Abstract
F-type lectins are phylogenetically widespread but selectively distributed fucose-binding lectins with L-fucose- and calcium-binding sequence motifs and an F-type lectin fold. Bacterial F-type lectin domains frequently occur in tandem with various protein domains in diverse architectures, indicating a possible role in directing enzyme activities or other biological functions to distinct fucosylated niches. Here, we report the biochemical characterization of a Streptosporangium roseum protein containing an F-type lectin domain in tandem with an NPCBM-associated domain and a family GH 29A alpha-l-fucosidase domain. We show that the F-type lectin domain of this protein recognizes fucosylated glycans in both α and β linkages but has high affinity for a Fuc-α-1,2-Gal motif and that the alpha-l-fucosidase domain displays hydrolytic activity on glycan substrates with α1-2 and α1-4 linked fucose. We also show that the F-type lectin domain does not have any effect on the activity of the cis-positioned alpha-l-fucosidase domain with the synthetic substrate, 4-Methylumbelliferyl-alpha-l-fucopyranoside or on inhibition of this activity by l-fucose or deoxyfuconojirimycin hydrochloride. However, the F-type lectin domain together with the NPCBM-associated domain enhances the activity of the cis-positioned alpha-l-fucosidase domain for soluble fucosylated oligosaccharide substrates. While there are many reports of glycoside hydrolase activity towards insoluble and soluble polysaccharides being enhanced by cis-positioned carbohydrate binding modules on the polypeptide, this is the first report, to our knowledge, of enhancement of activity towards aqueous, freely diffusible, small oligosaccharides. We propose a model involving structural stabilization and a bind-and-jump action mediated by the F-type lectin domain to rationalize our findings.
Keywords: alpha-l-fucosidase, carbohydrate-binding module, F-type lectin domain, fucose, Streptosporangium roseum
Introduction
F-type lectins are fucose-binding lectins with L-fucose (HX(26)RXDX(4)R/K) and calcium-binding sequence motifs and an F-type lectin fold (Bianchet et al. 2002). They are phylogenetically widespread but selectively distributed in viruses, bacteria and eukaryotes (Bishnoi et al. 2015), and have diverse functions ranging from innate immunity in fishes and amphibians to protoplast regeneration in the green alga, Bryopsis plumosa and virulence in the bacteria, Streptococcus pneumoniae and Streptococcus mitis (Vasta et al. 2017). Several eukaryotic F-type lectins (e.g., Anguilla anguilla agglutinin (AAA), Morone saxatilis agglutinin (MSA)) (Bianchet et al. 2002, 2010) and the F-type lectin domains (FLDs) of a couple of bacterial proteins (Streptococcus pneumoniae SP2159 and Streptococcus mitis lectinolysin) (Boraston et al. 2006; Farrand et al. 2008; Feil et al. 2012) have been biochemically and structurally characterized. Structurally, the FLD is a beta barrel with a jelly roll topology. The l-fucose binding site is a positively charged pocket at one end of the barrel with one histidine and two arginine residues that make hydrogen bonds with l-fucose. These basic residues are also part of the typical fucose binding FLD sequence motif, HX(26)RXDX(4)R/K (Bianchet et al. 2002, 2010; Boraston et al. 2006; Farrand et al. 2008).
FLDs are classified under the family “fucose binding lectins”, which is part of the only superfamily (galactose-binding domain like) of the fold “galactose-binding domain like” in SCOP, and together with discoidin domain and galactose-binding domain members under the coagulation factor 5/8 C-terminal (FA58C)/discoidin (PF00754) family in Pfam (Finn et al. 2014). In the Simple Modular Architecture Research Tool (SMART) database (Letunic et al. 2004), eukaryotic FLDs are classified under SMART accession number, SM00607 [the eel-Fucolectin Tachylectin-4 Pentraxin-1 domain (FTP)], and FLDs are also classified in the Carbohydrate-Active enZYmes (CAZy) database (Cantarel et al. 2009) as the carbohydrate binding module (CBM) family, CBM47 (http://www.cazy.org/CBM47).
Vasta et al. (Vasta et al. 2004; Odom and Vasta 2006) and we (Bishnoi et al. 2015) have previously analyzed the prevalence of FLDs in different life forms. These studies indicated that FLDs, while frequently existing as standalone lectin domains in eukaryotes, are almost always present in a variety of multi-domain architectural contexts in bacteria. The extent of diversity of co-occurring domains suggests that the sugar recognition capability of FLD is being harnessed in very different functional contexts in bacteria. Many of these FLD-containing polypeptides have co-occurring enzyme domains (Bishnoi et al. 2015) that include methyltransferase 21 and 23, lipase, and peptidase, and carbohydrate-active enzyme domains such as alpha-l-fucosidase, glycosyltransferase and cellulase. The latter proteins therefore have a domain architecture in which the FLD is positioned in cis (i.e., on the same polypeptide) with a carbohydrate-active enzyme domain. We hypothesized that cis positioning of the FLD with a carbohydrate-active enzyme domain might concomitantly result in a “cis effect”, i.e., the FLD might direct the activity of the cis positioned enzyme domain to carbohydrate substrates via its carbohydrate-binding ability and additionally or alternatively modulate the enzyme activity itself of the cis positioned domain.
We report here the biochemical characterization of a protein from an aerobic, Gram-positive free-living soil bacterium, Streptosporangium roseum DSM 43021 that belongs to the phylum Actinobacteria. This protein, hereafter referred to as SrFucNaFLD (GenBank accession number: ACZ87343.1), has an alpha-l-fucosidase domain (and an alpha-l-fucosidase C-terminal domain) in tandem with a small Novel Putative Carbohydrate Binding Module-associated domain (NPCBM-associated domain; also called NEW3 domain (Naumov 2004) and abbreviated as Na domain here) and an FLD. The Na domain has been previously reported to be associated with the Novel Putative Carbohydrate Binding Module (NPCBM domain; also called NEW2 domain (Naumov 2004, Rigden 2005)) on bacterial proteins that are galactosidases (Naumoff 2005).
Our study explores the glycan-binding specificity of the FLD, the kinetics and substrate specificity of the alpha-l-fucosidase domain, and the effect of the FLD on the activity of the cis positioned alpha-l-fucosidase domain. We find that cis positioning of the FLD enhances the activity of the alpha-l-fucosidase domain towards soluble fucosylated oligosaccharide substrates. We theorize that the observed “cis effect” is effected by structural stabilization as well as by functional targeting mediated by the lectin activity of the FLD.
Results
Cloning, expression and purification of SrFucNaFLD, mutants and domains
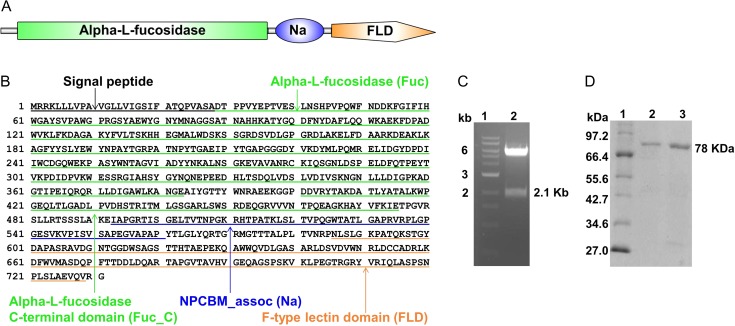
SrFucNaFLD (GenBank accession no: ACZ87343.1), as per the Conserved Domain Database (CDD), contains an N-terminal signal peptide followed by an alpha-l-fucosidase domain (Pfam family PF01120; smart00812), an alpha-l-fucosidase C-terminal domain (Pfam family PF16757), a small Na domain (Pfam family PF10633) and an FLD (Pfam family PF00754; smart00607) (Figure 1A and B). The FLD has a Ca2+ binding site as well as an FLD sequence motif, HX(27)RXDX(4)R, with residues, H623, R651 and R658, which are predicted to be critical for l-fucose binding. We successfully cloned and expressed SrFucNaFLD with a C-terminal hexahistidine tag and enriched it to ~85% purity using metal ion affinity chromatography (Figure 1C and 1D). The protein yield was ~0.5 mg starting from 400 mL of SrFucNaFLD-pET-28a(+) transformed E. coli BL21(DE3) culture. Recombinant SrFucNaFLD was also visualized by western analysis with an anti-C-terminal 6XHis tag antibody (data not shown).
Fig. 1.
(A) Domain architecture of SrFucNaFLD. (B) Protein sequence of S. roseum alpha-l-fucosidase in FASTA format depicting the amino acid sequence of the domains – alpha-l-fucosidase (Fuc), NPCBM_assoc (Na) and F-type lectin domain (FLD) within the polypeptide. (C) Digestion of recombinant clone with NcoI and XhoI demonstrating fallout of SrFucNaFLD gene from pET-28a(+); Lane 1: DNA marker from NEB; Lane 2: Positive clone with SrFucNaFLD insert (D) Coomassie blue-stained SDS-polyacrylamide gel with imidazole eluate fractions of a Ni-NTA metal ion affinity column, showing purified SrFucNaFLD protein; Lane 1: Protein marker from NEB; Lane 2 and 3: purified SrFucNaFLD protein.
We performed site-directed mutagenesis and made three SrFucNaFLD mutants, which had Ala in place of H623, R651 or R658 of the FLD sequence motif (Supplementary data, Figure S1), and the corresponding triple mutant (SrFucNaFLDH623AR651AR658A; referred to as SrFucNaFLDTM from this point onwards) (data not shown). We also generated a mutant, SrFucD244ANaFLD, in which the residue D244 of the alpha-l-fucosidase domain, which is equivalent to the catalytic nucleophile in Thermotoga maritima alpha-l-fucosidase (Sulzenbacher et al., 2004), was mutated to Ala (Supplementary data, Figure S1). Additionally, we made the mutant, SrFucD244ANaFLDH623AR651AR658A, which has mutations in both alpha-l-fucosidase domain and FLD (Supplementary data, Figure S1). We successfully expressed and purified all SrFucNaFLD mutants, similar to wildtype SrFucNaFLD (Supplementary data, Figure S1).
Apart from wildtype and mutant SrFucNaFLD, we also successfully expressed and purified recombinant proteins with C-terminal hexahistidine tags corresponding to just the alpha-l-fucosidase and alpha-l-fucosidase C-terminal domains (hereafter referred to as SrFuc), the alpha-l-fucosidase and alpha-l-fucosidase C-terminal domains together with the Npcbm_associated domain (hereafter referred to as SrFucNa), the Npcbm_associated domain (hereafter referred to as SrNa), the FLD (hereafter referred to as SrFLD), and the Npcbm_associated domain together with the FLD (hereafter referred to as SrNaFLD). We enriched SrFuc and SrFucNa to ~85%, and SrNaFLD, SrNa and SrFLD to ~90% using metal ion affinity chromatography, as assessed by visual examination of Coomassie blue-stained SDS-polyacrylamide gels (Supplementary data, Figure S2). We obtained protein yields of ~0.45 mg, ~0.6 mg, ~4 mg, ~3.3 mg and ~0.8 mg for purified SrFuc, SrFucNa, SrNaFLD, SrFLD and SrNa, respectively, starting from 150 mL of BL21(DE3) cultures transformed with the respective plasmids.
Using size exclusion chromatography, we determined that SrFLD exists as species of various sizes in solution (Supplementary data, Figure S3). Using Superdex 200 gel matrix, we extrapolated the molecular mass of the SrFLD species with the highest elution volume to be 9.3 kDa (as opposed to the theoretically calculated molecular mass of 17.1 kDa for the SrFLD monomer) using the linear equation between log (molecular mass) and Ve/Vo (ratio of elution volume to void volume) that we obtained with standard proteins of known molecular mass. We confirmed by SDS gel electrophoresis that the protein fractions corresponding to the SrFLD species with highest elution volume (peak 4) and other SrFLD species (peaks 1, 2 and 3) indeed had intact protein of expected size (Supplementary data, Figure S3). The results of our size exclusion chromatography therefore implied anomalous mobility. We obtained similar elution profiles with Sephacryl S200 gel matrix (composed of allyl dextran and N,N-methylene bisacrylamide) that lacks any carbohydrate moiety (data in accompanying manuscript, Mahajan and Ramya 2018). Our results indicated that (1) the anomalous mobility is not due to any carbohydrate-mediated binding of the protein to the gel matrix but is perhaps due to the protein assuming a non-spherical shape, and (2) the protein exists as a heterogeneous population of various oligomers in solution. We also performed intact mass analysis by mass spectrometry because we could not accurately assign oligomeric status by size exclusion chromatography. Using MALDI-MS as well as ESI-MS of SrFLD in solution, we identified masses corresponding to SrFLD oligomers; with MALDI-MS, we found the masses, 16.8 kDa, 33.7 kDa, 50.5 kDa, 67.3 kDa and 84.2 kDa, which correspond to the masses of SrFLD’s monomer, dimer, trimer, tetramer and pentamer, respectively (data in accompanying manuscript, Mahajan and Ramya 2018). Similarly, using MALDI-MS, we determined that SrNaFLD also oligomerizes in solution (with intact masses of 27.6 kDa and 55.2 kDa, corresponding to monomer and dimer, respectively) (Supplementary data, Figure S4).
