Abstract
Global control of hepatitis C virus (HCV) depends on development of a prophylactic vaccine. We studied escape for cross-genotype–reactive neutralizing antibody AR4A, providing valuable information for HCV vaccine design. We cultured HCV core-NS2 recombinants H77 (genotype 1a)/JFH1 or the highly antibody-susceptible hypervariable region 1 (HVR1)–deleted variants H77/JFH1∆HVR1 and J6(genotype 2a)/JFH1∆HVR1 in Huh7.5 cells with AR4A. Long-term AR4A exposure of H77/JFH1 and H77/JFH1∆HVR1 did not yield resistance. However, J6/JFH1∆HVR1 developed the envelope-E2 substitutions I696T or I696N, which reduced AR4A binding (I696N > I696T). I696N conferred greater AR4A resistance than I696T in J6/JFH1∆HVR1, whereas the reverse was observed in J6/JFH1. This was because I696N but not I696T conferred broadly increased antibody neutralization susceptibility to J6/JFH1. I696N and I696T abrogated infectivity of H77/JFH1 and broadly increased neutralization susceptibility of S52 (genotype 3a)/JFH1. In conclusion, I696 is in the AR4A epitope, which has a high barrier to resistance, thus strengthening the rationale for its inclusion in rational HCV vaccine designs.
Keywords: HCV, HVR1, hypervariable region 1, vaccine, immune evasion, antibody escape, liver disease
A hepatitis C virus (HCV) vaccine is urgently needed. The AR4A human monoclonal antibody targets a cross-genotype conserved HCV envelope epitope with potential relevance for vaccine development. We identify AR4A resistance substitutions and demonstrate a high barrier to AR4A resistance.
With >71 million individuals chronically infected with hepatitis C virus (HCV) worldwide, approximately 400 000 people die annually from HCV-related liver diseases [1, 2]. Novel direct-acting antiviral (DAA) therapies have cure rates of >95% [2, 3]. However, several factors, including treatment cost, high prevalence of occult infection, low treatment adherence in resource-poor parts of the world, and lack of protection against reinfection underline the necessity of vaccine development [3, 4].
HCV is an enveloped positive-stranded RNA virus of the Flaviviridae family, with 6 clinically relevant genotypes [1, 5, 6]. The HCV genome encodes a single polyprotein, which is processed into 3 structural proteins (core, E1, and E2), p7, and nonstructural proteins (NS2–5B). Neutralizing antibodies (NAbs) against HCV target the envelope protein complex E1/E2, which consequently is of primary interest in vaccine development [7, 8].
Studies have shown correlation between NAbs and clearance of acute HCV infection in humans [9, 10]. In addition, passive immunization with NAbs provided partial or even complete protection in chimpanzees [11–13] or human-liver-chimeric mice [14–17]. However, HCV heterogeneity remains a vaccine design obstacle [11]. In addition, HCV rapidly accumulates mutations [18], allowing escape from the humoral immune response. Therefore, successful HCV vaccine development depends on identifying cross-genotype conserved epitopes with high barriers to resistance.
By isolating NAbs from HCV-infected patients, we classified 5 antigenically distinct antigenic regions (termed AR1–5) and showed that AR3-, AR4-, and AR5-specific NAbs are potent in vitro across HCV genotypes, although variation was observed [19, 20]. This variation in cross-genotype susceptibility against AR3–5 antibodies decreased when the E2 motif hypervariable region 1 (HVR1) was deleted, indicating remarkable epitope conservation [21]. Interestingly, the AR4-specific NAb, AR4A, was unique in displaying virtually no cross-genotype variation for HVR1-deleted HCV [21]. Also, AR4A outperformed other promising NAbs, AR3A and AR3B, in protecting a genetically humanized mouse model from HCV challenge [22]. In vivo efficacy of AR4A was confirmed in another mouse challenge study [23]. Finally, AR4A is able to overcome the inherent HCV resistance against neutralization by autologous serum [24] and has high synergy with other potent NAbs [25, 26].
Taking advantage of the high neutralization susceptibility of HVR1-deleted HCV [21, 27, 28], we recently identified high-level AR5A resistance substitutions in cell-culture-infectious, JFH1-based core-NS2 recombinants H77/JFH1 (genotype 1a) and J6/JFH1 (genotype 2a) [29], which suggested that epitope conservation is a poor predictor of the barrier to resistance [29]. When using the same methods to study AR4A resistance, we were unable to induce escape in H77/JFH1 and observed only low-level resistance in J6/JFH1. Our results indicate that the AR4A epitope has a high barrier to resistance and would be a promising inclusion in the design of future HCV vaccine antigens.
METHODS
Antibodies and Reagents
Human monoclonal antibodies AR3A, AR4A, and AR5A generated from a genotype 1a–infected patient were produced as described previously [19, 20]. 9E10 antibody was kindly provided by Charles Rice [30]. AP33 antibody was kindly provided by Arvind Patel [31]. Plasmids with the core-NS2 sequence from genotypes 1a (H77), 2a (J6), or 3a (S52) and untranslated regions plus the NS3-NS5B region from genotype 2a (JFH1), with or without HVR1, were described previously (S52/JFH1∆HVR1 has the HVR1-deletion adaptive substitution A369V, and H77/JFH1∆HVR1 has the HVR1-deletion adaptive substitutions N476D and S733F) [27, 30, 32, 33], as well as a further cell culture–adapted mutant, H77/JFH1ΔHVR1/N417D/N532D, which has 2 additional HVR1-deletion adaptive substitutions, N417D and N532D, in addition to N476D and S733F [29]. J6 and J6∆HVR1 E1/E2 expression plasmids were previously described [34]. Point mutations were introduced into plasmids by conventional cloning techniques. The HCV sequence of each plasmid was confirmed by sequencing the final DNA preparation (Macrogen).
Cell Culture of Huh7.5 Cells
Huh7.5 cells [35], provided by Charles Rice, were cultured in Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen) supplemented with 10% of heat-inactivated fetal bovine serum, penicillin 100 U/mL, and streptomycin 100 μg/mL (Gibco/Invitrogen) with 5% CO2 at 37°C. Cells were split every 48–72 hours.
