Abstract
Background
At present, there is no effective targeted therapy for esophageal squamous cell carcinoma (ESCC), and it is urgent to find new targets for the treatment of ESCC. TRAF4 has been regarded as a cause of carcinogenesis due to overexpression in many cancer types and participation in multiple signaling pathways. However, there are few studies on TRAF4 in ESCC worldwide. Its expression in ESCC and whether it affects the prognosis of patients still remain unclear.
Material/Methods
We detected the expressions of TRAF4, ki-67, and p53 in 100 cases of ESCC and 80 cases of adjacent normal esophageal squamous epithelium tissues by immunohistochemical technique. We further explored the relationship between TRAF4 and ESCC and its prognosis through statistical analysis.
Results
TRAF4 was highly expressed in ESCC tissues and was mainly expressed in the cytoplasm. Overexpression of TRAF4 in ESCC was also associated with high expression of ki-67 and p53 (P<0.05). We also found that patients with high expression of TRAF4 had significantly lower OS than in patients with low TRAF4 expression (P<0.05). Overexpression of TRAF4 was an independent risk factor affecting the prognosis of patients (P<0.05).
Conclusions
We found that TRAF4 was highly expressed in ESCC tissues and was mainly expressed in the cytoplasm of cancer cells. Overexpression of TRAF4 was an independent risk factor affecting the overall prognosis of patients. The results indicated that TRAF4 may become a new target for the treatment of ESCC in the future.
MeSH Keywords: Esophageal Neoplasms, Ki-67 Antigen, Prognosis, TNF Receptor-Associated Factor 4, Tumor Suppressor Protein p53
Background
According to epidemiological statistics, China is a high-incidence area of esophageal squamous cell carcinoma, which is more common in men over 40 years old. Treatment of esophageal cancer is mainly early surgical treatment, but many patients diagnosed at later stages have high morbidity and mortality rates and have lost the opportunity for early surgery, and there is still no effective targeted therapy [1]. Esophageal cancer is more common in people with adenocarcinoma in Europe and the Americas. Most studies on esophageal cancer are mainly based on adenocarcinoma. It is urgent to find new targets for the treatment of esophageal squamous cell carcinoma.
Tumor necrosis factor receptor-related factor (TRAF) is an important intracellular signaling proteins of tumor necrosis factor receptor (TNFR), which activates the transcription factor NF-κB and JNK pathway by adjusting a variety of signal transduction pathways, such as the IL-1/Toll-like receptors, so as to regulate cell proliferation, differentiation, apoptosis, immune and inflammatory reaction and so on [2–4]. In 1994, Rothe et al. first discovered 2 proteins specific to TNFR binding in mouse cytotoxic T cell lines and named them TNFR1 and TNFR2. So far, 7 different TRAFs have been found [5]. These molecules share some structural properties. It mediates the interaction of TRAFs molecules, receptors, and corresponding signal proteins in cells through the homologous domain of its own carboxyl radicals. The signal is transmitted downstream through the interaction between DNA and protein through the ring finger/zinc finger domain contained in the amino terminal [6]. Among many family molecules, TRAF4 is the only one found in the nucleus, which may be related to some primary tumors and metastases. However, whether TRAF4 is mainly expressed in the nucleus or cytoplasm in tumor tissues has been controversial [7]. Unlike other family members that regulate immune and inflammatory responses, TRAF4 has been shown to be involved in the development and progression of multiple tumors [8,9]. TRAF4 is also expressed during embryonic development and is a key molecule in the process of individual development [10]. In recent years, studies around the world have found that TRAF4 is often highly expressed in breast cancer, lung cancer, colorectal cancer, and other tumors, and also affects the survival and prognosis of patients [11].
