Summary
The cysteine prodrug N-acetyl cysteine (NAC) is widely used as a pharmacological antioxidant and cytoprotectant. It has been reported to lower endogenous oxidant levels and to protect cells against a wide range of pro-oxidative insults. As NAC itself is a poor scavenger of oxidants, the molecular mechanisms behind the antioxidative effects of NAC have remained uncertain. Here we show that NAC-derived cysteine is desulfurated to generate hydrogen sulfide, which in turn is oxidized to sulfane sulfur species predominantly within mitochondria. We provide evidence suggesting the possibility that sulfane sulfur species produced by 3-mercaptopyruvate sulfurtransferase and sulfide:quinone oxidoreductase are the actual mediators of the immediate antioxidative and cytoprotective effects provided by NAC.
Introduction
N-acetyl cysteine (NAC) is one of the most widely used antioxidants in the context of clinical studies, animal and cell culture experiments. Many conclusions have been based on the outcome of experiments using NAC. When treatment with NAC is found to inhibit a particular cellular process or response, it is typically concluded that ‘reactive oxygen species’ (ROS) play an active role in it. For example, NAC was used to address the mechanism of action of anti-tumor drugs such as β-lapachone (Park et al., 2014), taurolidine (Rodak et al., 2005) and cisplatin (Wang et al., 2014). Furthermore, numerous insights concerning the involvement of ROS in tumor initiation, progression and metastasis have been obtained with the help of NAC (Irani et al., 1997; Le Gal et al., 2015; Ma et al., 2009; Piskounova et al., 2015; Radisky et al., 2005; Sayin et al., 2014; Schafer et al., 2009). Amongst many other applications, NAC has also been employed in addressing the physiological roles of mitochondrial ROS (Chouchani et al., 2016).
Despite its widespread use, it is not obvious how NAC acts as an antioxidant. On the one hand, it is often assumed that NAC scavenges oxidants directly through its thiol group, but this seems to be very unlikely in the light of kinetic data. The rate constants for the reaction of NAC with relevant physiological oxidants such as H2O2 and O2.- are too low (0.16 M-1s-1 and 68 M-1s-1 at pH 7.4 and 37°C) to make a significant contribution to oxidant scavenging (Benrahmoune et al., 2000; Winterbourn and Metodiewa, 1999).
On the other hand, it is often assumed that the antioxidative agency of NAC can be explained by its ability to act as a source of cysteine (Cys) for increased glutathione (GSH) biosynthesis. This explanation appears to apply to situations of severe GSH depletion, e.g. as induced in the liver by paracetamol overdose, a major indication for the clinical use of NAC (Green et al., 2013). The tracing of 34S-labeled NAC in Cys/Met-deprived cells confirmed its incorporation into newly synthesized GSH (Ono et al., 2017). Although it makes perfect sense that cells use NAC-derived Cys for GSH biosynthesis when the need for Cys is not matched by its supply, the question remains if the biosynthetic use of Cys generally or typically explains the effects afforded by NAC. It seems that it has rarely been tested if the increase (or maintenance) of GSH levels is actually required for the rescuing effect afforded by NAC. Some studies have observed that NAC failed to restore decreased GSH levels, but nevertheless provided cytoprotection (Patriarca et al., 2005). An in vivo labeling study in mice concluded that NAC is not utilized as a direct precursor of GSH synthesis (Zhou et al., 2015), in line with the suggestion that the rates of NAC uptake and deacetylation may not be adequate to maintain Cys levels for GSH biosynthesis (Raftos et al., 2007).
Overall, there does not seem to exist a consensus explanation for the cytoprotective and antioxidative agency of NAC, as noted previously (Parasassi et al., 2010; Samuni et al., 2013). Hence, it remains unclear how the large number and wide-ranging variety of studies reporting the ability of NAC to rescue cells from pro-oxidative and electrophilic insults can be explained. In this paper, we offer a new and unexpected perspective on the antioxidative prowess of NAC.
This study was motivated by our earlier observation that feeding NAC to flies induced oxidation of mitochondrial, but not cytosolic, roGFP2-based probes, in various tissues (Albrecht et al., 2011). We found that this phenomenon also holds true in human cells and we now show that NAC-induced roGFP2 oxidation is caused by sulfane sulfur species (hydropersulfides). We demonstrate that NAC is desulfurated to H2S and subsequently oxidized to sulfane sulfur species in mitochondria. We provide evidence suggesting that sulfane sulfur is a key mediator of the anti-oxidative and cytoprotective effects of NAC and that its generation is dependent on the activities of sulfane sulfur producing enzymes 3-mercaptopyruvate sulfurtransferase (MST) and/or sulfide:quinone oxidoreductase (SQR).
Results
NAC catabolism induces rapid oxidation of mitochondrial, but not cytosolic, roGFP2
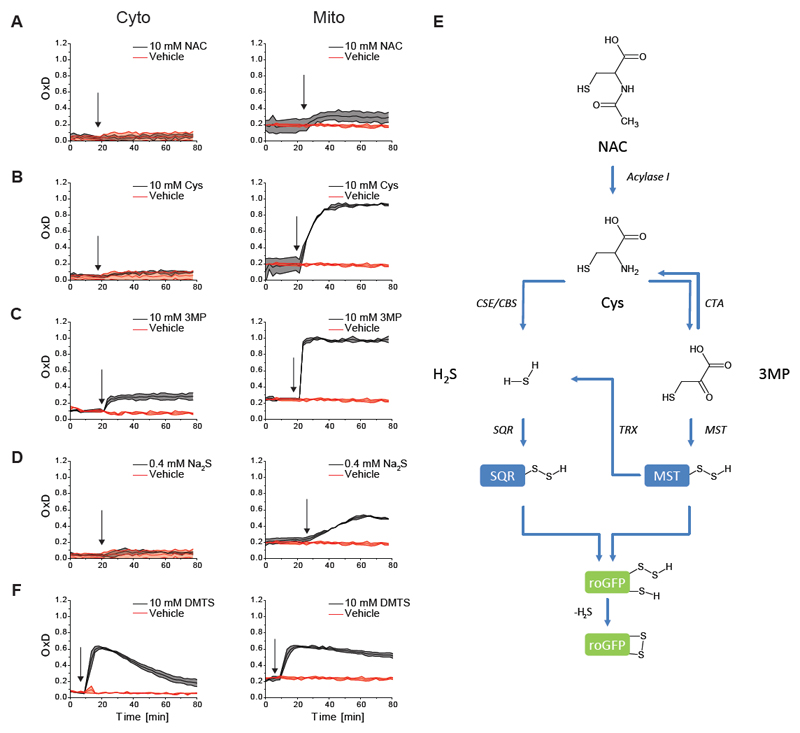
Motivated by our earlier Drosophila experiments (Albrecht et al., 2011), we started out by asking if NAC treatment would induce oxidation of the genetically encoded redox probe roGFP2 in cultured human cells. Indeed, roGFP2 expressed in the mitochondrial matrix (mt-roGFP2) rapidly responded to NAC, while cytosolic roGFP2 did not. The magnitude of the response was modest (an increase in the degree of probe oxidation from ~20 to 30%), yet highly reproducible (Figure 1A). In contrast to NAC, N-acetyl alanine (NAA) did not elicit any response, showing that the thiol group is crucial for the effect (Figure S1A). As NAC is known to be de-acetylated to cysteine (Cys) inside cells (Uttamsingh et al., 1998), we asked if direct provisioning of Cys would lead to the same phenomenon. Indeed, Cys applied at the same concentration was much more effective in inducing mt-roGFP2 oxidation (Figure 1B), suggesting that an increase in intracellular Cys levels is responsible for the NAC-induced phenomenon. Total glutathione levels were not affected by NAC treatment (Figure S1B), suggesting that the phenomenon is unrelated to glutathione synthesis.
Figure 1. NAC catabolism induces rapid oxidation of mitochondrial, but not cytosolic, roGFP2.
(A) Treatment of cells with NAC triggers partial oxidation of mitochondrially localized roGFP2, but not of cytosolic roGFP2.
(B-D) The same phenomenon can be provoked more strongly by treating with Cys (B), 3MP (C) and the H2S donor Na2S (D).
(E) These findings led to the working hypothesis that NAC, Cys, 3MP and H2S trigger roGFP2 oxidation through production of mitochondrial hydropersulfides.
(F) Exogenous application of the sulfane sulfur compound DMTS, which leads to the intracellular formation of persulfides, triggers roGFP2 oxidation in both compartments.
All curves represent the mean of three biological repeats, each carried out in technical quadruplicates, the error bars denoting the standard deviation (SD). OxD: degree of probe oxidation. The arrow indicates the time point of addition of the compound.