Transfection of Huh7.5 Cells
Huh7.5 cells were plated at 4 × 105 cells per well in 6-well plates and incubated overnight. HCV RNA was generated by T7-mediated in vitro transcription of XbaI-linearized plasmids. Huh7.5 cells were transfected using Lipofectamine 2000 (Invitrogen). Six hours after transfection, Huh7.5 cells were trypsinized and reseeded into 4 wells of 24-well plates at a cell density of 8 × 104 cells/well, along with plating in 6-well chamber slides for assessing the percentage of infected cells, as described elsewhere [29]. The percentage of infected cells was assessed at the 4 time points, using the primary mouse antibody anti-HCV NS5A 9E10 and the secondary antibody Alexa Fluor 488 goat anti-mouse immunoglobulin G (heavy and light chains; Invitrogen), as previously described [33]. Viral spread was monitored every 24 hours, and supernatants were harvested, sterile filtered, and stored at −80°C. Virus titers were determined as described previously [36, 37].
HCV Escape Assay
Huh7.5 cells were infected with the indicated virus at a multiplicity of infection of 0.001. Virus infection was monitored by immunostaining every 2–3 days as described above. After infection reached 0.1%–1% of the cells, the indicated concentrations of AR4A were added to each well following cell splitting. Cell supernatant was collected and filtered when infection reached 80%–90% of the cells. Direct Sanger sequencing of envelope sequences from cell culture–derived HCV RNA was performed by nested long reverse transcription–polymerase chain reaction procedures, as previously described [29].
Antibody Neutralization
This was done as previously described [21]. We plated 6 × 103 Huh7.5 cells per well in poly-D-lysine 96-well plates and incubated the cells for 24 hours. A virus dose resulting in a final readout of 30–150 focus-forming units of HCV in the control were incubated in quadruplicates with a dilution series of monoclonal antibody and relevant control antibody. Virus-antibody mixes along with 8 replicates of virus only were incubated for 1 hour at 37°C, added to Huh7.5 cells, and incubated for 4 hours at 37°C. Subsequently, the cells were washed, and we added fresh medium and incubated the cells for 48 hours. Cells were fixed and stained with 9E10 antibody, as described previously [33]. The data were normalized with 8 replicates of virus only and were analyzed using 3- or 4-parameter curve fitting in GraphPad Prism.
Immunostaining of HCV-Infected Cells for Evaluating AR4A Binding
Huh7.5 cells were plated at 4 × 105 cells per well in 6-well plates 24 hours before transfection, and transfected with HCV RNA as described above. Twenty-four hours after transfection, 10000 cells per well were plated in 8-well slides and incubated for 24 hours. Cells were fixed with paraformaldehyde at room temperature for 15 minutes. Anti-E1/E2 antibody (AR4A) [20] and anti-E2 antibody (AP33) [31] were used as primary antibodies at 1 µg/mL with a combination of Alexa Fluor 594 goat anti-mouse immunoglobulin G (heavy and light chains; Invitrogen) and Alexa Fluor 488 goat anti-human (Invitrogen) as secondary antibodies, using a Hoechst (Molecular Probes) nuclear counterstain. Images were acquired using a Zeiss Axio Observer Z1.
Flow Cytometry of AR4A Binding
Huh7.5 cells were transfected with HCV RNA as described above, or 293T cells were transfected with J6 E1/E2 expression plasmids in a similar fashion. A total of 48 hours after transfection, cells were treated with ethylenediaminetetraacetic acid and subjected to 2 phosphate-buffered saline washes between the following steps. Cells were fixed in 4% formaldehyde for 10 minutes and incubated for 1 hour with primary antibodies against E1/E2 (AR4A) [20] and E2 (AP33) [31], both at 1 µg/mL. This was followed by 1-hour incubation with the secondary antibodies Alexa Fluor 488 anti-human (ThermoFisher) and allophycocyanin anti-mouse (APC; Biolegend). Cells were resuspended and analyzed on a Becton Dickinson LSR Fortessa equipped with 405-, 488-, and 640-nm lasers and suitable filter sets. Data were collected using FACSDiva 8 (Becton Dickinson) and were analyzed with FlowJo 10 software (FlowJo). For data analysis, live cell populations were gated using front- and side-scatter dot plots and were subsequently analyzed using AP33 (APC) and AR4A (Alexa Fluor 488) density plots. The population of double-positive cells was used to calculate mean fluorescence intensity (MFI) and normalized AR4A fluorescence histograms.
RESULTS
Alanine Substitutions That Decrease AR4A Binding to H77 E1/E2 Resulted in Viral Fitness Loss of H77/JFH1
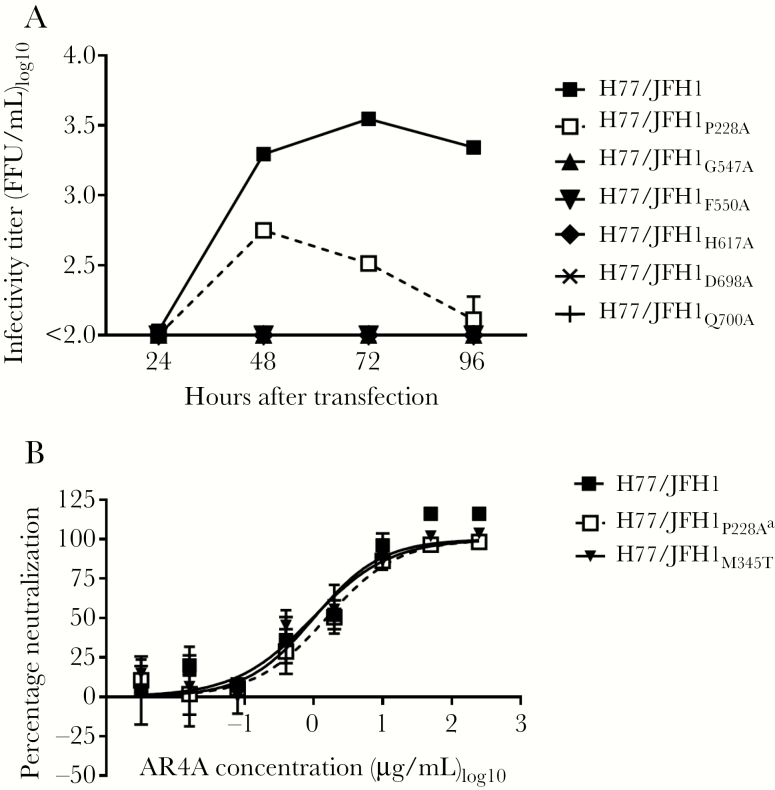
We previously showed that the envelope substitutions P228A, G547A, F550A, H617A, D698A, and Q700A reduced AR4A antibody binding to H77 E1/E2, using alanine scanning [20] (all positions are relative to the H77 reference genome [GenBank accession no. AF009606]). These substitutions were recently shown to have minor effects on the binding of antibodies targeting conformational E2 epitopes, except for F550A and H617A, which both globally destabilized E1/E2 [38]. To identify potentially viable AR4A-resistant viruses, the alanine substitutions were introduced into H77/JFH1 recombinants. Transfecting Huh7.5 cells, we found that P228A greatly reduced fitness, whereas the remaining 5 substitutions resulted in infectivity loss (Figure 1A). A first-passage virus stock was generated for H77/JFH1P228A, and the envelope genes were sequenced, revealing the additional substitution M345T (E1; T1374C). In a separate study, we found that H77/JFH1M345T has increased fitness (Velazquez-Moctezuma et al, unpublished data). A virus stock of H77/JFH1M345T without additional envelope substitutions was included when testing AR4A neutralization susceptibility to control for any effect of M345T. Neither P228A nor M345T affected AR4A susceptibility (Figure 1B).