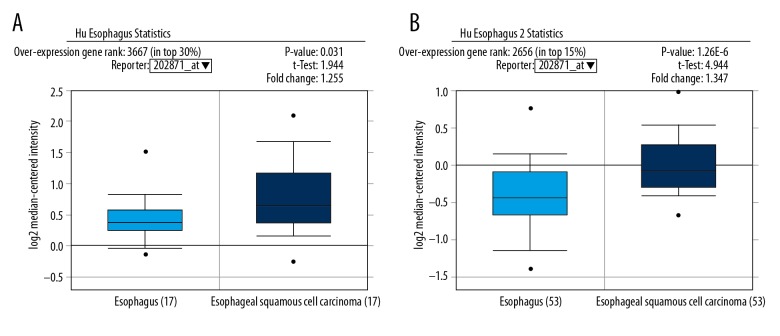
We predicted the expression of TRAF4 gene in esophageal cancer in advance through bioinformatics analysis. Our results suggest that the expression of TRAF4 in esophageal cancer and normal tissues may be different (Figure 1A, 1B). Therefore, we used immunohistochemistry to detect the distribution and expression of TRAF4 in esophageal squamous cell carcinoma and adjacent tissues, and performed statistical analysis of the correlation between OS and clinical features. To investigate the expression and association of TRAF4 in esophageal squamous cell carcinoma, we also studied the relationship between TRAF4, ki-67, and p53 expression.
Figure 1.
Oncomine database prediction results. (A) TRAF4 is related to the ESCC organization (P=0.031). (B) TRAF4 is related to the ESCC organization (P<0.001).
Material and Methods
Patients and specimens
We collected 100 tissue samples of esophageal cancer patients from Anhui Provincial Hospital from January 2008 to December 2012, and made tissue chips. Specimens were obtained from 74 male and 26 female patients, ages 48–82 years (mean age 65.3 years), all based on pathological confirmation of a clear diagnosis of ESCC. None of the patients had received radiotherapy, chemotherapy, or immunotherapy. These tissues were from patients undergoing esophagectomy. Among them, 80 patients had corresponding specimens of cancer and adjacent normal esophageal squamous epithelium tissues, numbered 1–80. The remaining 20 cases had only esophageal cancer specimens, numbered 80–100. The clinicopathological data on sex, age, tumor size, lymphatic metastasis, and TNM stage of all patients were collected from the case management system of the medical records room of Anhui Provincial Hospital. Follow-up lasted until July 2017. The study was approved by the Anhui Provincial Hospital Ethics Committee and all patients signed a written informed consent form.
Tissue chip manufacturing steps
The pathological tissue wax block was selected from the Pathology Department, and the conventional pathological sectioning was followed by HE staining. We invited the pathologist to locate the tumor tissue in the wax block. We used a US Beecher array instrument to make a hole (2 mm in diameter) on the blank wax block, and then accurately removed the desired tissue at the corresponding position of the tissue wax block according to the precise range drawn on the HE-stained section. After that, we put the desired tissue in the hole of the blank wax block, the number of the tissue placed in each hole was recorded in detail, and the above operation was repeated. Each specimen was taken from a single piece of cancer and adjacent tissues (3–5 cm from the cancer tissue) to make an array block. We used a microtome to slice continuously. After that, routine HE staining was performed, and 2 pathologists checked whether each of the chips was consistent with the design. Then, we assessed the qualified slice lines by immunohistochemistry.
Immunohistochemistry
The levels of TRAF4 were assessed by immunohistochemistry. Mouse anti-human TRAF4 monoclonal antibody was purchased from Santa Cruz (USA) diluted 1: 100. The immunohistochemistry kit and DAB chromogenic reagent are all products of Zymed Co. (USA). Immunohistochemical staining was carried out according to the instructions of the SP kit. The tissue chips were oven-dried overnight at 70°C. After routine dewaxing and hydration, 3% hydrogen peroxide was used to block endogenous peroxidase for 15 min. After rinsing with PBS, we then put it in citrate buffer, performed antigen retrieval by microwave or high pressure for 15 min, cooled it at room temperature, and treated it with normal goat blocking serum for 15 min at room temperature, and then an antibody was added at room temperature for 2 h at 4°C overnight. Then, we added biotin-labeled secondary antibody, incubate it for 15 min at room temperature, stained it with DAB, rinse it thoroughly with water, and stopped the reaction with hematoxylin. Positive and negative controls were set during the staining process, and the negative control was replaced with PBC instead of the primary antibody. The same immunohistochemical method was used to detect the expression of ki-67 and p53 in cancer and adjacent tissues.
