Abstract
There is limited information on the antigen specificity and functional potential of the influenza virus–specific CD4+ T-cell repertoire in humans. Here, enzyme-linked immunospot assays were used to examine circulating CD4+ T-cell specificities for influenza virus directly ex vivo in healthy adults. Our studies revealed CD4+ T-cell reactivity to multiple influenza virus proteins, including hemagglutinins, neuraminidases, M1 proteins, and nucleoproteins. Unexpectedly, the immunodominance hierarchies and functional potential of cells reactive toward influenza A virus were distinct from those toward influenza B virus. We also identified influenza virus–specific cells producing granzyme B. Our findings revealed individual and virus-specific patterns that may differentially poise humans to respond to infection or vaccination.
Keywords: Influenza virus, CD4+ T cells, cell mediated immunity, influenza vaccine
Human influenza virus–specific CD4+ T cells from a large cohort were quantified. CD4+ T-cell reactivity was exceptionally broad, and, unexpectedly, the immunodominance and functional potential of CD4+ T cells reactive toward influenza A and B viruses were strikingly distinct.
CD4+ T cells have multiple functions in protective immunity against influenza. These activities include assisting in the production of high-affinity, class-switched neutralizing and protective antibodies, as well as secreting cytokines that aid in recruitment and activation of innate immune effector cells. CD4+ T cells also promote the expansion, development of the effector function, and establishment of memory CD8+ T cells and may directly mediate cytolysis of infected cells [1–3]. These functions all may potentiate anti-influenza immunity, promoting neutralization of virus or accelerating viral clearance upon infection.
Despite the importance of CD4+ T cells in protection from influenza, our understanding of the breadth, specificity, and functionality of CD4+ T cells in humans with complex influenza exposure histories is limited. Previous studies were limited by small sample numbers [4], expansion of CD4+ T cells prior to response characterization [5], and preselection of potential epitopes by use of predictive algorithms [6] or particular major histocompatibility complex types [7] that may bias accurate estimates of the influenza virus–specific CD4+ T-cell repertoire in typical adults. In addition, CD4+ T cells have often been quantified by using the production of interferon γ (IFN-γ) as a sole indicator of CD4+ T-cell frequency [4–6], potentially underestimating the functional potential of the influenza virus–reactive cells. A striking deficit also exists in our understanding of CD4+ T-cell reactivity to influenza B virus, which causes substantial disease worldwide [8].
To better understand the influenza virus–specific memory CD4+ T cells available for recall in the general population, we analyzed reactivity directly ex vivo from peripheral blood mononuclear cells (PBMCs) isolated from a large cohort of healthy adults by using enzyme-linked immunospot (ELISPOT) assays, enumerating CD4+ T-cell reactivity by use of overlapping peptide libraries representing the entire translated sequence of 12 different influenza virus–derived proteins. These data quantifying the circulating CD4+ T-cell repertoire specific for influenza viral proteins at both the population level and among individual subjects provide the insight needed to begin to identify correlates of protection against acute infection and to enable the prediction of vaccine responses and the implementation of novel vaccination strategies.
MATERIALS AND METHODS
Synthetic Peptides and Libraries
17-mer peptides overlapping by 11 amino acids encompassing the entire translated sequences of the viral proteins were obtained from the Biodefense and Emerging Infections Research Repository, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The peptide arrays included A/New Caledonia/20/99(H1N1) hemagglutinin (sH1; NR-2602) and neuraminidase (N1; NR-2606), A/California/04/09(H1N1) hemagglutinin (pH1; NR-15433), A/New York/384/2005(H3N2) hemagglutinin (H3; NR-2603) and neuraminidase (N2; NR-2608), A/New York/348/2003(H1N1) nucleocapsid protein (NP; NR-2611) and matrix protein (M1; NR-2613), A/New York/444/2001 nonstructural protein 1 (NS1; NR-2612), and B/Florida/04/2006 hemagglutinin (HA-B; NR-18972), neuraminidase (NA-B; NR-19254), nucleocapsid protein (NP-B; NR-36045), and matrix protein (M1-B; NR-36046). A negative peptide pool encompassing 111 peptides from Sin Nombre Virus (NM H10) glycoprotein precursor protein (NR-4764) was also used. All peptides from a given protein were combined, with each peptide present at a final concentration of 1 μM in assays.
