Abstract
We recently identified a single potently neutralizing monoclonal antibody (mAb), mAb114, isolated from a human survivor of natural Zaire ebolavirus (EBOV) infection, which fully protects nonhuman primates (NHPs) against lethal EBOV challenge. To evaluate the ability of vaccination to generate mAbs such as mAb114, we cloned antibodies from NHPs vaccinated with vectors encoding the EBOV glycoprotein (GP). We identified 14 unique mAbs with potent binding to GP, 4 of which were neutralized and had the functional characteristics of mAb114. These vaccine-induced macaque mAbs share many sequence similarities with mAb114 and use the same mAb114 VH gene (ie, IGHV3-13) when classified using the macaque IMGT database. The antigen-specific VH-gene repertoire present after each immunization indicated that IGHV3-13 mAbs populate an EBOV-specific B-cell repertoire that appears to become more prominent with subsequent boosting. These findings will support structure-based vaccine design aimed at enhanced induction of antibodies such as mAb114.
Keywords: Ad5 immunization, cynomolgus macaques, ebolavirus, NPC1, receptor binding domain
Ebolavirus was the cause of the 2014 epidemic in West Africa, and it is associated with a high mortality rate (25%–90%) [1, 2]. Vaccines have shown efficacy in protecting macaques against challenge with Zaire ebolavirus (EBOV) [3–8]. In the absence of vaccination, several reports have shown the potential for passive administration of monoclonal antibody (mAb) cocktails in the treatment of macaques with ongoing EBOV infection [9–12]. We recently reported that administration of a single EBOV survivor antibody, mAb114, protected macaques from lethal challenge with EBOV when given as late as 5 days after inoculation [13].
The process through which EBOV enters target cells is a unique and complex multistep process. After attachment to the plasma membrane, the virus is taken up by macropinocytosis and transported into the target cell lysosomal compartment [14–18]. Lysosomes are characterized by low pH and by the presence of acid-dependent cellular proteases, including cathepsin L and B [19]. These proteases remove approximately 310 amino acids from the EBOV glycoprotein (GP) [20–22], expose a previously hidden receptor binding domain (RBD), and allow GP to engage its receptor, the Niemann-Pick disease, type C1 (NPC1) protein [22–24]. The mechanism of mAb114-mediated neutralization has been shown to be dependent on potent binding to the cleaved GP1 core (Supplementary Figure 1A) and strong competition with NPC1 for binding to cleaved GP [25].
In this report, we sought to determine the ability of vaccination to generate mAb114-like antibodies in a macaque vaccinated with deoxyribonucleic acid (DNA) and adenovirus type 5 (Ad5)-vectored vaccines encoding for the EBOV GP by amplification of immunoglobulin (Ig) gene transcripts from single-cell sorted EBOV GP-specific memory B cells and evaluated binding, neutralization, and functional characteristics of the cloned antibodies.
METHODS
Ethics Statement
The study was approved by the Vaccine Research Center Institutional Animal Care and Use Committee.
Vaccination and Sample Preparation
Monoclonal antibodies were cloned from a cynomolgus macaque (macaca fascicularis) that was primed with DNA and boosted 3 times with Ad5 EBOV GP. Prime consisted of 1 mg of DNA, whereas for boosting, 1011 (first and second boost) and 1012 (third boost) adenoviral particles were injected. All injections were performed intramuscularly. Blood was collected approximately 1 month after each vaccination. Peripheral blood mononuclear cells (PBMCs) were purified from Ficoll-Paque PLUS (GE Healthcare) ethylenediaminetetraacetic acid-treated blood and temporarily frozen until further experiments were performed.
Generation of Ebolavirus-Specific Probes and Conjugation for Flow Cytometry
The pCAGGS-GCN4-Avi vector was created by synthesizing DNA (Integrated DNA Technologies) encoding for a GCN4 site followed by an Avitag peptide (underlined) and His tags (MKQIEDKIEEILSKIYH IENEIARIKKLIGEVASSSGLNDIFEAQKIEWHEAHHHH HHG) and cloned into the pCAGGS expression vector using EcoRI and SmaI (New England Biolabs). A transmembrane-deleted EBOV GPΔTM (Δ657–676) and EBOV GPΔMUC (Δ309–505, Δ657–676) were amplified by polymerase chain reaction (PCR) using 5’ATGGTACCTAAATGGGCGTTACAGGA and CGACGCGTTCCAATACCTGCCGGT. The PCR products were subsequently cloned into pCAGGS-GCN4-Avi using KpnI and MluI (New England Biolabs). The EBOV Avitag proteins were expressed in HEK293T cells and purified as previously described [24, 25]. Site-specific biotinylation of purified proteins was performed by using the BirA enzyme kit (Avidity) following manufacturer’s instructions. Unbound or excess biotin was later removed by using Zeba spin desalting columns (Thermo Fisher Scientific). Finally, biotinylated full-length GP and GPΔ−MUC were conjugated at a 1:1 molar ratio to ExtrAvidin-phycoerythrin (PE) (Sigma-Aldrich) and streptavidin-APC (Life Technologies), respectively.
