Abstract
Non-ischemic dilated cardiomyopathy (DCM) has been recognized as a heritable disorder for over 25 years, yet clinical genetic testing is non-diagnostic in >50% of patients, underscoring the ongoing need for DCM gene discovery. Here, whole exome sequencing uncovered a novel molecular basis for idiopathic end-stage heart failure in two sisters who underwent cardiac transplantation at three years of age. Compound heterozygous recessive mutations in TAF1A, encoding an RNA polymerase I complex protein, were associated with marked fibrosis of explanted hearts and gene-specific nucleolar segregation defects in cardiomyocytes, indicative of impaired ribosomal RNA synthesis. Knockout of the homologous gene in zebrafish recapitulated a heart failure phenotype with pericardial edema, decreased ventricular systolic function, and embryonic mortality. These findings expand the clinical spectrum of ribosomopathies to include pediatric DCM.
Introduction
Dilated cardiomyopathy (DCM) is a progressive myocardial disorder characterized by left ventricular dilation and systolic dysfunction. Diagnosis is typically made in the fifth decade of life (1), but penetrance is variable and end-stage DCM in both adults and children is the most common indication for cardiac transplantation (2,3). There are diverse etiologies for the final common pathway of myopathic heart failure and premature death in DCM, yet a majority of patients have idiopathic disease, including 66% of childhood cases (4). Recognition of idiopathic DCM as a familial disorder (1) has been the impetus for discovering the molecular genetic bases of disease by candidate gene, locus mapping and, most recently, whole exome-sequencing strategies. This research has successfully identified over fifty DCM-associated genes (5,6), yet the diagnostic yield for current gene panel testing is only 37% (7). Moreover, the spectrum and frequency of mutations in known DCM genes differ among adults, children, and infants, suggesting unique pathogenic mechanisms within each age group. For example, truncating mutations in TTN have been reported to underlie 14–25% of DCM cases (7,8), yet they were absent in patients two years of age or younger owing to delayed penetrance (7). By contrast, RBM20 mutations are enriched in children 2–18 years of age (7,9). Exome sequencing provides a powerful platform for novel disease gene discovery, particularly in sporadic DCM of childhood attributable to mutations of major effect that disrupt fundamental myocellular functions.
Here, we report a unique family with DCM in a sibling pair, both of whom developed decompensated heart failure requiring cardiac transplantation by 3 years of age. Exome sequencing of the sibling-parent quartet uncovered compound heterozygous mutations in TAF1A, implicating an unsuspected candidate gene for DCM. Consistent with known TAF1A function, electron microscopy identified a gene-specific defect in cardiomyocyte nucleoli from explanted hearts, indicative of impaired ribosomal RNA (rRNA) synthesis. taf1a knockout in zebrafish recapitulated a lethal heart failure phenotype. These findings reveal ribosomopathy as a cause for early-onset heart failure.
Results
Clinical presentation
The proband presented at 2 6/12 years of age with transient left hemiplegia and subcortical ischemia in the right middle cerebral artery territory. Her developmental and past medical history were unremarkable except for failure to thrive (weight and height <3rd percentile). Echocardiography revealed DCM with severe four-chamber enlargement and decreased left ventricular ejection fraction (EF = 20%; normal ≥50%) in the absence of symptoms or physical findings of heart failure. While no intracardiac thrombus was identified, thromboembolism was the presumed etiology of her stroke. Electrocardiography demonstrated normal sinus rhythm, left atrial enlargement, and non-specific ST-T wave changes. An underlying cause for her DCM was not identified, despite an extensive diagnostic workup including metabolic testing, karyotyping, chromosomal microarray, and genetic testing with a 10-gene DCM panel. She remained stable for 10 months on an outpatient heart failure regimen of carvedilol, enalapril, digoxin, furosemide, spironolactone, and aspirin (10). However, her left ventricular function and growth failure did not improve and, following hospitalization for decompensated systolic heart failure and atrial flutter, she underwent cardiac transplantation at 3 4/12 years of age.