Figure 1.
Envelope substitutions shown to reduce AR4A binding at the protein level resulted in highly attenuated viruses. A, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated H77/JFH1 recombinants. Supernatants were collected at the indicated time points, and HCV infectivity titers were determined. The lower level of quantification was 100 focus-forming units (FFU)/mL. An independent duplicate transfection yielded similar results. B, First passages of the indicated viruses were subjected to a dilution series of antibody AR4A, from 250 µg/mL to 0.0032 µg/mL. The virus/antibody mixes, along with virus only, were added to Huh7.5 cells for 4 hours prior to wash and addition of fresh medium. Following 48 hours of infection, the cells were immunostained, and the number of FFU per well was counted as described in Methods. Error bars represent standard error of the mean of 4 replicates normalized to 8 replicates of virus only. The data were analyzed using 3-parameter curve fitting to obtain a sigmoidal dose-response curve. Independent neutralization experiments yielded similar results. aThe indicated virus had the additional E1 substitution M345T.
H77/JFH1 and H77/JFH1∆HVR1 Spread in the Presence of AR4A Without Acquiring Antibody Resistance
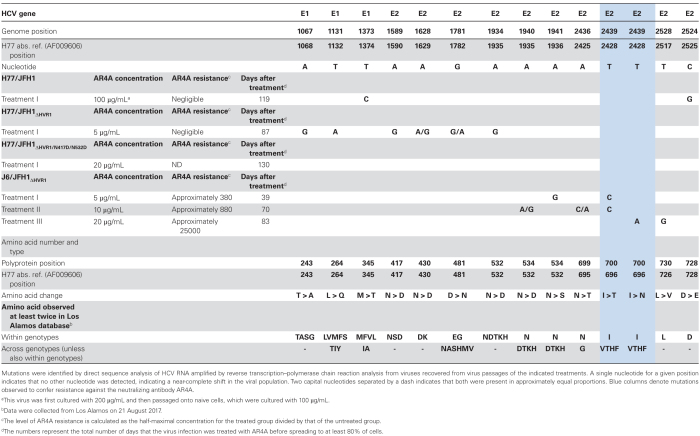
To induce AR4A-specific resistance substitutions, we cultured Huh7.5 cells infected with the recombinant virus H77/JFH1 with 200 or 100 µg/mL AR4A (Supplementary Figure 1A and 1B and Table 1). Virus spread in the AR4A-treated culture was significantly delayed. Once the virus spread at day 119 after treatment, we generated a virus stock and sequenced the envelope genes of recovered viruses, revealing the substitutions M345T (E1; T1374C) and D728E (E2; C2525G; Table 1). Testing AR4A susceptibility (Supplementary Figure 1C and Table 1), we found that the treated virus (half-maximal inhibitory concentration [IC50], 0.48 µg/mL) and the parental virus (IC50, 0.36 µg/mL) were similarly susceptible.
Table 1.
Coding Mutations Observed in the Hepatitis C Virus (HCV) Envelope Proteins Following AR4A Treatment
Next, we attempted to induce resistant mutants of the HVR1-deleted virus H77/JFH1ΔHVR1, using 5 µg/mL AR4A, in 3 separate cultures (Supplementary Figure 2A and Table 1). Only virus from treatment I spread following 87 days of treatment. In treatments II and III, we were unable to detect infected cells 94 days after treatment. A first-passage treatment I virus stock was generated from supernatant collected at the peak of infection. The envelope sequences had the substitutions T243A (E1; A1068G), L264Q (E1; T1132A), N417D (E2; A1590G), and N532D (E2; A1935G; Table 1). N417D and N532D are previously reported adaptive substitutions for H77/JFH1ΔHVR1 [29]. Testing AR4A susceptibility, we observed that virus from treatment I (IC50, 0.011 µg/mL) was similarly susceptible as parental virus (IC50, 0.0045 µg/mL; Supplementary Figure 2B and Table 1).
Finally, we cultured a high-fitness HVR1-deleted variant, H77/JFH1ΔHVR1/N417D/N532D [29], with 20 µg/mL AR4A (Supplementary Figure 2C and 2D). Although the virus spread under AR4A selective pressure, the envelope sequences had not acquired novel substitutions. This suggests that the presence of the 2 adaptive substitutions N417D and N532D [29] was sufficient to permit H77/JFH1ΔHVR1 to spread in the presence of AR4A.
Culturing J6/JFH1∆HVR1 With AR4A Resulted in Escape and High-Level Antibody Resistance
We have previously reported that J6/JFH1 can spread without acquiring resistance, because of high fitness and high resistance to NAbs [29], possibly through a high capacity for cell-to-cell spread as described for SA13/JFH1 [39]. Instead, it is possible to induce J6/JFH1 relevant escape substitutions by culturing J6/JFH1ΔHVR1 with antibodies [29]. Here, we treated cultures with 5, 10, and 20 µg/mL of AR4A (treatments I, II, and III, respectively; Supplementary Figure 3A and Table 1). All displayed significantly delayed spread. Using culture supernatants collected at the peak of infection, we generated first-passage virus stocks and tested AR4A neutralization susceptibility (Supplementary Figure 3B). Viruses from treatment I (IC50, 0.065 µg/mL) and treatment II (IC50, 0.15 µg/mL) were approximately 600 times more resistant than the parental virus (IC50, 0.00017 µg/mL). Virus from treatment III (IC50, 4.4 µg/mL) was approximately 40 times more resistant than virus from treatments I and II and approximately 25000 times more resistant than untreated J6/JFH1∆HVR1. Envelope sequencing identified the dominant substitutions N532S (E2; A1936G) and I696T (E2; T2428C) in virus from treatment I, I696T (E2; T2428C) in virus from treatment II, and I696N (E2; T2428A) and L726V (E2; T2517G; Table 1) in virus from treatment III. This implicated position I696 in AR4A resistance.
