Abstract
South Asia has a complex history of migrations and is characterized by substantial pigmentary and genetic diversity. For this reason, it is an ideal region to study the genetic architecture of normal pigmentation variation. Here, we present a meta-analysis of two genome-wide association studies (GWASs) of skin pigmentation using skin reflectance (M-index) as a quantitative phenotype. The meta-analysis includes a sample of individuals of South Asian descent living in Canada (N = 348), and a sample of individuals from two caste and four tribal groups from West Maharashtra, India (N = 480). We also present the first GWAS of iris color in South Asian populations. This GWAS was based on quantitative measures of iris color obtained from high-resolution iris pictures. We identified genome-wide significant associations of variants within the well-known gene SLC24A5, including the nonsynonymous rs1426654 polymorphism, with both skin pigmentation and iris color, highlighting the pleiotropic effects of this gene on pigmentation. Variants in the HERC2 gene (e.g., rs12913832) were also associated with iris color and iris heterochromia. Our study emphasizes the usefulness of quantitative methods to study iris color variation. We also identified novel genome-wide significant associations with skin pigmentation and iris color, but we could not replicate these associations due to the lack of independent samples. It will be critical to expand the number of studies in South Asian populations in order to better understand the genetic variation driving the diversity of skin pigmentation and iris color observed in this region.
Keywords: skin pigmentation, iris color, genome-wide association study, South Asia
Introduction
The South Asian continent is characterized by extensive linguistic and genetic diversity. Four different linguistic families (Indo-European, Dravidian, Tibeto-Burman, and Austro-Asiatic) are present in this region. Indo-European languages are spoken in the North and Central regions and Dravidian speakers are concentrated in the Southern states of India. The Tibeto-Burman and Austro-Asiatic languages have a more restricted geographic distribution, that is, the Northeast and Central India (Cordaux et al. 2004; Reddy et al. 2010; Chaubey et al. 2011). Recent genetic studies have shown that the genetic diversity in India fits very well with a model of mixture between two ancestral populations, the Ancestral Northern Indians (ANI), which are genetically close to Middle Easterners, Central Asians and Europeans, and Ancestral South Indians (ASI) (Reich et al. 2009). ANI ancestry is higher in Indo-European speakers than in Dravidian speakers, and is also higher in upper castes than in lower or middle caste groups (Reich et al. 2009). Not surprisingly, given the aforementioned linguistic and genetic diversity, a substantial amount of variation in pigmentary phenotypes, such as skin and hair pigmentation and iris color has been described in South Asia (Das and Mukherjee 1963; Jaswal 1979, 1983; Basu Mallick et al. 2013; Edwards et al. 2016; Jonnalagadda, Norton, et al. 2016; Jonnalagadda, Ozarkar, et al. 2016; Mishra et al. 2017; Norton et al. 2016). Some of the recent studies have tested the association of variants in pigmentation candidate genes with pigmentary traits in South Asian populations (Basu Mallick et al. 2013; Edwards et al. 2016; Jonnalagadda, Norton, et al. 2016; Mishra et al. 2017; Norton et al. 2016). However, to date only one genome-wide association study (GWAS) of skin pigmentation has been carried in South Asian populations. Stokowski et al. (2007) analyzed a sample of individuals of South Asian descent living in the United Kingdom, and reported associations of variants within the SLC24A5, SLC45A2, and TYR genes with skin-reflectance measures.
In this article, we present a meta-analysis of two GWAS of skin pigmentation using skin reflectance (M-index) as a quantitative phenotype. The meta-analysis includes a sample of individuals of South Asian descent living in Canada, and a sample of individuals from two caste and four tribal groups from West Maharashtra, India. We also present the first GWAS of iris color in South Asian populations, which is based on quantitative measures of iris color obtained from high-resolution iris pictures of the individuals of South Asian ancestry recruited in Canada. Iris color was quantified using the L*, a*, and b* coordinates of the CIELab color space, and we also measured the difference in iris color between the pupillary and ciliary regions of the iris (e.g., iris heterochromia).
Materials and Methods
Samples
South Asian Sample from Canada
Between 2012 and 2014, 348 healthy volunteers of South Asian ancestry participated in a research study on human pigmentation variation. All participants ranged between 18 and 35 years of age and were recruited using online and print advertisements directed toward the University of Toronto student community. A personal questionnaire was administered to each participant to determine their age, sex, self-described eye color, and whether or not they had been diagnosed with any pigmentation-related diseases or disorders. Biogeographical ancestry was determined using information from the personal questionnaire, which inquired about the ancestry, place of birth, and first language of each participant’s maternal and paternal grandparents. Individuals who stated that all of their grandparents originated in Pakistan, India, Bangladesh, or Sri Lanka were categorized as South Asian. When information about the grandparents was not known, the self-described ancestry of both parents was used to assess biogeographical ancestry.
