Abstract
Chlorarachniophyte and cryptophyte algae are unique among plastid-containing species in that they have a nucleomorph genome: a compact, highly reduced nuclear genome from a photosynthetic eukaryotic endosymbiont. Despite their independent origins, the nucleomorph genomes of these two lineages have similar genomic architectures, but little is known about the evolutionary pressures impacting nucleomorph DNA, particularly how their rates of evolution compare to those of the neighboring genetic compartments (the mitochondrion, plastid, and nucleus). Here, we use synonymous substitution rates to estimate relative mutation rates in the four genomes of nucleomorph-bearing algae. We show that the relative mutation rates of the host versus endosymbiont nuclear genomes are similar in both chlorarachniophytes and cryptophytes, despite the fact that nucleomorph gene sequences are notoriously highly divergent. There is some evidence, however, for slightly elevated mutation rates in the nucleomorph DNA of chlorarachniophytes—a feature not observed in that of cryptophytes. For both lineages, relative mutation rates in the plastid appear to be lower than those in the nucleus and nucleomorph (and, in one case, the mitochondrion), which is consistent with studies of other plastid-bearing protists. Given the divergent nature of nucleomorph genes, our finding of relatively low evolutionary rates in these genomes suggests that for both lineages a burst of evolutionary change and/or decreased selection pressures likely occurred early in the integration of the secondary endosymbiont.
Keywords: nucleotide substitution, mutation rate, genetic diversity, secondary endosymbiosis, evolution
Introduction
Endosymbiosis has defined eukaryotic evolution, generating both mitochondria and plastids, which arose by the primary endosymbiosis of an alpha-proteobacterium and a cyanobacterium, respectively (Gray 2012; Archibald 2015a). Primary plastids have subsequently spread to diverse lineages via eukaryote–eukaryote endosymbioses (Gould et al. 2008; Archibald 2009), giving rise to organelles with one or two additional membranes (Cavalier-Smith 2003). Consequently, nearly all eukaryotes contain multiple genetic compartments, each with its own distinct genome.
Chlorarachniophyte and cryptophyte algae exemplify this point. These two lineages gained photosynthesis through the secondary endosymbiosis of a green alga and a red alga, respectively, and still retain the nuclear genomes of their eukaryotic endosymbionts (Douglas et al. 2001; Gilson et al. 2006; Archibald 2007). Despite their different evolutionary origins, the available nucleomorph genome sequences of chlorarachniophytes and cryptophytes have many similar features. They are made up of three linear chromosomes, are generally AT-rich, and encode ∼500 proteins, 31 or fewer of which function in the plastid (Douglas et al. 2001; Gilson et al. 2006; Archibald 2007). Although our understanding of nucleomorph genome architecture has improved in recent years (Tanifuji et al. 2011, 2014; Moore et al. 2012; Suzuki et al. 2015), little is known about the mutation rate of nucleomorph DNA (nmDNA), particularly how it compares to those of the mitochondrial, plastid, and host nuclear DNA (mtDNA, ptDNA, and nuDNA).
Estimating mutation rate is not easy. But insights into this fundamental parameter can be gained by measuring the rate of synonymous nucleotide substitution (dS) between distinct species or strains (Kimura 1983). Under the assumption of neutrality, the relative values of dS for different genomes within a cell should reflect the relative mutation rates of those genomes (Kimura 1983). Indeed, relative substitution rate data have revealed that for most animals the mitochondrial mutation rate is much higher than that of the nucleus (Brown et al. 1979; Nabholz et al. 2008; James et al. 2016), whereas for land plants the opposite is true (Wolfe et al. 1987; Drouin et al. 2008; Richardson et al. 2013; Zhu et al. 2014). Substitution rate analyses of ptDNA have shown that land plant plastid genomes typically have higher and lower mutation rates, respectively, than their mitochondrial and nuclear counterparts (Wolfe et al. 1987; Drouin et al. 2008; Richardson et al. 2013; Zhu et al. 2014). But in plastid-bearing protists from diverse lineages, the ptDNA tends to have a lower dS than the mtDNA and nuDNA from the same cell, implying a low plastid mutation rate (Smith 2015). Together, these data have improved our understanding of organelle genetics (Sloan and Taylor 2012) and helped spur hypotheses on molecular evolution (Lynch et al. 2006).