Glycan binding studies of SrFucNaFLD, mutants and domains
Hemagglutination and Hemagglutination Inhibition Assays
Considering the oligomeric status of SrFLD and SrNaFLD in solution, we used an agglutination assay with human type-O erythrocytes to determine if SrFucNaFLD has any lectin property. SrFucNaFLD agglutinated human type-O erythrocytes and we found the lowest effective concentration to be 7.7 nM (Supplementary data, Figure S5). No hemagglutination was observed in the absence of SrFucNaFLD and a positive hemagglutination result was observed upon addition of the known F-type lectin, AAA (from Vector labs) (Supplementary data, Figure S5). Further, we found that hemagglutination by SrFucNaFLD was inhibited by the monosaccharide, l-fucose, but remained unaffected by the monosaccharides, d-fucose, d-glucose, d-galactose, d-mannose, d-arabinose, l-arabinose, d-ribose, 2-deoxy-d-galactose, d-lyxose, d-xylose, N-acetyl-d-glucosamine, N-acetyl-d-galactosamine and sialic acid. This indicated the l-fucose binding specificity of SrFucNaFLD. We also found that hemagglutination by SrFucNaFLD was inhibited by the glycoprotein, mucin, but not inhibited by the oligosaccharides, lactose and raffinose, or by the polysaccharides, fucoidan and arabinogalactan (Supplementary data, Figure S5). We inferred that fucosylated glycans on mucin contribute to its ability to inhibit hemagglutination by SrFucNaFLD. l-Fucose and mucin inhibited hemagglutination by SrFucNaFLD with minimum inhibitory concentrations of 1.25 mM and 0.023 ng/mL, respectively (Supplementary data, Figure S5). We therefore established through this assay that SrFucNaFLD is a lectin with specificity for l-fucose.
We also found that SrFLD (inconsistently; twice out of five trials) and SrNaFLD (consistently) agglutinated erythrocytes, but SrNa (consistently) did not (Supplementary data, Figure S6). This suggested that the FLD of SrFucNaFLD mediates hemagglutination and is required and sufficient for the lectin property displayed by SrFucNaFLD. The mutants, SrFucNaFLDH623A, SrFucNaFLDR651A and SrFucNaFLDR658A did not agglutinate erythrocytes (Supplementary data, Figure S6). This confirmed that all the three residues of the FLD sequence motif, H623A, R651A, R658A, are critical for fucose binding.
ELLA
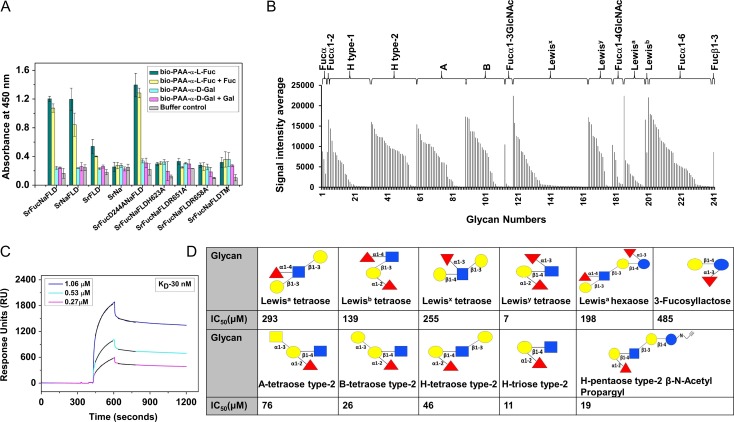
We used ELLA to confirm that the FLD of SrFucNaFLD is the domain with fucose-specific lectin property. We found that SrFucNaFLD, SrNaFLD and SrFLD bound to biotin-PAA-α-l-fucose, and not to biotin-PAA-α-d-galactose (Figure 2A). The intensity of binding to biotin-PAA-α-l-fucose was only marginally affected by 100 mM l-fucose, suggesting high binding strength (Figure 2A). Binding to biotin-PAA-α-l-fucose was not affected by >100 mM EDTA, suggesting that it is Ca2+ independent (Supplementary data, Figure S7). SrFucD244ANaFLD bound to biotin-PAA-α-l-fucose at a wide range of pH from pH 5.6 to pH 9.2 but not at pH 4.5 or lower (Supplementary data, Figure S8). There was no binding shown by SrNa to biotin-PAA-α-l-fucose or to biotin-PAA-α-d-galactose, confirming that the Na domain does not contribute to the lectin property of SrFucNaFLD (Figure 2A). Besides SrNaFLD and SrFLD, SrFucD244ANaFLD (with a mutation in the active site of the alpha-l-fucosidase domain) also bound to biotin-PAA-α-l-fucose, ruling out any role of the alpha-l-fucosidase and alpha-l-fucosidase C-terminal domains in lectin function. There was no binding by proteins with mutations in the FLD sequence motif, i.e., SrFucNaFLDH623A, SrFucNaFLDR651A, SrFucNaFLDR658A and SrFucNaFLDTM, to biotin-PAA-α-l-fucose or to biotin-PAA-α-d-galactose, confirming that these residues are critical for lectin function (Figure 2A).
Fig. 2.
(A) ELLA of immobilized SrFucNaFLD, SrNaFLD, SrNa, SrFLD, SrFucD244ANaFLD, SrFucNaFLDH6232A, SrFucNaFLDR651A, SrFucNaFLDR658A and SrFucNaFLDTM proteins using biotinylated PAA-α-l-fucose and PAA-α-d-galactose probes. (B) Binding profile of SrFucD244ANaFLD to fucosylated glycans of different glycan categories on the CFG glycan array. (C) SPR sensorgrams representing binding of SrFucNaFLD to PAA-linked fucose coated on a CM5 sensor chip. Three different concentrations of SrFucNaFLD (1.06–0.27 μM) are shown here. The black lines over each sensorgram are the association and dissociation fit curves. (D) IC50 values of inhibition of binding of SrFucD244ANaFLD to PAA-fucose by different glycans, as obtained from SPR analysis. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) system (Varki et al. 2015).
Glycan array analysis
We used the Consortium of Functional Glycomics glycan arrays to comprehensively identify glycan ligands of SrFucNaFLD. Considering that the active alpha-l-fucosidase domain present in SrFucNaFLD may cleave l-fucose off fucosylated glycans on the glycan microarray and confound data analysis, we subjected SrFucD244ANaFLD and SrNaFLD to glycan array analysis.
SrFucD244ANaFLD and SrNaFLD bound to l-fucose and l-fucose-containing glycans on the array with signal intensities <1500 (for SrFucD244ANaFLD) and <700 (for SrNaFLD) (Figure 2B, Supplementary data, Figure S9, Tables SI, SII). Both SrFucD244ANaFLD and SrNaFLD bound to both α- and β-linked l-fucose and to fucosylated glycans of different categories, suggesting that the FLD of SrFucNaFLD has a broad ligand specificity (Figure 2B, Supplementary data, Figure S9, Tables SI, SII). We observed higher signal intensities of binding for SrNaFLD to H type-2 glycans and difucosylated glycans (Lewisb and Lewisy) as compared to H type-1 glycans and monofucosylated glycans (Lewisa and Lewisx) (Supplementary data, Figure S9, Table SII).
We also subjected SrFucD244ANaFLDH623AR651AR658A and SrNa to glycan array analysis (Supplementary data, Figure S10, S11; Tables SIII, SIV). There was no significant glycan binding of these proteins to any fucosylated or non-fucosylated glycan on the array. This reiterated our conclusion from the ELLA and hemagglutination assays that the FLD and the residues, H623, R651 and R658, of the FLD sequence motif are indeed required and critical for l-fucose binding.
SPR studies
We used SPR to characterize the kinetics of SrFucD244ANaFLD binding to biotin-PAA-α-l-fucose immobilized via neutravidin on a CM5 chip (Figure 2C). We used SrFucD244ANaFLD instead of SrFucNaFLD (as done for glycan array analysis) to preclude the simultaneous use of immobilized biotin-PAA-α-l-fucose as substrate by the alpha-l-fucosidase domain. We found that the kon and koff values were in the range of 14.99 × 103 ± 4.80 × 103 M−1 s−1 and 2.84 × 10−4 ± 0.24 × 10−4 s−1, respectively, for several concentrations of SrFucD244ANaFLD. We attempted to calculate the dissociation constant (KD) for several concentrations of SrFucD244ANaFLD but the data from different concentrations of SrFucD244ANaFLD could not be fitted with global analysis. Further, due to fast dissociation rates, also reported in the literature for other lectin–ligand interactions, we were able to use only a narrow range of the data for individual curve fitting. Using a simple 1:1 interaction model and individual fitting of each data set, we calculated the average KD to be 31.54 ± 11.44 nM (Figure 2C). Considering the narrow range of data used for fitting as well as the potential for protein oligomerization and the heterogeneity of oligomers in solution, we note that this be regarded as an apparent dissociation constant.
We also performed SPR inhibition assays using various concentrations of competing ligands in solution to assess binding of SrFucD244ANaFLD to several fucosylated glycans, and we calculated IC50 values from these experiments (Figure 2D). We found that Lewisy antigen and Blood group H antigen triose type-2 were the most efficient in competing with immobilized biotin-PAA-α-l-fucose to bind to SrFucD244ANaFLD, with IC50 values of 7 μM and 11 μM, respectively. Both these glycans have an H type-2 moiety (Figure 2D). Blood group H antigen pentaose type-2 propargyl and blood group H antigen tetraose type-2, which also possess the H type-2 moiety, also inhibited binding but with higher IC50 values of 19 μM and 46 μM, respectively. Blood group B type-2 tetrasaccharide and blood group A type-2 tetrasaccharide also had higher IC50 values (26 μM and 76 μM respectively), suggesting that the galactose or N-acetylgalactosamine linked to the subterminal galactose might be a cause of steric hindrance (Figure 2D). Lewisb tetraose, which has an H type-1 moiety with an α-1,2-linked fucose, displayed little inhibition, indicating that SrFucD244ANaFLD discriminates against fucosylated glycans with type-1 sequences (Figure 2D). Glycans such as Lewisx tetraose and Lewisa tetraose which have α-1,3- or α-1,4-linked fucose also displayed little inhibition (Figure 2D). Our SPR experiments thus established that SrFucD244ANaFLD binds with high affinity to glycans with an H type-2 moiety.
Bioinformatics analysis of the alpha-l-fucosidase domain of SrFucNaFLD
The alpha-l-fucosidase domain of SrFucNaFLD was sequence aligned with representative members of GH29 alpha-l-fucosidase family – AlfA, AlfB and AlfC from Lactobacillus casei, BT_2192 and BT_2970 from Bacteroides thetaiotaomicron, TM306 from T. maritima, AfcB from Bifidobacterium bifidum JCM 1254, Blon_2336 from Bifidobacterium bifidum JCM 1254, Blon_0248 and Blon_0426 from Bifidobacterium longum ATCC15697, ALfuk1 from Paenibacillus thiaminolyticus, BFO_2737 from Tannerella forsythia ATCC 43037, FucA1 from Xanthomonas campestris ATCC 33913, FUCA1 from Canis lupus familiaris, AlfA from Dictyostelium fasciculatum, FCO1 from Fusarium oxysporum, FCO1 from Fusarium graminearum PH-1, α-1,3/1,4-l-fucosidase from Streptomyces sp. 142, FucA1 from Sulfolobus solfataricus P2, AtFuc1 from Arabidopsis thaliana, FucA1 and FucA2 from Homo sapiens, FucA1 from Rattus norvegicus, and Mfuc1, Mfuc2, Mfuc3, Mfuc4, Mfuc5, Mfuc6 and Mfuc7 from uncultured bacteria (Supplementary data, Figure S12).
We found that the alpha-l-fucosidase domain of SrFucNaFLD showed significant similarity to GH29 family members. Further, the Asp residue that acts as the catalytic nucleophile in the double displacement reaction was conserved in the alpha-l-fucosidase domain of SrFucNaFLD (Supplementary data, Figure S12).
