downregulation of XBP1. Ten μg total protein from C57epi cells uninfected/infected with C. muridarum for 48 hours and treated/nontreated with inhibitor of IRE1α RNase activity were prepared for Western blot analysis. Blot was probed with primary antibody against spliced XBP1. Lane 1: protein molecular weight marker. Lane 2 sample from noninfected C57epi cells at 48 hours. Lane 3: sample from C57epi cells infected with MoPn at MOI 5 for 48 hours. Lane 4: sample from C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of IRE1α RNase activity for 48 hours. C, Western blot of eIF2α phosphorylation during Chlamydia infection. Ten μg total protein from C57epi cells infected with C. muridarum (MoPn) for 24, 48, and 72 hours were prepared for Western blot analysis. Blot was probed with primary antibody against phosphorylated eIF2α. Lane 1: protein molecular weight marker. Lanes 2–4: samples from noninfected C57epi cells at 24, 48, and 72 hours. Lanes 5–7: samples from C57epi cells infected with C. muridarum (MOI 1) at 24, 48, and 72 hours. Lanes 8–10: samples from C57epi cells infected with C. muridarum (MOI 5) at 24, 48, and 72 hours. Abbreviations: bp, base pairs; C57epi cells, mouse oviduct epithelial cells; eIF2α, eukaryotic initiation factor 2-α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRE1α, inositol-requiring enzyme-1α; MOI, multiplicity of infection; MoPn, mouse pneumonitis; mRNA, messenger RNA; NI, noninfected; PCR, polymerase chain reaction; RNase, ribonuclease; Thap, thapsigargin; XBP1, X-box binding protein 1.
UPR Signaling Pathways as a Source of Extracellular Host Energy During Chlamydia Infection
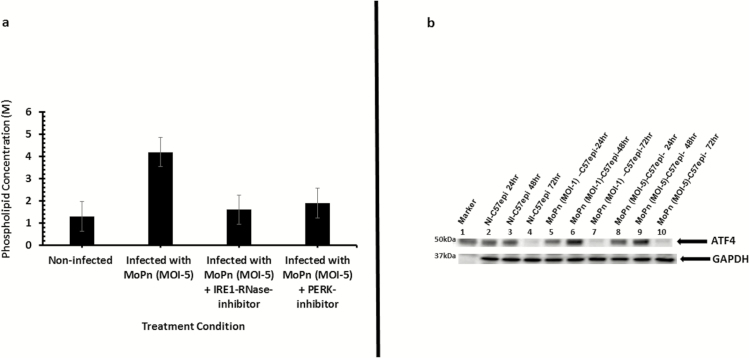
Chlamydial infection is characterized by a burst of supplementary host cell ATP production that is fostered by an upregulation of glucose transporters [7]. We tested the hypothesis that the substrate-level phosphorylation ATP generation step of the PERK pathway of UPR is exploited by chlamydia to promote its energy and nutrient parasitism, which supports the intracellular inclusion development. Because hexokinase II is upregulated as a crucial enzyme in the glycolytic pathway for rapid ATP production during substrate-level phosphorylation [39], we measured its expression as a marker of host cell energy boost during chlamydial infection. As shown in Figure 3A, there was an upregulation of hexokinase II during chlamydial infection. The upregulation of hexokinase II depended on the activation of PERK and IRE1α arms of the UPR because the inhibition of PERK kinase or IRE1α RNase activity resulted in the downregulation of hexokinase II (Figure 3A and 3B). Also, cellular glucose transporter-1 (Glut1) was upregulated during chlamydial infection, and the inhibition of the IRE1α RNase pathway resulted in its downregulation (Figure 3B). The results suggested that UPR increases host cell glucose utilization and ATP synthesis by substrate-level phosphorylation during chlamydial infection.
Figure 3.