Substitutions at Position I696 Conferred AR4A Resistance to J6/JFH1∆HVR1
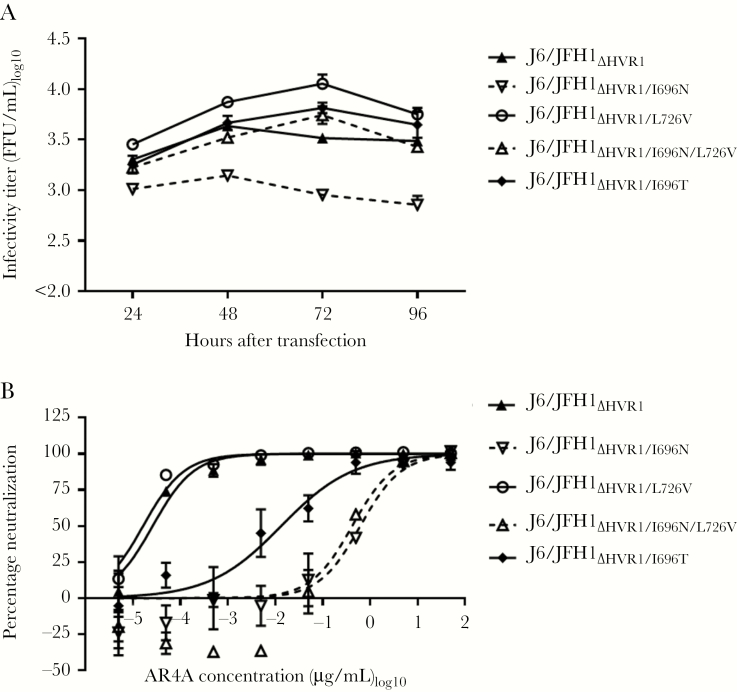
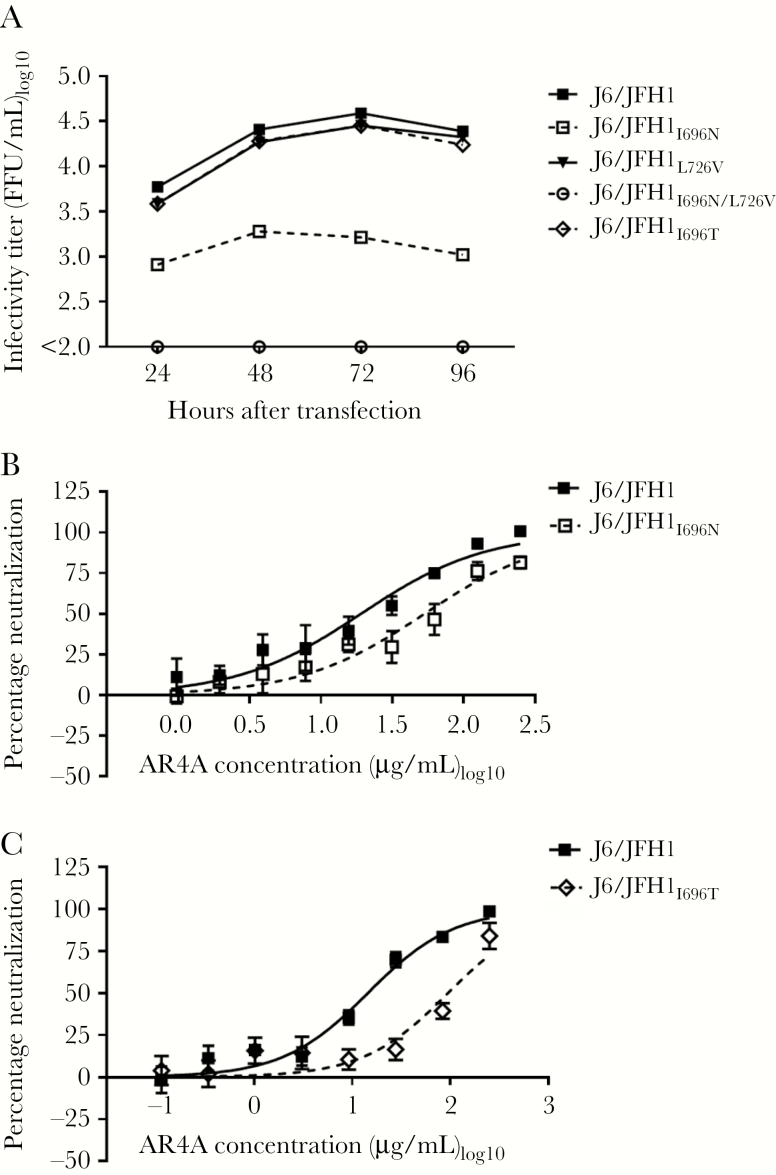
Substitutions I696T, I696N, and L726V or the combination I696N/L726V were introduced into J6/JFH1ΔHVR1. Transfecting Huh7.5 cells with these recombinants, we found that I696N greatly decreased virus fitness. L726V increased fitness, and virus with the combination I696N/L726V had titers slightly higher than J6/JFH1ΔHVR1 (Figure 2A). Virus titers were unaffected by I696T. We generated first-passage stocks of all 4 viruses and determined their envelope sequences. Only J6/JFH1ΔHVR1/I696T acquired the substitution, A217G (E1; C994G). Testing AR4A neutralization susceptibility, we found that J6/JFH1ΔHVR1/L726V (IC50, 0.000016 µg/mL) was similarly susceptible, compared with J6/JFH1ΔHVR1 (IC50, 0.000026 µg/mL). J6/JFH1ΔHVR1/I696N (IC50, 0.62 µg/mL) and J6/JFH1ΔHVR1/I696N/L726V (IC50, 0.41 µg/mL) were approximately 10000 less susceptible to AR4A. Finally, J6/JFH1ΔHVR1/I696T (IC50, 0.011 µg/mL) was approximately 420 times less susceptible, compared with J6/JFH1ΔHVR1 (Figure 2B). Thus, we showed that I696N conferred high-level AR4A resistance to J6/JFH1ΔHVR1, while L726V compensated virus fitness loss. I696T conferred slightly lower AR4A antibody resistance, compared with I696N, but did not affect virus fitness.
Figure 2.