As endogenous Cys levels are known to be tightly controlled (Stipanuk et al., 2006), we reasoned that the additional Cys provided by the de-acetylation of NAC may be subject to rapid catabolic conversion. Cys catabolism is known to proceed in three possible directions: The first possibility is the oxidation of Cys by cysteine dioxygenase to yield cysteine sulfinic acid (CSA) (Stipanuk et al., 2009). The second possibility is transamination (or oxidative deamination) of Cys to the corresponding α-keto acid, which is 3-mercapto-pyruvate (3MP) (Singh and Banerjee, 2011b). The third possibility is desulfuration of Cys (by cystathionine γ-lyase, CSE, and/or cystathionine β-synthase, CBS) to liberate reduced sulfur as H2S (Kabil et al., 2014). We therefore asked if the products of these three alternative catabolic transformations can mimic the effect produced by NAC and Cys. CSA did not elicit any probe response (Figure S1C), nor did other downstream products of oxidative Cys catabolism, including cysteate, sulfite or sulfate (Figure S1D-F). In stark contrast, the products of the other two catabolic pathways, i.e. those retaining sulfur in the reduced state, namely 3MP and H2S (the latter provided by the H2S donor Na2S) did fully reproduce the phenomenon, and both were even more effective than Cys in doing so (Figure 1C-D). The slow-releasing H2S donor GYY4137 had the same effect as Na2S (Figure S1G).
Together, these results suggested to us the possibility that NAC-induced roGFP2 oxidation is caused by sulfane sulfur (S0) species created inside mitochondria, based on the following consideration (summarized in Figure 1E): It is known that H2S is very efficiently oxidized by sulfide:quinone oxidoreductase (SQR) to yield hydropersulfides inside the mitochondrial matrix (Libiad et al., 2014). It is also known that per- and polysulfides are highly efficient in oxidizing roGFP2, at least in vitro (Greiner and Dick, 2015; Greiner et al., 2013). Thus, we postulated that hydropersulfides created by SQR oxidize roGFP2 within the mitochondrial matrix. In addition, MST, which is dually localized to both cytosol and mitochondria (Frasdorf et al., 2014), is known to use 3MP to create hydropersulfides, which may either directly contribute to roGFP2 oxidation, or be reduced to yield H2S, and thereby contribute to SQR-mediated hydropersulfide production. In addition, some 3MP may also be converted back to Cys by cysteine aminotransferase (CTA) (Singh and Banerjee, 2011a), also fueling Cys-dependent H2S generation. However, our observation that 3MP, but not Cys or H2S, induced a substantial cytosolic roGFP2 response (Figure 1C, left panel) concurs with the notion that cytosolic MST generates roGFP2-oxidizing persulfides from 3MP in the cytosol.
If endogenously produced S0 species are responsible for roGFP2 oxidation then one would also expect exogenously applied S0 species to trigger roGFP2 oxidation. To this end, we treated cells with dimethyl trisulfide (DMTS) which contains zero-valent sulfur and is expected to react with endogenous thiols (predominantly GSH) to generate hydropersulfides. Indeed, the roGFP2 probe responded to DMTS in both cytosol and mitochondria (Figure 1F), confirming that exogenously applied S0 species can induce roGFP2 oxidation inside living cells. Treatment of cells with the inorganic polysulfide salts Na2S2, Na2S3 and Na2S4 (Figure S1H-J) or with diallyl trisulfide (Figure S1K), known to react with GSH to yield persulfide intermediates (Liang et al., 2015), led to the same observation.
In summary, treatment of cells with the Cys prodrug NAC triggers mt-roGFP2 oxidation and this phenomenon can be mimicked with Cys and two specific downstream products of cysteine catabolism, H2S and 3MP. These findings led us to a working hypothesis (Figure 1E), amenable to further testing, namely that NAC treatment triggers endogenous S0/hydropersulfide formation, predominantly in the mitochondrial matrix.
NAC, Cys and 3MP trigger the generation of H2S and persulfides
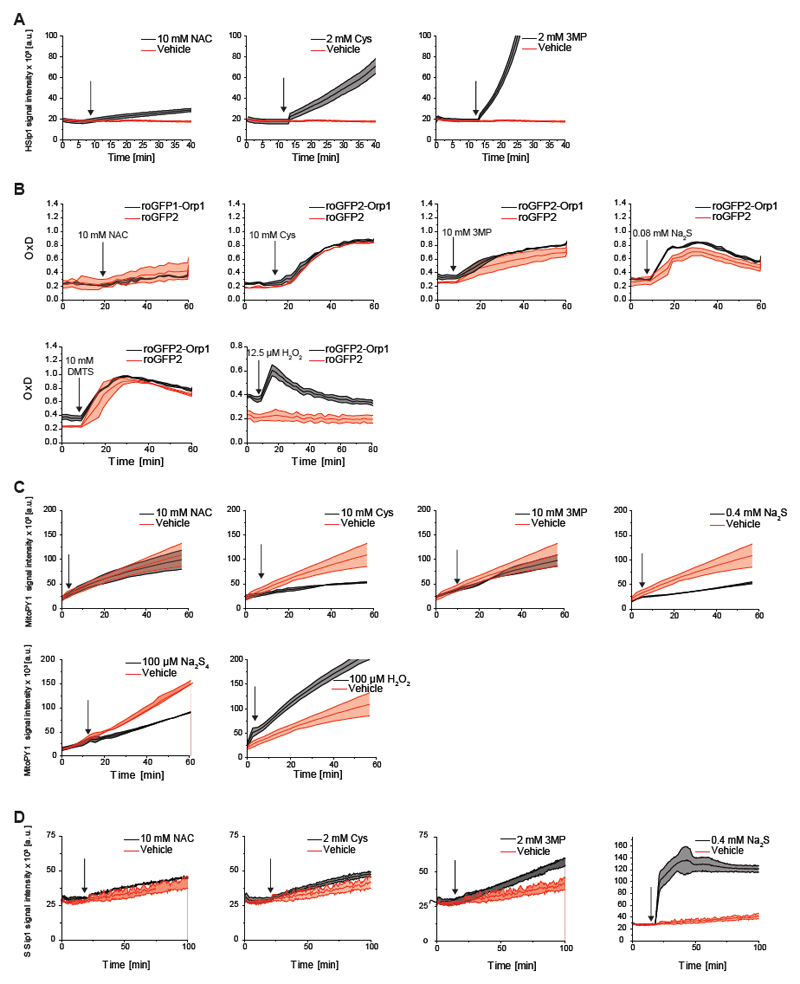
The working hypothesis (Figure 1E) predicted that NAC, Cys and 3MP treatments trigger endogenous H2S production. Indeed, all three compounds induced an increase in endogenous H2S levels, as detected by the H2S probe HSip-1 DA (Sasakura et al., 2011) (Figure 2A), in proportion to their potency in causing roGFP2 oxidation (Figure 1A-C).
Figure 2. NAC, Cys and 3MP trigger the generation of H2S and persulfides.
(A) NAC, Cys and 3MP trigger endogenous H2S generation, as detected by the probe HSip-1 DA.
(B) The mt-roGFP2 response is not significantly influenced by the translational fusion to Orp1, confirming that the observed response is H2O2-independent.
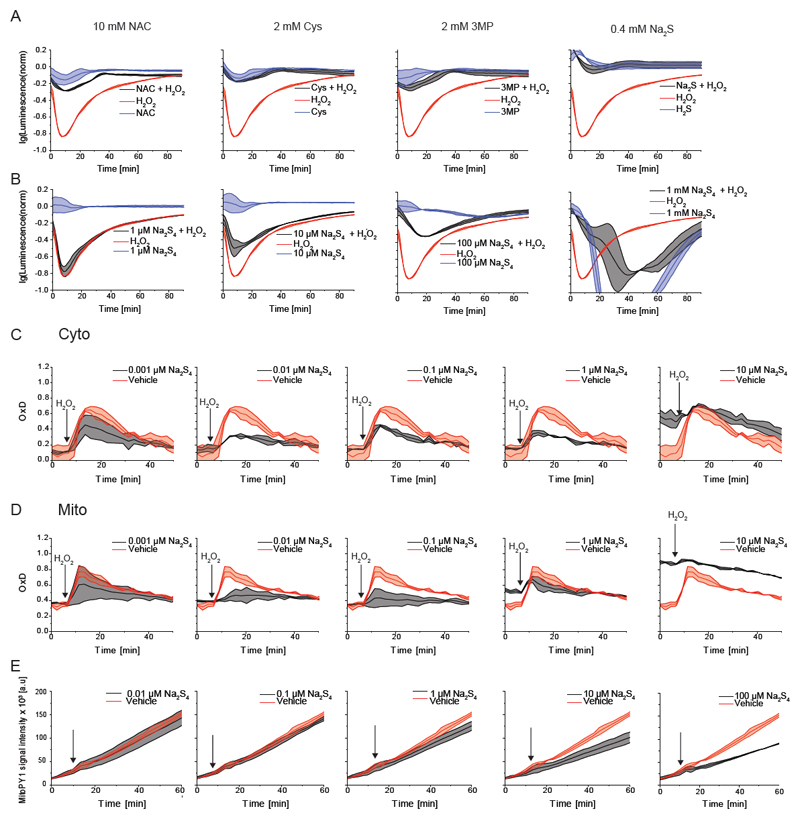
(C) The boronate based mitochondrially targeted chemical H2O2 probe mitoPY1 does not detect increased H2O2 generation in response to NAC, Cys, 3MP, Na2S or Na2S4, but rather a decrease. The last panel shows the response to H2O2 (positive control). The penultimate panel is also part of Fig. 5E.