Isolation of PBMCs From Human Blood Specimens
Following approval from the Division of Microbiology and Infectious Diseases (DMID) and the University of Rochester Research Subjects Review Boards (protocols 07-009 and 14-0064), blood specimens were obtained from a group of 45 healthy subjects 20–68 years of age who had provided informed consent. Plasma was removed, and PBMCs were isolated and frozen in fetal calf serum (Gibco) containing 10% dimethyl sulfoxide (Sigma-Aldrich) at a concentration of 15 million cells/mL. After thawing and overnight rest in culture, cells were washed and depleted of CD8+ and CD56+ T cells, using MACS microbeads per the manufacturer’s instructions (Miltenyi Biotec).
ELISPOT Assay
ELISPOT assays were performed as previously described [9]. Briefly, CD8- and CD56-depleted PBMCs were cultured with peptides on plates coated with 10 μg/mL anti-human IFN-γ (clone 1-D1K) or anti-human interleukin 2 (IL-2; clone MT2A91/2C95) for 36 hours at 37°C in 5% CO2 or on plates coated with 15 μg/mL anti-human granzyme B (GzmB; clone GB10) for 60 hours at 37°C in 5% CO2. After incubation, plates were washed and incubated with biotinylated anti-human IFN-γ (2 μg/mL; clone 7-B6-1), IL-2 (1 μg/mL; clone MT8G10), or GzmB (2 μg/mL; clone GB11) in wash buffer with 10% fetal calf serum for 2 hours. All antibodies were purchased from MabTech. The plates were washed and developed to detect mediators as previously described [9]. Quantification of cytokine-secreting cells was performed with an Immunospot reader (series 5.2), using Immunospot software, v5.1.
Statistics
Data are presented as the frequency of mediator-producing cells per million CD8- and CD56-depleted PBMCs. Means are depicted by a horizontal line, with statistical differences between protein conditions calculated using the Kruskal-Wallis test with the Dunn post hoc test. A P value of <.05 was considered statistically significant.
RESULTS
Because of gaps in our understanding of the circulating influenza virus–specific CD4+ T-cell repertoire, we designed a strategy anticipated to provide a generalizable estimate of the range in abundance and specificity of human CD4+ T cells reactive with influenza viral proteins and the diversity in CD4+ T-cell reactivity among individuals. Because the protein specificity of influenza virus–reactive CD4+ T cells could be strongly influenced by factors such as the frequency and type of influenza virus encounters (ie, infection vs vaccination), and because of the importance of influenza virus protein specificity in responses to infection [10] and vaccination [9], we sought to comprehensively evaluate antigen specificity and ascertain whether there were any detectable protein-dependent patterns in CD4+ T-cell reactivity.
A large cohort of 45 healthy adult subjects aged 22–68 years with no recorded recent history of influenza virus infection or vaccination was sampled (Supplementary Figure 1). PBMCs depleted of CD8+ T cells and natural killer cells were tested for reactivity to hemagglutinin, neuraminidase, NP, NS1, and M1 from circulating influenza viruses, using complete pools of overlapping peptides representing the entire translated sequence of each viral protein as stimulating antigens. ELISPOT assays were used to quantify reactivity directly ex vivo, scoring the abundance of influenza virus–reactive cells producing IFN-γ, IL-2, or GzmB (Figure 1). These mediators were chosen to quantify reactivity because IFN-γ is a key T-helper type 1 (Th1) effector cytokine, IL-2 has important functions in CD8+ T-cell proliferation and memory differentiation [11] and in the differentiation of cytotoxic CD4+ T cells [12], while GzmB is an important effector molecule for cytotoxicity [2, 12].