A, Images of Western blot analysis of hexokinase II expression during Chlamydia infection. Ten μg total protein from C57epi cells uninfected/infected with C. muridarum (MoPn) for 48 hours and treated/nontreated with inhibitor of PERK kinase activity were prepared for Western blot analysis. Blot was probed with primary antibody against hexokinase II. Lane 1: protein molecular weight marker. Lane 2: sample from noninfected C57epi cells at 48 hours. Lane 3: sample from C57epi cells infected with MoPn at MOI 5 for 48 hours. Lane 4: sample from C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of PERK kinase activity for 48 hours. B, Images of Western blot analysis of Glut1 and hexokinase II expression during Chlamydia infection. C57epi cells infected with C. muridarum (MoPn) induced the upregulation of key proteins in the IRE1α arm of UPR pathways, including Glut1 and hexokinase II, and the inhibition of IRE1α RNase activity, resulting in their downregulation. Ten μg total protein from C57epi cells uninfected/infected with C. muridarum for 48 hours and treated/nontreated with inhibitor of IRE1α RNase activity were prepared for Western blot analysis. Blot was probed with primary antibody against Glut1 or hexokinase II. Lane 1: protein molecular weight marker. Lane 2 sample from noninfected C57epi cells at 48 hours. Lane 3: sample from C57epi cells infected with MoPn at MOI 5 for 48 hours. Lane 4: sample from C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of IRE1α RNase activity for 48 hours. Abbreviations: C57epi cells, mouse oviduct epithelial cells; Glut1, glucose transporter-1; IRE1α, inositol-requiring enzyme-1α; MOI, multiplicity of infection; MoPn, mouse pneumonitis; NI, noninfected; PERK, protein kinase RNA-activated–like ER kinase; RNase, ribonuclease; UPR, unfolded protein response.
UPR Signaling Pathways as a Source of Extra Host Nutrients Parasitized by Chlamydia
The endonuclease activity of activated IRE1α generates a potent transcription factor sXBP1 [19] that upregulates enzymes in the lipid biosynthesis pathway [21, 40]. Chlamydia acquire certain essential phospholipids such as phosphatidylcholine [2], cholesterol, and sphingomyelin [3] from host cells because it cannot make them [41]. We hypothesized that activation of the IRE1α pathway of UPR will benefit chlamydia as a source of essential lipids. To verify this hypothesis, the upregulation of phospholipids in C57epi cell cultures infected with chlamydia was analyzed. As shown in Figure 4A, the infection of C57epi cells significantly upregulated phospholipids production at 48 hours (P > .005). In addition, we analyzed the expression levels of ATF4, a transcription factor that activates genes involved in amino acid import, glutathione biosynthesis, and resistance to oxidative stress [15].The results showed transcription factor ATF4 was upregulated during infection (Figure 4B). The activation of ATF4 peaked at 48 hours, which corresponds to the time Chlamydia is completing its developmental cycle [42]. The results indicated that UPR is a major source of essential lipids and nutrients for chlamydia development.
Figure 4.
A, Bar chart of lipid content of C. muridarum MoPn infected and noninfected C57epi cells. The infection of C57epi cells with MoPn (MOI 5) resulted in phospholipid upregulation as measured by the increase in choline content of infected and noninfected cells. B, Western blot analysis of ATF4 activation during Chlamydia infection. Ten μg total protein from C57epi cells infected with C. muridarum (MoPn) for 24, 48, and 72 hours were prepared for Western blot analysis. Blot was probed with primary antibody against ATF4. Lane 1: protein molecular weight marker; Lanes 2–4: samples from noninfected C57epi cells at 24, 48, and 72 hours. Lanes 5–7: samples from C57epi cells infected with C. muridarum (MOI 1) at 24, 48, and 72 hours. Lanes 8–10: samples from C57epi cells infected with C. muridarum (MOI 5) at 24, 48, and 72 hours. Abbreviations: ATF4, activating transcription factor 4; C57epi cells, mouse oviduct epithelial cells; MOI, multiplicity of infection; MoPn, mouse pneumonitis; NI, noninfected.