(D) NAC, Cys, 3MP and H2S trigger endogenous sulfane sulfur generation, as detected by the sulfane sulfur probe SSip-1 DA.
All curves represent the mean of three biological repeats, each carried out in technical quadruplicates, the error bars denoting the standard deviation (SD). OxD: degree of probe oxidation. The arrow indicates the time point of addition of the compound.
The working hypothesis (Figure 1E) further predicted that NAC, Cys, 3MP and H2S trigger endogenous S0 (hydropersulfide) production. We already established that roGFP2 can act as an indicator of S0 species, both in vitro (Greiner and Dick, 2015; Greiner et al., 2013) and in cellulo (Figure 1F), but we still needed to exclude the possibility that the observed roGFP2 oxidation is actually caused by mitochondrially generated H2O2. To this end, we compared the response of mito-roGFP2 and mito-roGFP2-Orp1. The translational fusion of the thiol peroxidase Orp1 greatly accelerates roGFP2 responses to H2O2 (Gutscher et al., 2009), but does not influence polysulfide-mediated roGFP2 oxidation (Muller et al., 2017). We found the two probes to exhibit almost identical responses (Figure 2B), strongly suggesting that mt-roGFP2 oxidation induced by NAC, Cys, 3MP or H2S treatment is not caused by H2O2. Moreover, the boronate-based mitochondrial H2O2 probe mitoPY1 (Dickinson et al., 2013) did not exhibit any enhanced oxidation in response to NAC, Cys, 3-MP or Na2S treatment, but rather showed the opposite response, namely an attenuation of basal probe oxidation (caused by endogenous H2O2), most prominently for Cys, H2S and Na2S4, in line with the expected anti-oxidative properties of these compounds (Figure 2C).
Finally, we aimed to confirm endogenous S0 generation more directly, and independently of roGFP2-based measurements. The general S0 probe SSip-1 DA (Takano et al., 2017) showed increased S0 formation in response to NAC, Cys, 3MP and H2S (Figure 2D), in proportion to mt-roGFP2 oxidation. In conclusion, several independent lines of evidence show that NAC, Cys and 3MP treatments cause H2S and S0 generation, in line with the working hypothesis (Figure 1E).
NAC-induced H2S production involves CSE and MST
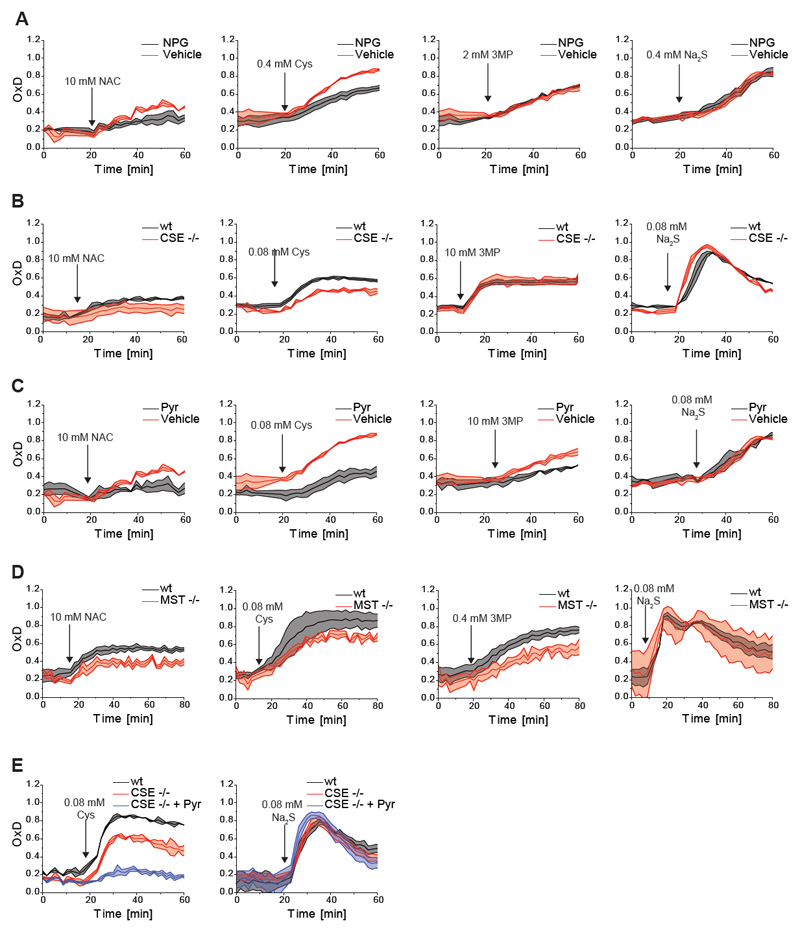
To further investigate the validity of the working hypothesis, we then asked if CSE contributes to NAC/Cys-induced H2S generation and mt-roGFP2 oxidation. Pre-treatment of cells with N-propargyl glycine (NPG), an inhibitor of CSE (Asimakopoulou et al., 2013; Sun et al., 2009), slowed down NAC- and Cys-induced H2S production (Figure S2A) and also attenuated the NAC- and Cys-induced mt-roGFP2 responses (Figure 3A, left panels). In contrast, NPG treatment did not influence 3MP- and Na2S-induced mt-roGFP2 responses (Figure 3A, right panels), as expected. Using CRISPR/Cas9 we then disrupted the CSE genomic locus (Figure S2B). Similar to NPG treatment, deletion of CSE attenuated NAC- and Cys-induced H2S generation (Figure S2C) and likewise attenuated the NAC- and Cys-induced mt-roGFP2 response (Figure 3B, left panels), but not the 3MP- and H2S-induced mt-roGFP2 responses (Figure 3B, right panels). These results indicated that CSE partially contributes to the phenomenon under study.
Figure 3. NAC-induced H2S production involves CSE and MST.
(A) N-propargyl glycine (NPG) partially inhibits NAC- and Cys-induced mt-roGFP2 oxidation, but not 3MP- or Na2S-induced mt-roGFP2 oxidation.
(B) Genetic disruption of CSE expression partially inhibits NAC- and Cys-induced mt-roGFP2 oxidation, but not 3MP- or Na2S-induced mt-roGFP2 oxidation.
(C) Pyruvate partially inhibits NAC-, Cys- and 3MP-induced mt-roGFP2 oxidation, but not Na2S-induced mt-roGFP2 oxidation.
(D) Genetic disruption of MST expression partially inhibits NAC-, Cys- and 3MP-induced mt-roGFP2 oxidation, but not Na2S-induced mt-roGFP2 oxidation.
(E) Pyruvate treatment of CSE KO cells leads to a near-complete suppression of Cys-induced mt-roGFP2 oxidation, while Na2S-induced mt-roGFP2 oxidation is not significantly affected.
All curves represent the mean of three biological repeats, each carried out in technical quadruplicates, the error bars denoting the standard deviation (SD). OxD: degree of probe oxidation. The arrow indicates the time point of addition of the compound.
To address the role of MST, for which no selective inhibitors are available, we first tested the influence of pyruvate, a non-selective inhibitor of MST (Porter and Baskin, 1996). Pyruvate treatment lowered NAC-, Cys- and 3MP-induced mt-roGFP2 responses, but not the response to H2S (Figure 3C), supporting the interpretation that the observed influence of pyruvate on the mt-roGFP2 response is caused by MST inhibition. Using CRISPR/Cas9 we then disrupted the MST genomic locus (Figure S2D). NAC and Cys induced H2S generation was not altered in MST KO cells, but 3MP induced H2S generation was partially suppressed (Figure S2E). Similar to pyruvate treatment, the deletion of MST lowered NAC-, Cys- and 3MP-induced mt-roGFP2 responses, but not the response to H2S (Figure 3D). The incomplete abolition of 3MP-induced H2S production and mt-roGFP2 oxidation in MST KO cells may be explained by the conversion of 3MP to Cys by CTA (Singh and Banerjee, 2011a), thereby allowing MST-independent H2S generation from 3MP. Indeed, when pyruvate was applied to CSE KO cells, the Cys-induced mt-roGFP2 response was almost completely abolished, but not the response to H2S (Figure 3E), suggesting that CSE and MST together are largely responsible for facilitating NAC/Cys-induced mt-roGFP2 oxidation.