Detailed relative rate analyses of nmDNA have not yet been carried out, even though such data could provide valuable insights into eukaryotic mutation rates and genome evolution (Smith et al. 2012). Here, we do just that. Using transcriptome sequence data from various isolates of the chlorarachniophyte Bigelowiella natans and the cryptophyte Hemiselmis andersenii, we provide the first relative mutation rate estimates for nucleomorph, mitochondrial, plastid, and host nuclear genomes from a single cell. We find that within B. natans and H. andersenii, dS is similar for the nucleomorph and nuclear DNAs, suggesting that the host and endosymbiont nuclear genomes have similar mutation rates. For both lineages, dS in the plastid genome is low relative to the nuclear and nucleomorph genomes, implying that the ptDNA has a low relative mutation rate. The mitochondrial rates showed the largest variability: elevated in B. natans and very low in H. andersenii. Overall, these mutation rate ratios provide insights into the evolutionary forces at play in endosymbiont genomes of photosynthetic eukaryotes and how they compare to those of the host.
Materials and Methods
All sequence data were downloaded from the National Center for Genome Resources (NCGR) MMETSP project page (http://data.imicrobe.us/project/view/104). The following B. natans samples were used: MMETSP0045 (strain CCMP2755), MMETSP1052 (strain CCMP623), MMETSP1054 (strain CCMP1259), MMETSP1055 (strain CCMP1258), and MMETSP1358 (strain CCMP1242). The following H. andersenii samples were used: MMETSP0043 (strain CCMP644), MMETSP1041 (strain CCMP439), MMETSP1042 (strain CCMP1180), and MMETSP1043 (strain CCMP441).
A reciprocal best BLAST hit (RBH) approach was used to identify putative orthologs between strains (Altschul et al. 1990; Tatusov et al. 1997; Bork et al. 1998; Camacho et al. 2009). An e-value cut-off of 1E-50 was used for nuclear genes. An additional step was required to identify nucleomorph genes prior to the RBH analysis. MMETSP sequences were used as query against either a curated set of known nucleomorph or nuclear genes in a competitive BLAST (e-value 1E-10), with the resulting files being processed via a custom Python script to compare alignment scores and assign sequences as nucleomorph or nuclear. The nucleomorph sequences were then used for RBH analysis with an e-value cut-off of 1E-20. As part of the pipeline to prepare files for input to the PAML package, custom Python scripts were written to 1) compile files of pairwise RBH for all strains into a single file, 2) retrieve relevant gene and protein sequences from assembly files from each strain into a single file per gene, 3) filter alignment files to exclude non block-selected residues, and 4) filter amino acid alignment files for improperly identified orthologs using a percent identity cut-off of 95% (Python Programming Language, www.python.org). MAFFT was used for multiple sequence alignment with maxiterate = 1,000 (Katoh et al. 2002). Clustal format alignment files were converted to FASTA format with seqret (Peter Rice; EMBOSS). The Gblocks software was used to identify aligned blocks of amino acids (options: -b4 = 4 -k = y) (Castresana 2000). Codon alignments were produced using pal2nal.pl (options: -output paml -blockonly -nogap -nomismatch) (Mikita et al. 2006).
Since the MMETSP pipeline used poly-A selected mRNA libraries, very few plastid- and mitochondrial-derived sequences are present in the assembled data sets. While a small number of plastid RBH orthologs were identified, a different strategy was needed to find mitochondrial genes. MMETSP raw RNAseq reads were thus mapped to the B. natans and H. andersenii mitochondrial genomes using Bowtie2 (Langmead and Salzberg 2012). Regions of interest were identified visually using the Integrated Genomics Viewer (Robinson et al. 2011). A Python script was written to retrieve the nucleotide sequence of regions of interest and incorporate polymorphism information for each strain directly from a SAMtools pileup file (Li et al. 2009). A similar strategy was used to retrieve rRNA subunit sequences.
Substitution rates were estimated using the CODEML and BASEML programs of PAML4.8 for the analysis of protein-coding sequences and noncoding sequences, respectively (Yang 2007). For CODEML the F3 x 4 codon model was used (Goldman and Yang 1994), while for BASEML the HKY85 nucleotide substitution model was used (Tamura and Nei 1993). A Python script was written to filter dS values of any gene with dS >1 under the assumption that the strains being compared are closely related, and therefore, an elevated dS value likely indicates an error in alignment or ortholog prediction. Following this filtering step, the script calculates the mean, median, and SD of all remaining dS values. Synonymous and nonsynonymous substitution values were averaged for all pairwise comparisons of each gene and those reported for each genome are the average across all genes. Statistical analyses were performed in R (R Core Team, 2015).