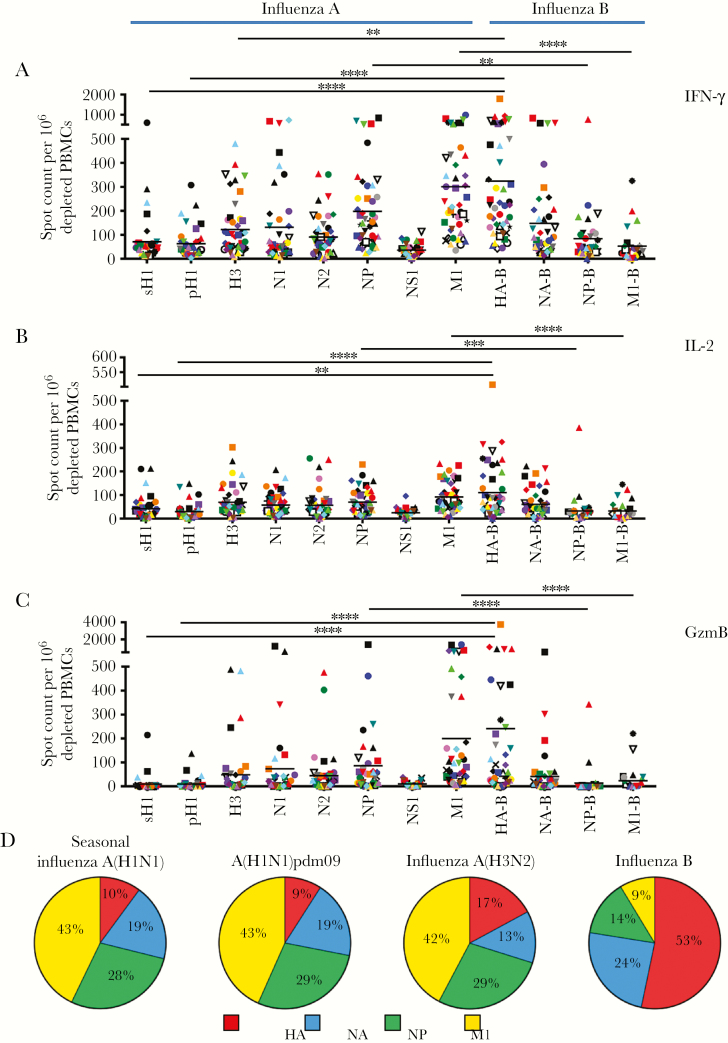
Figure 1.
A–C, Influenza virus–reactive CD4+ T cells secreting interferon γ (IFN-γ; A), interleukin 2 (IL-2; B), and granzyme B (GzmB; C) have diverse antigen specificity. Enzyme-linked immunospot (ELISPOT) analyses were performed following restimulation with peptide pools spanning the entire coding sequences of the sH1, pH1, H3, N1, N2, NP, NS1, M1, HA-B, NA-B, NP-B, and M1-B proteins (see Materials and Methods for descriptions of proteins). Responses are shown as the spot count per 106 CD8- and CD56-depleted peripheral blood mononuclear cells (PBMCs) with background subtracted. Donors are indicated by unique symbols (Supplementary Figure 1), with mean values designated by black lines. **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001, by the Kruskal-Wallis test with the Dunn post hoc test, for differences between influenza A virus and corresponding influenza B virus proteins. D, Differences in immunodominance hierarchies among CD4+ T cells specific to influenza A and influenza B viruses. Each slice of the pie depicts the relative fraction of the CD4+ T-cell response dedicated to hemagglutinin (red), neuraminidase (blue), nucleoprotein (green), and matrix protein (yellow), based on IFN-γ ELISPOT values. The total number of cytokine spots for the indicated proteins within each virus ranged from approximately 600–700 spots per 106 CD8- and CD56-depleted PBMCs. A(H1N1)pdm09, 2009 pandemic influenza A(H1N1) virus.