Determination of immunohistochemical results
We used positive range and intensity to analyze TRAF4 expression. The staining intensity scores were as follows: 0 (negative staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish brown), 3 (strong staining, brown). We used positive staining cells as a percentage of field of view to assess positive staining range: 0 (negative), 1 (positive rate <25%), 2 (positive rate 26–50%), 3 (positive rate 51–75%), and 4 (positive rate 75–100%). The final score was determined by multiplying the intensity score and the positive range. By observing at least 5 high-power fields and scores, a total score >4 was considered high expression. Ki-67 and p53 expression was quantified by using a visual grading system [12]. In this experiment, the p53 and ki-67 positive criteria were judged as: 0 (negative staining), 1 (<10% positive for diseased cells), 2 (10~50% positive for diseased cells), and 3 (>50% positive for diseased cells); 0 and 1 points are negative, and 2 and 3 points are positive [13]. All results were evaluated by 2 pathologists who were blinded to the experiment.
Statistical analysis
Statistical analyses were performed with SPSS version 22.0 software (IBM, Armonk, NY). Differences between groups were compared using the χ2 test. The relationship between TRAF4 expression and clinicopathologic characteristics and prognosis were explored by Kaplan-Meier survival analysis and log-rank test. Correlation analysis between the 2 variables was performed by using Spearman’s rank correlation analysis, and among multiple factors using Cox regression. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression and localization of TRAF4 in esophageal cancer and adjacent tissues
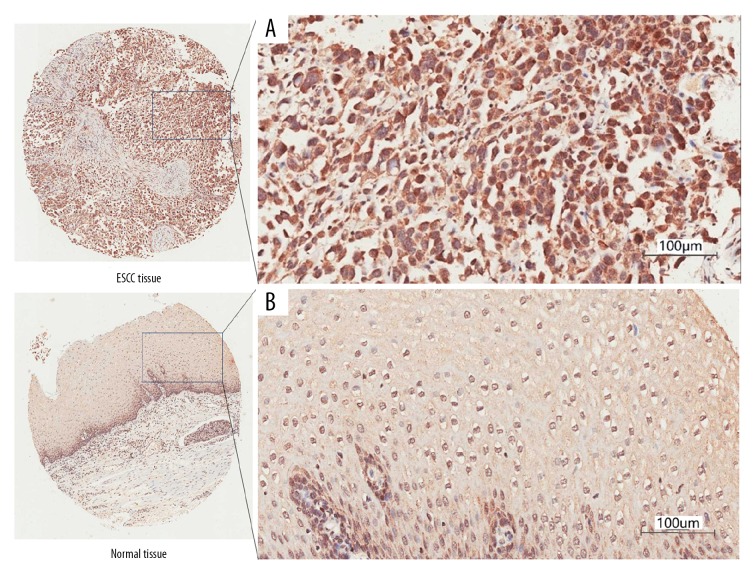
After immunohistochemical staining of the tissue microarray, we obtained 80 pairs of cancer tissues and corresponding adjacent normal esophageal squamous epithelium tissues, as well as 20 separate esophageal cancer tissues. During the operation, there were no squamous epithelium in 5 adjacent tissues. The results showed that although TARF4 is expressed in the nucleus and cytoplasm of esophageal cancer cells at the same time, but it is mainly expressed in the cytoplasm (85% vs. 44%). The expression of TRAF4 in the cytoplasm of esophageal cancer cells was significantly higher than in adjacent normal tissues (85/100 vs. 16/75, χ2=71.2, P<0.01 Table 1A, Figure 2A, 2B). In addition, the expression of TRAF4 in the nucleus of esophageal cancer cells was significantly higher than in adjacent normal tissues (44/100 vs. 6/75, χ2=27.2, P<0.01, Table 1B). The results show that TRAF4 is highly expressed in ESCC, and is mainly expressed in cytoplasm.
Table 1.
(A) Difference of TRAF4 expression level in cytoplasm between esophageal squamous cell carcinoma and adjacent normal tissues. (B) Difference of TRAF4 expression level in nucleus between esophageal squamous cell carcinoma and adjacent normal tissues.
| (A) | TRAF4 expression level in cytoplasm | Sum | High rate | |
|---|---|---|---|---|
| High | Low | |||
| ESCC tissues | 85 | 15 | 100 | 85% |
| Adjacent normal tissues | 16 | 59 | 75 | 21% |
| (B) | TRAF4 expression level in nucleus | Sum | High rate | |
| High | Low | |||
| ESCC tissues | 44 | 56 | 100 | 44% |
| Adjacent normal tissues | 6 | 69 | 75 | 8% |
Figure 2.