Antigen-Specific Single Memory B-Cell Sorts
The PBMCs were thawed and stained with the following antihuman-conjugated mAbs cross-reacting with macaque species: CD3-BV421 (BD Biosciences), CD4/8/14-BV421 (BioLegend), CD20-BV605, CD27-BV710 (both from BioLegend), IgG-Alexa680 (Custom-conjugated in house at the Vaccine Research Center), and IgM-FITC (BD Biosciences). Dead cells were stained with 0.25 μL/mL 7-aminoactinomycin D (Life Technologies). Zaire ebolavirus-specific memory B cells were identified by staining with 300 ng each of GP full-length-PE and GPΔ-MUC-APC (prepared in house, please see above). Samples were acquired on a FACS Aria II machine interfaced to the FacsDiva software (BD Biosciences), and EBOV-specific memory B cells were single-cell sorted into 96-well plates and frozen at −80°C until further experiments were performed. Flow cytometry analyses were performed by using the FlowJo software (Tree Star, Inc). The gating strategy and the definition of the B-cell subsets analyzed are summarized in Supplementary Figure 1B.
Amplification of VDJ/VJ Genes by Single-Cell Polymearse Chain Reaction, Cloning Into Immunoglobulin Expression Vectors, and Monoclonal Antibody Production
One 96-well plate with EBOV-specific single memory B cells was thawed, and ribonucleic acid was reverse-transcribed into complementary DNA with random hexamers (GeneLink) by using the Superscript III kit (Invitrogen) following manufacturer’s instructions. Nested PCR was then performed by using primers and procedures previously reported by Sundling et [26] al for the amplification of VDJ/VJ genes from single-cell sorted rhesus macaque (macaca mulatta) B cells. After single-pass sequencing of the second-round PCR products, the VDJ and VJ gene lineage for each sequence was determined by alignment to known macaque or human V, D, and J genes using IMGT/V-QUEST (http://www.imgt.org). Sequence-specific cloning primers were then designed for all productive and unique matched heavy and light chain sequences, and first-round PCR products were reamplified using the Pfu DNA Polymerase kit (Promega) following manufacturer’s instructions. Cloning of the final PCR products into rhesus macaque Igγ1, Igκ, or Igλ expression vectors was then performed [27]. Antibodies, including mAb114, were produced by transient transfection of Expi293F cells (Invitrogen) with 293fectin (Invitrogen) as described previously [25].
In Vitro Neutralization
Monoclonal antibodies were assessed for neutralization potency using a single-round infection assay with EBOV GP (Mayinga variant)-pseudotyped lentivirus particles expressing a luciferase reporter gene as previously described [4]. Final analyzed mAb concentrations were from 10 to 1 × 10−4 μg/mL except for ma-D08 from 6.25 to 1 × 10−4 μg/mL. VRC01 was used as irrelevant negative mAb control, and mAb114 was used as positive control [13]. Half-maximal inhibitory concentration (IC50) was calculated by 4-PL fit using the GraphPad Prism.
Production of GPTHL
Cleaved GP (GPTHL) was produced using thermolysin as previously described [24].
Enzyme-Linked Immunosorbent Assay
Immunoglobulin G binding to EBOV GP was measured by enzyme-linked immunosorbent assay (ELISA) as previously described [13] using 100 ng/well purified GΔTM, GPΔMUC, or GPTHL [24]. Anti-EBOV GP IgG half-maximal effective concentration (EC50) was calculated by 4-PL fit using the GraphPad Prism software.
Immunoprecipitation and Western Blotting
For each mAb, protein G agarose (Sigma-Aldrich) (100 μL packed resin) was incubated with 5 μg of IgG in 2 bead volume of phosphate-buffered saline (PBS) + 0.02% Tween 20 (Sigma-Aldrich) for 30 minutes at room temperature with end-over-end agitation. Unbound IgG was removed by washing the resin 3 times in 2 resin volumes of PBS + 0.02% Tween 20 + 0.1% bovine serum albumin ([BSA] Sigma-Aldrich). Resin was resuspended in 3 volumes of PBS + 0.02% Tween 20 + 0.2% BSA and 75 μL distributed into separate tubes for binding with GPΔMUC or GPTHL, respectively. After the addition of the protein, one-tenth volume was removed for the input blot, and the remainder was incubated for 1 hour at room temperature with end-over-end agitation. The resin was finally washed 3 times with 6 volumes of PBS + 0.02% Tween 20, and immune-complexes were detached by elution with 75 μL IgG elution buffer (Pierce). The supernatant and input were analyzed by Bio-Safe Coomassie G-250 Stain (Bio-Rad) and by Western blot for GP1 using an antirabbit polyclonal sera previously described [24] followed by a donkey antirabbit-horseradish peroxidase (HRP) (GE Healthcare) and by using Bio-Safe Coomassie G-250 Stain, respectively.