The results of these studies (Figure 1) demonstrated that most healthy adults had a broad influenza virus–specific CD4+ T-cell repertoire, with IFN-γ– and IL-2–producing CD4+ T cells reactive against many influenza A and B virus proteins. Within this diverse and highly variable influenza virus–specific CD4+ T-cell repertoire, an overarching hierarchy in CD4+ T-cell reactivity to different influenza viral proteins was observed. For influenza A virus (Figure 1), the most dominant reactivity was detected for epitopes derived from NP and M1, with more modest responses toward H3 and neuraminidase and generally the weakest responses toward H1 and NS1. Strikingly, marked differences in the immunodominance pattern were observed when reactivity to influenza B virus was quantified (Figure 1). Unlike influenza A virus–specific responses, anti–HA-B specific CD4+ T cells dominated the influenza B virus–specific response, with abundant neuraminidase-specific responses also detectable in many subjects. Reactivity to influenza B virus M1 and NP was modest. When IL-2 was used to assess reactivity to influenza viral proteins, it generally mirrored the hierarchies detected with IFN-γ, with a greater frequency of IFN-γ–producing cells for all influenza virus protein specificities. This Th1 bias supports the likelihood that, as adults, most of the subjects recruited for this study had been infected with influenza virus during their lifetime.
Figure 1D demonstrates the striking differences in the immunodominance hierarchies among influenza A and B virus–reactive cells. The average influenza virus protein–specific ELISPOT responses to the strain of viruses indicated above each panel were summed and a pie diagram constructed, where each slice of the pie depicts the relative fraction of the response dedicated to hemagglutinin, neuraminidase, NP, and M1 for each virus. It is clear that influenza B virus hemagglutinin–reactive CD4+ T cells (indicated in red) far outnumbered the other influenza B virus specificities, while M1 (indicated in yellow) dominated influenza A virus–reactive cells.
There has been an increasing appreciation of the importance of cytotoxic CD4+ T cells in protection from influenza, both within infected lungs in animal models [13] and in peripheral blood during human challenge studies [14]. We therefore sought to directly quantify the cytotoxic potential of influenza virus–specific CD4+ T cells, using GzmB ELISPOT assays (Figure 1C). We speculated that cytotoxic functionality might be enriched in CD4+ T-cell specificities directed against viral antigens at high abundance after infection, when inflammatory mediator production and viral protein expression are elevated. However, in contrast to the diverse viral CD4+ T-cell specificities detectable by IFN-γ and IL-2 production, our studies revealed that GzmB-reactive cells were largely restricted to HA-B and M1, with a lower frequency of responses directed against influenza A virus NP– and NA–derived epitopes. NP-B–, H1-, and M1-B–reactive CD4+ T cells from most subjects did not produce detectable GzmB, and, surprisingly, NS1-reactive cells, elicited only by infection, also produced very little GzmB.