Immunohistochemical staining of esophageal cancer and adjacent tissues at 200× magnification. (A) TRAF4 was overexpressed in the cytoplasm of esophageal cancer cells. (B) TRAF4 is under-expressed in the adjacent normal tissues.
Relationship between high expression of TRAF4 in cytoplasm of esophageal carcinoma and expression of ki-67 and p53
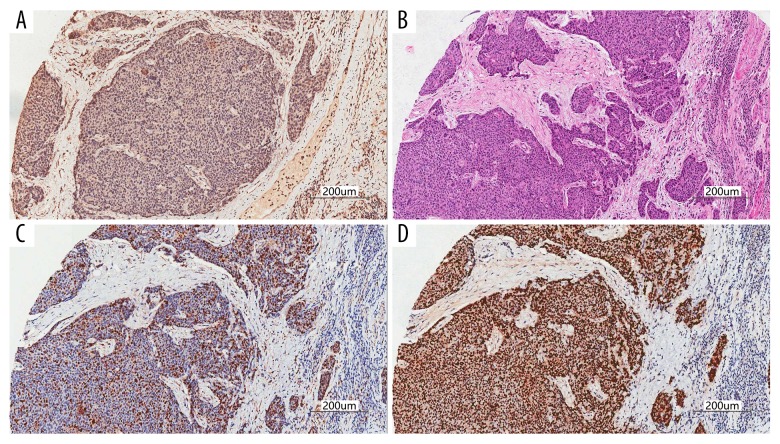
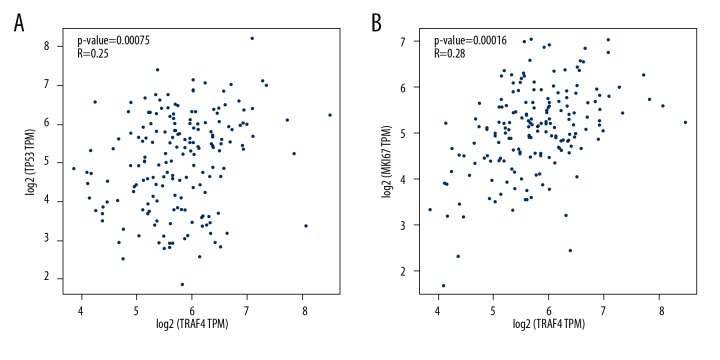
To explore the relationship between the expression of TRAF4 and ki-67 and P53 in esophageal cancer tissues, we serially sliced the tissue chip to obtain high-quality serial sections for immunohistochemistry under the same conditions. Immunohistochemical staining results showed that TRAF4 was highly expressed in ESCC tissues, and ki-67 and p53 were highly expressed in serial sections of the same site (Figure 3A–3D). We performed Spearman correlation analysis, and the results showed that high expression of TRAF4 in esophageal cancer tissues is often accompanied by high expression of ki-67 (r=0.512, P<0.01, Figure 3C) and P53 (r=0.234, P<0.05, Figure 3D). Our bioinformatics prediction by using the GEPIA website (TCGA data) also supports our results (Figure 4A, 4B).
Figure 3.
We performed immunohistochemistry and HE staining of ESCC sections at the same position under the same conditions. (A) TRAF4 was overexpressed in esophageal cancer cells. (B) Under the HE staining, the esophageal cancer tissue was deeply stained and the cell morphology was abnormal. (C) ki-67 was overexpressed in esophageal cancer cells. (D) p53 is overexpressed in esophageal cancer cells.
Figure 4.
Retrospective analysis of the database. (A) The expression of TRAF4 in esophageal cancer was associated with p53. (B) The expression of TRAF4 in esophageal cancer was associated with ki67.
We further explored the relationship between the expression of TARF4, ki-67, and p53 and clinicopathological features in esophageal cancer tissues. High expression of TRAF4 in ESCC tissues was associated with sex (P=0.048). The high expression of ki-67 in esophageal cancer tissues is usually accompanied by lymphatic metastasis and higher TNM staging. High expression of p53 was also positively correlated with TNM staging. However, we did not find a relationship between the expression of TRAF4 in ESCC tissues and age, tumor size, lymph node metastasis, TNM stage, or pathological stage (Table 2).
Table 2.