The younger sister also developed idiopathic DCM and underwent cardiac transplantation for end-stage heart failure at a similar age. Following her sister’s diagnosis, a screening echocardiogram at 13 months of age revealed mild-moderate left atrial enlargement but normal left ventricular chamber size and systolic function (EF = 60%). A surveillance study at 2 6/12 years of age was diagnostic for DCM, with moderate LV enlargement, moderate-severe LV systolic dysfunction (EF = 27%), moderate mitral valve regurgitation, and severe left atrial enlargement. Electrocardiography demonstrated normal sinus rhythm, left ventricular hypertrophy, and non-specific ST-T wave changes. Past medical history was similarly unremarkable and overt signs and symptoms of heart failure were absent. She was small but within normal limits for age (weight 25th percentile; height 10th percentile). The patient was placed on the same pharmacologic therapy as her sister and had a gradual decline in EF. Six months after diagnosis, she sustained cardiac arrest during a viral illness. Due to refractory junctional tachycardia and hemodynamic instability following resuscitation, she was placed on mechanical circulatory support as a bridge to cardiac transplantation, which occurred at 3 1/12 years of age.
Both sisters, ages 9 and 6 years, have maintained normal cardiac allograft function on chronic, steroid-free immunosuppressive therapy. They are physically active and attend school, but have global delays in cognitive and fine motor development. While they remain small for age (weight = 6th/<3rd percentiles and height = 27th/12th percentiles, respectively), no extra-cardiac disorders have developed to suggest a syndromic form of DCM. There is no history of heart failure in the extended family. Screening echocardiograms in the sisters’ mother (age 26; EF 68%) and father (age 33; EF 55–60%) excluded asymptomatic DCM, although the father had borderline left ventricular chamber enlargement (cross-sectional short-axis dimensions 1–2 mm above upper limits of normal).
Whole exome sequencing
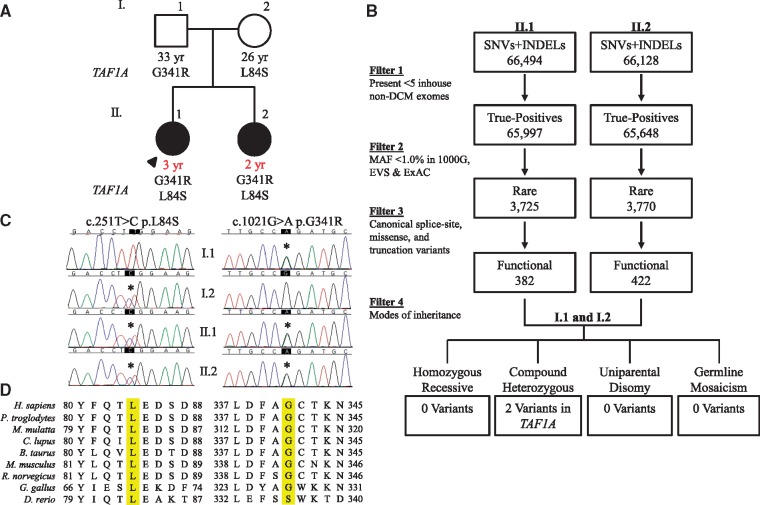
Chromosomal microarray analysis excluded chromosomal aneuploidy in the proband. To identify the genetic basis of DCM in the sibling pair, exome sequencing was performed on the family quartet and an iterative variant filtering scheme was employed, culminating in identification of TAF1A, encoding TATA-box binding protein associated factor 1A, as the sole candidate gene (Fig. 1A–C). The compound heterozygous TAF1A missense variants, c.251T > C, p.L84S and c.1021G > A, p.G341R (NM_005681.3), were rare, predicted to be damaging by in silico tools, and altered highly conserved amino acids within the protein product (Fig. 1D and Table 1).
Figure 1.
Identification of recessive TAF1A mutations in a sibling pair with DCM. (A) Family pedigree. Square, male; circle, female; solid, affected; open, unaffected; black font, age at screening echocardiography; red font, age at diagnosis; arrow, proband. (B) An iterative filtering scheme of whole exome sequencing variant calls identified a single candidate gene, TAF1A. (C) Sanger sequencing verified compound heterozygous missense mutations. (D) Conservation of L84, G341, and surrounding residues.
Table 1.