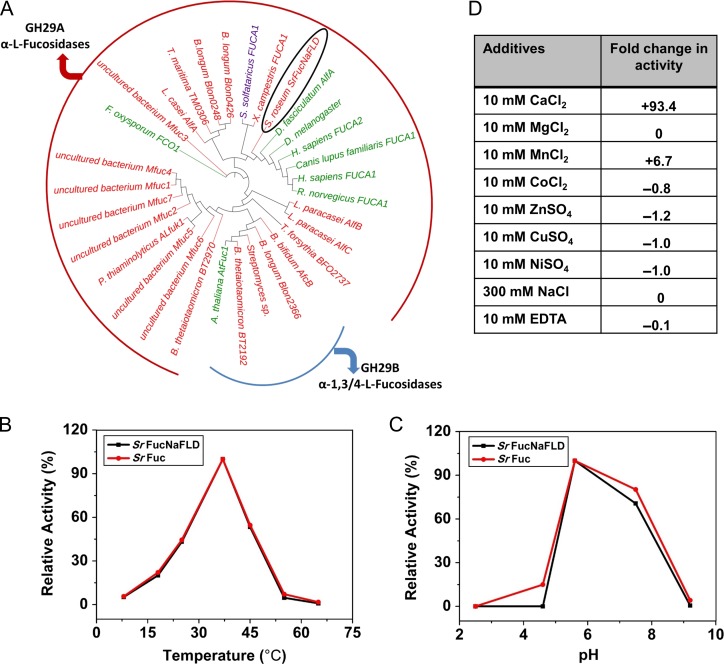
The family GH29 has two subfamilies – GH29-A and GH29-B. We generated a phylogenetic tree of the alpha-l-fucosidase domain of SrFucNaFLD together with representative members of both these subfamilies. We found separate clustering of GH29-A and GH29-B members (Figure 3A). We observed that the alpha-l-fucosidase domain of SrFucNaFLD was present in the GH29-A clade (Figure 3A). This suggested that the alpha-l-fucosidase domain of SrFucNaFLD might share the properties of GH29-A subfamily members.
Fig. 3.
(A) Phylogenetic tree of SrFuc domain along with representative GH29-A and B alpha-l-fucosidase family members. Branch nodes and labels are designated in red for bacterial members, in green for eukaryotic members and in purple for the archaeal member. SrFucNaFLD is circled in black. (B) Enzymatic activity of SrFuc protein at different temperatures. Relative activity (%) values were calculated with respect to the highest value (enzyme activity at 37°C). (C) Enzymatic activity of SrFuc protein at different pH. Relative activity (%) values were calculated with respect to the highest value (enzyme activity at pH 5.6) (D) Enzymatic activity of SrFuc protein in the presence of metal ions, NaCl and EDTA. ND: not detected. Fold change in activity was calculated with the formula, (M–C)/C where M refers to activity in presence of metal ions and C refers to activity in a control tube lacking metal ions.
Alpha-l-fucosidase activity of SrFuc and SrFucNaFLD
We assayed the alpha-l-fucosidase activity of purified SrFuc and SrFucNaFLD at different temperatures and at various pH using the substrate 4-Methylumbelliferyl-α-l-fucopyranoside, by measuring the fluorescence of the product, 4-Methylumbelliferone (4-MU). We chose to assay both SrFuc and SrFucNaFLD in order to understand the effect of FLD on the enzyme activity of the cis-positioned alpha-l-fucosidase domain. SrFuc and SrFucNaFLD were both optimally active at 37°C and at pH 5.6 to 7.5 (Figure 3B and C). Some alpha-l-fucosidase activity was retained at pH 9.2, but no significant activity was found at pH 2.6 and 4.5 (Figure 3C).
We also assayed the alpha-l-fucosidase activity of SrFuc in the presence of various metal ions. We found no substantial effect on alpha-l-fucosidase activity upon the addition of 10 mM Mg2+, 10 mM EDTA or 300 mM NaCl (Figure 3D). We found that Ca2+ and Mn2+ enhanced enzyme activity, with Ca2+ especially having a tremendous effect, inducing ~93-fold change in activity (Figure 3D). Zn2+, Co2+, Cu2+ and Ni2+ inhibited activity (Figure 3D). We would like to note that we performed these assays with the C-terminally hexahistidine-tagged proteins. There is a possibility that the inhibition of enzyme activity observed with Zn2+, Co2+, Cu2+ and Ni2+ is effected by metal ion coordination by the hexahistidine tag and subsequent alteration in enzyme structure and function.
Kinetics of alpha-l-fucosidase activity of SrFuc and SrFucNaFLD
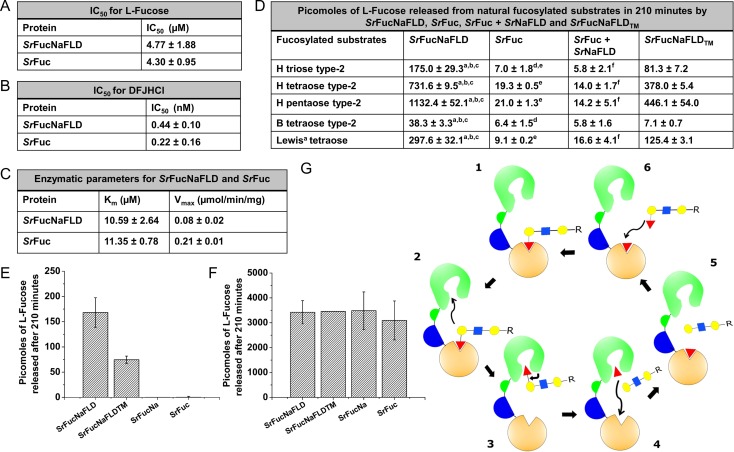
The alpha-l-fucosidase activity of SrFuc and SrFucNaFLD with the substrate, 4-Methylumbelliferyl-α-l-fucopyranoside, followed Michaelis Menten kinetics (Figure 4C, Supplementary data, Figure S13). We calculated the enzyme kinetic parameters - substrate concentration at half maximal velocity, Km and maximal velocity, Vmax (Figure 4C). There was no significant difference in the Km values of SrFuc and SrFucNaFLD for 4-Methylumbelliferyl-α-l-fucopyranoside (P = 0.86, two-tailed paired t-test). We observed significant batch-to-batch variations in Vmax, and we believe that this is due to differences in the fraction of inactive enzyme in the protein preparations. Thus, our experiments suggested that FLD has no significant effect on the enzymatic activity of the cis-positioned alpha-l-fucosidase domain with respect to enzyme kinetics with the substrate, 4-Methylumbelliferyl-α-l-fucopyranoside.
Fig. 4.
(A) IC50 of inhibition of SrFucNaFLD and SrFuc by l-fucose. (B) IC50 of inhibition of SrFucNaFLD and SrFuc by DFJHCl. (C) Vmax and Km values of SrFucNaFLD and SrFuc for the synthetic substrate, 4-Methylumbelliferyl-α-l-fucopyranoside. Values are mean ± standard error of two independent experiments. (D) l-fucose released by SrFucNaFLD, SrFuc, SrFuc + SrNaFLD and SrFucNaFLDTM from natural fucosylated substrates. aP < 0.05, t-test with SrFuc. bP < 0.05, t-test with SrFuc + SrNaFLD. cP > 0.05, t-test with SrNaFLDTM. dP < 0.05, t-test with SrFuc + SrNaFLD. eP > 0.05, t-test with SrNaFLDTM. fP > 0.05, t-test with SrNaFLDTM. SrFucNaFLD, SrFuc and SrFucNaFLDTM used in the assays were prepared simultaneously to eliminate batch-to-batch variations. (E) Histogram showing l-fucose released by SrFucNaFLD, SrFuc, SrFucNa and SrFucNaFLDTM from H tetraose type-2. SrFucNaFLD, SrFuc, SrFucNa, and SrFucNaFLDTM used in the assay were prepared simultaneously to eliminate batch-to-batch variations. (F) Histogram showing l-fucose released by SrFucNaFLD, SrFuc, SrFucNa and SrFucNaFLDTM, from the substrate, 4-Methylumbelliferyl-α-l-fucopyranoside. The values plotted are averaged from independent experiments conducted with multiple batches of protein (n = 3 for SrFucNaFLD, n = 4 for SrFuc and n = 7 for SrFucNa). (G) Schematic representation of proposed “bind and jump” model: (1) Binding of fucosylated oligosaccharide to the FLD (domain depicted in orange) of SrFucNaFLD. (2) Jumping of fucosylated oligosaccharide from the FLD to the alpha-l-fucosidase domain (depicted in green) of SrFucNaFLD. (3) Cleavage of l-fucose from the oligosaccharide by the alpha-l-fucosidase domain. (4) and (5) Binding of released l-fucose to the FLD. (6) Replacement of the l-fucose in the FLD binding pocket by a new fucosylated oligosaccharide due to higher binding affinity of the latter. (The SrFucNaFLD protein is depicted as a cartoon image with four domains – the alpha-l-fucosidase domain and the alpha-l-fucosidase C-terminal domain in green, the Na domain in blue and the FLD in orange. Monosaccharide symbols in the cartoon depiction of the fucosylated oligosaccharide follow the SNFG (Symbol Nomenclature for Glycans) system (Varki et al. 2015)).
Inhibition of alpha-l-fucosidase activity by l-fucose and deoxyfuconojirimycin hydrochloride
The product of alpha-l-fucosidase activity, l-fucose, and the compound, deoxyfuconojirimycin are known to be inhibitors of alpha-l-fucosidase activity (Alam and Balsubramanian 1978; Winchester et al. 1990; Berteau et al. 2002). We performed enzyme inhibition studies of SrFuc and SrFucNaFLD with various concentrations of l-fucose and deoxyfuconojirimycin hydrochloride (DFJHCl) in the presence of 25 μM 4-Methylumbelliferyl-α-l-fucopyranoside, and calculated IC50 values of inhibition.
l-Fucose, in the micromolar range, inhibited alpha-l-fucosidase activity (Figure 4A, Supplementary data, Figure S14). We observed similar IC50 values of l-fucose inhibition for SrFucNaFLD and SrFuc (P = 0.85, two-tailed paired T-test) (Figure 4A). We therefore found no effect of FLD on the cis-positioned alpha-l-fucosidase domain with respect to inhibition of alpha-l-fucosidase activity by l-fucose.
Nanomolar concentrations of DFJHCl inhibited alpha-l-fucosidase activity (Figure 4B, Supplementary data, Figure S15). The IC50 values for DFJHCl inhibition of SrFuc and SrFucNaFLD were also similar (P = 0.56, paired T-test) between SrFucNaFLD and SrFuc (Figure 4B). This suggests that the FLD has no effect on the cis-positioned alpha-l-fucosidase domain with respect to inhibition of alpha-l-fucosidase activity by DFJHCl.
Alpha-l-fucosidase activity of SrFucNaFLD and SrFuc on fucosylated oligosaccharides
We measured the alpha-l-fucosidase activity of SrFucNaFLD and SrFuc with unlabeled fucosylated oligosaccharide substrates using an alpha-l-fucosidase endpoint assay. This assay used l-fucose dehydrogenase (FDH) to convert the free l-fucose that was released from fucosylated oligosaccharides by alpha-l-fucosidase activity to l-fuconolactone with the concomitant formation of NADPH. The assay further used the enzyme, diaphorase, to oxidize NADPH to NADP and reduce resazurin to fluorescent resorufin. We cloned the FDH required for this assay from Pseudomonas sp., and heterologously expressed and purified it (Supplementary data, Figure S16).
We assayed alpha-l-fucosidase activity using blood group antigens (H triose type-2, H tetraose type-2, H pentaose type-1, H pentaose type-2 propargyl, A tetraose type-2 and B type 2 tetraose), Lewis antigens (Lewisa, Lewisx, Lewisb and Lewisy) and human milk oligosaccharides (2′-fucosyllactose, 3-fucosyllactose, Lacto-N-fucopentaose-III and Lacto-N-difucohexaose-II) as enzyme substrates. Due to high levels of contaminating free l-fucose, we were unable to determine if 3-fucosyllactose [Galβ1-4(Fucα1-3)Glc], Lewisb tetraose [Fucα1-2Galβ1-3(Fucα1-4)GlcNAc], Lewisx tetraose [Galβ1-4(Fucα1-3)GlcNAcβ1-3Gal], Lewisy tetraose [Fucα1-2Galβ1-4(Fucα1-3)GlcNAc], Lacto-N-difucohexaose II [Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4(Fucα1-3)Glc] and A tetraose type-2 [GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc] could be hydrolyzed by SrFucNaFLD or SrFuc.
Of the remaining glycans, we found that SrFucNaFLD utilized the substrates, 2′-fucosyllactose [Fucα1-2Galβ1-4Glc] (data not shown), H triose type-2 [Fucα1-2Galβ1-4GlcNAc], H tetraose type-2 [Fucα1-2Galβ1-4GlcNAcβ1-3Gal], H pentaose type-2 propargyl [Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4Glc-NAc-CH2CCH], and Lewisa tetraose [Galβ1-3(Fucα1-4)GlcNAcβ1-3Gal] (Figure 4D). SrFucNaFLD weakly utilized H pentaose type-1 [Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc] (data not shown), and B tetraose type-2 [Galα1-3(Fucα1-2)Galβ1-4GlcNAc] (Figure 4D) as substrates.
The results of these assays indicated that the alpha-l-fucosidase domain of SrFucNaFLD accepts both α1-2 and α1-4 linked fucosylated glycans as substrates. This is similar to the substrate preference of most GH29-A subfamily members. Further, our results suggested that the alpha-l-fucosidase activity of SrFucNaFLD increases with increasing length of fucosylated oligosaccharide; i.e., the order of preference for H type-2 oligosaccharides is H pentaose type-2 > H tetraose type-2 > H triose type-2. Our results also suggested that SrFucNaFLD prefers H type-2 to B type-2 oligosaccharides as substrate.