NAC-induced persulfide generation involves SQR and MST
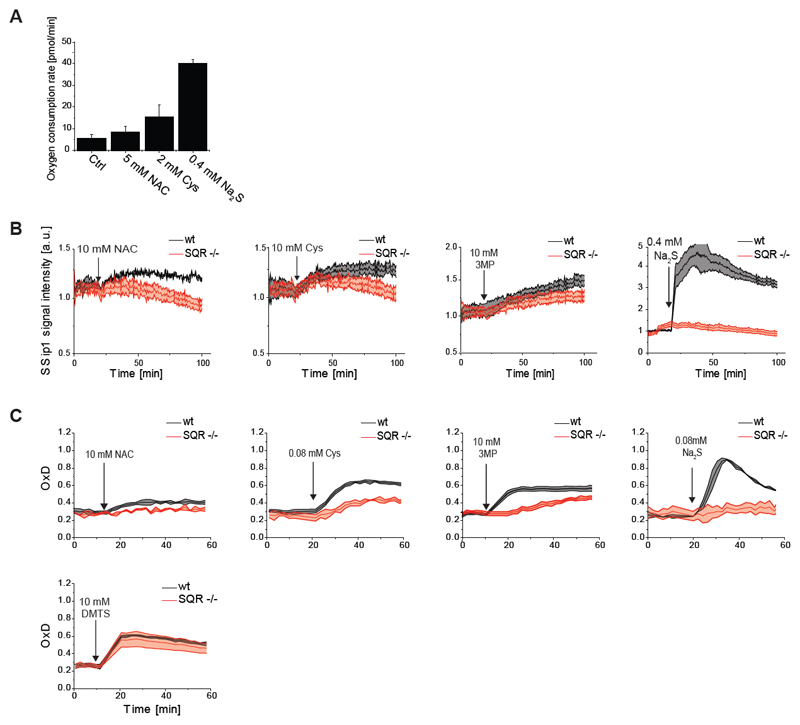
Given the evidence for H2S generation (Figure 2) and for the involvement of H2S-generating enzymes (Figure 3), we next addressed the question of how H2S is converted to S0 species. The primary pathway of intracellular H2S removal depends on SQR (Libiad et al., 2014). SQR activity is coupled to O2 consumption, as it feeds electrons extracted from H2S into the quinone pool of the respiratory chain. Indeed, NAC, Cys, and H2S treatment increased cellular O2 consumption, in proportion to the potency of the compounds in causing mt-roGFP2 oxidation (Figure 4A). If mt-roGFP2 oxidation is largely SQR-dependent (as predicted by the working hypothesis) it should depend on the activity of respiratory complexes III and IV, but not on complexes I, II or V. Indeed, mitochondrial uncoupling (Figure S3A) or inhibition of complex III or IV (Figure S3B), but not inhibition of complexes I or V (Figure S3C), substantially diminished NAC-induced mt-roGFP2 oxidation. Using CRISPR/Cas9 we then disrupted the SQR genomic locus (Figure S3D). S0 formation in response to NAC, Cys, and 3MP, measured with SSip-1 DA, was suppressed in the absence of SQR (Figure 4B). As expected, H2S-induced S0 generation was completely abolished in SQR deficient cells (Figure 4B, ultimate panel).
Figure 4. NAC-induced persulfide generation involves SQR and MST.
(A) NAC, Cys and Na2S treatment leads to increased cellular oxygen consumption.
(B) Disruption of SQR expression suppresses NAC-, Cys- and 3MP-induced sulfane sulfur generation, and completely abolishes H2S-induced sulfane sulfur generation, as detected by SSip-1 DA.
(C) Disruption of SQR expression suppresses NAC-, Cys- and 3MP-induced mt-roGFP2 oxidation, and completely abolishes H2S-induced mt-roGFP2 oxidation, but does not influence DMTS induced mt-roGFP2 oxidation.
All curves represent the mean of three biological repeats, each carried out in technical quadruplicates, the error bars denoting the standard deviation (SD). OxD: degree of probe oxidation. The arrow indicates the time point of addition of the compound.
Correspondingly, deletion of SQR attenuated NAC-, Cys- and 3MP-dependent mt-roGFP2 oxidation and completely prevented H2S-induced mt-roGFP2 oxidation (Figure 4C). The suppression of 3MP-induced mt-roGFP2 oxidation (Figure 4C, antepenultimate panel) suggests that a significant proportion of the hydropersulfides formed by MST is reduced to H2S to be subsequently oxidized by SQR. In contrast, the response to exogenously supplied S0 species (DMTS) was not significantly affected by SQR deletion, as expected (Figure 4C, ultimate panel). The same results were obtained with two SQR knockout cell lines independently generated with different guide RNAs (Figure S3D). Taken together, these results strongly suggest that SQR is a major generator of persulfides in the context of NAC or Cys treatment.
NAC induced generation of persulfides may provide protection against ‘oxidative stress’
The finding that NAC triggers the formation of hydropersulfides raised the question if hydropersulfide generation contributes to the cytoprotective (anti-oxidative) properties of NAC. If indeed persulfides mediate NAC-induced cytoprotection, then Cys, 3MP and H2S should mimic NAC in providing protection against oxidants, as they also induce S0 generation. To test this idea, we performed a real-time assay of intracellular reducing capacity that correlates with cell viability (based on a luciferase pro-substrate activated by intracellular reduction). A pronounced bolus of H2O2 (1.25 mM in a volume of 50 μL containing 10.000 cells; corresponding to 4×1012 H2O2 molecules per cell) caused a substantial, yet transient, drop in intracellular reducing capacity. This drop was not only prevented by pre-treatment with 10 mM NAC, but also by pretreatment with Cys (2 mM), 3MP (2 mM) or Na2S (0.4 mM) (Figure 5A). Thus, all four persulfide-generating compounds protected cells against acute peroxide stress. If persulfides are indeed responsible for the observed protection, then the direct provisioning of per/polysulfides should provide similar cytoprotective effects. To this end we tested the effect of increasing concentrations of Na2S4 in the same assay. Increasing Na2S4 concentrations indeed led to improved protection, but the highest concentration had the opposite effect, causing a major disruption in reducing capacity and cell viability (Figure 5B).
Figure 5. NAC induced generation of persulfides determines protection against oxidative stress.
(A) The cytoprotective effect of NAC against acute peroxide stress is mimicked by Cys, 3MP and Na2S.
(B) Exogenous provisioning of Na2S4 provides protection against acute peroxide stress within a certain concentration range (around 100 μM), but not at higher concentrations (1 mM).
(C-D) The oxidation of the H2O2 probe roGFP2-Orp1 is suppressed by of exogenously applied Na2S4, when applied at low concentrations (C). A very similar effect is observed with mitochondrial roGFP2-Orp1 (D).
(E) The response of the chemical H2O2 probe MitoPY1 is suppressed by exogenously applied Na2S4 in a concentration dependent manner.
All curves represent the mean of three biological repeats, each carried out in technical quadruplicates, the error bars denoting the standard deviation (SD). OxD: degree of probe oxidation. The arrow indicates the time point of addition of the compound.
To further test the idea that low concentrations of persulfides bring down endogenous H2O2 levels, we monitored the response of the H2O2 probe Orp1-roGFP2 in cells exposed to a modest bolus of H2O2 (25μM H2O2 in a volume of 120 μL containing 20.000 cells; corresponding to 9×1010 H2O2 molecules per cell). As expected, the H2O2 bolus led to an increase in probe oxidation, which reached a maximum and was followed by a reductive recovery phase. A 7-min pretreatment with low concentrations of Na2S4 (0.01 and 0.1 μM) substantially suppressed H2O2-induced probe oxidation in the cytosol, while higher concentrations (1 and 10 μM) promoted H2O2-independent probe oxidation (Figure 5C). A similar pattern was seen for the mitochondrial probe (Figure 5D).
Finally, we looked at endogenously generated mitochondrial H2O2 (in the absence of H2O2 addition from outside) using the chemical H2O2 probe mitoPY1. The titration of Na2S4 showed a concentration-dependent suppression of the MitoPY1 response (Figure 5E), again showing the ability of exogenously applied sulfane sulfur compounds to lower endogenous oxidant concentrations.