Results and Discussion
Similar Relative Mutation Rates in Nucleomorph and Host Nuclear Genomes
Phylogenetic analyses have consistently shown that chlorarachniophyte and cryptophyte nucleomorph gene sequences are highly divergent relative to their nuclear homologs in other species (Keeling et al. 1999; Archibald et al. 2001; Brinkmann et al. 2005; Marinov and Lynch 2016). These observations led to the assumption that nucleomorph genomes have high mutation rates, but concrete evidence supporting this prediction is lacking. Previous work comparing amino acid distances of plastid-targeted proteins encoded in the nucleus, nucleomorph, and plastid genomes of Guillardia theta and B. natans suggested that nucleomorph-encoded proteins are more divergent than nuclear- and plastid-encoded proteins (Patron et al. 2006). A similar trend was later seen when the amino acid sequences of 26 to 28 nucleus- and nucleomorph-encoded ribosomal proteins were compared, showing much higher substitution rates for the nucleomorph proteins (Hirakawa et al. 2014). Conversely, comparisons between two cryptophyte species (Rhodomonas salina and G. theta) and the red alga Cyanidioschyzon merolae provided evidence, albeit based on a small number of sequences, that cryptophyte genes in the nucleomorph are less divergent than those in the host nucleus (Patron et al. 2006). The same study also found nuclear and nucleomorph amino acid sequence distances between C. merolae and R. salina and G. theta to be larger than those between the two cryptophytes, implying that cryptophyte nucleomorph genomes are relatively stable. Thus, the current picture of evolutionary rates in nucleomorph genomes is complex and, at times, contradictory.
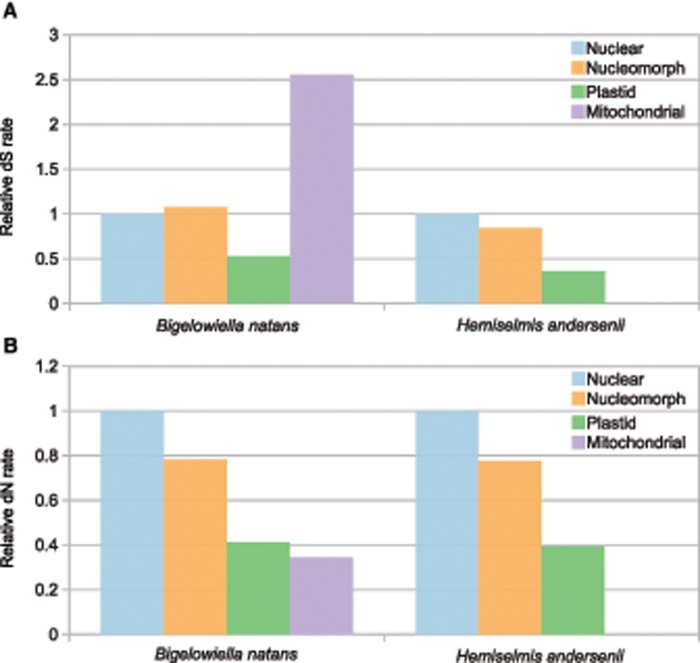
To better understand nucleomorph mutational pressures, we employed large transcriptome (RNA-Seq) data sets (Keeling et al. 2014) to measure synonymous and nonsynonymous substitution rates (dS and dN) by obtaining the average of pairwise comparisons among five strains of the chlorarachniophyte B. natans and four strains of the cryptophyte H. andersenii (table 1). Our substitution rate data encompassed ∼6,000 and 10,000 nuclear genes and 100 and 200 nucleomorph genes in B. natans and H. andersenii, respectively. We found nuclear and nucleomorph synonymous substitution rates to be similar in magnitude in both lineages (fig. 1). In H. andersenii, the average dS of the nuclear genes was slightly higher than that of the nucleomorph genes (4.80 x 10−3 vs. 4.08 x 10−3), while in B. natans the nuclear rate was slightly lower (4.99 x 10−3 vs. 5.36 x 10−3) (table 2). In both species, the SDs were large (i.e., greater than the mean), which was because most analyzed genes had a dS of zero, positively skewing the distributions. The mean dS values for nuclear and nucleomorph genomes were not significantly different in B. natans or H. andersenii (t-test P value = 0.4684 and 0.05686, respectively). Analysis of concatenated sequences gave a near-identical pattern with the nuclear dS being higher than the nucleomorph rates for both B. natans and H. andersenii (table 3). These data suggest that the host and endosymbiont nuclear genomes have experienced similar mutation rates and provide further support for the hypothesis that nucleomorph genomes are evolutionarily stable and no longer under strong mutational pressure resulting from ongoing reductive evolution.
Table 1.