SrFuc released low amounts of l-fucose from the substrates, B-tetraose type-2, H-triose type-2, H tetraose type-2, H pentaose type-2 propargyl, and Lewisa tetraose (Figure 4D). We found that the amount of l-fucose released by SrFucNaFLD was ~6 times that of SrFuc for B tetraose type-2, and ~25–53 times that of SrFuc for the remaining oligosaccharide substrates. We performed similar assays with SrFuc+SrNaFLD in order to determine if the difference in amounts of l-fucose released between SrFuc and SrFucNaFLD was due to a cis effect of the FLD on alpha-l-fucosidase activity. SrFuc+SrNaFLD behaved similar to SrFuc, releasing low amounts of l-fucose from H triose type-2, H tetraose type-2, H pentaose type-2, B tetraose type-2 and Lewisa tetraose (Figure 4D). Considering this result, we hypothesized that the FLD, via its lectin activity, might be enhancing the enzyme activity of the cis-positioned alpha-l-fucosidase domain for natural fucosylated oligosaccharide substrates. In order to test this hypothesis, we performed similar assays with SrFucNaFLDTM (which lacks the residues H623, R651 and R658 critical for the lectin function of the FLD). We found that the amounts of l-fucose released by SrFucNaFLDTM from the substrates, H triose type-2, H tetraose type-2, H pentaose type-2, and Lewisa tetraose, was ~2 times less than that released by SrFucNaFLD, and significantly higher (~7–31 times) than that released by SrFuc and SrFuc+SrNaFLD (Figure 4D). The amount of l-fucose released by SrFucNaFLDTM from B tetraose type-2 was however similar to that released by SrFuc and SrFuc+SrNaFLD (Figure 4D). This suggested that the lectin function of the FLD is not solely responsible for the cis effect observed in alpha-l-fucosidase activity of SrFucNaFLD towards the substrates, H triose type-2, H tetraose type-2, H pentaose type-2, and Lewisa tetraose. We theorized that the Na domain and/or the FLD might confer a structural advantage to the active site pocket of the alpha-l-fucosidase domain, thus making it more accessible to certain substrates. We therefore performed additional assays, comparing the alpha-l-fucosidase activity of SrFucNaFLD, SrFuc, SrFucNa and SrFucNaFLDTM using the substrate, H tetraose type-2 (Figure 4E). We found that SrFucNa behaved similar to SrFuc, with respect to release of l-fucose from H tetraose type-2 (Figure 4E). In contrast, SrFucNaFLD, SrFuc, SrFucNa and SrFucNaFLDTM behaved similarly with respect to release of l-fucose from the synthetic substrate, 4-Methylumbelliferyl-α-l-fucopyranoside, in a similar endpoint assay using FDH to detect l-fucose release (Figure 4F). This implied that the Na domain does not have any cis effect on the alpha-l-fucosidase domain and that only the FLD is responsible for the enhanced alpha-l-fucosidase activity observed with SrFucNaFLD on natural oligosaccharide substrates. Based on the results of the assays with SrFucNa and SrFucNaFLDTM, we inferred that the lectin activity and perhaps the structural placement of FLD are important for its cis effect on the alpha-l-fucosidase domain.
Discussion
l-Fucose (or 6-deoxy-galactose) is one of the few l-sugars present in nature and is found in low amounts in the biosphere but it is a key component of complex glycoconjugates occurring in various organisms ranging from bacteria to mammals (Becker and Lowe 2003; Ma et al. 2006). l-Fucose is generally situated at the non-reducing end of glycans, linked through an α-1,2 glycosidic bond to a d-galactosyl residue or through α-1,3, α-1,4 or α-1,6 glycosidic bonds to an N-acetyl-d-glucosamine residue (Ma et al. 2006; Stanley et al. 2009; Stanley and Cummings 2009). l-Fucose is frequently located at the outer terminal region of the glycan, exposed to the surrounding microenvironment, and recognized by various fucose-binding lectins or antibodies.
There are many fucose-binding lectins reported in literature with different specificities towards fucosylated glycans. They include F-type lectins such as AAA, which binds to H type-1 and Lewisa motifs (Baldus et al. 1996; Bianchet et al. 2002), Tachylectin-4 from horseshoe crab (Tachypleus tridentatus) which binds to 3-deoxy-l-fucose of E. coli O111 O-antigen (Saito et al. 1997), Streptococcus pneumoniae SP2159 FLD which recognizes ABH and Lewisy antigens (Boraston et al. 2006), and Streptococcus mitis lectinolysin which binds to Lewisb and Lewisy (Farrand et al. 2008). There are also lectins without an F-type lectin domain such as the lectins from Aleuria aurantia, Aspergillus oryzae and Rhizopus stolonifera, which bind to core α-(1-6)-linked l-fucose (Fukumori et al. 1990; Fujihashi et al. 2003; Oda et al. 2003; Wimmerova et al. 2003; Matsumura et al. 2007), Lotus tetragonolobus, which binds to Lewisx determinants and other fucosyl residues (VanEpps and Tung 1977), Ulex europaeus, which is specific towards H type-2 (Hindsgaul et al. 1982), Ralstonia solonacearum, which binds to blood group H and Lewis antigens (Sudakevitz et al. 2002), Aspergillus fumigatus, which binds to blood group epitopes and Lewis antigens, particularly Lewisy (Houser et al. 2013) and Burkholderia ambifaria, which binds to Fucα1-2Gal epitopes on plant cell wall fucosylated oligosaccharides (xyloglucans) and H type-2 and Lewisy human blood group oligosaccharides (Audfray et al. 2012).
Here, we have studied a microbial FLD with a CBM-like architecture. Streptosporangium roseum DSM43021 SrFucNaFLD has a non-enzymatic carbohydrate binding domain, FLD, associated with a carbohydrate-active enzyme domain, alpha-l-fucosidase. Besides this protein discussed in this study, S. roseum DSM43021 also has another alpha-l-fucosidase (Genbank accession no: ACZ90867.1), where the alpha-l-fucosidase domain is sandwiched between an N-terminal GH 65 family domain and a C-terminal Laminin_G_3 domain.
The SrFucNaFLD polypeptide has an alpha-l-fucosidase domain and an alpha-l-fucosidase C-terminal domain followed by an Na domain and an FLD. According to Pfam version 31.0 updated March 2017, of the 3097 sequences classified under PF01120 (alpha-l-fucosidase domain), 1747 sequences have a standalone alpha-l-fucosidase domain architecture whereas 710 sequences have the architecture of an (N-terminal) alpha-l-fucosidase domain (PF01120) and a (C-terminal) alpha-l-fucosidase C-terminal domain (PF16757). Moreover, of the 773 sequences classified under PF16757, only 7 sequences comprise a standalone alpha-l-fucosidase C-terminal domain. Among the remaining sequences, 710 sequences have just an alpha-l-fucosidase domain immediately N-terminal to the alpha-l-fucosidase C-terminal domain, and the other sequences have an alpha-l-fucosidase domain and other domain(s) as part of the domain architecture. SrFucNaFLD resembles the last mentioned category in domain architecture.
The Na domain in SrFucNaFLD is 30% similar to the Na domain of Streptomyces coelicolor A3(2). The Na domain is referred to as NPCBM-associated, NEW3 domain of alpha-galactosidase (Naumov 2004) and classified under PF10633 family in Pfam. It is mentioned as having similarity using HHSearch to the Pfam families DUF11 (PF01345) and CARDB (Cell Adhesion Related Domain found in Bacteria) (PF07705) superfamily. The Na domain has been found on bacterial proteins that are galactosidases and found associated with the NPCBM (NEW2) domain (PF08305) (Naumov 2004; Rigden 2005). A cursory analysis of the occurrence of the Na domain in polypeptides in the Pfam database shows that it is frequently located towards the C-terminal region of the polypeptide following a glycoside hydrolase domain and preceding the NPCBM domain. Interestingly, although the NPCBM domain itself is annotated as being found at the N-terminus of glycosylhydrolases, visual examination of the domain architectures of proteins with NPCBM domains indicates that only a few polypeptides possess an NPCBM domain at the N-terminal end of a Melibiase domain. The NPCBM domain seems to be commonly found in the C-terminus and not the N-terminus of the polypeptide; it is in fact the ultimate C-terminal domain of many of the polypeptides on which it is found.
We have demonstrated, using hemagglutination assays, ELLA, glycan microarray analysis, and SPR studies that the FLD in SrFucNaFLD is a Ca2+-independent fucose binding lectin with highest affinity for H type-2 motifs. We have demonstrated that the FLD is sufficient and required for l-fucose binding and that the residues of the FLD sequence motif, H623, R651 and R658 are critical for l-fucose binding. The Na domain displayed no binding to fucose or fucosylated glycoconjugates in ELLA and glycan array assays. Therefore, there currently exists no evidence implicating it as a carbohydrate binding domain.
The alpha-l-fucosidase domain in SrFucNaFLD is a GH29 family member by sequence similarity, and has both the conserved Asp residue which acts as a catalytic nucleophile and the Glu that acts as the acid/base residue responsible for the double displacement mechanism in the GH29 family of alpha-l-fucosidases (Supplementary data, Figure S17) (Tarling et al. 2003; Sulzenbacher et al. 2004; Cao et al. 2014). We demonstrated the alpha-l-fucosidase activity of the alpha-l-fucosidase domain of SrFucNaFLD using the synthetic substrate 4-Methylumbelliferyl-α-l-fucopyranoside, for which it has high affinity (Km ~10–11 μM). For comparison, the characterized human liver GH29 alpha-l-fucosidase has a Km of 0.43 mM for the same substrate (Alhadeff et al. 1975). We also studied inhibition of alpha-l-fucosidase activity by l-fucose and DFJHCl and established that the alpha-l-fucosidase domain of SrFucNaFLD can hydrolyze oligosaccharides with α1-4-linked (Lewisa tetraose) as well as α1-2-linked (H type-2) fucose. We could not verify if SrFucNaFLD could accept glycans having Fucα1-3 linkage as substrate.
We determined that SrFucNaFLD could hydrolyze complex blood group oligosaccharides using an FDH-based endpoint assay to measure l-fucose released by SrFucNaFLD from the oligosaccharides. However, the amount of l-fucose released was >10-fold lower than that released from the synthetic substrate, 4-Methylumbelliferyl-alpha-l-fucopyranoside. Considering that S. roseum is a soil-borne bacterium, we do not expect blood group oligosaccharides to be physiological substrates. It is likely that xyloglucans of plant cell walls are natural substrates of this alpha-l-fucosidase. Terminal fucose residues are present in XXFG nonasaccharides derived from the xyloglucan of many plants (McNeil et al. 1984), and a recent report demonstrated that the alpha-l-fucosidases (Mfuc1, 2, 4, 5 and 7) isolated from a soil metagenomic library use A. thaliana xyloglucan and Sambucus nigra xyloglucan as substrates (Lezyk et al. 2016). However, in assays with SrFuc, we could not detect any l-fucose release from DP7 and DP10 xyloglucans from apple (Sigma) (data not shown), although apple xyloglucans are known to contain l-fucose (Renard et al. 1992). We cannot rule out the possibility that other xyloglucan oligosaccharides or other fucosylated plant polysaccharides are the natural substrates of this enzyme.
GH29 alpha-l-fucosidase family is further classified into GH29-A and GH29-B subfamilies on the basis of sequence homology and substrate specificity (Ashida et al. 2009; Sakurama et al. 2012). Sequence based similarity and phylogenetic analysis as well as enzyme assays using natural oligosaccharides as substrates suggested that the alpha-l-fucosidase domain of SrFucNaFLD belongs to the GH29-A subfamily of alpha-l-fucosidases. GH29-A members have broad substrate specificity and accept fucose in α-1,2-, α-1,3- and α1,4- linkages, whereas GH29-B members usually accept only α-1,3- and α1,4-linked fucosides as substrates. For instance, the GH29B alpha-l-fucosidase from Bacteroidetes thetaiotamicron hydrolyses 3′-FL or Lewisx antigen (Guillotin et al. 2014), the GH29-B alpha-l-fucosidase from Bifidobacterium bifidum hydrolyzes 3′-FL, α-1,3- and α1,4- linked fucosyl residues from Lewisa, Lewisb, Lewisx and Lewisy antigens and lacto-N-fucopentaose II and III (Ashida et al. 2009), and the GH29A human fucosidases (FUCA1 and FUCA2) hydrolyze various α-(1-2), α-(1-3), α-(1-4), and α-(1-6) linked natural fucosylated glycoconjugates (Dawson and Tsay 1977; DiCioccio et al. 1982; Liu et al. 2009). The GH29A alpha-l-fucosidase from the pathogenic fungus, Fusarium graminareum, hydrolyses 2′-FL and xyloglucan (complex fucosylated oligosaccharide of plant cell wall) (Cao et al. 2014).