Summary of TAF1A variants
Cardiac pathology
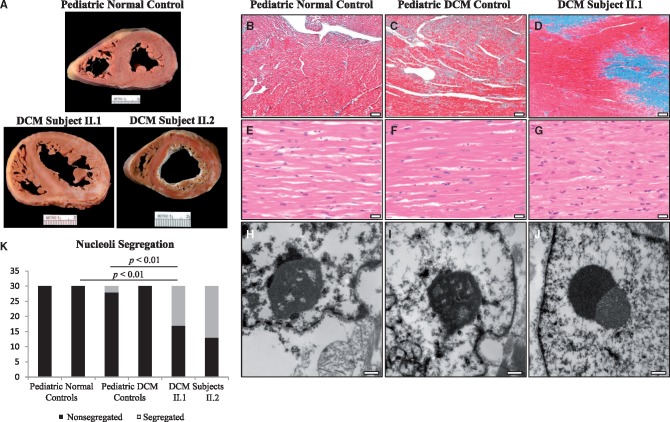
Examination of explanted hearts from the affected sisters revealed gross pathological features characteristic of end-stage DCM. Myocardial mass was increased (heart weight: II.1, 95.2 g; II.2, 66.5 g; expected, approximately 50 g) in conjunction with severe interstitial fibrosis evident in the gross specimens (Fig. 2A). Light microscopy and Masson trichrome staining demonstrated large patches of replacement-type interstitial fibrosis (Fig. 2D), distinct from the pericellular fibrosis seen in pediatric DCM controls (Fig. 2B and C). Hematoxylin and eosin staining revealed severe cardiomyocyte hypertrophy in the affected sibling pair (Fig. 2G) compared to moderate cardiomyocyte hypertrophy in the pediatric DCM controls (Fig. 2E and F). Because TAF1A is part of selectivity factor I (14), necessary for ribosomal RNA synthesis within nucleoli, nucleolar morphology was assessed by electron microscopy of explanted heart tissues from the affected sisters. Nucleolar segregation was identified in approximately 50% of cardiomyocytes examined (Fig. 2J and K), together with cytoplasmic aggregates coinciding with sarcomere degeneration (Supplementary Material, Fig. S1). These gene-specific findings were absent in pediatric normal and DCM controls (Fig. 2H,I,K; Supplementary Material, Fig. S1).
Figure 2.
TAF1A-associated pathological and ultrastructural alterations. (A) Cross section of explanted heart from a normal male control, age 6, and the affected sibling pair, both 3 years of age at time of cardiac transplantation. Increased myocardial mass and extensive fibrosis was seen in both siblings. (B–D) Representative photomicrographs of Masson’s trichrome stained left ventricular myocardial tissue from a normal male control, age 13, a female DCM control, age 9, and the proband. Replacement fibrosis was observed, unique to the affected sibling pair. Scale bars, 200 µm (40X magnification). (E–G) Hematoxylin and eosin staining revealed marked cardiomyocyte hypertrophy in both siblings. Scale bars, 20 µm (400X magnification). (H–J) Transmission electron microscopy revealed nucleolar segregation that was unique to the affected sibling pair. Scale bars, 400 nm (30,000X magnification). (K), Nucleolar segregation quantification. In total, 60 nucleoli/group were examined (30 nucleoli/individual) and classified as segregated or non-segregated based on morphological appearance.
Zebrafish modeling
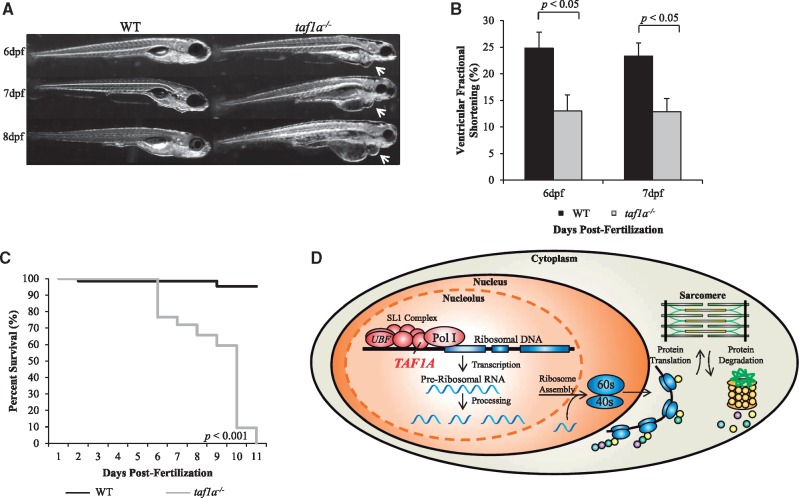
To model the recessive, presumed loss-of-function TAF1A mutations, stable knockout of the single taf1a zebrafish homolog was performed, resulting in a frameshift mutation leading to a premature stop codon (Supplementary Material, Fig. S2). Homozygous mutant zebrafish embryos recapitulated a heart failure phenotype beginning at 6 days post-fertilization, with progressive pericardial edema and a significant decrease in ventricular fractional shortening compared to WT embryos (Fig. 3A and B). Knockout of taf1a resulted in early lethality, with embryonic death occurring 6 to 11 days post-fertilization (Fig. 3C).