Strain Numbers, Sampling Locations, and Transcriptome Sequence Data Sets for the Bigelowiella natans and Hemiselmis andersenii Strains Used in This Study
| Organism | MMETSP Data Set | CCMP Strain | Location | Latitude:Longitude |
|---|---|---|---|---|
| B. natans | 0045 | 2755 | NW of Bermuda | 34.71 : −66.5 |
| 1052 | 623 | Gulf of Maine | 43 : −69 | |
| 1054 | 1259 | Gulf of Maine | 43 : −69 | |
| 1055 | 1258 | Gulf of Maine | 43 : −69 | |
| 1358 | 1242 | NW of Bermuda | 34.72 : −66.5 | |
| H. andersenii | 0043 | 644 | Long Island, Bahamas | 23 : −75 |
| 1041 | 439 | Panama city, Florida | 29.8 : −85.67 | |
| 1042 | 1180 | Gulf of Mexico | 21 : −93 | |
| 1043 | 441 | Long Island, Bahamas | 23 : −75 |
MMETSP, Marine Microbial Eukaryote Transcriptome Sequencing Project; CCMP, Provasoli-Guillard Center for Culture of Marine Phytoplankton (now the National Center for Marine Algae and Microbiota [NCMA]).
Fig. 1.
—Relative average synonymous (A) and nonsynonymous (B) substitution rates (dS and dN) between nuclear, nucleomorph, plastid, and mitochondrial genomes of the chlorarachniophyte Bigelowiella natans and the cryptophyte Hemiselmis andersenii. Values were calculated relative to the nuclear substitution rate. Substitution rates are based on averages of pairwise comparisons between five B. natans strains and four H. andersenii strains. Missing bars indicate a value of zero or data not available.
Table 2.
Average Synonymous (dS) and Nonsynonymous (dN) Substitution Values between Five Strains of Bigelowiella natans and between Four Strains of Hemiselmis andersenii
| dS Values (×10−3) | nuDNA | nmDNA | ptDNA | mtDNA | Relative Rate Ratios (nu:nm:pt:mt) |
|---|---|---|---|---|---|
| B. natans | 4.985 (10.47) | 5.362 (17.13) | 2.623 (4.28) | 12.726 (18.06) | 1:1.08:0.53:2.55 |
| H. andersenii | 4.824 (16.58) | 4.078 (13.24) | 1.735 (4.97) | 0 | 1:0.85:0.36:0 |
| dN Values (×10−3) | nuDNA | nmDNA | ptDNA | mtDNA | Relative Rate Ratios (nu:nm:pt:mt) |
| B. natans | 2.21 (5.46) | 1.7294 (6.42) | 0.9117 (3.41) | 0.7615 (1.96) | 1:0.78:0.41:0.35 |
| H. andersenii | 1.981 (6.81) | 1.539 (4.77) | 0.781 (2.6) | 0 | 1:0.78:0.39:0 |
nu, nuclear; nm, nucleomorph; pt, plastid; mt, mitochondrial.
Note.—SDs are shown in brackets.
Table 3.
Synonymous (dS) and Nonsynonymous (dN) Substitution Values of Concatenated Gene Sequences from the Nuclear (nu), Nucleomorph (nm), Plastid (pt), and Mitochondrial (mt) DNAs of Bigelowiella natans and Hemiselmis andersenii
| dS Values (×10−3) | nuDNA | nmDNA | ptDNA | mtDNA | Relative Rate Ratios (nu:nm:pt:mt) |
|---|---|---|---|---|---|
| B. natans | 5.23 (1.94) | 4.88 (1.85) | 2.86 (2.02) | 12.94 (10.08) | 1:0.93:0.55:2.47 |
| H. andersenii | 5.117 (0.45) | 4.45 (1.24) | 3.483 (1.40) | 0 | 1:0.87:0.68:0 |
| dN values (×10−3) | nuDNA | nmDNA | ptDNA | mtDNA | Relative Rate Ratios (nu:nm:pt:mt) |
| B. natans | 2.26 (0.58) | 1.69 (0.52) | 1.19 (1.46) | 0.78 (0.64) | 1:0.75:0.53:0.35 |
| H. andersenii | 2.1 (0.20) | 1.767 (0.56) | 1.75 (0.88) | 0 | 1:0.84:0.83:0 |
Note.—SDs are shown in brackets.
We also investigated nuclear and nucleomorph substitution rates using a branch-based model, implemented through CODEML (Yang 2007). The observed Nu:Nm synonymous-site substitution rate ratios derived from a branch-based model were very similar to those obtained from the pairwise analyses: 1:1.18 for B. natans and 1:0.82 for H. andersenii.
Pairwise comparisons of strains taken from the same or different sampling locations (table 1) showed different nucleus versus nucleomorph substitution rate patterns. When looking only at B. natans and H. andersenii strains from different locations, the nucleus-to-nucleomorph dS ratio was >1, whereas for strains from the same location the ratio was <1 (fig. 2). In other words, strains from the same location showed higher relative levels of nucleotide divergence in their nmDNA than those from different locations. However, since these substitution rate differences were not statistically significant (supplementary table 1, Supplementary Material online) and little is known about the population structures and recombination frequencies of these algae, we cannot draw meaningful conclusions from this comparison. Nevertheless, the nucleus-to-nucleomorph dS ratios for strains from the same or different locations mirrored the trends when all the strains were compared, which is consistent with the idea that nuclear and nucleomorph genomes are evolving at similar rates.