Enzyme-Linked Immunosorbent Assay-Based Antibody Competition With mAb114
The degree of antibody competition with mAb114 was determined by ELISA. mAb114 was biotinylated using the EZ-Link NHS-PEO solid-phase biotinylation kit (Pierce). Labeled mAb114 was tested by ELISA for binding to GPΔMUC to determine the optimal concentration to achieve 70%–80% maximal binding. The biotin-labeled mAb114 was then used as a probe to assess whether its binding (measured using streptavidin-HRP) was inhibited by preincubation of GPΔMUC-coated wells with the unlabeled macaque mAbs. Percentage of inhibition was calculated by subtracting the optical density values of biotinylated mAb114 in the presence of nonbiotinylated mAb114 at a concentration of 8 μg/mL and of biotinylated mAb114 in the presence of each of the macaque mAbs also at 8 μg/mL.
Biolayer Interferometry Antibody Cross-Competition Assay
Antibody cross-competition was determined by biolayer interferometry using a FortéBio Octet HTX instrument. A total of 15 µg/mL GPΔMUC protein was loaded onto biosensors using amine coupling (AR2G; FortéBio) for 600 seconds. mAb114, 13C6 (IBT), KZ52 (IBT), NPC1-dC, and isotype negative control were diluted to 35 µg/mL in PBST-BSA (1× PBS + 1% BSA + 0.01% Tween). Binding of competitor and analyte mAbs were each assessed for 300 seconds. The assay was performed in duplicate with agitation at 1000 rpm at 30°C. The percentage inhibition (PI) was calculated by the equation: PI = 100 − [(probing mAb binding in the presence competitor mAb)/(probing mAb binding in the absence of competitor mAb)] × 100.
Fab Fragment Preparation and Electron Microscopy
Fab fragments were produced using the Fab Preparation Kit (Pierce) following manufacturer’s instructions. GPΔMUC-mAb complexes were prepared incubating ~9 nM of GP produced in 293GnTi−/− with 40 nM Fab in a buffer containing 20 mM HEPES and 150 mM NaCl at pH 7.4 and incubating for 30 minutes at room temperature before flash freezing with liquid nitrogen. For electron microscopy, samples were adsorbed at a concentration of 0.03 mg/mL to glow-discharged carbon-coated copper grids and stained with 0.75% uranyl formate. Images were recorded on an FEI T20 microscope equipped with a 2k × 2k Eagle CCD camera using SerialEM [28]. Particle picking and 2-dimensional classification were performed with EMAN2 [29]. Representative class averages were then selected.
Binding Kinetics at Neutral and Low pH and Competition With Niemann-Pick Disease, Type C1-Domain C
Binding kinetics of Fab derived from mAbs to GPΔMUC or GPTHL and competition of mAbs and the C-domain from the Ebola receptor protein Niemann-Pick C1 (NPC1-dC) for binding to GPTHL [30] were determined based on biolayer interferometry using a fortéBio Octet HTX instrument as previously described [13, 25]. The NPC1-dC was produced as previously described [25].
Statistical Analyses
Paired Student t test was used to determine the statistical significance on the differences in binding potency to GPΔTM and GPΔMUC observed for the mAbs.
RESULTS
Vaccination Elicits Neutralizing Antibodies That Target the GP1 Core Similarly to mAb114
Protective vaccination, with DNA and adenoviral vectors encoding EBOV GP, has been demonstrated in cynomolgus macaques [3, 4, 31, 32]. Therefore, we stained and single-cell sorted EBOV-specific memory B cells from a cynomolgus macaque that was primed with DNA and boosted 3 times with Ad5 EBOV GP, by using GP-specific probes (Supplementary Figure 1B). We successfully PCR-amplified 56 productive matched heavy and light chains sequences and 30 unique sequence pairs from single B cells. Initial screening of supernatants from cells expressing the cloned mAb constructs revealed 14 mAbs with a potent EBOV GP-specific binding (EC50 <10 µg/mL; data not shown).