UPR Signaling Pathways Produce Molecules That Prevent Apoptosis and Enhance Host Cell Survival
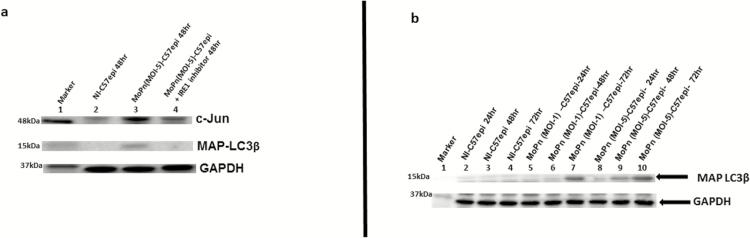
The mechanism by which chlamydial infection causes host cell resistance to apoptosis [35] (which allows for the initial host survival to ensure inclusion development) is unknown. We investigated the hypothesis that UPR produces key cell survival molecules during chlamydial infection by activation of the c-Jun N-terminal kinase pathway during chlamydial infection. As shown in Figure 5A, c-Jun, a component of the activating protein 1 (AP1) family of transcription factors that are antiapoptotic [24], and MAP-LC3, which promotes autophagy [23] to eliminate cell wastes as a cell survival strategy, were highly upregulated during chlamydial infection. The c-Jun and MAP-LC3 were produced in the IRE1α arm of UPR, as shown by the ability of IRE1α inhibitor to block their upregulation during chlamydial infection (Figure 5A and 5B). Thus, UPR induced during chlamydial infection contributes to the antiapoptotic phenotype of infected cells.
Figure 5.
A, Images of Western blot analysis of c-Jun and MAP-LC3β expression during Chlamydial infection. C57epi cells infected with C. muridarum (MoPn) induced the upregulation of key proteins in the UPR pathways, including c-Jun and MAP-LC3β and inhibition of IRE1 RNase activity result in their downregulation. Ten μg total protein from C57epi cells uninfected/infected with C. muridarum for 48 hours and treated/nontreated with inhibitor of IRE1α RNase activity were prepared for Western blot analysis. Blot was probed with primary antibody against c-Jun, Glut1, hexokinase II, and MAP-LC3β. Lane 1: protein molecular weight marker. Lane 2: sample from noninfected C57epi cells at 48 hours. Lane 3: sample from C57epi cells infected with MoPn at MOI 5 for 48 hours; Lane 4: Sample from C57epi cells infected with MoPn at MOI 5 in the presence of inhibitor of IRE1α RNase activity for 48 hours. B, Western blot of MAP-LC3β. Ten μg total protein from cells infected with C. muridarum (MoPn) for 24, 48, and 72 hours were prepared for Western blot analysis. Blot was probed with primary antibody against MAP-LC3β. Lane 1: protein molecular weight marker. Lanes 2–4: samples from noninfected C57epi cells at 24, 48, and 72 hours. Lanes 5–7: samples from C57epi cells infected with C. muridarum (MOI 1) at 24, 48, and 72 hours. Lanes 8–10: samples from C57epi cells infected with C. muridarum (MOI 5) at 24, 48, and 72 hours. Abbreviations: C57epi cells, mouse oviduct epithelial cells; Glut1, glucose transporter-1; IRE1α, inositol-requiring enzyme-1α; MAP-LC3β, microtubule-associated protein 1 light chain 3; MOI, multiplicity of infection; MoPn, mouse pneumonitis; NI, noninfected; RNase, ribonuclease.