The relationship between TRAF4, ki-67 and p53 expression and the clinicopathologic features in 100 ESCC patients(cases).
| Sum | TRAF4 | χ2 | P | Ki-67 | χ2 | P | p53 | χ2 | P | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | High | Low | |||||||||
| Gender | Male | 74 | 66 | 8 | 3.917 | 0.048 | 46 | 28 | 0.162 | 0.688 | 47 | 27 | 0.277 | 0.599 |
| Female | 26 | 19 | 7 | 15 | 11 | 15 | 11 | |||||||
| Age (years) | ≤65 | 51 | 43 | 8 | 0.038 | 0.845 | 32 | 19 | 0.133 | 0.715 | 33 | 18 | 0.323 | 0.57 |
| >65 | 49 | 42 | 7 | 29 | 20 | 29 | 20 | |||||||
| Tumor size (cm) | ≤5 | 56 | 46 | 10 | 0.005 | 0.944 | 31 | 25 | 0.353 | 0.553 | 36 | 20 | 0.668 | 0.414 |
| >5 | 29 | 24 | 5 | 18 | 11 | 16 | 13 | |||||||
| Lymphatic metastasis | Yes | 54 | 49 | 5 | 3.21 | 0.073 | 39 | 15 | 5.65 | 0.017 | 34 | 20 | 0.006 | 0.94 |
| No | 45 | 35 | 10 | 22 | 23 | 28 | 17 | |||||||
| TNM stages | I/II | 46 | 37 | 9 | 1.76 | 0.189 | 22 | 24 | 9.416 | 0.002 | 33 | 13 | 4.734 | 0.03 |
| III | 50 | 45 | 5 | 39 | 11 | 25 | 25 | |||||||
| Pathological grading | I/II | 72 | 61 | 11 | 0.016 | 0.901 | 40 | 32 | 3.204 | 0.073 | 43 | 29 | 0.566 | 0.452 |
| III | 28 | 24 | 4 | 21 | 7 | 19 | 9 | |||||||
Relationship between expression of TRAF4 in esophageal cancer tissues and patient prognosis
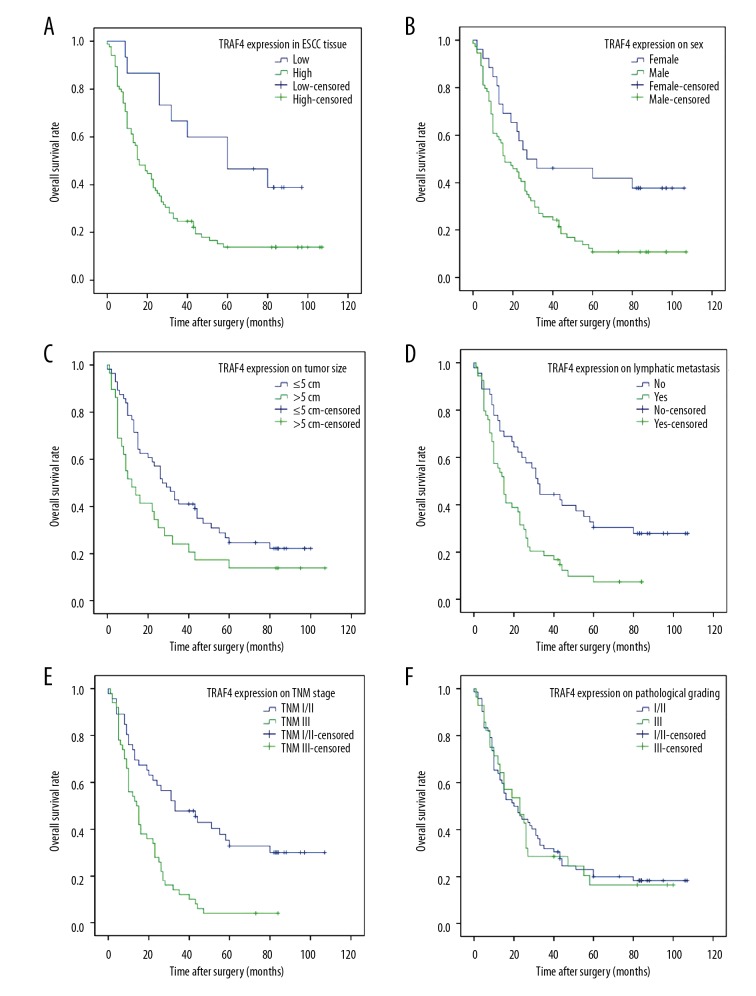
We found that the overall survival (OS) of patients with high expression of TRAF4 was significantly lower than that of patients with low expression of TRAF4 by Kaplan-Meier survival analysis and log-rank statistical test (95%CI 23.337–37.763 vs. 44.409–78.546, P<0.01, Figure 5A). Patient sex, tumor size, lymph node metastasis, and TNM staging were also associated with OS (P<0.05, Figure 5B–5E). However, there was no association between patient pathological staging and patient OS (P>0.05, Figure 5F). Our further Cox multivariate analysis suggested that overexpression of TRAF4 was closely associated with patient prognosis, and it was an independent risk factor for OS in patients with esophageal squamous cell carcinoma (Table 3).