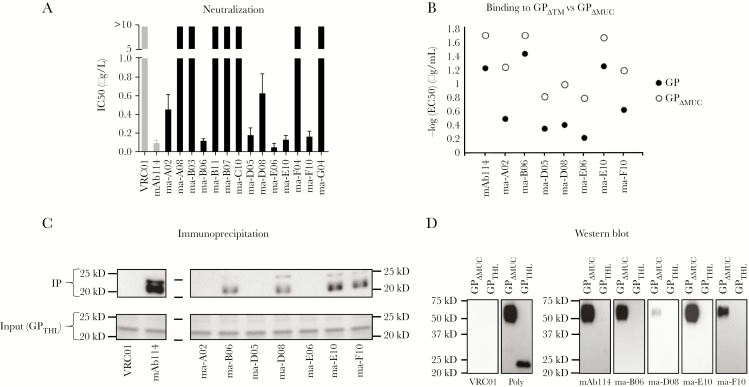
Because mAb114 is a potently neutralizing antibody [13], we first tested the purified 14 GP-specific mAbs for neutralizing activity using an in vitro pseudovirus assay. Seven of the 14 antibodies showed neutralization with similar IC50 compared with mAb114 (Figure 1A and Supplementary Figure 1C). The EBOV GP contains a large mucin-like domain (MLD) that is dispensable during infection. The binding affinity of mAb114 is slightly lower in the presence of the MLD (Figure 1B and Supplementary Figure 2). We noted that 7 of the neutralizing antibodies showed a similar decreased binding affinity when the MLD is present (Figure 1B and Supplementary Figure 2).
Figure 1.
Neutralization and binding characteristics of vaccination-induced macaque monoclonal antibodies (mAbs). (A) Bar chart representing the values of half-maximal inhibitory concentration ([IC50] μg/mL) calculated after in vitro neutralization assay using Zaire ebolavirus (EBOV) glycoprotein (GP)-pseudotyped-virus performed with the 14 mAbs that were cloned. In gray are negative controls VRC01 and mAb114 used as a reference. Seven of 14 mAbs showed ability to neutralize in vitro. The IC50 values were calculated from a total of 3 experiments run in triplicate. (B) Dot plots show the −log half-maximal effective concentration ([EC50] μg/mL) representing binding of the 7 neutralizers identified in A to GP (filled circles) and GPΔMUC (empty circles). All of the mAbs tested bound GPΔMUC with greater potency compared with GP (P = .0097). The EC50 values were calculated from a total of 2 experiments run in duplicate. (C) Immunoprecipitation of the 7 neutralizers performed with GPTHL. Four of seven mAbs were able to precipitate GPTHL. As control, input GPTHL is shown on a Coomassie-stained gel. (D) Western blot on the 4 mAbs identified in C performed with GPΔMUC and GPTHL. For mAb114, none of the mAbs tested were able to bind GPTHL by Western blot. As a positive control for GPTHL, binding of polyclonal serum is shown.
Endosomal protease cleavage of the EBOV GP removes the MLD and the “glycan cap” region, leaving a minimal GP1 core. A defining feature of mAb114 is binding to the GP1 core [25]. Therefore, we tested the capacity of the 7 neutralizing mAbs to bind after GP cleavage with thermolysin (GPTHL); a cleavage that faithfully mimics GP cleavage by cathepsins [21]. Four mAbs (ma-B06, ma-D08, ma-E10, and ma-F10) showed reactivity to GPTHL both by immunoprecipitation and by direct ELISA (Figure 1C and Supplementary Figure 3A). In addition to conformational contacts within the GP1 core, mAb114 makes contacts with the glycan cap [25]. We tested binding of mAb114 and the GP1 core-directed mAbs to GPΔMUC and GPTHL under denaturing and reducing conditions by Western blot. We observed that mAb114 reacted to GPΔMUC but not to GPTHL (Figure 1D), suggesting that the epitope within the glycan cap is conformation-independent. Each of the macaque GP1 core-binding antibodies reacted in a similar manner to mAb114. These data indicate that the vaccine-induced mAbs contact the glycan cap and GP1 core in a manner similar to mAb114.
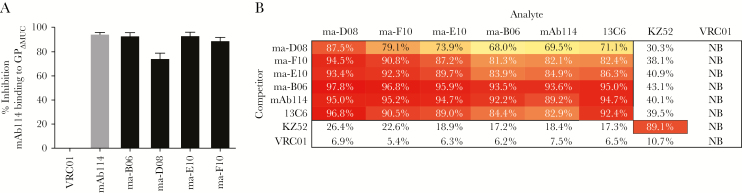
Vaccine-Induced GP1 Core-Directed Antibodies Compete With mAb114
To determine whether the 4 vaccine-induced GP1 core-directed mAbs target a similar region on GP as targeted by mAb114, we assessed direct competition for binding to GPΔMUC by ELISA (Supplementary Figure 3B). All 4 mAbs blocked mAb114 binding to a high degree (range, 80%–100%) (Figure 2A). We next used biolayer interferometry to analyze cross-competition among mAb114, the 4 GP1 core-directed mAbs, and 2 additional mAb controls, 13C6 and KZ52. 13C6 binds to the glycan cap and competes with mAb114 for binding to GP, likely due to steric hindrance [10, 25]. KZ52 binds to the base of GP and does not compete with mAb114 [25, 33]. Biolayer interferometry confirmed that the GP1 core-directed mAbs compete with mAb114 for binding to GP and revealed cross-competition with each other (Figure 2B). In addition, mAb114 and the macaque mAbs each showed competition with 13C6 but not with KZ52 (Figure 2B). Altogether, these results suggest that these vaccine-induced antibodies target a region on the GP1 core that partially or completely overlaps with the mAb114 epitopes.