Figure 3.
taf1a−/− zebrafish recapitulate a heart failure phenotype. (A) Lateral view of WT and taf1a−/− embryos at 6, 7, and 8 days post-fertilization, demonstrating pericardial edema that was absent in WT fish. (B) taf1a−/− embryos exhibited a significant reduction in ventricular fractional shortening compared to WT (n = 5 per group, P<0.05). No difference was observed between taf1a+/− and WT embryos (data not shown). (C) Kaplan-Meier survival curve demonstrates complete lethality in taf1a−/− fish by 11 days post-fertilization (WT = 65, taf1a−/− =64; P < 0.001). (D) Ribosome biogenesis. Ribosomal DNA is transcribed within the nucleolar compartment of the nucleus. Pre-ribosomal RNA is processed into mature ribosomal RNA, and then assembled into ribosomal subunits before being exported to the cytoplasm for protein translation. In cardiomyocytes, nascent proteins are continuously integrated into the existing sarcomere. Loss-of-function of TAF1A and impaired rRNA synthesis could result in an imbalance between protein synthesis and degradation.
Discussion
TAF1A and nucleolar stress
TAF1A functions in recruitment of RNA polymerase I to ribosomal DNA promoters, the essential first step in ribosome biogenesis (Fig. 3D) (14). Synthesis and processing of the rRNA 45S precursor occur within the nucleolus, comprised of granular, fibrillar, and dense fibrillar components (15). Transcription of pre-rRNA occurs on the borders between the fibrillar and dense fibrillar centers, while processing of pre-rRNA and initial ribosome subunit assembly occurs within the dense fibrillar center (15). During active ribosome synthesis, the three nucleolar components are dispersed, with the fibrillar and dense fibrillar components surrounded by the granular component (16). rRNA transcriptional arrest results in nucleolar segregation, where the fibrillar and granular components disengage and form juxtaposed structures, indicative of nucleolar stress (16). rRNA transcriptional arrest occurs through inhibition of RNA polymerase I, achieved pharmacologically by treatment with actinomycin D or doxorubicin (16,17). Notably, doxorubicin is a common cause of chemotherapy-induced DCM (18). Moreover, depletion of RRN3 in mouse embryonic fibroblasts, an interacting partner of TAF1A, results in disrupted nucleolar structure with segregation of the nucleolar components (19). Electron microscopy of explanted heart tissue from the affected sister-pair revealed prototypic nucleolar segregation, indicative of perturbed TAF1A-dependent rRNA synthesis and nucleolar stress. TAF1A-dependent cytoplasmic aggregation was also observed near sites of sarcomere degeneration, suggesting a defect in the critical balance between protein synthesis and degradation in the heart (Supplementary Material, Fig. S1) (20). Indeed, defects in protein degradation are known contributors to cardiac disease and heart failure (20), yet the essential role of the nucleolus in cardiovascular disease and morphologic changes as an early marker of myocellular stress have only recently been recognized (21).
Ribosomopathy as a cause of heart failure
Ribosomopathies are a group of genetic disorders that originate from defects in ribosome biogenesis, a ubiquitous process that occurs in all tissues, yet ribosomopathies are known to result in unique phenotypes confined to specific organs (22). Similarly, our data show that genetic disruption of TAF1A, a ubiquitous protein integral to the RNA polymerase I complex and ribosomal RNA synthesis, leads to a cardiac-specific phenotype. While cardiomyocytes have limited capacity to regenerate by cell division (21), constitutive replacement of most myocellular proteins occurs every 5-10 days (23). We postulate that physiologic mechanical and metabolic demands on the heart render it vulnerable to low rRNA synthetic reserve, particularly during active cardiac growth in development and postnatal life (Fig. 3D). Indeed, TAF1A mutations in the sibling pair led to end-stage DCM necessitating cardiac transplantation by 3 years of age. Validation of a defect in RNA synthesis will require further mechanistic studies. While deficiency of ribosomal genes has been associated with cardiomyopathy in Drosophila melanogaster (24), to our knowledge this is the first report of ribosomopathy as a cause for human heart failure. Identification of additional DCM-associated mutations in TAF1A or related genes would be required to further establish this genotype–phenotype relationship. Accordingly, we performed mutation scanning of 347 unrelated patients with DCM (85% adult-onset), but did not identify other rare or unique TAF1A coding variants that segregated with disease.