Figure 5.
Kaplan-Meier analysis of overall survival (OS) of patients with esophageal squamous cell carcinoma (ESCC). (A) OS curve of patients with ESCC based on TRAF4 expression in tumor tissues; (B) OS curve of patients with ESCC based on sex; (C) OS curve of patients with ESCC based on tumor size; (D) OS curve of patients with ESCC based on lymphatic metastasis. (E) OS curve of patients with ESCC based on TNM stages; (F) OS curve of patients with ESCC based on pathological grading.
Table 3.
Univariate and multivariate analysis of factors associated with OS.
| OS | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | P | HR | 95% CI | P | ||
| TRAF4 expression | High | 30.55 | 23.337–37.763 | 0.004 | 2.789 | 1.221–6.356 | 0.015 |
| Low | 61.478 | 44.409–78.546 | |||||
| Sex | Male | 29.176 | 22.050–43.094 | 0.006 | 0.485 | 0.232–1.012 | 0.054 |
| Female | 54.594 | 37.973–71.216 | |||||
| Tumor size (cm) | ≤5 | 41.924 | 32.521–51.336 | 0.043 | 0.681 | 0.401–1.156 | 0.681 |
| >5 | 28.276 | 15.772–40.779 | |||||
| Lymphatic metastasis | Yes | 22.247 | 16.319–28.174 | 0.001 | 4.155 | 0.984–17.543 | 0.053 |
| No | 48.306 | 36.494–60.118 | |||||
| TNM stages | I/II | 49.992 | 37.903–62.081 | <0.001 | 0.099 | 0.023–0.417 | 0.002 |
| III | 19.000 | 14.111–23.889 | |||||
Discussion
Since it was first discovered in human breast cancer tissues in 1995, TRAF4 has been regarded as an oncogene due to overexpression in many cancer types and is thought to be involved in carcinogenesis by participating in multiple signaling pathways. Camilleri et al. performed immunohistochemical analysis of pathological specimens from 623 patients with 20 different tumor types, and found that TRAF4 was overexpressed in various tumor cells such as breast cancer and lung adenocarcinoma, liver cancer, and colon cancer, including lung adenocarcinoma. Overexpression of TRAF4 was observed in 60% of tissues (82 of 137). TRAF4 may play an important role in most tumor proliferation signaling pathways [14]. In China, the prevalent type of esophageal cancer is squamous cell carcinoma [15]. However, there are few studies on TRAF4 overexpression in esophageal squamous cell carcinoma and the relationship with patient prognosis. In the present immunohistochemistry study of 100 patients with esophageal cancer, we found that TRAF4 is mainly localized in the cytoplasm, and only a small amount is expressed in the nucleus. By comparing 80 pairs of cancer tissues with corresponding adjacent tissues, we found that the expression of TRAF4 in esophageal squamous cell carcinoma was significantly higher than that in normal squamous epithelium. This indicates that overexpression of TRAF4 is closely related to the occurrence of esophageal squamous cell carcinoma.