Figure 2.
Direct competition with mAb114 and cross competition between the macaque GP1 core-directed monoclonal antibodies (mAbs). (A) Bar chart representing the percentage of binding inhibition of biotinylated mAb114 to GPΔMUC in the presence of each of the 4 putative mAb114-like antibodies. All mAbs showed a high degree of competition with mAb114 for binding to GPΔMUC (calculated at the concentration of 8 μg/mL). The gray bar shows the maximal degree of self-competition of mAb114. Values were calculated from a total of 2 experiments run in duplicate. (B) Biolayer interferometry showing percentage of interference across mAbs for binding to GPΔMUC. All the putative mAb114-like antibodies competed with mAb114 and to each other for binding to GPΔMUC. In addition, for mAb114, all the macaque mAbs partially competed with 13C6. This experiment was performed twice. Values are shown from 1 representative experiment. Abbreviation: NB, not binding.
Macaque GP1 Core-Targeted Monoclonal Antibodies Bind Glycoprotein at Low pH and Compete With NPC1 for Receptor Binding
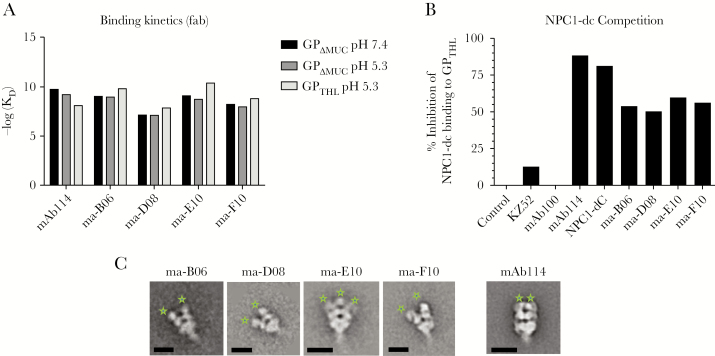
We previously showed that mAb114 has stable high-affinity binding at low pH [25]. This allows mAb114 to remain bound to cleaved GP in low pH compartments where receptor engagement occurs [24, 25]. Therefore, we measured the affinity of binding to GPΔMUC and GPTHL using biolayer interferometry at physiologic and low pH (Figure 3A and Supplementary Figure 3C). Although the affinity of mAb114 decreased slightly after cleavage, the macaque GP1 core mAb affinities slightly increased and ranged from 13.8 (ma-D08) to 0.4 nM (ma-E10) (Figure 3A and Supplementary Figure 3C).
Figure 3.
Kinetics of the macaque GP1 core antibodies at neutral and low pH, Niemann-Pick disease, type C1-domain C (NPC1-dC) competition for binding to GPTHL and electron microscopy. (A) Bar chart on the binding kinetics of the GP1 core antibodies compared with mAb114. −log (KD) values were calculated by biolayer interferometry performed with Fab binding to GPΔMUC at neutral and low pH (black and dark gray bars, respectively) as well as to GPTHL at low pH only (light gray bars). All monoclonal antibodies (mAbs) showed similar kinetics compared with mAb114 with ma-D08 showing slightly lower values. Fab fragments derived from papain digestion of mAbs were used for measuring kinetics. This experiment was performed twice. Values are shown from 1 representative experiment. (B) Bar chart on competition of the GP1 core antibodies with NPC1-dC compared with mAb114 performed by biolayer interferometry using whole IgG binding to GPTHL. All mAbs showed similar competition with NPC1-dC for binding to GPTHL. Human immunodeficiency virus GP120 (indicated as “Control”) and 2 additional mAbs (KZ52 and mAb100) that are known to bind the base of GPTHL far from the receptor binding domain (RBD) were used as negative controls. NPC1-dC was used to assess the maximal degree of self-competition. This experiment was also performed twice, and values are shown from 1 representative experiment. (C) Electron microscopy reveals that all of the RBD blockers approach glycoprotein (GP) vertically but with a slightly more open angle compared with mAb114 (Fab indicated with green stars). The scale bars are 10 nm.
The defining characteristic of mAb114 is competition with the Ebola receptor protein, NPC1, for binding to GPTHL [25, 34]. Therefore, we asked whether ma-B06, ma-D08, ma-E10, and ma-F10 competed with NPC1 for binding to GPTHL. We observed that all of the mAbs competed with NPC1 for binding to GPTHL (Figure 3B) and similarly to mAb114, they might neutralize infection by blocking GP interaction with its receptor.