Fig. 2.
—Average synonymous and nonsynonymous substitution rates (dS and dN) of nuclear, nucleomorph, plastid, and mitochondrial genomes for strains sampled at the same or different locations. Note the different y-axis ranges. (A) Pairwise comparison of Bigelowiella natans strains. (B) Pairwise comparison of Hemiselmis andersenii strains. Missing bars indicate a value of zero or data not available.
It needs to be emphasized that we cannot be certain that the strains used to measure dS in B. natans and H. andersenii are from distinct lineages or populations; indeed, many of the genes we analyzed from both B. natans and H. andersenii showed no substitutions across the entire aligned regions. If the B. natans and H. andersenii strains used in our relative rate analyses do come from the same population, we have measured and compared the relative levels of silent-site nucleotide diversity (πsilent) among the four genomes. reflects the effective population size and the mutation rate (Neμ). In this case, the effective population size of the various genetic compartments (also called the effective number of genes, Ng) should not be dramatically different. The nuclear genome of B. natans is haploid and that of H. andersenii is likely haploid as well (Hirakawa and Ishida 2014). The nucleomorph, mitochondrial, and plastid genomes in explored chlorarachniophytes and cryptophytes are polyploid and presumably uniparentally inherited (Hirakawa and Ishida 2014), but the latter two DNAs (and potentially the former) are effectively haploid (Palmer 1987). Thus, our comparative work should still provide key insights into the underlying mutation pressures impacting these genomes.
Are Chlorarachniophyte and Cryptophyte Nucleomorph Genomes Evolving at Different Rates?
The nucleomorphs of chlorarachniophytes and cryptophytes evolved from lineages of primary photosynthetic algae acquired in separate endosymbiotic events (Gould et al. 2008; Archibald 2015b). As noted earlier, comparisons of nucleomorph genomes within and between these two lineages have uncovered many remarkable examples of convergent evolution (Tanifuji et al. 2011, 2014; Moore et al. 2012; Suzuki et al. 2015). But there have also been differences in the way reductive evolution has shaped chlorarachniophyte and cryptophyte nucleomorph genomes, such as giving rise to contrasting gene and intron contents (Douglas et al. 2001; Gilson et al. 2006; Lane et al. 2007; Moore et al. 2012; Tanifuji et al. 2014). Given the similarities in nucleomorph genome architecture and levels of reductive evolution, one might expect them to display evidence of similar evolutionary pressures.
Despite having a near-identical average nucleus-to-nucleomorph dS ratio, B. natans and H. andersenii differed in the distribution of dS values between the nucleus and nucleomorph (supplementary fig. 1, Supplementary Material online). A Kolmogorov–Smirnov (KS) test comparing the nucleomorph and nuclear dS values for each lineage showed a significant difference for B. natans (KS test P value = 1.734e-05) but not for H. andersenii (KS test P value = 0.8612). This implies that there are some underlying differences in the evolutionary rates of B. natans nuclear and nucleomorph genes. The raw substitution rate values are not directly comparable between B. natans and H. andersenii because the divergence times between the various strains are unknown, but it is noteworthy that dS of the nuclear genome was more similar between the two lineages (0.0049 vs. 0.0048) than that of the nucleomorph (0.0054 vs. 0.0041). Although speculative, the higher nucleomorph dS for B. natans relative to H. andersenii could be indicative of a slightly higher mutation rate in the former. These trends are consistent with previous work showing that for B. natans amino acid sequence divergence was higher for certain nucleomorph- versus nucleus-encoded proteins, but for the cryptophyte G. theta divergence levels were similar in both compartments (Patron et al. 2006).
Analyses of B. natans ribosomal RNA (rRNA) sequences also suggest that its nucleomorph genome is evolving slightly faster than the nuclear genome: substitution rates of the concatenated rRNA genes from the nucleomorph were nearly double those of the nucleus (0.00044 vs. 0.00024) (supplementary table 2, Supplementary Material online). Conversely, not a single nucleotide substitution was found in the nuclear rRNA genes of H. andersenii, whereas the nucleomorph rRNA-coding regions had an overall substitution rate of 0.00035. Rates of nonsynonymous substitution (dN) across the nuclear and nucleomorph genomes of B. natans and H. andersenii did not follow the same pattern as the synonymous and rRNA substitution rate data. For both B. natans and H. andersenii, dN for the nucleomorph was lower than that of the nucleus, and this was true when looking at averages among genes and concatenated data sets (tables 2 and 3). This pattern could reflect high levels of purifying selection in nucleomorph genomes, potentially pointing to large evolutionary changes having taken place in the distant past. However, this idea is not necessarily supported by the comparison of the average dN and average dS because mean values can flatten the possible variation of the per-gene dN/dS ratio (see supplementary fig. S2, Supplementary Material online, for a cumulative distribution plot of dN).