The substitutions I696T and I696N conferred AR4A resistance to J6/JFH1ΔHVR1. A, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated recombinants harboring I696T, I696N, L726V or the combination I696N/L726V. Error bars represent standard errors of the mean (SEMs). Supernatants were collected at the indicated time points, and hepatitis C virus infectivity titers were determined. Lower level of quantification was 100 focus-forming units (FFU)/mL. B, First passages of the viruses were subjected to a dilution series of AR4A, from 50 µg/mL to 0.000005 µg/mL, and neutralization was assessed and analyzed as described in Figure 1, except we used 4-parameter curve fitting to obtain a sigmoidal dose-response curve. Error bars represent SEMs. Independent neutralization experiments yielded similar results.
I696T Conferred Higher AR4A Resistance Than I696N for J6/JFH1 Retaining HVR1
Substitutions I696T, I696N, and L726V or I696N/L726V were introduced into J6/JFH1. Transfecting Huh7.5 cells, we observed that in contrast to J6/JFH1ΔHVR1, virus titers were not measurable for J6/JFH1I696N/L726V. I696N alone greatly decreased virus viability, while I696T did not (Figure 3A). We generated first-passage stocks and confirmed the envelope sequences. Testing AR4A neutralization susceptibility, we found that J6/JFH1I696N (IC50, 53 μg/mL) was approximately 3 times less susceptible, compared with J6/JFH1 (IC50, 19 μg/mL; Figure 3B). J6/JFH1I696T (IC50, 95 μg/mL) was approximately 7 times less susceptible, compared with J6/JFH1 (IC50, 14 μg/mL; Figure 3C).
Figure 3.
The substitutions I696N and I696T conferred AR4A resistance to the parental recombinant virus J6/JFH1. A, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated recombinants with single or combined mutations. Supernatants were collected at the indicated time points, and hepatitis C virus infectivity titers were determined. The lower level of quantification was 100 focus-forming units (FFU)/mL. B and C, First passages of the indicated viruses were subjected to a dilution series of AR4A, from 250 µg/mL to 0.9 µg/mL (B) or from 250 µg/mL to 0.1 µg/mL (C), and neutralization was assessed and analyzed as described in Figure 1, except we used 4-parameter curve fitting to obtain a sigmoidal dose-response curve. Error bars represent standard errors of the mean. Independent neutralization experiments yielded similar results.
AR4A Binding to J6 E1/E2 Was Reduced More by I696N Than by I696T
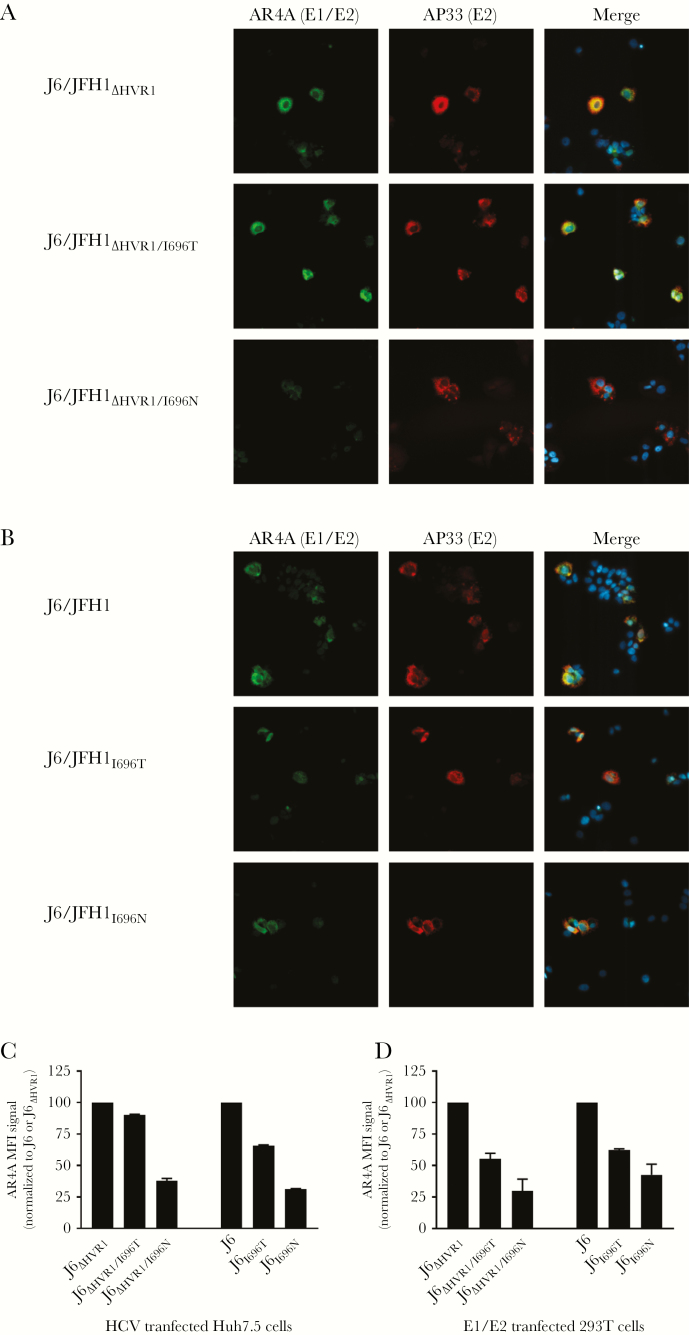
To assess the effects of I696T and I696N on AR4A binding to E1/E2, we performed immunostaining of Huh7.5 cells transfected with J6/JFH1ΔHVR1, J6/JFH1ΔHVR1/I696T, or J6/JFH1ΔHVR1/I696N recombinants, using AR4A and the control E2 antibody, AP33 (Figure 4A). AP33 signal appeared unaltered, indicating similar levels of E2. I696N had a decreased AR4A signal, compared with the parental virus, whereas I696T had no or marginal effect. Immunostaining of Huh7.5 cells transfected with J6/JFH1, J6/JFH1I696T, and J6/JFH1I696N confirmed these findings in the corresponding viruses with HVR1 (Figure 4B).
Figure 4.
AR4A binding to HCV J6 E1/E2 was reduced more by I696N than by I696T. A, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated J6/JFH1ΔHVR1 recombinants. Cells were fixed 48 hours after transfection and stained using the primary antibodies AR4A (anti-E1/E2) and AP33 (anti-E2) as described in Methods. Cell nuclei were visualized with Hoechst. B, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated J6/JFH1 recombinants and stained as described above. C, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated J6/JFH1 recombinants with and without HVR1. Cells were prepared for flow cytometry by releasing them into suspension, fixing them, and staining them using the primary antibodies AR4A (anti-E1/E2) and AP33 (anti-E2) as described in Methods. The AR4A mean fluorescence intensity (MFI) was calculated for the double-positive cell population as shown in Supplementary Figure 4 and normalized to either J6 or J6ΔHVR1, as appropriate. The bars represent the means (±SDs) of the 2 independent experiments. D, 293T cells were transfected with the indicated J6 E1/E2 expression vectors with and without HVR1. Cells were prepared for flow cytometry by releasing them into suspension, fixing them, and staining them using the primary antibodies AR4A (anti-E1/E2) and AP33 (anti-E2) as described in Methods. The AR4A MFI was calculated for the high-AR4A double-positive cell population as shown in Supplementary Figure 5 and normalized to either J6 or J6ΔHVR1, as appropriate. The bars represent the means (±SDs) of the 2 independent experiments.