A structural feature of mAb114 interaction with GP is binding with a near-vertical angle of approach within the chalice of GP [25, 34] and 3 Fabs per trimer. We used negative-stain electron microscopy and single-particle analysis to compare the mode of binding of each Fab to GPΔMUC to that of the Fab114. We noted that at least 2 Fabs bound to each GP trimer (Figure 3C). It is interesting to note that although each of the vaccine-induced RBD-blocking Fabs bound to the trimer within the chalice, they each had a more open angle when compared with mAb114 (Figure 3C). Taken together, these data indicate that immunization is capable of inducing mAb114-like antibodies.
Vaccine-Induced Receptor Binding Domain-Blocking Antibodies Originate From a Similar VH Gene Lineage as mAb114
Because the Ig variable region gene families influence the gross binding characteristics of antibodies [35], we asked whether the vaccine-induced RBD-blocking antibodies would originate from B cells carrying similar gene families. In contrast with the other neutralizing antibodies that we cloned, all of the RBD-blocking antibodies originate from the macaque IGHV3-13 and the IGLV1S2 families. It is interesting to note that the IGHV3-13 is the same gene family from which mAb114 originated in human (Figure 4A). Because our previous reports suggested that the heavy chain alone is sufficient for binding of mAb114 to GP [13, 25], we focused our analyses on the mAb heavy chains. We noted that the complementarity determining regions (CDRs) 1 and 2 on the antibody heavy chains in the macaque RBD blockers shows a high degree of homology between each other (Figure 4B), further suggesting that these antibodies bind a very similar region on GP. On the other hand, the mAb114 heavy chain CDR 1 and 2 differ significantly from the 4 macaque antibodies (Figure 4B). This may be due to differences between human and macaque Ig alleles as well as to differences in somatic hypermutations (SHM) [36]. These differences might contribute to the slightly differing angles of approach that we observed between the macaque mAbs and the human mAb114 (Figure 3C).
Figure 4.
Immunoglobulin VH-gene sequence analyses and somatic hypermutations (SHM) on the 7 macaque neutralizing antibodies compared with mAb114. (A) Alignment of immunoglobulin VH-gene sequences for the 7 macaque neutralizers point out that all of the receptor binding domain (RBD) blockers originated from the IGHV3-13*01 family, the same of mAb114 itself as well as a similar degree of SHM between the macaque RBD blockers and mAb114. All of the macaque RBD blockers had an overall lower degree of SHM compared with the other neutralizers, which were mapped to bind the glycan cap, suggesting that the RBD blockers might be more recent than other antibodies such as glycan cap binders. Among all of the RBD blockers, ma-D08 appeared to be the closest to germline in terms of VH-gene region homology. (B) Protein sequence alignment between mAb114 and the the vaccine-induced macaque RBD blockers further highlights the relatedness of these monoclonal antibodies (mAbs) as well as (C) the similar type of affinity maturation that they have undergone during selection, which was observed by comparing the mAb protein sequence to their predicted unmutated common ancestors (UCAs). The red boxes indicate the complementarity determining regions (CDR), whereas underlined in red are the amino acid mutations compared with the UCS. Abbreviations: D, deletions; I, insertions; S, substitutions.
We next focused on the overall heavy chain level of B-cell maturation and noted that the amount of SHM was similar to that of mAb114 (86.1%–92.2% vs 87.4% VH-region identity) as a result of the alignment to the macaque versus human reference databases (Figure 4A). Furthermore, when the CDR sequences for each mAb were compared with their respective inferred unmutated common ancestors, similar numbers and putative types of SHM were also observed (Figure 4B). We noted that the extent of SHM in the RBD blockers was lower than that seen in the 3 other neutralizing mAbs (86.1%–92.2% vs 73.6%–77.5% VH region identity) (Figure 4A and Supplementary Figures 2 and 3A). This suggests that these antibodies might have developed before the RBD blockers because SHM accumulate after each cell division [35].
Macaque IGHV3-13 Monoclonal Antibodies Populate the Antigen-Specific B-Cell Repertoire Progressively During the Course of Vaccination
mAb114 was isolated from a human survivor who may have experienced multiple antigen exposures, and the mAbs presented here were isolated from a macaque that received multiple vaccine-derived antigen exposures. Therefore, we hypothesized that sustained antigen exposure might enhance the generation of mAb114-like antibodies. To test this hypothesis, we analyzed the macaque VH-gene repertoire present after each immunization on PBMC collected 1 month after each vaccination. As expected, we found that EBOV+ memory B cells increase with time (Figure 5). In addition, we were able to identify precursors for each of the 3 VH3-13 mAbs from the second boost and noted that after each antigen encounter, the Ig variable region continues to be refined by SHM (Supplementary Figure 4). In line with this, the shape of the total IgG+ B-cell population versus total IgM+ B cells changes with time becoming more stringent during secondary immune responses, indicating memory B cell recall rather than engagement of newly formed naive B cells (Figure 5). Finally, single-cell sorts and PCR on EBOV+ B cells revealed that the frequency of macaque IGHV3-13 B cells although absent after prime, progressively increased during the course of vaccination (Figure 5). Although all of VH3-13 B cells generated in this study may not be mAb114-like, these data suggest that sustained antigen exposure increases the proportion of VH3-13 B cells, and, to be effective at inducing mAb114-like mAbs, vaccination strategies may need repeated exposures to antigen.