Green- and Red-Algal Derived Secondary Plastid Genomes Have Low Relative Mutation Rates
Land plant plastid genomes tend to have higher synonymous substitution rates than their mitochondrial counterparts (Wolfe et al. 1987; Sloan et al. 2012), but in unicellular eukaryotes, the pattern seems to be reversed. Nearly all plastid-bearing protists examined thus far, including red algae, green algae, and species with secondary red-algal plastids, have a mitochondrion-to-plastid dS ratio ≥1, and in many cases dS in the ptDNA is lower than that of the nuDNA (Smith and Keeling 2012; Smith 2015). The reasons for these differences in relative mutation rate of mtDNA versus ptDNA are poorly understood, but are likely connected (at least in part) to differences in DNA maintenance and repair machinery (Moriyama et al. 2014; Zhang et al. 2016).
No one, however, has explored relative-rates in the organelle genomes of species with secondarily derived green algal plastids, such as B. natans, making the data presented here more interesting. Indeed, consistent with previous studies of protists, we found low relative rates of synonymous substitution in the ptDNA of B. natans: dS of the plastid was, on average, about half that of the nucleus and nucleomorph and one-fifth that of the mitochondrion (fig. 1). An almost identical trend was observed for H. andersenii for which dS of the ptDNA was ∼0.5 times that of the nuclear and nucleomorph DNAs.
Substitution rates in the rRNA-coding regions from the B. natans plastid were also low (between a half and one-twentieth of those of the nucleus, nucleomorph, mitochondrion), again, indicating that the plastid genome has a lower mutation rate than its neighboring genomes. Unfortunately, a similar comparison was not possible for H. andersenii as plastid rRNA data were not available. Low relative rates of nonsynonymous substitution were also observed in the plastid genomes of B. natans and H. andersenii (fig. 1), mirroring the dS data. Altogether, these results provide additional evidence that plastid genomes have low relative mutation rates across a wide diversity of photosynthetic organisms, now including species with green-algal derived secondary plastids, and reinforce the notion that land plants are outliers in terms of their relative plastid mutational rate patterns.
Extreme Differences in mtDNA Substitution Rates between Chlorarachniophytes and Cryptophytes
The nucleomorph and plastid genomes of chlorarachniophytes and cryptophytes are derived from green and red algae, respectively—organisms for which we have some expectations of relative substitution rates based on previous studies (Popescu and Lee 2007; Hua et al. 2012; Smith et al. 2012). However, the mitochondrial genomes of chlorarachniophytes and cryptophytes belong to the host lineages for which there is limited knowledge, as exemplified by the fact that the first nuclear genome sequences from these two lineages were described recently (Curtis et al. 2012). The mtDNAs of B. natans and H. andersenii have also been sequenced (Kim et al. 2008; Tanifuji et al. 2016), but until now little was known about substitution rates in these genomes. It has been proposed that mitochondrial genome architecture may be related to mutational patterns (Smith and Keeling 2015). Mitochondrial genomes are known to be highly variable in architecture, which is also true for plastid genomes, but to a lesser degree (Smith and Keeling 2015). Accordingly, mutation rate estimates for mitochondrial genomes are more variable and have a broader range than those of plastid genomes, which, as already mentioned, is thought to be related to differences in the underlying DNA maintenance and repair pathways of mitochondria relative to plastids (Smith and Keeling 2015).
Our analyses of the H. andersenii mtDNA revealed not one nucleotide substitution among the four isolates, contrasting the high levels of substitution in the B. natans mitochondrial genome, and suggesting that these two lineages have contrasting mtDNA mutational patterns. This finding is made more remarkable by the fact that dS for the nucleus, nucleomorph, and plastid genomes were very similar between B. natans and H. andersenii (tables 2 and 3). The lack of observed mtDNA sequence divergence in H. andersenii can be interpreted in various ways. The mitochondrial mutation rate for H. andersenii might be genuinely lower than those of the nucleus, nucleomorph, and plastid. Alternatively, we might not have sampled enough mitochondrial genes to get an accurate picture of the synonymous substitution rate. And there is always the possibility that the synonymous sites we analyzed are not neutral, and are under selective constraints. Why this would be true for the mtDNA of H. andersenii and not the other genomes of this organism, or for the mtDNA of B. natans, is not obvious.