Discussion
In this study we report that treatment of cells with NAC triggers the formation of the roGFP2 disulfide bond (C147–C204) when roGFP2 is expressed in the mitochondrial matrix. We present multiple lines of evidence suggesting that NAC-induced roGFP2 oxidation is caused by enzymatically generated sulfane sulfur species. Most likely, roGFP2 is first hydropersulfidated on one of its two vicinal cysteines (C147 or C204). The inner sulfur atom of the roGFP2-bound hydropersulfide then invites a nucleophilic attack by the neighboring thiol group, hence forming the intramolecular disulfide and releasing H2S as a leaving group.
At present, the exact mechanism by which roGFP2 becomes hydropersulfidated remains unknow. We present evidence suggesting that SQR-derived hydropersulfides play a major role in the process, as H2S-induced roGFP2 oxidation is completely abolished in SQR KO cells. SQR is known to generate an enzyme-bound hydropersulfide which donates sulfur predominantly to GSH, generating GSSH (Landry et al., 2017). It is however not clear if non-enzymatic transpersulfidation from GSSH to roGFP2 can be efficient in producing roGFP2 persulfidation. This is perhaps unlikely, since unhindered hydropersulfides seem to prefer attack at the inner sulfur atom, to release H2S, which is a better leaving group (Filipovic et al., 2017). Transpersulfidation (i.e. the nucleophilic attack of a thiol on the outer sulfur atom of the hydropersulfide) may instead depend on steric and inductive support provided by specialized enzymes. It has been speculated that members of the thiosulfate sulfur transferase (TST) family (which can use thiosulfate and/or GSSH to generate an enzyme-bound hydropersulfide) transfer persulfides to other proteins (Mishanina et al., 2015). Whether TSTs or other enzymes play a role in roGFP2 hydropersulfidation (and hence disulfide formation) remains to be investigated.
Irrespective of the exact mechanism of roGFP2 hydropersulfidation, we conclude that roGFP2 can be used to detect changes in hydropersulfide generation in living cells and in real-time. The observed responses of roGFP2 to NAC, Cys and H2S were restricted to the mitochondrial matrix, apparently because persulfides are predominantly generated in this compartment, and/or the presence of transpersulfidating enzymes. Nevertheless, 3MP also caused cytosolic roGFP2 oxidation (Figure 1C), probably because of the presence of a cytosolic MST population. At present, it is not known if MST can directly transfer hydropersulfides to roGFP2. These findings have general implications for the use of roGFP2-based redox probes: as sulfane sulfur species can oxidize roGFP2-Orp1 in an Orp1-independent manner, it is important to compare fused and unfused roGFP2 probes in side-by-side experiments to distinguish between Orp1-dependent and Orp1-independent roGFP2 oxidation events (as demonstrated in Fig. 2B). A recent in vitro comparison of roGFP2 based probes has come to the same conclusion (Muller et al., 2017).
In our study we investigated the pathways by which NAC triggers sulfane sulfur generation. We focused on the pathways previously shown to connect Cys metabolism to hydropersulfide generation and implicated CSE, SQR and MST in the process. However, we cannot exclude the possibility that additional pathways can contribute to NAC-induced hydropersulfide generation. We tried to create SQR/MST double knock-out cells to see if they would still support NAC/Cys-induced persulfide generation, but we were not able to obtain such cells, which do not appear to be viable. Of note, a recent paper proposes the existence of a hitherto unrecognized pathway that leads from Cys to persulfides, depending on a ‘cysteine persulfide synthetase’ activity ascribed to mitochondrial cysteinyl-tRNA synthetase (CARS2) (Akaike et al., 2017). If CARS2 contributes to NAC-induced sulfane sulfur generation remains to be investigated.
Having found that NAC treatment leads to the intracellular generation of hydropersulfides, the question arises if sulfane sulfur species (hydropersulfides or hydropersulfide-derived species) contribute to the cytoprotective and antioxidative effects of NAC. We found that all three persulfide-generating catabolites downstream of NAC (Cys, 3MP and H2S) mimicked NAC in that they also protected cells against acute peroxide stress. This finding suggested the possibility that acute stress protection is at least partially based on persulfide generation.
We therefore asked if the direct provisioning of a sulfane sulfur species to cells would mimic the antioxidative effects of NAC/Cys/3MP/H2S, perhaps in a more potent manner. To deliver sulfane sulfur to cells, we used polysulfides. Both inorganic (Na2S2, Na2S3, Na2S4) and organic (DMTS, DATS) polysulfides were confirmed to induce intracellular roGFP2 oxidation in a compartment-independent manner. It is well known that solubilized polysulfides are intrinsically unstable and subject to rapid disproportionation into multiple species (Filipovic et al., 2017). Inside cells they are also subject to reduction. Hence, the approach of adding polysulfides to cells is not well defined in terms of the products and their distribution. Nevertheless, polysulfides can be expected to generate intermediary hydropersulfide species when they react with intracellular thiols, as for example shown for the reaction between GSH and DATS (Liang et al., 2015).
Testing sodium tetrasulfide within a wide range of concentrations, we observed a pronounced antioxidative effect at concentrations far below those that induce detectable roGFP2 oxidation: Na2S4 suppressed H2O2-induced oxidation of the roGFP2-Orp1 probe at low concentrations, potentially indicating direct and efficient H2O2 scavenging by sulfane sulfur species. A similar result was obtained with a chemical probe that monitors endogenous mitochondrial H2O2 levels.
These findings are in line with the idea that low-molecular-weight hydropersulfides (e.g. GSSH) can serve as antioxidants and electrophile scavengers. Hydropersulfides can be considered 'hyperactivated thiols' (Francoleon et al., 2011), because of the so-called α-effect, which is the increase in reactivity (acidity, nucleophilicity) due to lone electron pairs on the atom in the adjacent alpha position (Edwards and Pearson, 1962). The α-effect makes the outer sulfur atom of the persulfide highly reactive towards oxidants and electrophiles (Cuevasanta et al., 2015). Likewise, persulfides readily reduce one-electron oxidants to form the resonance stabilized perthiyl radical (Bianco et al., 2016), which may be efficiently removed by radical recombination. Superior antioxidant properties of small-molecule hydropersulfides in a direct comparison to corresponding thiols have been observed previously (Ida et al., 2014). Hence, hydropersulfides appear to be more efficient scavengers of radicals, peroxides and electrophiles than thiols.
It is possible that the observed antioxidative effects of polysulfide treatment are also caused by sulfane sulfur species other than persulfides. Further (limited) oxidation of sulfane sulfur species can lead to products that are highly efficient in trapping peroxyl radicals, specifically polysulfide-1-oxides (Chauvin et al., 2016), suggesting the possibility that these could also play a biological role as antioxidants. It seems that fully reduced organosulfur must be oxidized to make more efficient antioxidants, from the thiol (-2) to the per/polysulfide (-1/0) and then perhaps to the subsequent oxidation number, as in polysulfide-1-oxides (+1).
Taken together, our findings suggest that hydropersulfides (and/or other S0 species derived from hydropersulfides) can act as direct oxidant scavengers already at low concentrations (as reflected by the suppression of H2O2-induced roGFP2-Orp1 or MitoPY1 oxidation), and additionally as inducers of protein hydropersulfidation, which however seems to require higher concentrations (as reflected by the induction of roGFP2 oxidation). It seems likely that hydropersulfide-induced roGFP2 oxidation is indicative of the concomitant hydropersulfidation of many other proteins. Proteins with vicinal thiols (like roGFP2) can rapidly convert hydropersulfide groups to disulfides, but other proteins may acquire the hydropersulfide as a more stable modification. Indeed, protein hydropersulfidation seems to be relatively common as a detectable posttranslational modification (Doka et al., 2016; Gao et al., 2015; Longen et al., 2016; Wedmann et al., 2016).
Protein hydropersulfidation may also contribute to cytoprotection because it can be expected to protect protein thiols against irreversible oxidation: while unprotected protein thiols can form irreversibly oxidized products (sulfinic and sulfonic acids), the corresponding protein persulfide oxidation products (perthiosulfinic and perthiosulfonic acid) can still be reduced to regenerate the unmodified thiol (Millikin et al., 2016; Ono et al., 2014). In other words, critical protein thiols can be protected against hyperoxidation by their prior hydropersulfidation, because the outer (highly reactive) sulfur atom of a hydropersulfide can be sacrificed (i.e. hyperoxidized) to effectively safeguard the inner sulfur atom.
Our findings may relate to various previous observations. Previous experiments indicated that the cytoprotective effects of H2S (Ahmad et al., 2016; Kimura and Kimura, 2004; Wen et al., 2013) may be mediated by sulfane sulfur species. For example, the cytoprotective action of H2S on cells under ischemia was abrogated upon depletion of SQR (Hine et al., 2015), implicating a role for the mitochondrial H2S oxidation pathway in cytoprotection. A study investigating the role of H2S in cardioprotection found that NAC replicates the effects of the H2S donor NaHS, when used at ≈1000-fold higher concentrations (Li et al., 2015). Moreover, it has been noted that exposure of cells to H2O2 induces the increased expression of both the cystine xCT transporter (Lewerenz et al., 2013) and CSE (Wang et al., 2012), compatible with the idea that increased persulfide generation from Cys and/or Cys2 is a physiologically relevant oxidative stress response (Millikin et al., 2016).