To better quantify the effects of I696T and I696N on AR4A binding, we performed flow cytometry of Huh7.5 cells transfected with these same recombinants (Figure 4C; one of 2 independent experiments shown in Supplementary Figure 4). I696T reduced AR4A binding less than I696N both in the absence and the presence of HVR1. To assess AR4A binding to E1/E2 in the absence of other HCV proteins, we transfected 293T cells with the analogous E1/E2 expression vectors (Figure 4D; one of 2 independent experiments shown in Supplementary Figure 5), which confirmed the finding.
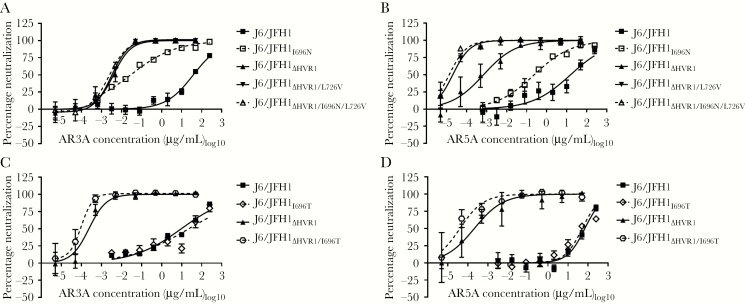
I696N Conferred Broadly Increased Neutralization Susceptibility in an HVR1-Dependent Manner, Whereas I696T Did Not
To investigate how I696T conferred higher AR4A resistance than I696N to HCV retaining HVR1, we tested the neutralization susceptibility of J6/JFH1 and J6/JFH1∆HVR1 recombinants with these substitutions, using AR3A and AR5A [19, 20]. J6/JFH1I696N (IC50, 0.028 μg/mL) was approximately 1300 times more susceptible to AR3A, compared with J6/JFH1 (IC50, 38 μg/mL; Figure 5A), and approximately 90 times more susceptible to AR5A (IC50, 0.22 μg/mL), compared with J6/JFH1 (IC50, 20 μg/mL; Figure 5B). In the absence of HVR1, I696N did not increase HCV susceptibility to neutralization (Figure 5A and 5B). The substitution I696T did not significantly affect neutralization susceptibility in J6/JFH1, with or without HVR1 (Figure 5C and 5D). This indicated that I696N broadly increased neutralization susceptibility in an HVR1-dependent manner, whereas I696T did not.
Figure 5.
I696N greatly increased the antibody susceptibility of J6/JFH1 retaining HVR1 against the neutralizing antibodies AR3A and AR5A, whereas I696T did not. First passages of the indicated viruses were subjected to a dilution series of antibodies AR3A (A and C) or AR5A (B and D), and neutralization was assessed and analyzed as described in Figure 1, except we used 4-parameter curve fitting to obtain a sigmoidal dose-response curve. Error bars represent standard errors of the mean. Independent neutralization experiments yielded similar results.
Substitutions at Position I696 Affected Fitness and Antibody Susceptibility in Genotypes 1a and 3a
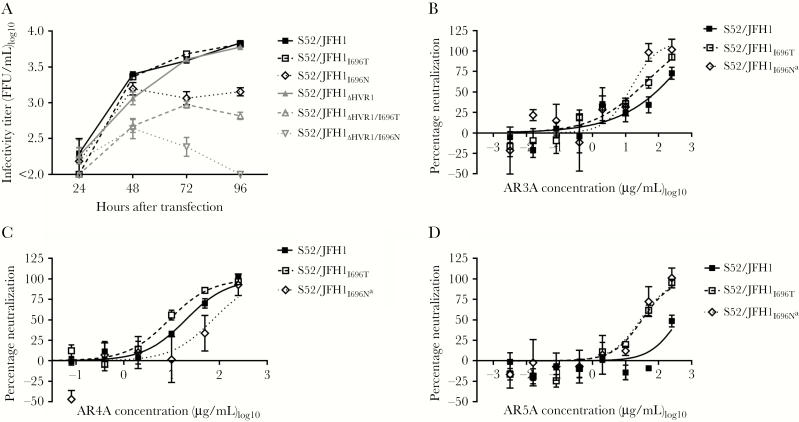
We introduced I696T and I696N in core-NS2 recombinants of genotype 1a (H77/JFH1) and 3a (S52/JFH1), with and without HVR1, and transfected Huh7.5 cells. I696T and I696N greatly decreased the fitness of both H77/JFH1 and H77/JFH1ΔHVR1 (Supplementary Figure 6). This was also the case for S52/JFH1ΔHVR1 (Figure 6A). For S52/JFH1, I696N greatly decreased the fitness, while I696T did not (Figure 6A). We generated first-passage virus stocks of S52/JFH1I696T and S52/JFH1I696N. Sequencing revealed that S52/JFH1I696N had acquired the substitution V553A (E2; C1999T), whereas S52/JFH1I696T did not have additional substitutions. Next, we tested neutralization susceptibility against AR3A (Figure 6B), AR4A (Figure 6C), and AR5A (Figure 6D). While both I696T and I696N increased susceptibility against AR3A and AR5A, we observed an approximately 4-fold increase of AR4A-specific resistance for S52/JFH1I696N.
Figure 6.
Substitutions at position I696 had varied effects on virus fitness and antibody susceptibility of hepatitis C virus (HCV) genotype 3a recombinants. A, Huh7.5 cells were transfected with in vitro–transcribed RNA of the indicated S52/JFH1 recombinants (genotype 3a). Supernatants were collected at the indicated time points, and HCV infectivity titers were determined. The lower level of quantification was 100 focus-forming units (FFU)/mL. B–D, First passages of the indicated viruses were subjected to a dilution series of AR3A (B), AR4A (C), and AR5A (D), from 250 µg/mL to 0.0032 µg/mL, and neutralization was assessed and analyzed as described in Figure 1, except we used 4-parameter curve fitting to obtain a sigmoidal dose-response curve. Error bars represent standard errors of the mean. Independent neutralization experiments yielded similar results. aThe indicated virus had the additional E2 substitution V553A.