The Role of UPR in Chlamydia Replication and Disease
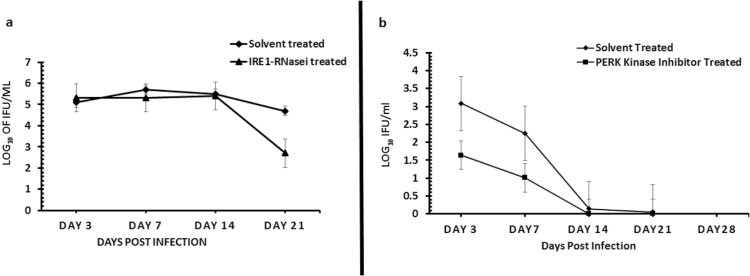
To directly investigate the role of UPR activation in Chlamydia pathogenesis, we tested the hypothesis that the blockade of the key signaling pathways of UPR will affect Chlamydia replication in vitro and in vivo. As shown in Figure 6, the inhibition of IRE1α RNase activity or PERK kinase activity significantly inhibits the formation Chlamydia inclusions in cultures of C57epi cells (P > .03 and .04, respectively). In vivo, the inhibition of IRE1α RNase activity or PERK kinase activity by intravaginal application of inhibitors to chlamydial-infected mice showed that the inhibition of IRE1α RNase activity resulted in a significant decrease in chlamydial shedding (P > .045). The inhibition of PERK kinase activity also showed a reduction trend in Chlamydia shedding, although statistically insignificant (P > .2) (Figure 7A and 7B). Furthermore, mice treated with the inhibitor of IRE1α RNase activity showed a 30% increase in fertility compared to 0% fertility in infected, nontreated mice (data not shown). The results indicated that UPR activation is essential for chlamydial replication and pathogenesis.
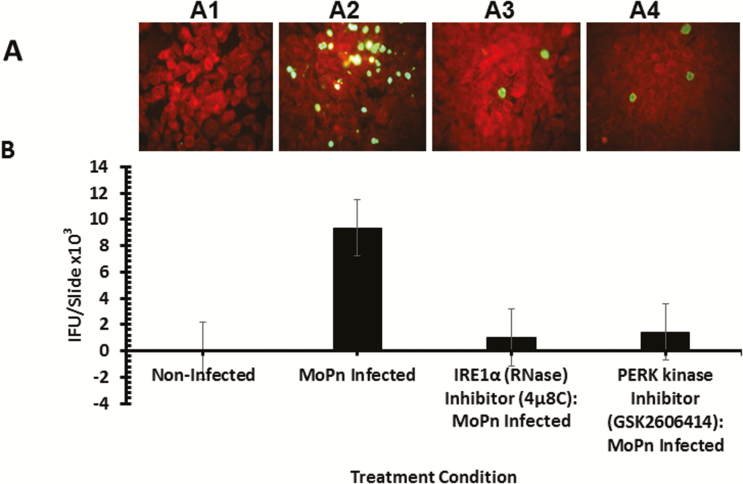
Figure 6.
Role of the IRE1α and PERK signaling pathways of UPR in Chlamydia replication in vitro. Fluorescence microscope images and bar chart of MoPn infectious-forming units recovered from C57epi cells treated with or without IRE1α RNase or PERK inhibitors. A, Fluorescence microscope images of noninfected C57epi cells (A1), C57epi cells infected with MoPn (A2), and C57epi cells infected with MoPn and treated with IRE1α RNase (A3) or PERK kinase (A4) inhibitor. B, Bar chart of MoPn infectious-forming units recovered per slide of noninfected, MoPn (MOI 5)–infected, and MoPn (MOI 5)–infected/treatment with IRE1α RNase or PERK kinase inhibitors. All cultures were incubated for 48 hours. Abbreviations: C57epi cells, mouse oviduct epithelial cells; IRE1α, inositol-requiring enzyme-1α; MOI, multiplicity of infection; MoPn, mouse pneumonitis; NI, noninfected; PERK, protein kinase RNA-activated–like ER kinase; RNase, ribonuclease; UPR, unfolded protein response.
Figure 7.
Role of the IRE1α (7A) and PERK (7B) signaling pathways of UPR in Chlamydia replication in vivo. Graph shows the number of inclusion-forming units recovered from mice infected with C. muridarum (MoPn) and treated/untreated with inhibitor of IRE1α RNase after a primary infection (7A) or PERK kinase after a secondary infection (7B) activity. The triangle symbol plot represents mice infected intravaginally with MoPn and treated intravaginally with inhibitor of IRE1α RNase or PERK kinase activity. The diamond symbol plot represents mice infected intravaginally with MoPn and treated intravaginally with 2% DMSO (solvent concentration used in the preparation of IRE1α RNase or PERK kinase inhibitor). The y-axis represents the number of inclusion-forming units recovered; the x-axis is the number of days postinfection. Abbreviations: DMSO, dimethylsulphoxide; IRE1α, inositol-requiring enzyme-1α; MoPn, mouse pneumonitis; PERK, protein kinase RNA-activated–like ER kinase; RNase, ribonuclease; UPR, unfolded protein response.