Although several GH29-A subfamily and GH29-B subfamily members (Arabidopsis thaliana, Genbank accession no: NP_180377; Streptomyces sp., Genbank accession no: AAD10477.1) including some co-occurring with coagulation factor FA58C domain (Bacteroidetes thetaiomitocron BT_2192, Genbank accession no: NP_811105; Bifidobacterium longum, Genbank accession no: WP_012578563; Bifidobacterium bifidum, Genbank accession no: WP_047270681) have been characterized, and a few characterized GH29 family alpha-l-fucosidases do have CBM-like architecture, there are no biochemical studies of the effect, if any, of an FLD on the enzymatic activity of a cis-positioned alpha-l-fucosidase domain.
Our studies indicated that cis-positioning of the FLD with respect to the alpha-l-fucosidase domain in SrFucNaFLD did not alter enzyme kinetics with respect to the synthetic substrate, 4-Methylumbelliferyl-α-l-fucopyranoside, or inhibition of alpha-l-fucosidase activity by l-fucose or DFJHCl. However, the alpha-l-fucosidase domain of S. roseum was found to display enhanced activity against fucosylated oligosaccharides when co-associated with the FLD in cis (on the same polypeptide) but not in trans (on different polypeptides). Interestingly, we observed a similar cis effect albeit of a significantly lesser magnitude against fucosylated oligosaccharides (other than B tetraose type-2) with SrFucNaFLDTM that lacks the His and Arg residues critically required for lectin function. This suggested that the lectin function of the FLD is not solely responsible for the cis effect observed in alpha-l-fucosidase activity of SrFucNaFLD for certain substrates. Assays performed with SrFucNa implied that the Na domain did not have any role in this cis effect. Therefore, we conclude that the enzyme activity of the alpha-l-fucosidase domain in SrFucNaFLD is enhanced by the lectin activity of the cis positioned FLD as well as by some structural advantage imparted by the cis positioned FLD.
Typically, the non-enzymatic carbohydrate binding module of a CBM binds to insoluble polysaccharides, and thereby increases their effective concentration near the co-associated enzymatic domain, and enhances catalytic efficiency (Abbott et al. 2008; Hoffmam et al. 2016). This “proximity effect” is related to the observation that CBMs can increase enzyme processivity (Zheng and Ding 2013). For instance, Clostridium stercorarium xylanase (Xyn10B) has a catalytic domain of glycosylhydrolase (GH10) and a CBM9 domain. Removal of the CBM9 domain from Xyn10B has been demonstrated to reduce hydrolysis of insoluble xylan and plant cell walls but not of soluble xylan (Ali et al. 2001). The fusion of a type II carbohydrate binding domain of Pseudomonas fluorescens subsp. cellulosa xylanase A (XYLACBD) and cellulase E (CELECBD) to the catalytic domain of Clostridium thermocellum endoglucanase (EGE) has also been demonstrated to increase the hydrolysis activity of endoglucanase (EGE) against the insoluble form of cellulose (Bolam et al. 1998).
Some CBMs have also been reported to have either an enhanced or a neutral effect on soluble polysaccharides in literature. Bacillus subtilis exo-acting β-fructosidase (SacC), which is co-associated with CBM66 (CBM family 66), specifically hydrolyses levan (a water soluble polysaccharide). It was observed that removal of Bacillus subtilis BsCBM66 from SacC resulted in a ∼100-fold reduction of exo-acting β-fructosidase activity against water soluble polysaccharide, levan (Cuskin et al. 2012). In contrast, a study of Clostridium thermocellum glycosylhydrolase GH5 showed that both standalone (rGH5) and CBM32-associated C. thermocellum glycosylhydrolase (rGH5-CBM32) were highly active towards konjac glucomannan (water soluble polysaccharide) (Mizutani et al. 2012).
In this study, the natural fucosylated glycans used as potential substrates of S. roseum alpha-l-fucosidase domain were soluble oligosaccharides. Considering the high rate of diffusion in aqueous media for such substrates, it is difficult to attribute the enhanced activity of the FLD-cis-positioned alpha-l-fucosidase domain to only a direct proximity effect.
The three-dimensional structure of the alpha-l-fucosidase domain of SrFucNaFLD is not available. However, three-dimensional structures of several GH29 alpha-l-fucosidase homologs solved by X-ray diffraction of crystals are publically available in the Protein Data Bank (PDB). These include apo- (PDB 1HL8 and 2ZWY), fucose-bound (PDB 1ODU) and several inhibitor-bound structures of Thermotoga maritima GH29A alpha-l-fucosidase TM0306 (TmFuc), open apo (4PSP), closed apo (4NI3) and open l-fucose-bound (4PSR) structures of Fusarium graminearum GH29A alpha-l-fucosidase (FgFuc), apo (4J27) and many inhibitor-bound structures of Bacteroides thetaiotaomicron GH29A alpha-l-fucosidase BT2970 (BtFuc2970), open (3MO4) and closed (3UES) structures of Bifidobacterium longum subsp. infantis GH29B alpha-l-fucosidase BiAfcB Blon2336 (BlFuc), and apo (3EYP), IPTG- (4OUE) and oNPTG-bound (4OZO) structures of Bacteroides thetaiotaomicron GH29B alpha-l-fucosidase BT2192 (BtFuc2192).
Of these alpha-l-fucosidases, two GH29A members, TmFuc and FgFuc have a domain architecture that is partially similar to that of SrFuc, i.e., the catalytic GH29 alpha-l-fucosidase domain is followed by an alpha-l-fucosidase C-terminal domain. The alpha-l-fucosidase C-terminal domain is an eight-β-stranded two-layer beta sandwich domain containing a Greek key motif and a small α-helix (Cao, H., et al., Sulzenbacher et al. 2004). This domain presumably safeguards a large hydrophobic patch on the surface of the catalytic alpha-l-fucosidase domain against aqueous exposure (Sulzenbacher, G., et al.). GH29B member, BlFuc lacks the alpha-l-fucosidase C-terminal domain but instead has a beta-sandwich domain predicted to be a CBM32 domain, with some homology to the domains in GH29B member, BtFuc2192 and GH29A member, BtFuc2970 (Sela, D.A., et al.).
The structure of GH29A member, TmFuc demonstrates that the catalytic alpha-l-fucosidase domain is a (β/α)8 barrel domain that resembles the TIM barrel except that helix 5 of the TIM barrel is replaced by a small loop region and helix 6 is replaced by a disordered region; several alpha helices, 310 helices and several surface loops decorate the core barrel structure (Sulzenbacher, G., et al.). Several amino acid residues in the active site pocket, including the catalytic nucleophile (Asp 224 in TmFuc) make hydrogen bonds with l-fucose and a hydrophobic concavity formed by aromatic amino acids enclose the C6 methyl group of l-fucose (Sulzenbacher, G., et al.). In the crystal structure of a covalent glycosyl-enzyme intermediate obtained with fucosyl fluoride, the acid/base catalyst (Glu 266 inTmFuc) is located 5.5 A away from the catalytic nucleophile, Asp 224, a distance optimal for the double displacement mechanism of retaining glycosidases (Sulzenbacher, G., et al.). TmFuc displays considerable active site conformational flexibility as demonstrated by the stabilization of disordered loop regions near the substrate binding site upon binding of certain inhibitors (van Bueren et al. 2007; Wu et al. 2010).
The structures of GH29A member, FgFuc (Cao, H., et al.) and GH29B member, BlFuc (Sakurama, H., et al., Sela et al. 2012) demonstrate the existence of two active site conformations – open and closed forms, confirming active site conformational flexibility. Open and closed conformations differ in the active site loop conformation and in the orientation of the acid/base catalyst Glu residue. The open form is the unreactive form; the loop in the active site is disordered with the acid/base catalyst (Glu-288 in FgFuc; Glu217 in BlFuc) oriented away from the active site pocket. In the closed form, the loop covers the active site pocket and the Glu residue is suitably positioned to act as an acid/base catalyst. Two loops show conformational changes – loop 1 with residues 287 to 290 (WERG) and loop 2 with residues 391 to 395 (GGSFT) in FgFuc, and loop 1 with residues 173 to 182 and loop 2 with residues 215 to 220 in BlFuc (Cao, H., et al.) (Sakurama, H., et al.).
Thus, the overall fold, active site pocket, and open and closed active site conformations are conserved in GH29A and GH29B members. However, some active site residues and the aglycone binding region vary, and there is considerable variability in the residues of the disordered loop regions.
The LNFP-II bound structure of GH29B member, BlFuc displays a Gal-binding site that involves a residue, Gly 173, of the mobile loop, besides the residues, Trp 213, Glu237 and Asp 283 (Supplementary Figure S17). Mutagenesis of these residues result in a reduction in enzyme activity for 3-fucosyllactose, suggesting that the Gal-binding site might be important for appropriate induced fit movement and positioning of the substrate in the active site.
Although the overall structure, loops involved in induced fit movement and l-fucose-binding site are common to both GH29A and GH29B members, the site in TmFuc that is equivalent to the Gal-binding site in BlFuc is occupied by the amino acid residue, Arg 254, which makes a hydrogen bond with l-fucose and is highly conserved in GH29A members (Supplementary Figure S17). In contrast, the Gal-binding site residues are invariant in GH29B members (Sakurama, H., et al.). This structural difference is potentially responsible for the difference in substrate specificity beween GH29A and GH29B members (Sakurama, H., et al.). The structure of oNPTG-bound GH29B member, BtFuc2192 also provides evidence for the Gal-binding site in GH29B members (Guillotin, L., et al.).
By virtue of its similarity to characterized GH29 members, it is highly likely that SrFuc also displays active site conformational flexibility. SrFuc is a GH29A family member and a sequence alignment with GH29 alpha-l-fucosidases indicates that it lacks the Gal-binding site residues and instead has the conserved Arg residue (equivalent to Arg 254 of TmFuc) (Supplementary Figure S17). Based on the structural information available for its homologs, the active site pocket of SrFuc is anticipated to house the l-fucose end of the oligosaccharide substrate. The l-fucose is expected to be cleaved off the subterminal galactose by the concerted action of the catalytic nucleophile (predicted to be Asp 244) and the acid/base catalyst (predicted to be Glu 298).
Therefore, in the SrFucNaFLD protein, both the FLD and the alpha-l-fucosidase domains can house fucosylated oligosaccharides in their active site/binding pocket with l-fucose located within these concavities and making significant interactions with amino acids in the pocket. Considering this and the absence of a Gal-binding site in the alpha-l-fucosidase domain of SrFuc, we propose a model that involves a structural advantage imparted by the FLD to alpha-l-fucosidase and a “bind-and-jump” event mediated by the FLD to rationalize our experimental results (Figure 4G).
We envision that a fucosylated oligosaccharide substrate might bind first to the FLD binding pocket of SrFucNaFLD, perhaps aided by relatively better accessibility (Figure 4G). As expected in a lectin-saccharide binding equilibrium, the SrFucNaFLD-fucose complex is in a state of constant association and dissociation. As a consequence of proximity (due to the presence of the FLD and the alpha-l-fucosidase domains on the same polypeptide) and perhaps relative structural orientation of the two domains in space, l-fucose dissociating form the oligosaccharide can “jump” into and associate with the alpha-l-fucosidase active site pocket where l-fucose is cleaved off the oligosaccharide (Figure 4G). Based on the available structural information, it is not likely to be sterically feasible for the alpha-l-fucosidase domain to reach into the binding cleft of the FLD and cleave the bound l-fucose off the subterminal galactose without a “jump” event. We thus theorize that the FLD increases substrate accessibility through a “bind and jump” or an internal diffusion mechanism of ligands as reported for lectins in literature (Dam et al. 2009); i.e., the substrate might bind to the FLD first, akin to a typical CBM, but then jump to the alpha-l-fucosidase active site (Figure 4G). Additionally, the cleaved l-fucose released during the reaction might jump into and bind to the FLD binding pocket from the alpha-l-fucosidase domain. The cis positioning of the FLD might thus also serve to lower product inhibition (Figure 4G). Moreover, this l-fucose which is now bound to the FLD binding cleft may be easily replaced by another molecule of fucosylated oligosaccharide substrate due to the higher binding affinity of the FLD for the latter (Figure 4G).
We further envision that the FLD in SrFucNaFLD might positively stabilize the active site conformational flexibility of the cis positioned alpha-l-fucosidase domain. We speculate that the active site disordered loop in the apo enzyme might flap like a lid to “open” and “closed” conformations over the active site pocket and the presence of the cis-positioned FLD might stabilize this loop, allowing for better access of certain fucosylated oligosaccharides to the active site of the fucosidase domain. Alternatively or additionally, the cis proximity of the FLD polypeptide might further stabilize the disordered loop of the alpha-l-fucosidase domain in the substrate-bound enzyme and better orient the acid/base residue for catalysis. It is also possible that some other structural advantage is conferred on to the alpha-l-fucosidase by the cis-positioned FLD.