Our findings do not rule out the possibility that NAC can protect cells in several different ways, e.g. by directly feeding the biosynthesis of GSH, depending on conditions. Nevertheless, our results point towards a more general answer to the question of how NAC, whose thiol group exhibits low intrinsic reactivity towards oxidants, can act as an antioxidant inside cells. The answer appears to be the conversion of NAC-derived thiols into 'hyperactivated thiols', that is hydropersulfides, acting as direct oxidant scavengers and/or protective caps for critical protein thiols.
Significance
N-acetyl cysteine (NAC) is one of the most widely used antioxidants in the context of clinical studies, animal experiments and cell culture experiments. Many conclusions have been based on the outcome of NAC treatment experiments. Despite this, the mechanisms behind its antioxidant action remain poorly understood. Beyond its use in research, NAC has been promoted to the general public as a food supplement. Recent studies show that NAC can accelerate cancer progression in mice, making a better understanding of how NAC is acting inside cells even more critical. Here we delineate the catabolism of NAC inside cells and identify sulfane sulfur species as likely mediators of the acute antioxidative and cytoprotective effects of NAC. These findings indicate that NAC experiments need to be interpreted more carefully and further suggest new and refined strategies for pharmacological cytoprotection.
Star⋆Methods
Key Resources Table
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tobias P. Dick (t.dick@dkfz.de).
Experimental model and subject detail
Cell lines
The human lung adenocarcinoma cell line H838 (gender: male) was purchased from ATCC (CRL-5844) and cultivated in Dulbecco’s modified Eagle’s Medium (DMEM, Lonza) supplemented with 10% fetal calf serum (Gibco), 100 μg/ml streptomycin (Gibco) and 100 U/ml penicillin (Gibco) at 37°C in a humidified atmosphere with 5% CO2. The Phoenix Ampho packaging cell line was cultured in the same medium. All cell lines were confirmed to be free of mycoplasma, viral infections and contaminations with other cell lines by Multiplex Human Cell Line Authentication (Multiplexion, Heidelberg).
Method details
Generation of stably transduced H838 cells
RoGFP2 and roGFP2-Orp1 (with or without the mitochondrial targeting sequence) were introduced into cells using the retroviral expression vector pLPCX. Transfection of the Phoenix Ampho cells was performed by calcium phosphate precipitation. Supernatants for transduction of H838 cells were prepared 24 hours after transfection by passing through a 0.45 μm filter and supplementing with 8 μg/ml polybrene (Sigma). This procedure was repeated twice on two consecutive days. Stably transduced H838 cells were selected in the presence of 12.5 μg/ml puromycin (Sigma) 72 hours after transduction. Cells were then sorted by FACS to obtain a homogenous population of probe-expressing cells.
Generation of CRISPR/Cas9-mediated knockout cells
The following guide RNAs (gRNAs) were used in the study: For CSE: exon 5 (5’-GGAGACATTATTTTGGTCGTGG-3’) and exon 1 (5’-TTCCAACATTTGGCCACGCAGG-3’) (purchased from Sigma-Aldrich); for SQR: exon 8 (5’-GTCCTCAAGACCAGTCCTGTGG-3’) (Sigma-Aldrich) and exon 3 (5’-TGCCGTGGGACGACCAGATG-3’) (Genscript); for MST: exon 4 (5’-CGCGTTACCGTCTCGGGGCT-3’) (Genscript). The gRNAs were expressed using an U6gRNA-Cas9-2A-RFP (Sigma-Aldrich), or pSpCas9 BB-2A-Puro (PX459) v2.0 vector after transient transfection using Lipofectamine LTX following the manufacturer’s protocol. Depending on the vector, cells were selected either with 12.5 μg/ml puromycin (Sigma), or by FACS. Individual clones were grown after diluting cells to a concentration of 0.5 cells per 100 μl in 96 well plates. Knockout clones were identified by western blotting. Antibodies used were rabbit anti-CSE (abcam, ab151769) at a dilution of 1:500, mouse anti-SQR (abcam, ab71978) at a dilution of 1:500 and mouse anti-MST (Santa Cruz, sc-374326) at a dilution of 1:100. Rabbit anti-GAPDH (abcam, ab 9485) (1:10000) and mouse anti-β-actin (sigma # A5441) (1:2500) were used as loading controls.
Chemicals
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich. Polysulfide salts were obtained from Dojindo Molecular Technologies. 3-mercaptopyruvate was purchased from Santa Cruz. The source of pyruvate was MP Biomedicals. All chemicals were prepared in sterile PBS (Gibco) and, if necessary, the pH was adjusted to 7.4 using Trizma base. H2S stock solutions (± 100 mM) were prepared fresh daily from Na2S.9H2O, as described (Nagy et al., 2014). In brief, salt crystals were washed and then dissolved in ultrapure water and immediately transferred to ice. The concentration was determined by UV-Vis spectrophotometry, recording the absorbance at 230 nm (ε230HS− = 7700 M−1 cm−1), after diluting the solution to a concentration of approximately 50 μM. Working H2S solutions were prepared by diluting the stock solution into the working buffer immediately before use.
Fluorescence measurements of roGFP2 oxidation
The day before the measurement cells stably expressing the probe were seeded into a black flat-bottomed 96-well imaging plate (BD Falcon 353219) at a density of 166 x 103 cells/ml in DMEM. A non-transduced control was included on the same plate for background subtraction. Before the measurement the growth medium was removed, the cells washed with PBS and imaged in 120 μl imaging buffer (130 mM NaCl, 5 mM KCl, 10 mM D-glucose, 1 mM MgCl2, 1 mM CaCl2 and 20 mM HEPES). RoGFP2 oxidation was measured using a PHERAstar FS fluorescence plate reader (BMG Labtech), which allows the simultaneous detection of the two excitation maxima of roGFP2 (400 nm and 485 nm) when emission is monitored at 520 nm. The readout of the roGFP2 measurement was expressed as the degree of sensor oxidation (OxD) (see equation 1). In order to obtain the fluorescence intensity values for a fully oxidized and reduced probe, following the run each well was sequentially treated with N,N,N’,N’-tetramethylazodicarboxamide (diamide) to a final concentration of 2 mM and DTT to a final concentration of 10 mM. All treatments were carried out after 5 cycles and were performed in technical quadruplicates seeded in different quadrants of the imaging plate, so as to avoid position effects.
In this equation, I represents the fluorescence intensity detected at λ=520 nm after excitation at either 400 nm or 485 nm. The subscripts “ox”, “red” and “sample” indicate the fluorescence intensity measured for the fully oxidized (i.e. diamide-treated), the fully reduced (i.e. DTT-treated) controls and the sample, respectively.
Measurement of total glutathione levels
The concentration of total glutathione was determined using the DTNB recycling assay (Rahman et al., 2006). Cells for the assay were seeded at a density of 1 x 105 cells/ml in 10 cm dishes on the day prior to the measurement. Cells were collected by trypsinysation and an aliquot was taken for the determination of the packed cell volume (PCV) determined using PCV tubes (TPP). Cells were then lysed in 10% TCA (trichloroacetic acid). The lysate was clarified by centrifugation and the samples were diluted 1:100 in KPE buffer (100 mM K2HPO4, 100 mM KH2PO4 and 5 mM EDTA). 20μl of the resulting solution were transferred into a 96-well plate. The same volume of standards was employed. 120 μl of a 1:1 mixture of NADPH (AppliChem) and DTNB (both 1.11 mg/ml in KPE buffer) was then added to each well and the plate was placed into the plate reader set to detect absorbance at λ=412 nm. The measurement was allowed to run for a couple of cycles until a stable baseline was established for each sample and then paused in order to add 50 μl of the glutathione reductase (3.3 units/ml in KPE buffer, Sigma-Aldrich) to each well. The run was then continued until the absorbance values of the highest standard reached saturation. The absolute values of the concentrations were determined using the equation obtained from the linear regression analysis performed with a series of samples of known concentrations. In our case the range of concentrations was from 0.4 to 26 μM. The measurement was performed using biological triplicates, carried out in technical duplicates.
Measurement of mitochondrial H2O2 production
RoGFP2-independent assessment of mitochondrial H2O2 production was performed using the fluorescence probe MitoPY1 (Tocris). Cells were seeded into a black flat-bottomed 96-well imaging plate (BD Falcon 353219) at a density of 166 x 103 cells/ml in DMEM. Cells were loaded with 10 μM MitoPY1 in PBS for 1 h at 37°C. The plate was then washed twice with PBS and cells were imaged in PBS. The signal was acquired by monitoring the fluorescence at λex= 480 nm and λem= 520 nm. Experiments were performed in biological triplicates, using technical quadruplicates.