DISCUSSION
The detailed host–pathogen interactions and biochemical pathways that drive the successful intracellular parasitism and inclusion development by Chlamydia are incompletely defined. UPR is a cellular response to ER stress [8, 43], whose major signaling pathways may be exploited by energy and metabolically deficient pathogens to increase ATP and metabolites required for development, similar to rapidly dividing cancer cells [17]. Previous attempts to investigate chlamydial-induced UPR were inconclusive. Thus, the short-lived UPR induced by Simkania negevensis (a Chlamydia-related species), was considered unremarkable [44, 45]. Also, the UPR observed in Chlamydophila pneumoniae–infected, IFN-γ–treated cells was attributed to the cytokine [16, 45]. Thus, the ability of human or animal Chlamydia species to directly induce UPR was uncertain. Besides, the mechanisms by which the host cell augments ATP production, maintains the unidirectional flow of nutrients into the inclusion, and establishes an initial antiapoptotic state that allows for chlamydial development, had remained unclear. Our results indicate conclusively that Chlamydia induces the 3 arms UPR. We confirmed the endonuclease activity of activated IRE1α and the phosphorylation and upregulation of phosphorylated eIF2α on the PERK signaling pathway of UPR [13]. Also, the activation of IRE1α generates a potent transcription factor sXBP1 [19] that upregulates lipid biosyntheses [21, 40]. Chlamydial infection significantly upregulated phospholipid production and ATF4 (a transcription factor that activates genes involved in amino acid import), glutathione biosynthesis, and resistance to oxidative stress [15]. Thus, the activation of the IRE1α pathway benefits chlamydia as a source of essential lipids and metabolites. We demonstrated that the PERK pathway upregulated hexokinase II, a crucial enzyme in the glycolytic pathway for rapid ATP production during substrate-level phosphorylation [39], indicating that UPR increases host cell glucose utilization and ATP synthesis during chlamydial infection.
The host survival is essential for Chlamydia replication, and premature apoptosis prevents inclusion formation [46]. The IRE1α/TRAF2 complex formed through the activation of IRE1α [22] activates c-Jun (a component of the AP1 family of transcription factors that are antiapoptotic), which promotes cell survival through upregulation of the microtubule-associated protein 1 light chain 3 (MAP-LC3) (an inducer of autophagy to eliminate cell wastes as a cell survival strategy) [23] and downregulation of tumor suppressors [24]. We showed that the activation of IRE1α upregulated c-Jun and MAP-LC3, which benefits chlamydia because their antiapoptotic activities support host cell survival. In addition, activated ATF6α inhibits the PERK pathway through its upregulation of p58IPK, which reduces the phosphorylation of eIF2α and the expression of CHOP [28, 29]. The blocking of CHOP, a proapoptotic molecule, ensures the survival of ER-stressed cells [30]. Therefore, ATF6α is a necessary feedback molecule in ER homeostasis and cell survival during UPR. Our findings provide a compelling molecular and biochemical mechanism by which chlamydial infection causes host cell resistance to apoptosis [35]. Last, we directly demonstrated the role of UPR activation in Chlamydia pathogenesis by showing that the inhibition of IRE1α or PERK significantly inhibited the formation of Chlamydia inclusions in vitro, decreased chlamydial shedding in vivo, and partially restored fertility in infected mice.
In conclusion, we verified that UPR is a key mechanism by which Chlamydia derives it supplementary ATP and nutrients, as well as maintain host cell survival that ensure its productive development, leading to the various diseases. The induction of UPR during chlamydial infection may include the extra protein expression (eg, Chlamydia early genes [47]) or the unidirectional flow of ATP and nutrients into the inclusion that cause ER stress, triggering UPR. Alternatively, Chlamydia cytotoxin [48] may induce UPR through binding to BiP [49], like other bacterial inducers [33]. Chlamydia produces a cytotoxin thought to play a role in pathogenesis [48]. Our findings have important implications in therapeutic targeting of chlamydial activators of UPR signaling pathways and pathologic expression of the host’s UPR-related molecules in order to prevent chlamydial diseases.