DCM is characterized by marked genetic heterogeneity and a majority of causal genes are yet to be discovered. Consequently, the diagnostic yield of DCM gene panel testing (37%) falls short of other prototypic heritable cardiac disorders like hypertrophic cardiomyopathy (60%) and long QT syndrome (75%) (25). To this end, our report exemplifies a current limitation of standard DCM gene panel testing, and the added value of family-based whole exome sequencing on a research basis for ongoing discovery of DCM-associated genes (26–28).
Materials and Methods
Study subjects
Written informed consent was obtained under a research protocol approved by the Mayo Clinic Institutional Review Board and subjects were phenotypically classified by echocardiography. Diagnostic criteria for DCM were left ventricular diastolic and/or systolic short-axis chamber dimension Z-score ≥2.0, and left ventricular ejection fraction <50%.
Chromosomal microarray and copy number variant analysis
A custom 180K oligonucleotide microarray (Agilent) was utilized for chromosomal microarray on a DNA sample from the older affected sibling, with a genome-wide functional resolution of 100 kilobases (kb). Deletions ≥200 kb and duplications ≥500 kb were considered clinically relevant. Those below the size threshold were considered relevant if sufficient evidence supporting pathogenicity was present in Online Mendelian Inheritance in Man (OMIM) (http://ncbi.nlm.nih.gov/omim), PubMed (http://ncbi.nlm.nih.gov/pubmed), or ClinGen Dosage Sensitivity Map (http://ncbi.nlm.nih.gov/projects/dbvar/clingen/index.shtml). Higher resolution screening for copy number variants (CNVs) was accomplished by analyzing whole exome sequencing data with PatternCNV software (29). No CNVs were identified that segregated according to Mendelian modes of inheritance.
Whole exome sequencing and bioinformatics analysis
DNA was isolated from peripheral-blood white cells from the family quartet and subject to whole exome sequencing utilizing the Mayo Clinic Medical Genome Facility and Bioinformatics Core. The Agilent SureSelect Human All Exon 50 Mb capture kit (version 2.0; Agilent) was utilized for exome capture. Each sample was run on its own lane and 101 base pair, paired-end sequencing was performed on Illumina’s HiSeq2000 platform (Illumina, Inc.). Reads were aligned to the hg19 reference genome with Novoalign (http://novocraft.com) followed by sorting and marking of duplicate reads using Picard (http://picard.sourceforge.net). Local realignment of insertions/deletions and base quality score recalibration were performed using the Genome Analysis Toolkit (30). Single nucleotide variants and insertions/deletions were called across all four samples simultaneously using the Genome Analysis Toolkit’s UnifiedGenotyper with variant quality score recalibration (31). All four samples had over 97% of reads map to the hg19 reference genome and on average, 95% of the 50 Mb capture region had ≥20X coverage. Similarly, analysis of a comprehensive list of 55 known DCM genes (data not shown) revealed, on average, over 96% of known DCM genes had ≥20X coverage.
The resultant variant call format files were analyzed with QIAGEN’s Ingenuity® Variant AnalysisTM software (QIAGEN). Variants that mapped to the coding region and passed variant quality score recalibration were filtered to exclude false-positive platform-specific sequencing errors that were present in 5 or more in-house non-DCM controls. Variants were then filtered for rarity, excluding those with a minor allele frequency ≥1.0% in 1000 Genomes (whole genome sequencing data from 1,092 individuals) (32), the Exome Variant Server (whole exome sequencing data from 6,503 individuals) (http://evs.gs.washington.edu/EVS/), and the Exome Aggregation Consortium (WES data from 60,706 individuals) (33). Next, all canonical splice-site, missense, and truncation variants were retained. Inheritance modeling was then carried out, utilizing exome sequencing data from the unaffected parents to model all potential modes of inheritance, including homozygous recessive, compound heterozygous, uniparental disomy, and germline mosaicism.
Sanger sequencing was performed on DNA samples from the family quartet to confirm the TAF1A variants identified through whole exome sequencing. Primers utilized for PCR were: TAF1A L84S: F 5’-GCTGGCTTTGGTACTGG-3’, R 5’-TGAAGCTGTAGGCATAAACT-3’; TAF1A G341R: F 5’-GAACTGTTAAAACCATTGC-3’, R 5’-GTATGTTCTCCTTATGACT-3’.