Considering such a model, one would expect the glycan binding specificity of the FLD to affect the substrate preference of the alpha-l-fucosidase domain. In fact, H type-2 glycans are both ligands and substrates of the FLD and the alpha-l-fucosidase domain, respectively. From our limited panel of substrates used at a single high concentration, it is not clear if the FLD does indeed preferentially enhance alpha-l-fucosidase activity for its high affinity ligands. However, future studies employing more oligosaccharide substrates and FLDs with different glycan-binding specificities or different alpha-l-fucosidase domains or both will help shed light on this aspect. Although other lectins or carbohydrate binding modules could also likely substitute for the FLD, the FLD fold with the N-terminal and C-terminal ends of the polypeptide adjacent to each other (Bianchet et al. 2002; Vasta et al. 2004), might enable the required orientation in cis-positioning that facilitates internal diffusion of the ligand.
Considering our model, natural conditions such as ion and salt concentrations, pH and temperature, which might affect the individual domains differently, might additionally determine or modulate the cis effect of the FLD on the alpha-l-fucosidase domain. We determined that the alpha-l-fucosidase domain is optimally active in the pH range 5.6 to 7.5. The FLD is active at this range of pH, too (5.6–9.2). While the alpha-l-fucosidase enzymatic activity of SrFuc was greatly enhanced in the presence of 10 mM Ca2+ ions, the lectin property of FLD is Ca2+ independent, similar to the F-type lectins, AAA and MSA (Bianchet et al. 2002; Odom and Vasta 2006). We also found that the alpha-l-fucosidase enzymatic activity of SrFuc was enhanced by Mn2+ ions, unaffected by 10 mM Mg2+ and inhibited by Zn2+, Cu2+ and Ni2+. While we have not determined the effect of various ions and temperature on the lectin property of the FLD, it is intriguing to imagine the myriad possibilities for regulating enzyme activity by tweaking such conditions in a two-domain system of this kind.
To conclude, we have biochemically characterized the lectin property and the alpha-l-fucosidase activity of an S. roseum protein, and demonstrated that the FLD enhances alpha-l-fucosidase activity towards aqueous, freely diffusible, small oligosaccharides. We believe that our study offers a simple strategy for engineering alpha-l-fucosidases for increased activity that might be translated to other carbohydrate-active enzymes as well.
Material and Methods
Cloning, expression, and purification of recombinant SrFucNaFLD and mutants
The nucleotide sequence (2196 bp) coding for the protein sequence of SrFucNaFLD from S. roseum DSM 43021 (GenBank accession number ACZ87343.1) was codon optimized for expression in Escherichia coli, custom synthesized, and cloned into pUC57 vector (GenScript, Piscataway, NJ). The region of the nucleotide sequence coding for the mature polypeptide (minus the signal peptide of 28 amino acids) was amplified by PCR and cloned in the expression vector, pET-28a(+) using primers, 5′-CAT GCC ATG GAT ACG CCG CCG GTT TAT GAA C-3′ and 5′-CCG CTC GAG GCC ACG CAC TTG GAC TTC TGC-3′ so as to also encode a C-terminal hexahistidine tag in the expressed recombinant protein. The plasmid generated was confirmed by DNA sequencing, and transformed in E. coli BL21(DE3). Recombinant protein expression was induced in a secondary culture of BL21(DE3) grown in LB at 37°C with 0.1 mM IPTG (GoldBio) when the cell density corresponded to OD600 of 0.8 and thereafter incubated at 22°C with shaking for 12 h. The bacterial cell pellet was resuspended in lysis solution (300 mM sodium chloride, 20 mM Tris, pH 7.5, 1% N-Lauroylsarcosine sodium salt and 20 mM imidazole), lysed by sonication and purified by Ni-NTA metal ion affinity chromatography. The column was washed with 50 mM imidazole in Tris-buffered saline (TBS) (150 mM sodium chloride and 20 mM Tris, pH 7.5) and the protein eluted with 250 mM imidazole in TBS. Purified protein was dialyzed extensively against TBS. Protein purity was assessed by SDS-PAGE and western analysis was done with mouse anti-C-terminal 6xHis antibody (Invitrogen). Protein concentration was estimated by OD280 measurement as well as by Bradford assay.
Mutations, H623A, R651A, R658A and D244A were made in SrFucNaFLD using appropriate primers (5′-CAG GTT CAA CGA CCG CTA CCG CTG AAC CGG-3′ and 5′-CCG GTT CAG CGG TAG CGG TCG TTG AAC CTG-3′ for H623A; 5′-GTG TTG ACG TCT GGA ATG CCC TGG ACT GCT GTG-3′ and 5′-CAC AGC AGT CCA GGG CAT TCC AGA CGT CAA CAC-3′ for R651A; 5′-CTG GAC TGC TGT GCC GAT GCT CTG AAA GAC TTT TGG-3′ and 5′-CCA AAA GTC TTT CAG AGC ATC GGC ACA GCA GTC CAG-3′ for R658A; 5′-CGG ACA TTA TCT GGT GCG CTG GCC AAT GGG AAA AAC-3′ and 5′- GTT TTT CCC ATT GGC CAG CGC ACC AGA TAA TGT CCG-3′ for D244A) and a site-directed mutagenesis kit (Novagen). All mutant proteins were expressed and purified by Ni-NTA metal ion affinity chromatography, similar to the wildtype SrFucNaFLD protein.
Cloning, expression, and purification of recombinant proteins comprising the alpha-l-fucosidase domain (SrFuc), alpha-l-fucosidase domain and Na domain (SrFucNa), Na domain and FLD (SrNaFLD), Na domain (SrNa), or FLD (SrFLD)
Constructs of all domains were cloned in pET-28a(+) to encode a C-terminal hexahistidine tag. SrFuc was cloned using primers, 5′-CAT GCC ATG GAT ACG CCG CCG GTT TAT GAA C-3′ and 5′-CCG CTC GAG ACC CGG CGT TTC GAT TTT AAA AAC-3′. SrNaFLD was cloned using primers, 5′-CAT GCC ATG GCG GTC CGT TCA CTG CTG CGC ACC-3′ and 5′- GCA GAA GTC CAA GTG CGT GGC CTC GAG CGG-3′. SrNa was cloned using primers, 5′-CAT GCC ATG GCG GTC CGT TCA CTG CTG CGC ACC-3′ and 5′-CAG CTC GAG GTT AAC GGT CAG CGG CAG TGC GGT-3′. SrFucNa was cloned using primers, 5′- CAT GCC ATG GAT ACG CCG CCG GTT TAT GAA C-3′ and 5′-CAG CTC GAG GTT AAC GGT CAG CGG CAG TGC GGT-3′. A structural alignment in the Fold and function assignment system (FFAS) server (Jaroszewski et al. 2005) of SrFLD along with available FLD structures was used in order to define the boundary of the FLD. SrFLD (with R584 as the starting N-terminal amino acid residue) was cloned using 5′-CAT GCC ATG GCG CGT CCG AAT CTG AGT CTG GG-3′ and 5′-GCA GAA GTC CAA GTG CGT GGC CTC GAG CGG-3′. All recombinant proteins were expressed in E. coli BL21(DE3) and purified by Ni-NTA metal ion affinity chromatography, similar to SrFucNaFLD, with a few variations as mentioned below. For expression of SrFuc, SrNaFLD, SrFucNa, SrNa and SrFLD constructs, transformed BL21(DE3) cells were grown at 37°C until their cell density corresponded to OD600 of 0.8, then induced with 1 mM IPTG, and incubated with continuous shaking at 200 rpm for 4 h after induction. The lysis buffer used was TBS with 0.5% N-Lauroylsarcosine sodium salt for SrFuc transformed cells, and TBS with 20 mM imidazole for SrNaFLD, SrNa and SrFLD transformed cells. The wash buffer used was 30 mM imidazole in TBS for SrFuc and SrFucNa, and 40 mM imidazole in TBS for SrNaFLD, SrNa and SrFLD.
Hemagglutination and hemagglutination inhibition assays
A 2% suspension of type-O human erythrocytes was made in TBS (20 mM Tris-HCl, 0.15 M NaCl buffer, pH 7.5). Then, 25 μL of 2-fold serially diluted purified wildtype or mutant SrFucNaFLD or its domains was mixed with 25 μL of this 2% suspension of erythrocytes in a U-shaped microtitre plate, and the mixture was incubated for 1 h at 37°C. The plate was observed for hemagglutination, and the reciprocal of the highest dilution of protein showing hemagglutination was recorded as the titer. For controls, commercially procured AAA or just TBS were used instead of lectin. For hemagglutination inhibition assays, 12.5 μL of 2-fold serially diluted 200 mM mono/oligosaccharide or 10 mg/mL polysaccharide/glycoprotein in TBS was mixed with 12.5 μL of SrFucNaFLD solution in TBS (titer of 4 agglutination units) in a U-shaped microtitre plate and incubated at 37°C for 45 min. Subsequently, 25 μL of the 2% suspension of erythrocytes was added, the mixture was incubated for 1 h at 37°C and the plate was observed for hemagglutination.
Enzyme-linked lectin assay (ELLA) using biotin-PAA-α-l-fucose
Wells of a MaxiSorp flat-bottom 96-well plate (Nunc) coated with wildtype or mutant SrFucNaFLD or its domains (100 μL of 2 μg/mL stock solutions in TBS) were blocked with 3% bovine serum albumin and incubated with one of the following solutions: biotin-PAA-α-l-fucose (Glycotech), biotin-PAA-α-l-fucose + 0.1 M l-fucose (Sigma), biotin-PAA-α-d-galactose, biotin-PAA-α-d-galactose + 0.1 M d-galactose (Sigma) or just buffer. Following incubation and washes, wells were incubated with HRP-conjugated streptavidin at room temperature for 1 h, followed by addition of TMB-ELISA substrate. The reaction was stopped by adding 2 M H2SO4 and the color developed monitored by measuring absorbance at 450 nm. The buffers, glycine-HCl (pH 2.5), citrate buffer (pH 4.5), MES (pH 5.6), TBS (pH 7.5), glycine-NaOH (pH 9.2) and TBS containing 100 mM EDTA were used instead of plain TBS to determine the effect of different pH and EDTA on glycan binding.
Glycan microarray analysis
Purified recombinant proteins, SrFucD244ANaFLD (200 μg/mL), SrNaFLD (200 μg/mL), SrNa (50 μg/mL) and SrFucD244ANaFLDH623AR651AR658A (200 μg/mL) were extensively dialyzed against TBS with 10 mM CaCl2, and were subjected to glycan microarray analysis following the standard procedure of the Protein-Glycan Interaction Core (H) at CFG (Consortium for Functional Glycomics, Emory University USA) to determine glycan binding specificity. CFG glycan microarrays versions 5.2 (with 203 fucosylated glycans and a total of 609 glycans) and 5.3 (with 200 fucosylated glycans and total of 600 glycans) were used. Mouse C-terminal 6xHis antibody and fluorescently labeled Alexa488 secondary anti-mouse antibody were used to detect binding of the proteins to the glycans on the CFG microarray, and fluorescence intensities were measured in a Perkin Elmer Scan Array scanner. The data was filtered to remove the highest and lowest spot of the six replicates. The glycan binding specificity of the proteins were determined by ranking the glycan binding data according to signal intensity, and categorizing fucosylated glycans (Supplementary data, Tables SI, SII, SIII, SIV) according to fucose linkages and glycan structural motifs. Some complex glycans with more than one glycan motif were classified into more than one category.
Surface Plasmon Resonance (SPR) studies of SrFucD244ANaFLD binding to fucosylated glycans
SPR experiments were performed on a BIAcore 3000 instrument (GE Healthcare) at 25°C in 10 mM HEPES buffer with 150 mM NaCl and 0.05% Tween-20, pH 7.5 at a flow rate of 10 μL/min. A research-grade CM5 sensor chip was used. The surfaces of flow channels (FC) 3 and 4 were activated with a 1:1 mixture of 0.1 M NHS (N-hydroxysuccinimide) and 0.1 M EDC (3-(N,N-dimethylamino)-propyl-N-ethylcarbodiimide) at a flow rate of 5 μL/min. Neutravidin (40 μg/mL in 10 mM sodium acetate, pH 5.0) was immobilized on FC3 and FC4. All the surfaces were blocked with 1 M ethanolamine, pH 8.0. Biotin-PAA-α-l-fucose, 200 μg/mL, was then captured on flow channel 4. FC3 was left blank to serve as a reference surface. To collect kinetic binding data, the SrFucD244ANaFLD protein (77.7 kDa) was injected (association 240 s, dissociation 600 s) over the two flow cells and binding measured as response units (RU) over time after blank subtraction (FC4–FC3). The flow channels were fully regenerated with 1 M l-fucose after each injection. The data was fitted to a simple 1:1 interaction model available within BIA evaluation software version 4.