H2S detection
H2S was monitored using the HSip-1 DA (Sasakura et al., 2011) probe. Cells were seeded in black flat-bottomed 96-well imaging plate (BD Falcon 353219) at a density of 166 x 103 cells/ml in DMEM. The following day HSip-1 DA in HBSS was added directly to the medium to a final concentration of 20 μM and incubated at 37°C for 30 min. Cells were then washed with HBSS and imaged in HBSS. Signal acquisition was performed using PHERAstar FS fluorescence plate reader (BMG Labtech) at λex= 480 nm and λem= 520 nm. Experiments were performed in biological triplicates, using technical quadruplicates.
Sulfane sulfur detection
Changes in intracellular sulfane sulfur levels were monitored with the SSip-1 DA probe (Takano et al., 2017). Cells were seeded in black flat-bottomed 96-well imaging plate (BD Falcon 353219) at a density of 166 x 103 cells/ml in DMEM. The following day the medium was discarded, cells washed with HBSS and loaded with SSip-1 DA in DMEM at a concentration of 5 μM for 1 h at 37°C. The cells were then washed twice with HBSS and the SSip-1 DA signal acquisition in HBSS was carried out by recording the fluorescence λex= 470 nm and λem= 520 nm. Experiments were performed in biological triplicates, using technical quadruplicates.
Measurement of the oxygen consumption rate
The oxygen consumption rate (OCR) was determined using a Seahorse XF96 analyzer (Agilent Technologies). One day prior to the measurement cells were seeded in a Seahorse assay 96-well plate at a density of 75 x 103 cells/ml in DMEM. On the day of the experiment DMEM was removed, the cells were washed twice with Seahorse XF assay medium supplemented with 1 mM pyruvate and 5 mM glucose and incubated in it for 1 h in at 37°C in a CO2-free incubator. Following the OCR determination, the same plate was used for protein quantification using the sulforhodamine B based in vitro toxicology assay kit (Sigma-Aldrich) according to the manufacturer’s instructions. OCR was normalized to protein levels. The experiment was performed in biological triplicates, each using technical hexaplicates.
Viability assay
Cell viability was assessed using the RealTime-Glo™ MT Cell viability assay (Promega) according to the manufacturer’s instructions. Cells for the experiment were seeded in complete DMEM at a density of 75x103 cells/ml in white clear flat-bottomed 96 well plates (Corning) in complete DMEM. Compounds, H2O2 and the assay reagents were added simultaneously immediately before monitoring the luminescence using the PHERAstar FS fluorescence plate reader (BMG Labtech). Experiments were performed in biological triplicates, using technical quadruplicates.
Quantification and statistical analysis
Unless otherwise indicated, all experiments in this study were carried out using biological triplicates, using technical quadruplicates each time, as indicated in the figure legends. The curves represent the mean and the error bars the standard deviation. No hypothesis test was used. All calculations were performed using OriginPro 2017G.
Supplementary Material
Supplemental Information includes 3 figures and can be found with this article online at
Acknowledgements
We thank Monika Langlotz (ZMBH Flow Cytometry & FACS Core Facility) for performing cell sorting. We acknowledge funding by BMBF (LungSysII), DFG (SPP1710 and SFB1036) and the European Commission (ERC 742039) to T.P.D.
Footnotes
Author contributions
Conceptualization, D.E. and T.P.D.; Methodology, D.E., K.H., Y.U. and T.P.D.; Formal Analysis, D.E.; Investigation, D.E.; Resources, Y.T., K.H. and Y.U.; Writing, D.E. and T.P.D.; Visualization, D.E.; Supervision, T.P.D.; Funding Acquisition, T.P.D.
Declaration of Interests
The authors declare no competing interests.
References
- Ahmad A, Olah G, Szczesny B, Wood ME, Whiteman M, Szabo C. AP39, A Mitochondrially Targeted Hydrogen Sulfide Donor, Exerts Protective Effects in Renal Epithelial Cells Subjected to Oxidative Stress in Vitro and in Acute Renal Injury in Vivo. Shock (Augusta, Ga) 2016;45:88–97. doi: 10.1097/SHK.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T, Ida T, Wei FY, Nishida M, Kumagai Y, Alam MM, Ihara H, Sawa T, Matsunaga T, Kasamatsu S, et al. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat Commun. 2017;8:1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht SC, Barata AG, Grosshans J, Teleman AA, Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Asimakopoulou A, Panopoulos P, Chasapis CT, Coletta C, Zhou Z, Cirino G, Giannis A, Szabo C, Spyroulias GA, Papapetropoulos A. Selectivity of commonly used pharmacological inhibitors for cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE) British journal of pharmacology. 2013;169:922–932. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benrahmoune M, Therond P, Abedinzadeh Z. The reaction of superoxide radical with N-acetylcysteine. Free Radic Biol Med. 2000;29:775–782. doi: 10.1016/s0891-5849(00)00380-4. [DOI] [PubMed] [Google Scholar]
- Bianco CL, Chavez TA, Sosa V, Saund SS, Nguyen QN, Tantillo DJ, Ichimura AS, Toscano JP, Fukuto JM. The chemical biology of the persulfide (RSSH)/perthiyl (RSS.) redox couple and possible role in biological redox signaling. Free Radic Biol Med. 2016;101:20–31. doi: 10.1016/j.freeradbiomed.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin JR, Haidasz EA, Griesser M, Pratt DA. Polysulfide-1-oxides react with peroxyl radicals as quickly as hindered phenolic antioxidants and do so by a surprising concerted homolytic substitution. Chem Sci. 2016;7:6347–6356. doi: 10.1039/c6sc01434h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532:112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Lange M, Bonanata J, Coitino EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. J Biol Chem. 2015;290:26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Lin VS, Chang CJ. Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat Protoc. 2013;8:1249–1259. doi: 10.1038/nprot.2013.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Science advances. 2016;2:e1500968. doi: 10.1126/sciadv.1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JO, Pearson RG. Factors Determining Nucleophilic Reactivities. J Am Chem Soc. 1962;84:16. [Google Scholar]
- Filipovic MR, Zivanovic J, Alvarez B, Banerjee R. Chemical Biology of H2S Signaling through Persulfidation. Chem Rev. 2017 doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoleon NE, Carrington SJ, Fukuto JM. The reaction of H(2)S with oxidized thiols: generation of persulfides and implications to H(2)S biology. Archives of biochemistry and biophysics. 2011;516:146–153. doi: 10.1016/j.abb.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Frasdorf B, Radon C, Leimkuhler S. Characterization and interaction studies of two isoforms of the dual localized 3-mercaptopyruvate sulfurtransferase TUM1 from humans. J Biol Chem. 2014;289:34543–34556. doi: 10.1074/jbc.M114.605733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, et al. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Heard KJ, Reynolds KM, Albert D. Oral and Intravenous Acetylcysteine for Treatment of Acetaminophen Toxicity: A Systematic Review and Meta-analysis. West J Emerg Med. 2013;14:218–226. doi: 10.5811/westjem.2012.4.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner R, Dick TP. Real-time assays for monitoring the influence of sulfide and sulfane sulfur species on protein thiol redox states. Methods in enzymology. 2015;555:57–77. doi: 10.1016/bs.mie.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Greiner R, Palinkas Z, Basell K, Becher D, Antelmann H, Nagy P, Dick TP. Polysulfides link H2S to protein thiol oxidation. Antioxidants & redox signaling. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine C, Harputlugil E, Zhang Y, Ruckenstuhl C, Lee BC, Brace L, Longchamp A, Trevino-Villarreal JH, Mejia P, Ozaki CK, et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160:132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science (New York, NY) 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Kabil O, Vitvitsky V, Banerjee R. Sulfur as a signaling nutrient through hydrogen sulfide. Annu Rev Nutr. 2014;34:171–205. doi: 10.1146/annurev-nutr-071813-105654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Kimura H. Hydrogen sulfide protects neurons from oxidative stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:1165–1167. doi: 10.1096/fj.04-1815fje. [DOI] [PubMed] [Google Scholar]