Figure 5.
Flow cytometry, immunoglobulin VH-gene sequence analyses, and somatic hypermutations (SHM) after each vaccination procedure. Flow cytometry analyses on the presence of Zaire ebolavirus (EBOV)+ memory B cell were performed on samples collected 1 month after each vaccination procedure. Increase of the frequency of EBOV+ memory B cells as well as increase in binding affinity (appearance of cells double positive for both glycoprotein [GP] and GPΔMUC) is evident during the course of vaccination. In line with this, sequence analyses on EBOV+ single-cell sorted cells showed a gradual increase of SHM (red portions of pie charts). IGHV3-13*01 B cells (green portions of pie charts) are absent after prime and progressively increase after each boost. The VH-gene region homology and the frequency of IGHV3-13*01 B cells was calculated based on productive unique heavy chain sequences obtained from each sort (starting from 96 cells). Abbreviations: Ad5, adenovirus type 5; DNA, deoxyribonucleic acid.
CONCLUSIONS
Preventive immunization of macaques with recombinant adenovirus vaccines encoding for the EBOV GP has been shown to be protective against lethal challenge with EBOV by CD8+ T cell-mediated virus clearance [4, 8]. The findings in this report indicate that immunization with GP encoding vectors induced antibodies with similar binding and functional properties as the protective antibody mAb114. Furthermore, repeated immunization with Ad5 increased the proportions of mAbs in the VH3-13 gene family. This suggests the possibility that the potency of immunization against EBOV might be enhanced with a multiboost strategy by possibly adding the protective function of a class of antibodies acting like mAb114 (that is by blocking the interaction between NPC1 and cleaved GP through stable binding to the RBD region at low pH) to that of CD8+ T cells.
It is interesting to note that mAb114-like antibody responses in both human and macaque originate from B cells harboring antibodies encoded by very closely related gene families. The relation between antibody specificity and Ig variable region gene families is well known in the field of human immunodeficiency virus (HIV) infection, and it is currently driving HIV vaccine research toward the design of specific B-cell lineage immunogens [37, 38]. The same direction is being explored for universal influenza vaccine design based on defined convergent (shared between different individuals) Ig gene signatures that have recently been described for vaccine-induced antibodies binding to the conserved region of the influenza hemagglutinin stem [39]. Thus, the definition of the B-cell ontogeny provided here for mAb114-like antibodies in EBOV will support structure-based vaccine design for enhanced triggering of germline-reverted mAb114-like antibody precursors. Selective or increased engagement of naive B cells precursors to mAb114-like antibodies might generate higher frequencies of memory B cells and allow quicker differentiation into mAb114-like antibody-secreting cells. Memory B cell-mediated serologic memory may persist over a long period of time, such as in the case of measles-specific memory B cells [40]. Further studies will need to be conducted to design and test modified vaccines as well as different vaccination strategies that enrich selection of B cells encoding for antibodies directed to blocking access to the RBD. In addition, modified vaccines and/or different vaccination strategies should also be evaluated for the generation of antibodies with multiple specificities, on their possible synergistic effects and on their degree of cross-reactivity to other ebolavirus species.
In addition, investigations into the degree of affinity maturation needed for the optimal neutralization and retention of binding in the low pH environments are needed. Although all 4 mAb114-like macaque mAbs that we isolated retain good binding and neutralization at low pH, we noted that ma-D08, which showed the lowest degree of binding and neutralization, also showed the lowest degree of SHM compared with the macaque VH3-13 germline (Figure 4A). This may indicate that SHM may indeed play a role for neutralization and binding. Continued maturation may be required to allow high-affinity recognition of the major epitope in the GP1 core for mAb114 binding, which is partially covered by the glycan cap and MLD, respectively [33, 41] (Supplementary Figure 1A), and may not be easily accessible to B cells.
In summary, we have shown that (1) mAbs generated through vaccination with EBOV GP encoding vectors in macaque present with binding and functional characteristics similar to mAb114, which was instead isolated from a human survivor; (2) these mAbs share many sequence similarities with mAb114 and may originate from same, or closely related, VH genes as mAb114; and (3) multiple exposures to antigen may be required for these antibodies to arise and fully mature. Overall, these findings will support structure-based vaccine design and/or multiboost vaccine strategies for enhanced triggering of mAb114-like antibodies.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Nicole Doria-Rose for useful discussion on the methods for isolating monoclonal antibodies, James Cunningham for support, Soo-mi Lee for help purifying probes, Ulrich Baxa for tips on sample preparation for electron microscopy, Kendra Leigh for critical review, and Brandon DeKosky, Amy Ransier, and Sam Darko for discussion on the sequencing data.