Again, dS in the B. natans mtDNA was 2.5–5 times higher than the averages of the other three genomes in this organism (table 2). This is in line with relative-rate data from other secondary red-algal containing lineages (i.e., haptophytes and heterokonts), where mitochondrial substitution rates were predicted to be 5–30 times those of the plastid and thought to be connected to mtDNA genomic contraction (Smith and Keeling 2012). However, despite having the highest relative synonymous substitution rate, the B. natans mtDNA had the lowest relative dN among the four genomes, potentially reflecting strong purifying selection within the mitochondrion. There is reason to believe that the B. natans mitochondrial genome has undergone rapid evolution: it uses alternate initiation codons, and displays a low degree of gene synteny and gene content within mtDNAs from other chlorarachniophyte species (Tanifuji et al. 2016). Thus, it is possible that, like our prediction for the nucleomorph genomes, rapid evolution of the mtDNA occurred long ago when selection pressures were diminished, such as during the initial phase of secondary endosymbiont integration. Overall, our data point to a trend of a high mutation rate in the B. natans mitochondrial genome that is countered by strong purifying selection.
Conclusions
Genome-wide substitution rate data were used to estimate relative mutation rates in all four genomes within chlorarachniophyte and cryptophyte algae. On average, the relative synonymous substitution rates were similar between the nuclear and nucleomorph genomes in these two algal lineages. This suggests that in B. natans and H. andersenii the nucleomorph and nuclear genomes have similar mutation rates—a finding that appears to be at odds with the fact that nmDNAs display high levels of sequence divergence among lineages (Keeling et al. 1999; Archibald et al. 2001; Brinkmann et al. 2005; Marinov and Lynch 2016). Given these seemingly conflicting data, we argue that rapid evolutionary changes occurred in these endosymbiotically derived genomes in the distant evolutionary past but are no longer underway. There is some evidence that the B. natans nucleomorph genome mutation rate is slightly elevated relative to the nuclear rate and potentially evolving more rapidly than the nucleomorph genome of the cryptophyte H. andersenii, but the difference in rates is small, and in some instances not statistically significant. This is in line with previous work (Patron et al. 2006) suggesting that cryptophyte nucleomorph genomes are evolving more conservatively than those of chlorarachniophytes. We have provided the first evidence that secondary green algal plastids have low relative substitution rates, following the patterns seen in secondary red plastids and primary plastids outside the land plant lineage (Smith 2015). Relative mitochondrial substitution rates in B. natans were high, while those in H. andersenii were low. Our results run counter to the assumption that the highly divergent nucleomorph genomes of chlorarachniophytes and cryptophytes have high mutation rates; the data are instead more consistent with a burst of nucleomorph genome sequence divergence taking place as their respective secondary endosymbionts were being “converted” into organelles, followed by a reduction in evolutionary rates. It will be important to compare substitution rates in a much larger diversity of chlorarachniophytes and cryptophytes to see if these patterns hold in all members of these lineages.
It should be stressed that synonymous sites can be under various selective constraints (Chamary et al. 2006; Meiklejohn et al. 2007), and factors such as compositional biases and codon usage can impact dS and the assumption of neutrality (Popescu and Lee 2007). In this context, it will be particularly interesting to have substitution rate data from the intergenic regions of the different genomes of these lineages for gauging the underlying mutational pressures. Finally, it remains to be determined whether the various B. natans and H. andersenii strains employed here represent members of distinct populations or individuals from the same population.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Associate Editor and three reviewers for helpful feedback on earlier versions of this manuscript. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellowship to C.J.G. (PDF 417789-2015), and NSERC Discovery Grants to D.R.S. (RGPIN 435173-2013) and J.M.A. (RGPIN 05871-2014).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Cavalier-Smith T, Maier U, Douglas S.. 2001. Molecular chaperones encoded by a reduced nucleus: the cryptomonad nucleomorph. J Mol Evol. 52(6):490–501. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2007. Nucleomorph genomes: structure, function, origin and evolution. Bioessays 29(4):392–402. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2009. The puzzle of plastid evolution. Curr Biol. 19(2):R81–R88. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2015a. Endosymbiosis and eukaryotic cell evolution. Curr Biol. 25(19):R911–R921. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2015b. Genomic perspectives on the birth and spread of plastids. Proc Natl Acad Sci U S A. 112(33):10147–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, et al. 1998. Predicting function: from genes to genomes and back. J Mol Biol. 283(4):707–725. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, et al. 2005. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol. 54(5):743–757. [DOI] [PubMed] [Google Scholar]
- Brown WM, George M, Wilson AC.. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 76(4):1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2003. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos Trans R Soc Lond B Biol Sci. 358(1429):109–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD.. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 7(2):98–108. [DOI] [PubMed] [Google Scholar]
- Curtis BA, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492(7427):59–65. [DOI] [PubMed] [Google Scholar]