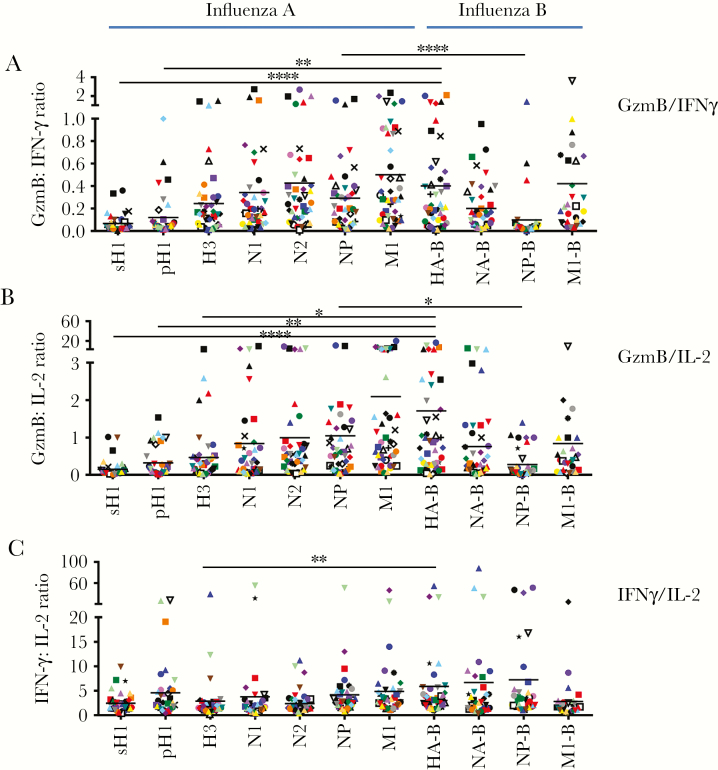
To further evaluate any protein-specific biases in GzmB production, the ratio of GzmB-producing cells to either IFN-γ– or IL-2–producing cells was examined, with the ratio of IFN-γ– to IL-2–producing cells serving as a comparator group (Figure 2). While the ratio of IFN-γ– to IL-2–producing cells among CD4+ T cells specific for influenza virus proteins was relatively comparable for all subjects (Figure 2C), quantification of the ratio of GzmB- to IFN-γ–producing cells (Figure 2A) and the ratio of GzmB- to IL-2–producing cells (Figure 2B) revealed that the frequency of GzmB-producing CD4+ T cells relative to the other mediators was not equivalent among CD4+ T cells of different viral specificities. The lowest frequency of CD4+ T cells producing GzmB relative to those producing either IFN-γ or IL-2 was detected in response to H1 and NP-B. These data support the existence of protein-specific biases in the functional potential of influenza virus–reactive cells.
Figure 2.
Protein-specific biases in the production of granzyme B (GzmB). Using the previously quantified cell frequencies as measured by enzyme-linked immunospot analysis, we calculated the ratio of GzmB-producing CD4+ T cells to interferon γ (IFN-γ)–producing CD4+ T cells (GzmB:IFN-γ ratio; A) and the ratio of GzmB-producing CD4+ T cells to interleukin 2 (IL-2)–producing CD4+ T cells (GzmB:IL-2 ratio; B) for each protein, with the ratio of IFN-γ–producing CD4+ T cells to IL-2–producing CD4+ T cells (IFN-γ:IL-2 ratio; C) serving as a comparator. While the IFN-γ:IL-2 ratio was relatively similar across all viral specificities, there was variability in the frequency of GzmB production among CD4+ T cells specific for different viral antigens. Donors are indicated by their symbols, with mean values from all subjects depicted with black lines. *P ≤ .05, **P ≤ .01, and ****P ≤ .0001, by the Kruskal-Wallis test with the Dunn post hoc test, for differences between influenza A virus and corresponding influenza B virus proteins.
DISCUSSION
The results presented here demonstrate the substantial diversity in the influenza virus–specific CD4+ T-cell repertoire in healthy adults, as well as the variability in the abundance and functional potential of cells specific for different influenza viral antigens. These studies reveal that many adults have robust and diverse influenza viral antigen specificities in their CD4+ T-cell compartment. While most individuals had Th1-biased CD4+ T-cell reactivity directed against many different influenza virus proteins, the production of GzmB by CD4+ T cells was largely restricted to those specific for HA-B and influenza A virus M1 and NP, with an almost undetectable number of granzyme-producing cells specific for H1 and NP-B.