However, the mechanism by which TRAF4 promotes tumorigenesis remains unclear. Liu et al. found that TRAF4 overexpressing HCC cell line promotes phosphorylation of Akt and induces HCC cell migration and invasion by activating the PI3K/Akt signaling pathway [16,17]. Yang et al. also found that TRAF4 promotes the growth, invasion, and migration of oral squamous cell carcinoma cells through the Wnt-β-catenin signaling pathway [18]. These studies indicate that TRAF4 participates in the development and progression of multiple tumors in a variety of ways. Therefore, in this study, we compared the expression of TRAF4, ki-67, and p53 in ESCC tissues in the same tissue site and under the same experimental conditions. The results show that high expression of TRAF4 in esophageal squamous cell carcinoma is accompanied by high expression of ki-67 and p53. Further big data correlation analysis also supports our experimental results. Ki-67 is a common indicator of clinical tumor pathology. A high ki-67-positive rate is often accompanied by accelerated tumor growth, poor tissue differentiation, and poor prognosis. Although the p53 gene is a tumor-suppressor gene, it is mutated in tumor tissues, thereby exhibiting overexpression of p53 protein in malignant tumors. Many studies have reported that high expression of ki-67 and p53 often indicates poor prognosis in patients. Therefore, high expression of TRAF4 may lead to the poor prognosis of patients by promoting the growth and proliferation of tumor cells and by inhibiting cell apoptosis. The statistical results also showed that the expression of TRAF4 was related to sex. The expression of TRAF4 in ESCC tissue of male patients was significantly higher than in female patients. This is also consistent with the epidemiological status of the majority of male patients with esophageal squamous cell carcinoma.
In recent years, some scholars have found that changing the expression level of TRAF4 by gene vector can affect the malignant behavior of cancer cells. For example, Jiang used small interfering RNA (siRNA) to silence the expression of TRAF4 gene in esophageal cancer cell Eca-109, and found that the inhibition rate, apoptosis rate, and caspase-3 activation rate of proliferation in the silent group were increased (P<0.05), and the proportion of cells in G0/G1 phase was significantly higher than in the control group (P<0.05) [19]. Yao also found that silencing TRAF4 inhibits osteosarcoma cell growth in vivo and in vitro, consistent with these findings [20]. In addition, some studies have found that downregulation of TRAF4 levels has an effect of inhibiting the progression of non-small cell lung cancer [21]. Coincidentally, Zhang found that the proliferation ability of MDA-MB-231 cells was enhanced by the overexpression of TRAF4 in breast cancer cells, and the proportion of cells in S phase was increased [22]. The above results indicate that targeting silencing of TRAF4 can inhibit the growth of malignant tumors, and the expression of TRAF4 can affect the occurrence and development of tumors.
Through further Kaplan-Meier and log-rank statistical tests, we found that the overall survival (OS) of patients with high expression of TRAF4 in the cytosol was significantly lower than that of patients with low expression of TRAF4, and the Cox multivariate analysis suggests that TRAF4 is an independent risk factor affecting overall patient survival. These results suggest that TRAF4 is involved in the development of esophageal squamous cell carcinoma. The high expression of TRAF4 suggests a poor overall prognosis. Targeted silencing of TRAF4 may benefit patients with advanced esophageal squamous cell carcinoma. In the future, TRAF4 gene knockout and inhibition of TRAF4 expression may promote apoptosis of esophageal cells, and inhibit proliferation and distant metastasis.
We were not able to deeply study the specific mechanism by which TRAF4 affects the prognosis of patients. In the future, we will collect specimens and expand the sample size of the experiment. Further research is needed on the mechanism of TRAF4 in the occurrence and development of ESCC.
Conclusions
We found that TRAF4 was highly expressed in ESCC tissues and was mainly expressed in the cytoplasm of cancer cells. The high expression of TRAF4 was also accompanied by high expression of ki-67 and p53. Patients with high expression of TRAF4 had significantly lower OS than patients with low TRAF4 expression. Overexpression of TRAF4 was an independent risk factor affecting the prognosis of patients, indicating that TRAF4 may become a new target for the treatment of ESCC in the future.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (No. 81472329)