SPR inhibition studies were done with 11 different fucosylated glycans (procured from Elicityl) – Lewisa tetraose (GLY054), Lewisb tetraose (GLY045), Lewisx tetraose (GLY050), Lewisy tetraose (GLY048), Lewisa hexaose or Lacto-N-Difucohexaose II (GLY055), 3-fucosyllactose (GLY060), blood group A type-2 tetraose (GLY035-2), blood group B type-2 tetraose (GLY038-2), blood group H type-2 tetraose (GLY032-2), blood group H type-2 triose (GLY031-2) and blood group H type-2 pentaose proparagyl (GLY033-2). For each inhibition assay, SrFucD244ANaFLD protein (~1 μM) was incubated with different concentrations of glycan for 45 min at room temperature. The mixture was then passed through FC3 and FC4 using the same parameters. For IC50 evaluation, the response obtained in the absence of any glycan was considered to be 100% and the relative binding was calculated for each concentration of glycan. Inhibition curves were obtained by plotting percent inhibition of binding versus the logarithmic value of glycan inhibitor concentration.
Multiple sequence alignment and phylogenetic tree generation
The tool, Tree-based Consistency Objective Function For alignment Evaluation (T-Coffee) (Notredame et al. 2000), was employed for generation of multiple sequence alignments of SrFuc along with characterized representatives of GH29 family members. The characterized GH29 representatives used for the analysis were AlfA (Genbank accession no: CAQ67115.1), AlfB (Genbank accession no: WP_016384650.1) and AlfC (Genbank accession no: WP_045625564.1) from Lactobacillus casei, BT_2192 (Genbank accession no: NP_811105.1) and BT_2970 (Genbank accession no: WP_008767257.1) from Bacteroides thetaiotaomicron, TM306 (Genbank accession no: WP_010865090.1) from T. maritima, AfcB (Genbank accession no: WP_047270681.1) from Bifidobacterium bifidum JCM 1254, Blon_2336 (Genbank accession no: WP_012578563.1) from Bifidobacterium bifidum JCM 1254, Blon_0248 (Genbank accession no: WP_012576690.1) and Blon_0426 (Genbank accession no: WP_012576844.1) from Bifidobacterium longumATCC15697, ALfuk1 from Paenibacillus thiaminolyticus (Genbank accession no: CBM40947.1), BFO_2737 (Genbank accession no: WP_014225862.1) from Tannerella forsythia ATCC 43037, FucA1 (Genbank accession no: WP_011038013.1) from Xanthomonas campestris ATCC 33913, FUCA1 (Genbank accession no: NP_001003250.1) from Canis lupus familiaris, AlfA (Genbank accession no: EGG13477.1) from Dictyosetlium fasciculatum, FCO1 (Genbank accession no: AFR68934.1) from Fusarium oxysporum, FCO1 (Genbank accession no: KNB14523.1) from Fusarium graminearum PH-1, α-1,3/1,4-l-fucosidase (Genbank accession no: KNB14523.1) from Streptomyces sp. 142, FucA1 (Genbank accession no: WP_009988419.1) from Sulfolobus solfataricus P2, AtFuc1 (Genbank accession no: NP_180377.2) from Arabidopsis thaliana, FucA1 (Genbank accession no: NP_000138.2) and FucA2 (Genbank accession no: CAB53746.1) from Homo sapiens, FucA1 (Genbank accession no: NP_036694.2) from Rattus norvegicus, and Mfuc1 (Genbank accession no: AIC77298.1), Mfuc2 (Genbank accession no: AIC77299.1), Mfuc3 (Genbank accession no: AIC77300.1), Mfuc4 (Genbank accession no: AIC77301.1), Mfuc5 (Genbank accession no: AIC77302.1), Mfuc6 (Genbank accession no: AIC77303.1) and Mfuc7 (Genbank accession no: AIC77304.1) from uncultured bacteria. A phylogenetic tree was constructed using the S. roseum alpha-l-fucosidase protein and the representative members of GH29 alpha-l-fucosidase family. The Neighbor-Joining (NJ) method (Saitou and Nei 1987) was used to generate a phylogenetic tree and evolutionary relationships were analyzed by Molecular Evolutionary Genetics Analysis (MEGA v6.0) (Tamura et al. 2013). The tree was viewed by using Interactive Tree Of Life (iTOL) server (Letunic and Bork 2011).
Alpha-l-fucosidase activity assays of SrFucNaFLD and SrFuc using the synthetic substrate, 4-Methylumbelliferyl-α-l-fucopyranoside
For studying the effect of temperature and pH, 1 μM SrFucNaFLD or SrFuc was mixed with 50 μM 4-Methylumbelliferyl-α-l-fucopyranoside (Sigma) in a 96 well black Maxisorp microtiter plate (Nunc), the assay was performed at 37°C in TBS at the temperatures – 8°C, 18°C, 25°C, 37°C, 45°C, 55°C, or 65°C, and the fluorescence of the product, 4-MU was detected using an excitation wavelength of 365 nm and an emission wavelength of 450 nm every 1 min for a period of 15 min. For studying the effect of metal ions, the alpha-l-fucosidase activity of SrFuc was assayed in the presence of 10 mM calcium chloride, magnesium chloride, manganese chloride, cobaltous chloride, copper sulfate, zinc sulfate, nickel sulfate, sodium chloride and ethylene diamine tetraacetate, and fluorescence readings were measured in kinetics mode every 1 min over a time period of 60 min.
For substrate kinetics, 4-Methylumbelliferyl-α-l-fucopyranoside was included at 2-fold serial dilutions starting from 200 μM and up to a concentration of 1.5 μM. The reaction was initiated by the addition of SrFucNaFLD or SrFuc to a final concentration of 0.25 μM and the fluorescence was monitored in a Synergy H1 hybrid plate reader for 60 min at intervals of 1 min at 37°C. Appropriate controls were included wherein TBS was used in place of enzyme or substrate. Using a standard curve of 4-MU fluorescence at various concentrations, the fluorescence readings were converted into nmol 4-MU formed per minute by SrFuc and SrFucNaFLD. Initial velocities were calculated from the slopes of the kinetics plots and used for generating the Michaelis-Menten plot. Sigma plot software was used for curve fitting and calculation of Km and Vmax values.
For studying the inhibition of alpha-l-fucosidase activity by l-fucose and deoxyfuconojirimycin hydrochloride (DFJHCl), an assay solution containing 0.25 μM SrFucNaFLD or SrFuc, 25 μM 4-Methylumbelliferyl-α-l-fucopyranoside and l-fucose (Sigma) (serially diluted 2-fold starting from 1 mM) or DFJHCl (serially diluted 10-fold starting from 500 μM) in TBS was used. Initial velocities were calculated for all serial dilutions of l-fucose and Sigma plot software was used for calculation of IC50 values.
Cloning, expression, purification and activity assay of Pseudomonas sp l-fucose dehydrogenase (FDH)
The nucleotide sequence (GenBank accession number D32042.1) coding for the mature polypeptide sequence of FDH was codon optimized for expression in E. coli, custom synthesized (GenScript, Piscataway, NJ), and cloned into pET-28a(+) to encode a C-terminal hexahistidine tag. Positive clones were confirmed by DNA sequencing. For recombinant protein expression, a secondary culture of E. coli BL21(DE3) cells transformed with the FDH-pET-28a(+) plasmid were grown at 37°C until the cell density corresponded to an OD600 of 0.8, then induced with 0.01 mM IPTG, and incubated with continuous shaking at 160 rpm for 16 h at 16°C. The bacterial cell pellet was resuspended in a TBS-based lysis solution and purified by Ni-NTA metal ion affinity chromatography using 40 mM imidazole in TBS as the wash buffer and 250 mM imidazole in TBS as the elution buffer. Purified protein was dialyzed extensively against TBS and assayed for FDH activity using the resazurin/diaphorase system (Guilbault and Kramer 1965), wherein the enzyme, diaphorase is used to convert resazurin into fluorescent resorufin, making use of the NADPH which is released by the action of FDH. The reaction was initiated by adding 50 μL of the purified FDH preparation to 50 μL TBS containing 0.1 M l-fucose, 20 mM NADP, 10 μM resazurin (Sigma) and 0.2 units/mL diaphorase (Sigma). Resorufin formed was detected by monitoring fluorescence at an excitation wavelength of 530 nm and an emission wavelength of 590 nm. The optimal FDH concentration for the assay was determined by performing the assay with 2-fold serial dilutions starting with 0.25 mg/mL FDH.
Alpha-l-fucosidase activity assays using fucosylated oligosaccharide substrates
The alpha-l-fucosidase activity of SrFucNaFLD, SrFuc, SrFucNa and SrFucNaFLDTM with unlabeled oligosaccharide substrates was assayed by using an FDH endpoint assay. The fluorescence-based FDH assay using the above-mentioned resazurin/diaphorase system was used to determine the amount of free l-fucose released from fucosylated oligosaccharides by alpha-l-fucosidase activity. The fucosylated glycans, blood group H type-2 triose (GLY031-2), blood group H type-2 tetraose (GLY032-2), blood group H type-2 pentaose (GLY033-1), blood group H type-2 pentaose proparagyl (GLY033-2), blood group A type-2 tetraose (GLY035-2), blood group B type-2 tetraose (GLY038-2), Lewisa tetraose (GLY054), Lewisx tetraose (GLY050), Lewisb tetraose (GLY045), Lewisy tetraose (GLY048), Lacto-N-Difucohexaose II (GLY055) and 3-Fucosyllactose (GLY060) from Bio-Indenta, and 2′-Fucosyllactose and Lacto-N-fucopentaose III from Sigma were used as substrates. A solution containing 4 μM of SrFucNaFLD, SrFuc, SrFucNa, SrFuc+SrNaFLD or SrFucNaFLDTM was mixed with 2 mM of fucosylated glycan and the reaction volume was made up to 50 μL by TBS buffer containing 10 mM of CaCl2, and was incubated at 37°C for 210 min. At the end of this time period, the reaction mix was mixed with an assay mix of 50 μL containing 0.2 μM FDH, 1 mM NADP, 10 μM resazurin and 0.2 Units/mL diaphorase in TBS and the resorufin fluorescence was monitored using an excitation wavelength of 530 nm and an emission wavelength of 590 nm every 1 min in kinetic mode in a Synergy H1 hybrid plate reader. Initial velocities of FDH enzyme activity were calculated from the fluorescence readings obtained during this enzyme assay. Using the Michaelis Menten plot of FDH made with various concentrations of free l-fucose, the l-fucose concentrations present in the alpha-l-fucosidase reactions after 210 min were calculated from the initial velocities. We used 210 min of reaction time for the alpha-l-fucosidase assays because the resazurin fluorescence in our preliminary experiments saturated around 200 min indicating no more l-fucose release after this time point. Subsequently, amounts of l-fucose present in the reaction volume were calculated, and represented as picomoles l-fucose released from the substrate by the alpha-l-fucosidase after 210 min.
The FDH concentration was optimized by trying different concentrations of FDH (micromolar to nanomolar range) and the concentration at which initial velocities could be measured accurately (0.1 μM) was used for the assays reported. With higher FDH concentration (>0.5 μM) we were unable to accurately measure initial velocity due to the reaction progressing too fast to measure using our assay set up.
We estimate the sensitivity limit of this endpoint alpha-l-fucosidase assay described above to be around 3–5 pmol released in 100 μL reaction volume based on the linear equation fitting the standard curve as well as based on the fluorescence detected in the standard curve of mM fucose versus initial velocity of FDH. We consider that any value less than 5 pmol released in reaction volume (prior to subtraction of buffer control) is activity not detected by our assay.
The standard curve of l-fucose concentration versus initial velocity of FDH was fitted to Michaelis Menten kinetics equation (using the whole range of concentrations) as well as to a linear equation (using only lower concentrations in the linear range). The linear fit was found to be better with lower residuals for the lower concentrations which were actually relevant for the study (since only fucose of such low concentrations were released in the endpoint assay) therefore this linear fit was used for the calculations.
Supplementary Material
Acknowledgements
The authors thank Dr. P. Babu and Dr. S. Gopalan for discussions relating to the research reported here, and the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics (NCFG) at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grant P41 GM103694) for the glycan array analysis. The authors acknowledge CSIR-IMTECH (manuscript communication number 035/2017) for the research facilities and infrastructure.
Abbreviations
- FLD
F-type lectin domain
- SPR
surface plasmon resonance
- Fuc
alpha-l-fucosidase
- Na
Npcbm_associated
- NPCBM
novel putative carbohydrate binding module
- CDD
conserved domain database
- iTOL
interactive tree of life
- MEGA
molecular evolutionary genetic analysis.
Funding
This work was supported by the Science and Engineering Research Board, Department of Science and Technology, Government of India (FAST-TRACK grant no. SR/FT/LS-87/2012 to R.T.N.C.). R.B. and S.M. acknowledge the University Grants Commission (UGC) and Department of Biotechnology (DBT), Government of India, respectively, for their fellowships.
Conflict of interest statement
The authors declare that there are no conflicts of interest relevant to the subject of this manuscript.
Authors' contributions
R.T.N.C. conceived the study, R.B. and S.M. performed the biochemical experiments. All authors participated in experimental design, data analysis and manuscript preparation.