- Douglas SE, et al. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410(6832):1091–1096. [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J.. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49(3):827–831. [DOI] [PubMed] [Google Scholar]
- Gilson PR, et al. 2006. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci U S A. 103(25):9566–9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang ZA.. 1994. Codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI.. 2008. Plastid evolution. Annu Rev Plant Biol. 59:491–517. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 4(9):a011403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Ishida KI.. 2014. Polyploidy of endosymbiotically derived genomes in complex algae. Genome Biol Evol. 6(4):974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Suzuki S, Archibald JM, Keeling PJ, Ishida K.. 2014. Overexpression of molecular chaperone genes in nucleomorph genomes. Mol Biol Evol. 31(6):1437–1443. [DOI] [PubMed] [Google Scholar]
- Hua J, Smith DR, Borza T, Lee RW.. 2012. Similar relative mutation rates in the three genetic compartments of Mesostigma and Chlamydomonas. Protist 163(1):105–115. [DOI] [PubMed] [Google Scholar]
- James JE, Piganeau G, Eyre-Walker A.. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, et al. 1999. The secondary endosymbiont of the cryptomonad Guillardia theta contains alpha-, beta-, and gamma-tubulin genes. Mol Biol Evol. 16(9):1308–1313. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12(6):e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, et al. 2008. Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae). BMC Genomics 9:215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Lane CE, et al. 2007. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci U S A. 104(50):19908–19913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. ; 1000 Genome Project Processing Subgroup. 2009. The sequence alignment/map format SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S.. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311(5768):1727–1730. [DOI] [PubMed] [Google Scholar]
- Marinov GK, Lynch M.. 2016. Conservation and divergence of the histone code in nucleomorphs. Biol Direct. 11(1):18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM.. 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23(6):259–263. [DOI] [PubMed] [Google Scholar]
- Mikita S, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CE, Curtis B, Mills T, Tanifuji G, Archibald JM.. 2012. Nucleomorph genome sequence of the cryptophyte alga Chroomonas mesostigmatica CCMP1168 reveals lineage-specific gene loss and genome complexity. Genome Biol Evol. 4(11):1162–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Tajima N, Sekine K, Sato N. 2014. Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae. Genome Biol Evol. 6(1):228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Glémin S, Galtier N.. 2008. Strong variations of mitochondrial mutation rate across mammals – the longevity hypothesis. Mol Biol Evol. 25(1):120–130. [DOI] [PubMed] [Google Scholar]
- Palmer JD. 1987. Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. Am Nat. 130:S6–29. [Google Scholar]
- Patron NJ, Rogers MB, Keeling PJ.. 2006. Comparative rates of evolution in endosymbiotic nuclear genomes. BMC Evol Biol. 6:46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu CE, Lee RW.. 2007. Mitochondrial genome sequence evolution in Chlamydomonas. Genetics 175(2):819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. 2015. R: a language and environment for statistical computing Vienna (Austria: ): R Foundation for Statistical Computing. http://www.R-project.org/. [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD.. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative Genomics Viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10(1):e1001241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Taylor DR.. 2012. Evolutionary rate variation in organelle genomes: the role of mutational processes In: Bullerwell CE, editor. Organelle genetics. Heidelberg (Germany: ): Springer-Verlag; p. 123–146. [Google Scholar]
- Smith DR. 2015. Mutation rates in plastid genomes: they are lower than you might think. Genome Biol Evol. 7(5):1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hua J, Lee RW, Keeling PJ.. 2012. Relative rates of evolution among the three genetic compartments of the red alga Pophyra differ from those of green plants and do not correlate with genome architecture. Mol Phylogenet Evol. 65(1):339–344. [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ.. 2012. Twenty-fold difference in evolutionary rates between the mitochondrial and plastid genomes of species with secondary red plastids. J Eukaryot Microbiol. 59(2):181–184. [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ.. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 112(33):10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Shirato S, Hirakawa Y, Ishida K.. 2015. Nucleomorph genome sequences of two chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata. Genome Biol Evol. 7(6):1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, Archibald JM, Hashimoto T.. 2016. Comparative genomics of mitochondria in chlorarachniophyte algae: endosymbiotic gene transfer and organelle genome dynamics. Sci Rep. 6:21016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanifuji G, et al. 2011. Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol Evol. 3:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanifuji G, et al. 2014. Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics 15:374.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ.. 1997. A genomic perspective on protein families. Science 278(5338):631–637. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM.. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 84(24):9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Coevolution between Nuclear-Encoded DNA Replication, Recombination, and Repair Genes and Plastid Genome Complexity. Genome Biol Evol. 8(3):622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Jain K, Mower JP.. 2014. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 31(5):1228–1236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.