Histochemistry and electron microscopy
Left ventricular tissue specimens, procured from the sisters’ explanted hearts at the time of transplantation (ages: 3 years), were formalin fixed and paraffin embedded by standard clinical laboratory protocols. Four additional specimens served as controls. Left ventricular tissue was obtained at autopsy from 6-year-old and 13-year-old males, both of whom died from non-cardiac causes and had no evidence of DCM. Explanted left ventricular tissue was available from 2 individuals with genetically-defined DCM who underwent cardiac transplantation for end-stage heart failure: a 9-year-old female with a missense mutation in LMNA, encoding lamin A/C, and a 12-year-old female with a missense mutation in RBM20, encoding RNA binding motif protein 20. Sections 5 µm thick were obtained from the paraffin tissue blocks, and hematoxylin and eosin (H&E) and Masson trichrome-stained slides were generated using standard clinical histochemistry protocols.
For electron microscopy, formalin fixed cardiac tissue was obtained from the paraffin tissue blocks and deparaffinized by standard clinical laboratory protocols. Tissues were rinsed 3 times in 0.1 M sodium phosphate buffer, post-fixed, and stained in 1% osmium tetroxide. Tissues were then rinsed 3 times in distilled water, en bloc stained in 2% aqueous uranyl acetate, and dehydrated in a graded series of ethanol followed by absolute acetone. Tissues were subsequently infiltrated and embedded in epoxy resin. Semi-thin (0.6 µm) sections for light microscopy were cut with an ultramicrotome and stained with toluidine blue. Thin sections, approximately 100 nm in thickness, were cut with an ultramicrotome, post-stained with 0.3% aqueous lead citrate, and examined in a FEI Tecnai G2 12 transmission electron microscope operated at 80 kV. Digital images were captured with an AMT 2k side mount CCD camera and AMT image capture software. For statistical analysis, 30 nucleoli/individual (60 nucleoli/group) were analyzed and classified as either non/segregated based upon morphological appearance. One individual from each group was randomly chosen for chi-squared analysis followed by multiple comparisons with Bonferroni correction (significance: P < 0.01).
Zebrafish modeling
Wild-type (WT) WIK zebrafish were utilized as controls and for generation of taf1a−/− fish to model the loss-of-function TAF1A mutations. Zebrafish were maintained under 14-h light and 10-h dark cycles at 28.5°C. For Transcription Activator-like Effector Nuclease (TALEN) assembly and mRNA injection, the gene sequence of taf1a was first verified in WT WIK zebrafish via PCR and Sanger sequencing. TALEN pairs targeting taf1a exon 1 were then designed in ZiFit (http://zifit.partners.org/ZiFiT/Disclaimer.aspx) (34). The GOLDEN GATE TALEN and TAL Effector kits were utilized for TALEN assembly (Addgene) (35). TALEN sequences were verified with Sanger sequencing, and mRNA was generated with the mMessage mMachine® T3 high yield capped RNA transcription kit (Ambion). 50 ng each of left- and right-arm TALEN capped mRNA were injected into zebrafish embryos at the 1- to 2-cell stage. The following sequences were targeted by TALEN: taf1a left arm 5’-GCACAGAGAAATGGAT-3’, taf1a right arm 5’–GTGTTTCTGCAATGTT–3’. Polymerase chain reaction (PCR) amplification and restriction enzyme (Eco53KI, New England Biolabs) digestion were performed to confirm successful mutagenesis and to identify heterozygous and homozygous mutants. The following primer pairs and conditions were utilized: taf1a exon 1: F 5’–TAGTGAGTCAATAGTGTTT–3’, R 5’–ACTTTACACGCATTCTGG–3’. The restriction enzyme Eco53KI generated PCR products of the following lengths: WT = 285 bp; taf1a+/− = 285 bp, 167 bp, and 118 bp; taf1a−/− = 167 bp, and 118 bp.