References
- Abbott DW, Eirin-Lopez JM, Boraston AB. 2008. Insight into ligand diversity and novel biological roles for family 32 carbohydrate-binding modules. Mol Biol Evol. 25:155–167. [DOI] [PubMed] [Google Scholar]
- Alam T, Balsubramanian AS. 1978. The purification, properties and characterization of three forms of alpha-L-fucosidase from monkey brain. Biochim Biophys Acta. 524:373–384. [DOI] [PubMed] [Google Scholar]
- Alhadeff JA, Miller AL, Wenaas H, Vedvick T, O’Brien JS. 1975. Human liver alpha-L-fucosidase. Purification, characterization, and immunochemical studies. J Biol Chem. 250:7106–7113. [PubMed] [Google Scholar]
- Ali MK, Hayashi H, Karita S, Goto M, Kimura T, Sakka K, Ohmiya K. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci Biotechnol Biochem. 65:41–47. [DOI] [PubMed] [Google Scholar]
- Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 19:1010–1017. [DOI] [PubMed] [Google Scholar]
- Audfray A, Claudinon J, Abounit S, Ruvoen-Clouet N, Larson G, Smith DF, Wimmerova M, Le Pendu J, Romer W, Varrot A et al. 2012. Fucose-binding lectin from opportunistic pathogen Burkholderia ambifaria binds to both plant and human oligosaccharidic epitopes. J Biol Chem. 287:4335–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus SE, Thiele J, Park YO, Hanisch FG, Bara J, Fischer R. 1996. Characterization of the binding specificity of Anguilla anguilla agglutinin (AAA) in comparison to Ulex europaeus agglutinin I (UEA-I). Glycoconj J. 13:585–590. [DOI] [PubMed] [Google Scholar]
- Becker DJ, Lowe JB. 2003. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 13:41R–53R. [DOI] [PubMed] [Google Scholar]
- Berteau O, McCort I, Goasdoue N, Tissot B, Daniel R. 2002. Characterization of a new alpha-L-fucosidase isolated from the marine mollusk Pecten maximus that catalyzes the hydrolysis of alpha-L-fucose from algal fucoidan (Ascophyllum nodosum). Glycobiology. 12:273–282. [DOI] [PubMed] [Google Scholar]
- Bianchet MA, Odom EW, Vasta GR, Amzel LM. 2002. A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 9:628–634. [DOI] [PubMed] [Google Scholar]
- Bianchet MA, Odom EW, Vasta GR, Amzel LM. 2010. Structure and specificity of a binary tandem domain F-lectin from striped bass (Morone saxatilis). J Mol Biol. 401:239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishnoi R, Khatri I, Subramanian S, Ramya TN. 2015. Prevalence of the F-type lectin domain. Glycobiology. 25:888–901. [DOI] [PubMed] [Google Scholar]
- Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, Gilbert HJ. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem J. 331(Pt 3):775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston AB, Wang D, Burke RD. 2006. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 281:35263–35271. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 37:D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Walton JD, Brumm P, Phillips GN Jr. 2014. Structure and substrate specificity of a eukaryotic fucosidase from Fusarium graminearum. J Biol Chem. 289:25624–25638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Flint JE, Gloster TM, Morland C, Basle A, Henrissat B, Coutinho PM, Strazzulli A, Solovyova AS, Davies GJ et al. 2012. How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc Natl Acad Sci USA. 109:20889–20894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam TK, Gerken TA, Brewer CF. 2009. Thermodynamics of multivalent carbohydrate-lectin cross-linking interactions: Importance of entropy in the bind and jump mechanism. Biochemistry. 48:3822–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Tsay G. 1977. Substrate specificity of human alpha-L-fucosidase. Arch Biochem Biophys. 184:12–23. [DOI] [PubMed] [Google Scholar]
- DiCioccio RA, Barlow JJ, Matta KL. 1982. Substrate specificity and other properties of alpha-L-fucosidase from human serum. J Biol Chem. 257:714–718. [PubMed] [Google Scholar]
- Farrand S, Hotze E, Friese P, Hollingshead SK, Smith DF, Cummings RD, Dale GL, Tweten RK. 2008. Characterization of a streptococcal cholesterol-dependent cytolysin with a Lewis y and b specific lectin domain. Biochemistry. 47:7097–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil SC, Lawrence S, Mulhern TD, Holien JK, Hotze EM, Farrand S, Tweten RK, Parker MW. 2012. Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity. Structure. 20:248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J et al. 2014. Pfam: The protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihashi M, Peapus DH, Kamiya N, Nagata Y, Miki K. 2003. Crystal structure of fucose-specific lectin from Aleuria aurantia binding ligands at three of its five sugar recognition sites. Biochemistry. 42:11093–11099. [DOI] [PubMed] [Google Scholar]
- Fukumori F, Takeuchi N, Hagiwara T, Ohbayashi H, Endo T, Kochibe N, Nagata Y, Kobata A. 1990. Primary structure of a fucose-specific lectin obtained from a mushroom, Aleuria aurantia. J Biochem. 107:190–196. [DOI] [PubMed] [Google Scholar]
- Guilbault GG, Kramer DN. 1965. Fluorometric procedure for measuring the activity of dehydrogenases. Anal Chem. 37:1219–1221. [DOI] [PubMed] [Google Scholar]
- Guillotin L, Lafite P, Daniellou R. 2014. Unraveling the substrate recognition mechanism and specificity of the unusual glycosyl hydrolase family 29 BT2192 from Bacteroides thetaiotaomicron. Biochemistry. 53:1447–1455. [DOI] [PubMed] [Google Scholar]
- Hindsgaul O, Norberg T, Le Pendu J, Lemieux RU. 1982. Synthesis of type 2 human blood-group antigenic determinants. The H, X, and Y haptens and variations of the H type 2 determinant as probes for the combining site of the lectin I of Ulex europaeus. Carbohyd Res. 109:109–142. [DOI] [PubMed] [Google Scholar]
- Hoffmam ZB, Zanphorlin LM, Cota J, Diogo JA, Almeida GB, Damasio AR, Squina F, Murakami MT, Ruller R. 2016. Xylan-specific carbohydrate-binding module belonging to family 6 enhances the catalytic performance of a GH11 endo-xylanase. N Biotechnol. 33:467–472. [DOI] [PubMed] [Google Scholar]
- Houser J, Komarek J, Kostlanova N, Cioci G, Varrot A, Kerr SC, Lahmann M, Balloy V, Fahy JV, Chignard M et al. 2013. A soluble fucose-specific lectin from Aspergillus fumigatus conidia–structure, specificity and possible role in fungal pathogenicity. PLoS One. 8:e83077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. 2005. FFAS03: A server for profile–profile sequence alignments. Nucleic Acids Res. 33:W284–W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2011. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32:D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezyk M, Jers C, Kjaerulff L, Gotfredsen CH, Mikkelsen MD, Mikkelsen JD. 2016. Novel alpha-L-fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PLoS One. 11:e0147438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Chen CS, Chang SS, Mong KK, Lin CH, Chang CW, Tang CY, Li YK. 2009. Identification of essential residues of human alpha-L-fucosidase and tests of its mechanism. Biochemistry. 48:110–120. [DOI] [PubMed] [Google Scholar]
- Ma B, Simala-Grant JL, Taylor DE. 2006. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 16:158r–184r. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Ramya T N C. 2018. Nature-inspired engineering of an F-type lectin for increased binding strength. Glycobiology. doi:10.1093/glycob/cwy082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Higashida K, Ishida H, Hata Y, Yamamoto K, Shigeta M, Mizuno-Horikawa Y, Wang X, Miyoshi E, Gu J et al. 2007. Carbohydrate binding specificity of a fucose-specific lectin from Aspergillus oryzae: A novel probe for core fucose. J Biol Chem. 282:15700–15708. [DOI] [PubMed] [Google Scholar]
- McNeil M, Darvill AG, Fry SC, Albersheim P. 1984. Structure and function of the primary cell walls of plants. Annu Rev Biochem. 53:625–663. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Fernandes VO, Karita S, Luis AS, Sakka M, Kimura T, Jackson A, Zhang X, Fontes CM, Gilbert HJ et al. 2012. Influence of a mannan binding family 32 carbohydrate binding module on the activity of the appended mannanase. Appl Environ Microbiol. 78:4781–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumoff DG. 2005. GH97 is a new family of glycoside hydrolases, which is related to the alpha-galactosidase superfamily. BMC Genomics. 6:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov DG. 2004. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Mol Biol (Mosk). 38:463–476. [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302:205–217. [DOI] [PubMed] [Google Scholar]
- Oda Y, Senaha T, Matsuno Y, Nakajima K, Naka R, Kinoshita M, Honda E, Furuta I, Kakehi K. 2003. A new fungal lectin recognizing alpha(1-6)-linked fucose in the N-glycan. J Biol Chem. 278:32439–32447. [DOI] [PubMed] [Google Scholar]
- Odom EW, Vasta GR. 2006. Characterization of a binary tandem domain F-type lectin from striped bass (Morone saxatilis). J Biol Chem. 281:1698–1713. [DOI] [PubMed] [Google Scholar]
- Renard CM, Lomax JA, Boon JJ. 1992. Apple-fruit xyloglucans: A comparative study of enzyme digests of whole cell walls and of alkali-extracted xyloglucans. Carbohyd Res. 232:303–320. [DOI] [PubMed] [Google Scholar]
- Rigden DJ. 2005. Analysis of glycoside hydrolase family 98: Catalytic machinery, mechanism and a novel putative carbohydrate binding module. FEBS Lett. 579:5466–5472. [DOI] [PubMed] [Google Scholar]
- Saito T, Hatada M, Iwanaga S, Kawabata S. 1997. A newly identified horseshoe crab lectin with binding specificity to O-antigen of bacterial lipopolysaccharides. J Biol Chem. 272:30703–30708. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sakurama H, Tsutsumi E, Ashida H, Katayama T, Yamamoto K, Kumagai H. 2012. Differences in the substrate specificities and active-site structures of two alpha-L-fucosidases (glycoside hydrolase family 29) from Bacteroides thetaiotaomicron. Biosci Biotechnol Biochem. 76:1022–1024. [DOI] [PubMed] [Google Scholar]
- Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 78:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Cummings RD. 2009. Structures common to different glycans In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. New York: Cold Spring Harbor, p. 175–198. [PubMed] [Google Scholar]
- Stanley P, Schachter H, Taniguchi N. 2009. N-glycans In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. New York: Cold Spring Harbor, p. 101–114. [PubMed] [Google Scholar]
- Sudakevitz D, Imberty A, Gilboa-Garber N. 2002. Production, properties and specificity of a new bacterial L-fucose- and D-arabinose-binding lectin of the plant aggressive pathogen Ralstonia solanacearum, and its comparison to related plant and microbial lectins. J Biochem. 132:353–358. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Bignon C, Nishimura T, Tarling CA, Withers SG, Henrissat B, Bourne Y. 2004. Crystal structure of Thermotoga maritima alpha-L-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J Biol Chem. 279:13119–13128. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarling CA, He S, Sulzenbacher G, Bignon C, Bourne Y, Henrissat B, Withers SG. 2003. Identification of the catalytic nucleophile of the family 29 alpha-L-fucosidase from Thermotoga maritima through trapping of a covalent glycosyl-enzyme intermediate and mutagenesis. J Biol Chem. 278:47394–47399. [DOI] [PubMed] [Google Scholar]
- van Bueren AL, Higgins M, Wang D, Burke RD, Boraston AB. 2007. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol. 14:76–84. [DOI] [PubMed] [Google Scholar]
- VanEpps DE, Tung KS. 1977. Fucose-binding Lotus tetragonolobus lectin binds to human polymorphonuclear leukocytes and induces a chemotactic response. J Immunol. 119:1187–1189. [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta GR, Ahmed H, Odom EW. 2004. Structural and functional diversity of lectin repertoires in invertebrates, protochordates and ectothermic vertebrates. Curr Opin Struct Biol. 14:617–630. [DOI] [PubMed] [Google Scholar]
- Vasta GR, Amzel LM, Bianchet MA, Cammarata M, Feng C, Saito K. 2017. F-type lectins: a highly diversified family of fucose-binding proteins with a unique sequence motif and structural fold, involved in self/non-self-recognition. Front Immunol. 8:1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmerova M, Mitchell E, Sanchez JF, Gautier C, Imberty A. 2003. Crystal structure of fungal lectin: Six-bladed beta-propeller fold and novel fucose recognition mode for Aleuria aurantia lectin. J Biol Chem. 278:27059–27067. [DOI] [PubMed] [Google Scholar]
- Winchester B, Barker C, Baines S, Jacob GS, Namgoong SK, Fleet G. 1990. Inhibition of alpha-L-fucosidase by derivatives of deoxyfuconojirimycin and deoxymannojirimycin. Biochem J. 265:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ho CW, Ko TP, Popat SD, Lin CH, Wang AH. 2010. Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range. Angew Chem Int Ed Engl. 49:337–340. [DOI] [PubMed] [Google Scholar]
- Zheng F, Ding S. 2013. Processivity and enzymatic mode of a glycoside hydrolase family 5 endoglucanase from Volvariella volvacea. Appl Environ Microbiol. 79:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.