DISCUSSION
AR4A targets a conserved epitope and has a superior protective effect in animal models [20, 21, 23]. Here, we used a newly established method to study AR4A escape and found that tested isolates were incapable of developing high-level AR4A resistance. In addition, we showed that position I696 is in the AR4A epitope. In addition, substitutions at this position were found to have isolate-dependent effects, including potential fitness loss and broadly increased neutralization susceptibility.
We analyzed alanine substitutions that we previously showed to have decreased AR4A binding to H77 E1/E2 [19, 20]. Infectivity of H77/JFH1 was abrogated except for P228A, which did not confer AR4A resistance. Although additional alanine substitutions recently reported to affect AR4A binding [38] could result in resistant viable viruses, the low infectivity of the alanine mutants tested here suggests that in vitro escape studies represent a more tenable approach to identify viable resistance substitutions.
Several studies have demonstrated a correlation between in vitro resistance substitutions and resistance substitutions found in vivo [40–44]. H77/JFH1 did not develop AR4A-specific resistance substitutions, even after long-term high-dose antibody culture. The virus developed the E1 substitution M345T that we have shown is adaptive, suggesting the virus evaded neutralization through improved fitness, as previously described for J6/JFH1 cultured with AR5A [29]. Similarly, upon culturing H77/JFH1ΔHVR1 in the presence of AR4A, we identified the substitutions N417D and N532D, which abolish E2 glycosylation sites N417 and N532 and have been implicated in increasing antibody susceptibility and virus fitness [45]. We have previously shown that N417 did not affect AR5A susceptibility for HVR1-deleted virus [29] and that N417D together with N532D greatly increased the fitness of H77/JFH1ΔHVR1 [29]. When culturing H77/JFH1ΔHVR1/N417D/N532D with AR4A, the virus spread without acquiring additional envelope substitutions. These results indicate that the increased fitness of H77/JFH1 and H77/JFH1ΔHVR1 permitted virus spread by rendering the antibody dose too low to prevent spread. We previously reported that, with comparable antibody doses, both H77/JFH1ΔHVR1 and J6/JFH1ΔHVR1 developed resistance substitutions against AR5A after <30 days of culture [29]. The fact that H77/JFH1 and H77/JFH1ΔHVR1 took 87–130 days to spread with AR4A (only acquiring increased fitness) supports that it is difficult for H77/JFH1 to develop AR4A-specific resistance substitutions.
Culturing J6/JFH1∆HVR1 in the presence of AR4A for 39–83 days, we identified substitutions at position I696 that increased AR4A resistance for J6/JFH1, with and without HVR1. This position is completely conserved within genotype 2 and 93% conserved across genotypes with 2.4% threonine (Los Alamos HCV sequence database). I696N greatly decreased AR4A neutralization susceptibility for J6/JFH1∆HVR1 but required the fitness-compensating substitution L726V, which decreased the fitness of J6/JFH1. I696T had a lower effect on AR4A binding than I696N, as shown by immunofluorescence and flow cytometry. This was reflected in the fact that I696T conferred less resistance than I696N against AR4A for J6/JFH1∆HVR1. Interestingly, I696T conferred greater resistance than I696N for J6/JFH1, likely because I696N broadly increased neutralization susceptibility for J6/JFH1 but not for J6/JFH1∆HVR1. I696T did not affect the fitness of J6/JFH1 with or without HVR1 and did not affect susceptibility to other NAbs for either virus, thus indicating that it is a de novo AR4A-specific resistance substitution.
In a previous complete alanine scanning mutagenesis study of recombinant E1/E2, alanine substitution at I696 had no effect on AR4A binding, whereas D698A and Q700A substitutions reduced AR4A binding [20, 38]. Combined, these results firmly establish that E2 positions 696–700 form part of the AR4A epitope, although AR4A co-crystallization of E1/E2 would be needed to fully elucidate the epitope. Our data complement recent findings of polymorphisms in E1/E2 that have been shown to broadly modulate NAb susceptibility, including susceptibility to AR4A [39, 46, 47].
Interestingly, we found that H77/JFH1 was nonviable when I696T or I696N were introduced. For S52/JFH1, we observed some effect on the viability of I696N but not of I696T. I696N decreased AR4A susceptibility, but both I696T and I696N broadly increased NAb susceptibility. Thus, our data indicate that substitutions at position I696 can have isolate-dependent effects on virus fitness and broad NAb susceptibility.
We previously reported that substitutions of L665 conferred high-level AR5A resistance in the 6 major HCV genotypes [29]. Here, we found that I696T conferred low-level AR4A resistance to J6/JFH1. Together with our inability to induce resistance in H77/JFH1 despite long-term cultures with antibody, this indicates that the barrier to resistance for AR4A is higher than that for AR5A. While the promise of epitope-targeted vaccine development is still being explored [48], 2 studies have shown some efficacy for linear HCV envelope peptides [49, 50]. However, these approaches cannot be readily adapted to NAbs targeting nonlinear conformational epitopes, such as AR4A. A better approach might be to emphasize the AR4A epitope in E1/E2 vaccine antigens, but improved structural understanding of the E1/E2 heterodimer is needed.
In conclusion, by culturing viruses in the presence of the vaccine-relevant NAb AR4A, we identified the resistance substitution I696T for J6/JFH1, whereas no resistance could be induced for H77/JFH1. Our findings indicate that AR4A targets a low-variation epitope with a high barrier to resistance, which in conjunction with high AR4A cross-genotype conservation makes the epitope a prime candidate for inclusion in HCV vaccine candidates.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Lubna Ghanem, Lotte Mikkelsen, Anne-Louise Sørensen (Copenhagen University Hospital, Hvidovre), Erick Giang, and Shaun Castillo (The Scripps Research Institute, La Jolla), for general laboratory support; Charles Rice (Rockefeller University, New York) and Arvind Patel (University of Glasgow), for providing reagents; and Bjarne Ørskov Lindhardt, Ove Andersen (Copenhagen University Hospital, Hvidovre), and Carsten Geisler (University of Copenhagen), for their support of the project.