- Landry AP, Ballou DP, Banerjee R. H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase. J Biol Chem. 2017;292:11641–11649. doi: 10.1074/jbc.M117.788547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyurek LM, Lindahl P, Nilsson J, et al. Antioxidants can increase melanoma metastasis in mice. Science translational medicine. 2015;7:308re308. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxidants & redox signaling. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang Y, Wei C, Bai S, Zhao Y, Li H, Wu B, Wang R, Wu L, Xu C. Mediation of exogenous hydrogen sulfide in recovery of ischemic post-conditioning-induced cardioprotection via down-regulating oxidative stress and up-regulating PI3K/Akt/GSK-3β pathway in isolated aging rat hearts. Cell & Bioscience. 2015;5:11. doi: 10.1186/s13578-015-0003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wu H, Wong MW, Huang D. Diallyl Trisulfide Is a Fast H2S Donor, but Diallyl Disulfide Is a Slow One: The Reaction Pathways and Intermediates of Glutathione with Polysulfides. Org Lett. 2015;17:4196–4199. doi: 10.1021/acs.orglett.5b01962. [DOI] [PubMed] [Google Scholar]
- Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J Biol Chem. 2014;289:30901–30910. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S, Richter F, Kohler Y, Wittig I, Beck KF, Pfeilschifter J. Quantitative Persulfide Site Identification (qPerS-SID) Reveals Protein Targets of H2S Releasing Donors in Mammalian Cells. Scientific reports. 2016;6 doi: 10.1038/srep29808. 29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Cavallin LE, Yan B, Zhu S, Duran EM, Wang H, Hale LP, Dong C, Cesarman E, Mesri EA, et al. Antitumorigenesis of antioxidants in a transgenic Rac1 model of Kaposi's sarcoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8683–8688. doi: 10.1073/pnas.0812688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikin R, Bianco CL, White C, Saund SS, Henriquez S, Sosa V, Akaike T, Kumagai Y, Soeda S, Toscano JP, et al. The chemical biology of protein hydropersulfides: Studies of a possible protective function of biological hydropersulfide generation. Free Radic Biol Med. 2016;97:136–147. doi: 10.1016/j.freeradbiomed.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishanina TV, Libiad M, Banerjee R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nature chemical biology. 2015;11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Schneider JF, Degrossoli A, Lupilova N, Dick TP, Leichert LI. Systematic in vitro assessment of responses of roGFP2-based probes to physiologically relevant oxidant species. Free Radic Biol Med. 2017;106:329–338. doi: 10.1016/j.freeradbiomed.2017.02.044. [DOI] [PubMed] [Google Scholar]
- Nagy P, Pálinkás Z, Nagy A, Budai B, Tóth I, Vasas A. Chemical aspects of hydrogen sulfide measurements in physiological samples. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840:876–891. doi: 10.1016/j.bbagen.2013.05.037. [DOI] [PubMed] [Google Scholar]
- Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic Biol Med. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Jung M, Zhang T, Tsutsuki H, Sezaki H, Ihara H, Wei FY, Tomizawa K, Akaike T, Sawa T. Synthesis of l-cysteine derivatives containing stable sulfur isotopes and application of this synthesis to reactive sulfur metabolome. Free Radic Biol Med. 2017;106:69–79. doi: 10.1016/j.freeradbiomed.2017.02.023. [DOI] [PubMed] [Google Scholar]
- Parasassi T, Brunelli R, Costa G, De Spirito M, Krasnowska E, Lundeberg T, Pittaluga E, Ursini F. Thiol redox transitions in cell signaling: a lesson from N-acetylcysteine. ScientificWorldJournal. 2010;10:1192–1202. doi: 10.1100/tsw.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Min KJ, Lee TJ, Yoo YH, Kim YS, Kwon TK. beta-Lapachone induces programmed necrosis through the RIP1-PARP-AIF-dependent pathway in human hepatocellular carcinoma SK-Hep1 cells. Cell death & disease. 2014;5:e1230. doi: 10.1038/cddis.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarca S, Furfaro AL, Domenicotti C, Odetti P, Cottalasso D, Marinari UM, Pronzato MA, Traverso N. Supplementation with N-acetylcysteine and taurine failed to restore glutathione content in liver of streptozotocin-induced diabetics rats but protected from oxidative stress. Biochim Biophys Acta. 2005;1741:48–54. doi: 10.1016/j.bbadis.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DW, Baskin SI. The effect of three alpha-keto acids on 3-mercaptopyruvate sulfurtransferase activity. Journal of biochemical toxicology. 1996;11:45–50. doi: 10.1002/(SICI)1522-7146(1996)11:1<45::AID-JBT6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftos JE, Whillier S, Chapman BE, Kuchel PW. Kinetics of uptake and deacetylation of N-acetylcysteine by human erythrocytes. The international journal of biochemistry & cell biology. 2007;39:1698–1706. doi: 10.1016/j.biocel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006;1:3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- Rodak R, Kubota H, Ishihara H, Eugster HP, Konu D, Mohler H, Yonekawa Y, Frei K. Induction of reactive oxygen intermediates-dependent programmed cell death in human malignant ex vivo glioma cells and inhibition of the vascular endothelial growth factor production by taurolidine. J Neurosurg. 2005;102:1055–1068. doi: 10.3171/jns.2005.102.6.1055. [DOI] [PubMed] [Google Scholar]
- Samuni Y, Goldstein S, Dean OM, Berk M. The chemistry and biological activities of N-acetylcysteine. Biochimica et biophysica acta. 2013;1830:4117–4129. doi: 10.1016/j.bbagen.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Sasakura K, Hanaoka K, Shibuya N, Mikami Y, Kimura Y, Komatsu T, Ueno T, Terai T, Kimura H, Nagano T. Development of a highly selective fluorescence probe for hydrogen sulfide. Journal of the American Chemical Society. 2011;133:18003–18005. doi: 10.1021/ja207851s. [DOI] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med. 2014;6:221ra215. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Banerjee R. PLP-dependent H(2)S Biogenesis. Biochimica et biophysica acta. 2011a;1814:1518–1527. doi: 10.1016/j.bbapap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Banerjee R. PLP-dependent H(2)S biogenesis. Biochim Biophys Acta. 2011b;1814:1518–1527. doi: 10.1016/j.bbapap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanuk MH, Dominy JE, Lee JI, Coloso RM. Mammalian cysteine metabolism: New insights into regulation of cysteine metabolism. J Nutr. 2006;136:1652s–1659s. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Ueki I, Dominy JE, Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino acids. 2009;37:55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Collins R, Huang S, Holmberg-Schiavone L, Anand GS, Tan CH, van-den-Berg S, Deng LW, Moore PK, Karlberg T, et al. Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S. J Biol Chem. 2009;284:3076–3085. doi: 10.1074/jbc.M805459200. [DOI] [PubMed] [Google Scholar]
- Takano Y, Hanaoka K, Shimamoto K, Miyamoto R, Komatsu T, Ueno T, Terai T, Kimura H, Nagano T, Urano Y. Development of a reversible fluorescent probe for reactive sulfur species, sulfane sulfur, and its biological application. Chemical communications (Cambridge, England) 2017;53:1064–1067. doi: 10.1039/c6cc08372b. [DOI] [PubMed] [Google Scholar]
- Uttamsingh V, Keller DA, Anders MW. Acylase I-catalyzed deacetylation of N-acetyl-L-cysteine and S-alkyl-N-acetyl-L-cysteines. Chem Res Toxicol. 1998;11:800–809. doi: 10.1021/tx980018b. [DOI] [PubMed] [Google Scholar]
- Vasas A, Doka E, Fabian I, Nagy P. Kinetic and thermodynamic studies on the disulfide-bond reducing potential of hydrogen sulfide. Nitric oxide : biology and chemistry. 2015;46:93–101. doi: 10.1016/j.niox.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Wang F, Liu S, Shen Y, Zhuang R, Xi J, Fang H, Pan X, Sun J, Cai Z. Protective effects of N-acetylcysteine on cisplatin-induced oxidative stress and DNA damage in HepG2 cells. Experimental and therapeutic medicine. 2014;8:1939–1945. doi: 10.3892/etm.2014.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Guo Z, Wang S. Cystathionine gamma-lyase expression is regulated by exogenous hydrogen peroxide in the mammalian cells. Gene expression. 2012;15:235–241. doi: 10.3727/105221613x13571653093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedmann R, Onderka C, Wei S, Szijarto IA, Miljkovic JL, Mitrovic A, Lange M, Savitsky S, Yadav PK, Torregrossa R, et al. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci. 2016;7:3414–3426. doi: 10.1039/c5sc04818d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Wang H, Kho SH, Rinkiko S, Sheng X, Shen HM, Zhu YZ. Hydrogen sulfide protects HUVECs against hydrogen peroxide induced mitochondrial dysfunction and oxidative stress. PLoS One. 2013;8:e53147. doi: 10.1371/journal.pone.0053147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Coles LD, Kartha RV, Nash N, Mishra U, Lund TC, Cloyd JC. Intravenous Administration of Stable-Labeled N-Acetylcysteine Demonstrates an Indirect Mechanism for Boosting Glutathione and Improving Redox Status. Journal of pharmaceutical sciences. 2015;104:2619–2626. doi: 10.1002/jps.24482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.