References
- 1.Siegel RL, Miller KD. Cancer statistics, 2015. Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Wan-qiao Z, et al. [Intracellular adaptor proteins TRAFs family]. Chinese Bulletin of Life Sciences. 2008;20(4):611–17. [in Chinese] [Google Scholar]
- 3.Ahmed F, Shiraishi T, Vessella RL. Tumor necrosis factor receptor associated factor-4: an adapter protein overexpressed in metastatic prostate cancer is regulated by microRNA-29a. Oncol Rep. 2013;30:2963–68. doi: 10.3892/or.2013.2789. [DOI] [PubMed] [Google Scholar]
- 4.Esparza EM. TRAF4 functions as an intermediate of GITR-induced NF-kappaB activation. Cell Mol Life Sci. 2004;61:3087–92. doi: 10.1007/s00018-004-4417-0. [DOI] [PubMed] [Google Scholar]
- 5.Rothe M, Wong SC, Henzel WJ. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–92. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 6.Takayanagi H, Ogasawara K, Hida S, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–5. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 7.Glauner H, Siegmund D, Motejadded H, et al. Intracellular localization and transcriptional regulation of tumor necrosis factor (TNF) receptor-associated factor 4 (TRAF4) Eur J Biochem. 2002;269:4819–29. doi: 10.1046/j.1432-1033.2002.03180.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhao ZJ, Ren HY, Yang F, et al. Expression, correlation, and prognostic value of TRAF2 and TRAF4 expression in malignant plural effusion cells in human breast cancer. Diagn Cytopathol. 2015;43:897–903. doi: 10.1002/dc.23330. [DOI] [PubMed] [Google Scholar]
- 9.Ren HY, Wang J, Yang F, et al. Cytoplasmic TRAF4 contributes to the activation of p70s6k signaling pathway in breast cancer. Oncotarget. 2015;6:4080–96. doi: 10.18632/oncotarget.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masson R, Régnier CH, Chenard MP, et al. Tumor necrosis factor receptor associated factor 4 (TRAF4) expression pattern during mouse development. Mech Dev. 1998;71:187–91. doi: 10.1016/s0925-4773(97)00192-5. [DOI] [PubMed] [Google Scholar]
- 11.Haiyang Z, Guowu Q, Xin W, et al. [Effect of RNA interference of TRAF4 expression on apoptosis and Wnt signaling pathway in rectal cancer cells]. Journal of Qingdao University. 2018;54(4):415–22. [in Chinese] [Google Scholar]
- 12.Penault-Llorca F, André F, Sagan C, et al. Ki67 expression and docetaxel efficacy in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009;27:2809–15. doi: 10.1200/JCO.2008.18.2808. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Wang LF, Tian HM, et al. [Expression of retinoic acid receptor-beta mRNA and p16, p53, Ki67 proteins in esophageal carcinoma and its precursor lesions]. Zhonghua Zhong Liu Za Zhi. 2005;27:152–55. [in Chinese] [PubMed] [Google Scholar]
- 14.Camilleri-Broët S, Cremer I, Marmey B, et al. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene. 2007;26:142–47. doi: 10.1038/sj.onc.1209762. [DOI] [PubMed] [Google Scholar]
- 15.Rubenstein JH. Epidemiology, diagnosis, and management of esophageal adenocarcinoma. Gastroenterology. 2015;149(2):302–17. doi: 10.1053/j.gastro.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu K, Wu X, Zang X, et al. TRAF4 regulates migration, invasion, and epithelial-mesenchymal transition via PI3K/AKT signaling in hepatocellular carcinoma. Oncol Res. 2017;25:1329–40. doi: 10.3727/096504017X14876227286564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao W, Wang X, Cai Q, et al. TRAF4 enhances osteosarcoma cell proliferation and invasion by Akt signaling pathway. Oncol Res. 2014;22:21–28. doi: 10.3727/096504014X14077751730351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Wei D, Wang W, et al. TRAF4 enhances oral squamous cell carcinoma cell growth, invasion and migration by Wnt-β-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8:11837–46. [PMC free article] [PubMed] [Google Scholar]
- 19.Ming J. [Effects of TRAF4 silencing on the proliferation, apoptosis and cell cycle of esophageal cancer cell line Eca-109]. Acta Medicinae Universitatis Scientiae et Technologiae Huazhong. 2016;45(4) [in Chinese] [Google Scholar]
- 20.Yao W, Wang X, Cai Q, et al. Knockdown of TRAF4 expression suppresses osteosarcoma cell growth in vitro and in vivo. Int J Mol Med. 2014;34:1655–60. doi: 10.3892/ijmm.2014.1948. [DOI] [PubMed] [Google Scholar]
- 21.Chen T, Gao F, Feng S, et al. MicroRNA-370 inhibits the progression of non-small cell lung cancer by downregulating oncogene TRAF4. Oncol Rep. 2015;34:461–68. doi: 10.3892/or.2015.3978. [DOI] [PubMed] [Google Scholar]
- 22.Xiao-li Z, Xiao-yi M. [Transfecting pcDNA3-TRAF4-DM-TRAF improves proliferation of human breast cancer cell MDA-MB-231]. Basic & Clinical Medicine. 2015;10:1382–86. [in Chinese] [Google Scholar]