Financial support. This work was supported by the Faculty of Health and Medical Sciences, University of Copenhagen (PhD stipend to R. V.-M.); the Danish Council for Independent Research, Medical Sciences (DFF postdoctoral grant 11-116529 to J. P. and advanced top researcher grant 0602-02366B to J. B.); the Lundbeck Foundation (research grants R100-A9689 [to J. P. and J. B.] and R108-A10894 and R221-2016-1455 [to J. B.]); the Novo Nordisk Foundation (grants NNF12OC0002037, NNF14OC0012533, and NNF17OC0029372 to J.B.); the Danish Council for Independent Research (grant 4004-00598 to J. B.); the National Institutes of Health (awards AI079031, AI106005, AI123365, and AI123861 to M. L.); and Novo Nordisk Foundation (2015 Novo Nordisk Prize to J. B.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Bukh J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol 2016; 65:S2–S21. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization (WHO). Hepatitis C. Fact sheet number 164. Geneva: WHO, 2017. [Google Scholar]
- 3. Bartenschlager R, Baumert TF, Bukh J, et al. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 2018; 248:53–62. [DOI] [PubMed] [Google Scholar]
- 4. Walker CM. Designing an HCV vaccine: a unique convergence of prevention and therapy?Curr Opin Virol 2017; 23:113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith DB, Becher P, Bukh J, et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 2016; 97:2894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simmonds P, Becher P, Bukh J, et al. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 2017; 98:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fauvelle C, Colpitts CC, Keck ZY, Pierce BG, Foung SK, Baumert TF. Hepatitis C virus vaccine candidates inducing protective neutralizing antibodies. Expert Rev Vaccines 2016; 15:1535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ball JK, Tarr AW, McKeating JA. The past, present and future of neutralizing antibodies for hepatitis C virus. Antiviral Res 2014; 105:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pestka JM, Zeisel MB, Bläser E, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A 2007; 104:6025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osburn WO, Snider AE, Wells BL, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology 2014; 59:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bukh J, Engle RE, Faulk K, et al. Immunoglobulin with high-titer in vitro cross-neutralizing hepatitis C virus antibodies passively protects chimpanzees from homologous, but not heterologous, challenge. J Virol 2015; 89:9128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morin TJ, Broering TJ, Leav BA, et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog 2012; 8:e1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farci P, Shimoda A, Wong D, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A 1996; 93:15394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prentoe J, Verhoye L, Velázquez Moctezuma R, et al. HVR1-mediated antibody evasion of highly infectious in vivo adapted HCV in humanised mice. Gut 2016; 65:1988–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanwolleghem T, Bukh J, Meuleman P, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology 2008; 47:1846–55. [DOI] [PubMed] [Google Scholar]
- 16. Meuleman P, Bukh J, Verhoye L, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology 2011; 53:755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 2012; 142:1279–87.e3. [DOI] [PubMed] [Google Scholar]
- 18. von Hahn T, Yoon JC, Alter H, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 2007; 132:667–78. [DOI] [PubMed] [Google Scholar]
- 19. Law M, Maruyama T, Lewis J, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 2008; 14:25–7. [DOI] [PubMed] [Google Scholar]
- 20. Giang E, Dorner M, Prentoe JC, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 2012; 109:6205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prentoe J, Velázquez-Moctezuma R, Foung SK, Law M, Bukh J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology 2016; 64:1881–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Jong YP, Dorner M, Mommersteeg MC, et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med 2014; 6:254ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O’Shea D, Law J, Egli A, et al. Prevention of hepatitis C virus infection using a broad cross-neutralizing monoclonal antibody (AR4A) and epigallocatechin gallate. Liver Transpl 2016; 22:324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pedersen J, Carlsen TH, Prentoe J, et al. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology 2013; 58:1587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carlsen TH, Pedersen J, Prentoe JC, et al. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology 2014; 60:1551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mankowski MC, Kinchen VJ, Wasilewski LN, et al. Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc Natl Acad Sci U S A 2018; 115:E82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prentoe J, Jensen TB, Meuleman P, et al. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol 2011; 85:2224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bankwitz D, Steinmann E, Bitzegeio J, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol 2010; 84:5751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Velázquez-Moctezuma R, Law M, Bukh J, Prentoe J. Applying antibody-sensitive hypervariable region 1-deleted hepatitis C virus to the study of escape pathways of neutralizing human monoclonal antibody AR5A. PLoS Pathog 2017; 13:e1006214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science 2005; 309:623–6. [DOI] [PubMed] [Google Scholar]
- 31. Owsianka A, Tarr AW, Juttla VS, et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol 2005; 79:11095–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheel TK, Gottwein JM, Jensen TB, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A 2008; 105:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gottwein JM, Scheel TK, Hoegh AM, et al. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 2007; 133:1614–26. [DOI] [PubMed] [Google Scholar]
- 34. Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J Virol 2014; 88:1725–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 2002; 76:13001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheel TK, Gottwein JM, Carlsen TH, et al. Efficient culture adaptation of hepatitis C virus recombinants with genotype-specific core-NS2 by using previously identified mutations. J Virol 2011; 85:2891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gottwein JM, Scheel TK, Callendret B, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 2010; 84:5277–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gopal R, Jackson K, Tzarum N, et al. Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog 2017; 13:e1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathiesen CK, Prentoe J, Meredith LW, et al. Adaptive mutations enhance assembly and cell-to-cell transmission of a high-titer hepatitis C virus genotype 5a Core-NS2 JFH1-based recombinant. J Virol 2015; 89:7758–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pantua H, Diao J, Ultsch M, et al. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol 2013; 425:1899–914. [DOI] [PubMed] [Google Scholar]
- 41. Keck ZY, Saha A, Xia J, et al. Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. J Virol 2011; 85:10451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keck ZY, Olson O, Gal-Tanamy M, et al. A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. J Virol 2008; 82:6067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gal-Tanamy M, Keck ZY, Yi M, et al. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc Natl Acad Sci U S A 2008; 105:19450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung RT, Gordon FD, Curry MP, et al. Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. Am J Transplant 2013; 13:1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Helle F, Vieyres G, Elkrief L, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 2010; 84:11905–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wasilewski LN, El-Diwany R, Munshaw S, et al. A hepatitis C virus envelope polymorphism confers resistance to neutralization by polyclonal sera and broadly neutralizing monoclonal antibodies. J Virol 2016; 90:3773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. El-Diwany R, Cohen VJ, Mankowski MC, et al. Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog 2017; 13:e1006235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Correia BE, Bates JT, Loomis RJ, et al. Proof of principle for epitope-focused vaccine design. Nature 2014; 507:201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pierce BG, Boucher EN, Piepenbrink KH, et al. Structure-based design of hepatitis C virus vaccines that elicit neutralizing antibody responses to a conserved epitope. J Virol 2017; 91:doi: 10.1128/JVI.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He L, Cheng Y, Kong L, et al. Approaching rational epitope vaccine design for hepatitis C virus with meta-server and multivalent scaffolding. Sci Rep 2015; 5:12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.