Notes
Disclaimer. The conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention(CDC)/the Agency for Toxic Substances and Disease Registry.
All animal protocols were approved by the CDC Institutional Animal Care and Use Committee (IACUC) under Protocol # 2605IGIMOUC-A1. The CDC IACUC is guided by Title 9, Chapter I, Subchapter A—Animal Welfare (USDA Regulations).
Financial support. This work was supported by the Centers for Disease Control and Prevention and Public Health Service grants (AI41231, GM 08248, RR03034, and 1SC1GM098197) from the National Institutes of Health.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Torrone E, Papp J, Weinstock H; Centers for Disease C, Prevention Prevalence of Chlamydia trachomatis genital infection among persons aged 14–39 years United States, 2007–2012. MMWR Morb Mortal Wkly Rep 2014; 63:834–8. [PMC free article] [PubMed] [Google Scholar]
- 2. Cox JV, Abdelrahman YM, Peters J, Naher N, Belland RJ. Chlamydia trachomatis utilizes the mammalian CLA1 lipid transporter to acquire host phosphatidylcholine essential for growth. Cell Microbiol 2016; 18:305–18. [DOI] [PubMed] [Google Scholar]
- 3. Hatch GM, McClarty G. Phospholipid composition of purified Chlamydia trachomatis mimics that of the eucaryotic host cell. Infect Immun 1998; 66:3727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elwell CA, Engel JN. Lipid acquisition by intracellular Chlamydiae. Cell Microbiol 2012; 14:1010–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stephens RS, Kalman S, Lammel C, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 1998; 282:754–9. [DOI] [PubMed] [Google Scholar]
- 6. Das UN. Pyruvate is an endogenous anti-inflammatory and anti-oxidant molecule. Med Sci Monit 2006; 12:RA79–84. [PubMed] [Google Scholar]
- 7. Ojcius DM, Degani H, Mispelter J, Dautry-Varsat A. Enhancement of ATP levels and glucose metabolism during an infection by Chlamydia. NMR studies of living cells. J Biol Chem 1998; 273:7052–8. [DOI] [PubMed] [Google Scholar]
- 8. Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014; 25:563–73. [DOI] [PubMed] [Google Scholar]
- 9. Blázquez A-B, Escribano-Romero E, Merino-Ramos T, Saiz J-C, Martín-Acebes MA. Stress responses in flavivirus-infected cells: Activation of unfolded protein response and autophagy. Front Microbiol 2014; 5:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han X, Zhou J, Zhang P, et al. IRE1α dissociates with BiP and inhibits ER stress-mediated apoptosis in cartilage development. Cell Signal 2013; 25:2136–46. [DOI] [PubMed] [Google Scholar]
- 11. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334:1081–6. [DOI] [PubMed] [Google Scholar]
- 12. Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2000; 2:326–32. [DOI] [PubMed] [Google Scholar]
- 13. Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000; 6:1099–108. [DOI] [PubMed] [Google Scholar]
- 14. Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 2011; 286:10939–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11:619–33. [DOI] [PubMed] [Google Scholar]
- 16. Shima K, Klinger M, Schütze S, et al. The role of endoplasmic reticulum-related BiP/GRP78 in interferon gamma-induced persistent Chlamydia pneumoniae infection. Cell Microbiol 2015; 17:923–34. [DOI] [PubMed] [Google Scholar]
- 17. Bi M, Naczki C, Koritzinsky M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 2005; 24:3470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joshi A, Newbatt Y, McAndrew PC, et al. Molecular mechanisms of human IRE1 activation through dimerization and ligand binding. Oncotarget 2015; 6:13019–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 2009; 186:323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006; 313:104–7. [DOI] [PubMed] [Google Scholar]
- 21. Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 2004; 167:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000; 287:664–6. [DOI] [PubMed] [Google Scholar]
- 23. Sun T, Li D, Wang L, et al. c-Jun NH2-terminal kinase activation is essential for up-regulation of LC3 during ceramide-induced autophagy in human nasopharyngeal carcinoma cells. J Transl Med 2011; 9:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hettinger K, Vikhanskaya F, Poh MK, et al. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ 2007; 14:218–29. [DOI] [PubMed] [Google Scholar]