The most unexpected aspect of our data is the marked difference in human CD4+ T-cell immunodominance and cell functionality present between the influenza A and influenza B viral proteins. While for influenza A virus, CD4+ T cells specific for internal virion proteins dominate the response, the reverse was seen for influenza B virus, where HA-B was immunodominant. These findings raised the possibility that influenza viral protein abundance within virions might differ. However, a similar abundance of these proteins was found within the influenza A and influenza B virions from which most inactivated vaccines are made [15]. To better understand the differences noted here between influenza A and B virus–specific immunity, additional knowledge of the persistence of influenza B virus antigens, the frequency of infection with these distinct viruses, and the inflammatory responses elicited by influenza A virus versus influenza B virus is necessary. Interestingly, the growing use of quadrivalent vaccines that contain twice the amount of influenza B virus hemagglutinin may serve to further accentuate the disparities in the influenza A virus– and influenza B virus–specific CD4+ T-cell repertoires noted in this study.
Given this diverse and variable circulating CD4+ T-cell repertoire in healthy adults, questions regarding how to best use vaccination to enhance CD4+ T-cell reactivity against influenza virus and provide broad and durable protective immunity are raised. Future studies are needed to improve our understanding of which CD4+ T-cell subsets are critical for influenza protection and to develop vaccination platforms that allow for targeted boosting of these specificities. Such knowledge is crucial for successful novel vaccination strategies that enhance cellular immunity, allowing for broader cross-reactive protection across human populations.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the NYICE clinical core for enrolling study participants and processing clinical samples, and the study participants for their willingness to contribute to scientific research.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (Centers of Excellence for Influenza Research and Surveillance contract HHSN272201400006C, contract HHSN272201200005C to A. J. S., and grant 1K08AI106954 to J. L. N.); and by the Doris Duke Charitable Foundation (Clinical Scientist Development Award 2015098 to J. L. N.).
Potential conflicts of interest. J. J. T. reports professional relationships with or consultancies for Sequiris, Novartis, GlaxoSmithKline, Pfizer, Merck, Vaxart, Vir Biotechnology, FluGen, Takeda, Medicago, Protein Sciences, Vaccitech, and Janssen. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 10th Annual Centers of Excellence for Influenza Research and Surveillance Network Meeting, Atlanta, Georgia, 16–19 July 2017; 9th Annual Centers of Excellence for Influenza Research and Surveillance Network Meeting, Memphis, Tennessee, 26–29 June 2016.
References
- 1. Sant AJ, Richards KA, Nayak J. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr Opin Immunol 2018; 53:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strutt TM, McKinstry KK, Marshall NB, Vong AM, Dutton RW, Swain SL. Multipronged CD4(+) T-cell effector and memory responses cooperate to provide potent immunity against respiratory virus. Immunol Rev 2013; 255:149–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zens KD, Farber DL. Memory CD4 T cells in influenza. Curr Top Microbiol Immunol 2015; 386:399–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 2010; 185:4998–5002. [DOI] [PubMed] [Google Scholar]
- 5. Hayward AC, Wang L, Goonetilleke N, et al. ; Flu Watch Group Natural T cell-mediated protection against seasonal and pandemic influenza. results of the flu watch cohort study. Am J Respir Crit Care Med 2015; 191:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Assarsson E, Bui HH, Sidney J, et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol 2008; 82:12241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uchtenhagen H, Rims C, Blahnik G, et al. Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat Commun 2016; 7:12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paul Glezen W, Schmier JK, Kuehn CM, Ryan KJ, Oxford J. The burden of influenza B: a structured literature review. Am J Public Health 2013; 103:e43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nayak JL, Richards KA, Yang H, Treanor JJ, Sant AJ. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J Infect Dis 2015; 211:1408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alam S, Knowlden ZA, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 2014; 88:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 2016; 16:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown DM, Lampe AT, Workman AM. The differentiation and protective function of cytolytic CD4 T cells in influenza infection. Front Immunol 2016; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol 2012; 86:6792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilkinson TM, Li CK, Chui CS, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012; 18:274–80. [DOI] [PubMed] [Google Scholar]
- 15. Hutchinson EC, Charles PD, Hester SS, et al. Conserved and host-specific features of influenza virion architecture. Nat Commun 2014; 5:4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.