Financial support. This work was supported by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases. This work has been funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health (NIH), under contract HHSN261200800001E. Leidos Biomedical Research, Inc. provided support in the form of a salary (to Y. T.). J. M. received grant support from NIH-5K08AI079381 and a Boston Children’s Hospital Faculty Development Award.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Kucharski AJ, Edmunds WJ. Case fatality rate for Ebola virus disease in West Africa. Lancet 2014; 384:1260. [DOI] [PubMed] [Google Scholar]
- 2. Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sullivan NJ, Geisbert TW, Geisbert JB, et al. . Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 2003; 424:681–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sullivan NJ, Geisbert TW, Geisbert JB, et al. . Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med 2006; 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marzi A, Engelmann F, Feldmann F, et al. . Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc Natl Acad Sci U S A 2013; 110:1893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis 2007; 196(Suppl 2):S430–7. [DOI] [PubMed] [Google Scholar]
- 7. Zhou Y, Sullivan NJ. Immunology and evolvement of the adenovirus prime, MVA boost Ebola virus vaccine. Curr Opin Immunol 2015; 35:131–6. [DOI] [PubMed] [Google Scholar]
- 8. Stanley DA, Honko AN, Asiedu C, et al. . Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat Med 2014; 20:1126–9. [DOI] [PubMed] [Google Scholar]
- 9. Qiu X, Audet J, Wong G, et al. . Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med 2012; 4:138ra81. [DOI] [PubMed] [Google Scholar]
- 10. Olinger GG Jr, Pettitt J, Kim D, et al. . Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A 2012; 109:18030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marzi A, Yoshida R, Miyamoto H, et al. . Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PLoS One 2012; 7:e36192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu X, Wong G, Audet J, et al. . Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014; 514:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corti D, Misasi J, Mulangu S, et al. . Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016; 351:1339–42. [DOI] [PubMed] [Google Scholar]
- 14. Misasi J, Sullivan NJ. Camouflage and misdirection: the full-on assault of Ebola virus disease. Cell 2014; 159:477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davey RA, Shtanko O, Anantpadma M, Sakurai Y, Chandran K, Maury W. Mechanisms of Filovirus entry. Curr Top Microbiol Immunol 2017; 411:323–52. [DOI] [PubMed] [Google Scholar]
- 16. Hunt CL, Kolokoltsov AA, Davey RA, Maury W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J Virol 2011; 85:334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 2010; 6:e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quinn K, Brindley MA, Weller ML, et al. . Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J Virol 2009; 83:10176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White JM, Schornberg KL. A new player in the puzzle of filovirus entry. Nat Rev Microbiol 2012; 10:317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005; 308:1643–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 2006; 80:4174–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Misasi J, Chandran K, Yang JY, et al. . Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol 2012; 86:3284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carette JE, Raaben M, Wong AC, et al. . Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011; 477:340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Côté M, Misasi J, Ren T, et al. . Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011; 477:344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Misasi J, Gilman MS, Kanekiyo M, et al. . Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 2016; 351:1343–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods 2012; 386:85–93. [DOI] [PubMed] [Google Scholar]
- 27. Saunders KO, Pegu A, Georgiev IS, et al. . Sustained delivery of a broadly neutralizing antibody in nonhuman primates confers long-term protection against simian/human immunodeficiency virus infection. J Virol 2015; 89:5895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 2005; 152:36–51. [DOI] [PubMed] [Google Scholar]
- 29. Tang G, Peng L, Baldwin PR, et al. . EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- 30. Miller EH, Obernosterer G, Raaben M, et al. . Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 2012; 31:1947–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sullivan NJ, Hensley L, Asiedu C, et al. . CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat Med 2011; 17:1128–31. [DOI] [PubMed] [Google Scholar]
- 32. Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. Development of a preventive vaccine for Ebola virus infection in primates. Nature 2000; 408:605–9. [DOI] [PubMed] [Google Scholar]
- 33. Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008; 454:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krishnan A, Miller EH, Herbert AS, et al. . Niemann-Pick C1 (NPC1)/NPC1-like1 chimeras define sequences critical for NPC1’s function as a flovirus entry receptor. Viruses 2012; 4:2471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson KJ, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol 2013; 4:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Wang JH. Generation and repair of AID-initiated DNA lesions in B lymphocytes. Front Med 2014; 8:201–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Medina-Ramírez M, Garces F, Escolano A, et al. . Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J Exp Med 2017; 214:2573–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jardine JG, Kulp DW, Havenar-Daughton C, et al. . HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016; 351: 1458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Joyce MG, Wheatley AK, Thomas PV, et al. . Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 2016; 166:609–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med 2007; 357:1903–15. [DOI] [PubMed] [Google Scholar]
- 41. Tran EE, Simmons JA, Bartesaghi A, et al. . Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol 2014; 88:10958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.