- 25. Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 2002; 3:99–111. [DOI] [PubMed] [Google Scholar]
- 26. Sato Y, Nadanaka S, Okada T, Okawa K, Mori K. Luminal domain of ATF6 alone is sufficient for sensing endoplasmic reticulum stress and subsequent transport to the Golgi apparatus. Cell Struct Funct 2011; 36:35–47. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem 2000; 275:27013–20. [DOI] [PubMed] [Google Scholar]
- 28. van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem 2003; 278:15558–64. [DOI] [PubMed] [Google Scholar]
- 29. Boriushkin E, Wang JJ, Li J, Jing G, Seigel GM, Zhang SX. Identification of p58IPK as a novel neuroprotective factor for retinal neurons. Invest Ophthalmol Vis Sci 2015; 56:1374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiong Z, Jiang R, Li X, Liu Y, Guo F. Different roles of GRP78 on cell proliferation and apoptosis in cartilage development. Int J Mol Sci 2015; 16:21153–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu M, Gelowani V, Eblimit A, et al. ATF6 is mutated in early onset photoreceptor degeneration with macular involvement. Invest Ophthalmol Vis Sci 2015; 56:3889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kohl S, Zobor D, Chiang WC, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet 2015; 47:757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes?Nat Rev Microbiol 2015; 13:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith JA, Khan M, Magnani DD, et al. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages. PLOS Pathog 2013; 9:e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Messinger JE, Nelton E, Feeney C, Gondek DC. Chlamydia infection across host species boundaries promotes distinct sets of transcribed anti-apoptotic factors. Front Cell Infect Microbiol 2015; 5:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Igietseme JU, He Q, Joseph K, et al. Role of T lymphocytes in the pathogenesis of Chlamydia disease. J Infect Dis 2009; 200:926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajaram K, Giebel AM, Toh E, et al. Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun 2015; 83:2870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cawley K, Deegan S, Samali A, Gupta S. Assays for detecting the unfolded protein response. Methods Enzymol 2011; 490:31–51. [DOI] [PubMed] [Google Scholar]
- 39. Pellegatti P, Raffaghello L, Bianchi G, Piccardi F, Pistoia V, Di Virgilio F. Increased level of extracellular ATP at tumor sites: in vivo imaging with plasma membrane luciferase. PLOS One 2008; 3:e2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008; 320:1492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci USA 2003; 100:6771–6. doi:10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 2005; 5:149–61. [DOI] [PubMed] [Google Scholar]
- 43. Naidoo N, Davis JG, Zhu J, et al. Aging and sleep deprivation induce the unfolded protein response in the pancreas: implications for metabolism. Aging Cell 2014; 13:131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehlitz A, Karunakaran K, Herweg JA, et al. The chlamydial organism Simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell Microbiol 2014; 16:1224–43. [DOI] [PubMed] [Google Scholar]
- 45. Derré I. Chlamydiae interaction with the endoplasmic reticulum: contact, function and consequences. Cell Microbiol 2015; 17:959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ying S, Pettengill M, Latham ER, Walch A, Ojcius DM, Häcker G. Premature apoptosis of Chlamydia-infected cells disrupts chlamydial development. J Infect Dis 2008; 198:1536–44. [DOI] [PubMed] [Google Scholar]
- 47. Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 2000; 37:913–25. [DOI] [PubMed] [Google Scholar]
- 48. Belland RJ, Scidmore MA, Crane DD, et al. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci USA 2001; 98:13984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paton AW, Beddoe T, Thorpe CM, et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 2006; 443:548–52. [DOI] [PubMed] [Google Scholar]