Because a taf1a antibody was not available to determine the knockdown efficiency in zebrafish, reverse transcription PCR was utilized. Three zebrafish embryos per micro-centrifuge tube were homogenized using a mortar and pestle (Fisher Scientific). Triplicates for each time point were genotyped. RNA was extracted using TRIzol® LS reagent (Sigma) according to the manufacturer’s instructions, and 500 ng of purified mRNA was converted to complementary DNA (cDNA) utilizing the SuperScriptTM III First-Strand Synthesis System (Invitrogen). The following primer pairs were utilized: taf1a exons 1-2: F 5’-TGAAAGTGAACATCGGCAAC-3’, R 5’- TCCGCAACCACAGACTGAT-3’; taf1a exons 7-8: F 5’- GGAGAATTACGCATACAACA-3’, R 5’-ACTCCTTTCCAATGTATCC-3’; gapdh: F 5’–CCACCCATGGAAAGTACAAG-3’, R 5’-CTCTCTTTGCACCACCCTTA-3’.
For phenotypic assessment of zebrafish, videos of anesthetized embryos were recorded to determine ventricular shortening fraction utilizing a Zeiss Axioplan two microscope (Carl Zeiss) and a Nikon COOLPIX 8700 digital camera (Nikon) as previously described (36). Briefly, anesthetized zebrafish embryos were positioned horizontally on a microscope slide in a thin layer of 3% methyl cellulose to obtain a lateral view of the heart. Quicktime (.MOV) video files were analyzed to identify the maximum ventricular diastolic (VD) and minimum ventricular systolic (VS) dimensions. Five WT and taf1a−/− zebrafish embryos were analyzed per group. Ventricular fractional shortening (VFS) was calculated by dividing the difference between ventricular width in diastole and systole by the ventricular width in diastole. For statistical analysis (P < 0.05), Fisher’s Exact T-test was utilized for ventricular fractional shortening, while log-rank test was utilized for Kaplan-Meier survival.
Supplementary Material
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by funding from the National Institutes of Health [R01 HL071225 to TMO, RO1 HL107304 to XX, T32GM072474 to PAL], American Heart Association [14PRE18070007 to PAL], and Mayo Clinic Center for Individualized Medicine to TMO.
References
- 1. Michels V.V., Moll P.P., Miller F.A., Tajik A.J., Chu J.S., Driscoll D.J., Burnett J.C., Rodeheffer R.J., Chesebro J.H., Tazelaar H.D. (1992) The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N. Engl. J. Med., 326, 77–82. [DOI] [PubMed] [Google Scholar]
- 2. Lund L.H., Edwards L.B., Kucheryavaya A.Y., Dipchand A.I., Benden C., Christie J.D., Dobbels F., Kirk R., Rahmel A.O., Yusen R.D.. et al. (2013) The registry of the international society for heart and lung transplantation: thirtieth official adult heart transplant report—2013; focus theme: age. J. Heart Lung Transplant, 32, 951–964. [DOI] [PubMed] [Google Scholar]
- 3. Dipchand A.I., Kirk R., Edwards L.B., Kucheryavaya A.Y., Benden C., Christie J.D., Dobbels F., Lund L.H., Rahmel A.O., Yusen R.D.. et al. (2013) The registry of the international society for heart and lung transplantation: sixteenth official pediatric heart transplantation report–2013; focus theme: age. J. Heart Lung Transplant, 32, 979–988. [DOI] [PubMed] [Google Scholar]
- 4. Towbin J.A., Lowe A.M., Colan S.D., Sleeper L.A., Orav E.J., Clunie S., Messere J., Cox G.F., Lurie P.R., Hsu D.. et al. (2006) Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA, 296, 1867–1876. [DOI] [PubMed] [Google Scholar]
- 5. Hershberger R.E., Hedges D.J., Morales A. (2013) Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat. Rev. Cardiol., 10, 531–547. [DOI] [PubMed] [Google Scholar]
- 6. Olson T.M., Chan D.P. (2013) Dilated cardiomyopathy In Allen H.D., Driscoll D.J., Shaddy R.E., Feltes T.F. (eds.), Moss and Adams' Heart Disease in Infants, Children, and Adolescents: Including the Fetus and Young Adult. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA, Vol 2, 8th ed., pp. 1235–1246. [Google Scholar]
- 7. Pugh T.J., Kelly M.A., Gowrisankar S., Hynes E., Seidman M.A., Baxter S.M., Bowser M., Harrison B., Aaron D., Mahanta L.M.. et al. (2014) The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet. Med., 16, 601–608. [DOI] [PubMed] [Google Scholar]
- 8. Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E.. et al. (2012) Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med., 366, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brauch K.M., Karst M.L., Herron K.J., de Andrade M., Pellikka P.A., Rodeheffer R.J., Michels V.V., Olson T.M. (2009) Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol., 54, 930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirk R., Dipchand A.I., Rosenthal D.N., Addonizio L., Burch M., Chrisant M., Dubin A., Everitt M., Gajarski R., Mertens L.. et al. (2014) The international society for heart and lung transplantation guidelines for the management of pediatric heart failure: executive summary. J. Heart Lung Transplant, 33, 888–909. [DOI] [PubMed] [Google Scholar]
- 11. Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 12. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pollar K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res., 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Comai L., Zomerdijk J.C., Beckmann H., Zhou S., Admon A., Tjian R. (1994) Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science, 266, 1966–1972. [DOI] [PubMed] [Google Scholar]
- 15. James A., Wang Y., Raje H., Rosby R., DiMario P. (2014) Nucleolar stress with and without p53. Nucleus, 5, 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernandez-Verdun D., Roussel P., Thiry M., Sirri V., Lafontaine D.L. (2010) The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA, 1, 415–431. [DOI] [PubMed] [Google Scholar]
- 17. Merski J.A., Daskal I., Busch H. (1976) Effects of Adriamycin on ultrastructure of nucleoli in the heart and liver cells of the rat. Cancer Res., 36, 1580–1584. [PubMed] [Google Scholar]
- 18. Singal P.K., Iliskovic N. (1998) Doxorubicin-induced cardiomyopathy. N. Engl. J. Med., 339, 900–905. [DOI] [PubMed] [Google Scholar]
- 19. Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Gröne H.J., Schütz G., Grummt I. (2005) Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell, 19, 77–87. [DOI] [PubMed] [Google Scholar]
- 20. Willis M.S., Patterson C. (2013) Proteotoxicity and cardiac dysfunction – Alzheimer’s disease of the heart? N. Engl. J. Med., 368, 455–464. [DOI] [PubMed] [Google Scholar]
- 21. Hariharan N., Sussman M.A. (2014) Stressing on the nucleolus in cardiovascular disease. Biochim. Biophys. Acta, 1842, 798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armistead J., Triggs-Raine B. (2014) Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett., 588, 1491–1500. [DOI] [PubMed] [Google Scholar]
- 23. Willis M.S., Schisler J.C., Portbury A.L., Patterson C. (2009) Build it up-tear it down: protein quality control in the cardiac sarcomere. Cardiovasc. Res., 81, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casad M.E., Abraham D., Kim I.M., Frangakis S., Dong B., Lin N., Wolf M.J., Rockman H.A. (2011) Cardiomyopathy is associated with ribosomal protein gene haplo-insufficiency in Drosophila melanogaster. Genetics, 189, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ackerman M.J., Priori S.G., Willems S., Berul C., Brugada R., Calkins H., Camm A.J., Ellinor P.T., Gollob M., Hamilton R.. et al. (2011) HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm, 8, 1308–1339. [DOI] [PubMed] [Google Scholar]
- 26. Theis J.L., Sharpe K.M., Matsumoto M.E., Chai H.S., Nair A.A., Theis J.D., de Andrade M., Wieben E.D., Michels V.V., Olson T.M. (2011) Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circ. Cardiovasc. Genet., 4, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Long P.A., Larsen B.T., Evans J.M., Olson T.M. (2015) Exome sequencing identifies pathogenic and modifier mutations in a child with sporadic dilated cardiomyopathy. J. Am. Heart Assoc., 4, e002443. doi: 10.1161/JAHA.115.002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Long P.A., Evans J.M., Olson T.M. (2015) Exome sequencing establishes diagnosis of Alström syndrome in an infant presenting with non-syndromic dilated cardiomyopathy. Am. J. Med. Genet. A, 167A, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C., Evans J.M., Bhagwate A.V., Prodduturi N., Sarangi V., Middha M., Sicotte H., Vedell P.T., Hart S.N., Oliver G.R.. et al. (2014) PatternCNV: a versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics, 30, 2678–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.. et al. (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The 1000 Genomes Consortium. (2012) An integrated map of genetic variation from 1092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sander J.D., Zaback P., Joung J.K., Voytas D.F., Dobbs D. (2007) Zinc Finger Targeter (ZiFit): an engineered zinc finger/target site design tool. Nucleic Acids Res., 35, W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. (2001) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res., 39, e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoage T., Ding Y., Xu X. (2012) Quantifying cardiac functions in embryonic and adult zebrafish. Method. Mol. Biol., 843, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.