Skin and hair pigmentation were quantitatively measured using the DSM II Dermaspectrometer (Cortex Technologies, Hadsund, Denmark). Measurements were taken three times on the inner skin of the upper right arm and reported as Melanin (M) index. If participants wore contact lenses they were asked to remove these before having iris photographs taken. High-resolution photographs of the right iris of each study subject were taken with a Fujifilm Finepix S3 Pro 12-megapixel DSLR mounted on a Nikkor 105-mm macro lens. To control for lighting and exposure, photographs were taken in the same room with a coaxial biometric illuminator to deliver a constant and uniform source of light to each iris at 5,500 K (D55 illuminant). All photographs were taken under the same setting conditions, with an aperture of f/19, exposure sensitivity (ISO) set at 200 and a shutter speed of 1/125 s. Photographs were then stored as both 12 megapixel jpeg and RAW formats for analysis. Iris pigmentation was digitally scored using a custom program designed to crop out both the pupil and sclera to retain only the iris. A wedge of the iris was then extracted, and color scores in CIELab coordinates were calculated from the pupillary and ciliary zones. In addition to the L*, a*, and b* coordinates for the iris wedge, the program calculated the parameter delta, which describes color differences in the pupillary and ciliary regions of the iris. Detailed information about this program has been described in Edwards et al. (2016).
South Asian Sample from West Maharashtra, India
The samples from India represent six tribe and caste populations collected from West Maharashtra, which have been described in a prior study (Jonnalagadda, Ozarkar, et al. 2016). Each participant’s age, sex, native place, and clan was recorded along with 5–8 ml whole blood, which was collected in EDTA vials. Genomic DNA was extracted using phenol–chloroform extraction method (Sambrook et al. 1989), and was checked for its quality on a 1% agarose gel. The DNA samples were quantified using the Eppendorf BioPhotometer plus. The final number of samples genotyped was 480.
Constitutive skin pigmentation was measured quantitatively using the DSM II Colormeter (Cortex Technology, Denmark) and recorded in the form of Melanin Index (MI) measures with higher MI values representing darker pigmentation. Three measurements were recorded on the inner surface of both upper arms and were averaged to yield a mean MI value for each study participant.
Genotyping, Phasing, and Imputation
South Asian Sample from Canada
Genotyping was carried out with Illumina’s Infinium Multi-Ethnic Global Array (MEGA) at the Clinical Genomics Centre (Mount Sinai Hospital, Toronto, Ontario, Canada) using standard protocols. The MEGA array, which includes ∼1.7 million markers, was designed to capture common genome variation in diverse population groups. Four samples were included as blind duplicates, and the concordance rate was in all samples >99.99%. We used the program GenomeStudio to carry out the basic QC steps recommended by Illumina. After this initial QC step, ∼1.4 million were retained for further analyses. The number of autosomal markers included was ∼1.36 million. We performed additional QC steps to remove samples and markers, according to the following criteria. Sample QC involved: 1/removal of samples with missing call rates <0.9, 2/removal of samples that were outliers in Principal Component Analysis (PCA) plots, 3/removal of samples with sex discrepancies, 4/removal of samples that were outliers for heterozygosity, and 5/removal of related individuals (pi-hat > 0.2). Likewise, marker QC involved: 1/removal of markers with genotype call rate <0.95, 2/removal of markers with Hardy–Weinberg P values <10−6, 3/removal of Insertion/Deletion (Indel) markers, 4/removal of markers with allele frequencies <0.01, 5/removal of markers not present in the 1000 Genomes reference panel, or that do not match on chromosome, position, and alleles, 6/removal of A/T or G/C SNPs with MAF >40% in the 1000 Genomes South Asian reference samples, and 7/removal of SNPs with allele frequency differences >20% between the study sample and the 1000 Genomes South Asian reference sample. After these QC steps, we retained 333 samples and 640,625 markers. After performing the QC steps described earlier, the samples were phased using the program SHAPEIT2 and imputed at the Sanger Imputation Service, using the Positional Burrows-Wheeler Transform (PBWT) algorithm (Durbin 2014) and the samples of the 1000 Genomes as reference haplotypes.
South Asian Sample from West Maharashtra, India
Genotyping was carried out with Applied Biosystem’s Axiom TM Precision Medicine Research Array (PMRA) at the Imperial Life Sciences Pvt Ltd. Laboratory (Gurgaon, Haryana, India) using standard protocols. The PMRA array includes ∼900,000 markers and was designed to capture common genome variation in diverse population groups. The program Axiom Analysis Suite was used to carry out basic QC steps. After this initial QC step, ∼522,125 polymorphic markers and 478 samples were retained for further analyses.
We performed additional QC steps to remove samples and markers, according to the following criteria. Sample QC involved: 1/removal of samples with sex discrepancies 2/removal of samples that were outliers for heterozygosity, 3/removal of samples with missing call rates < 0.95, 4/removal of related individuals (pi-hat > 0.25), 5/removal of samples that were outliers in Principal Component Analysis (PCA) plots. Marker QC involved: 1/removal of markers with genotype call rate < 0.95, 2/removal of markers with Hardy–Weinberg P values < 10−6, 3/removal of markers with minor allele count < 4, 4/removal of Insertion/Deletion (Indel) markers, 5/removal of markers not present in the 1000 Genomes reference panel, or that do not match on chromosome, position, and alleles, 6/removal of A/T or G/C SNPs with MAF > 40% in the 1000 Genomes South Asian reference samples, and 7/removal of SNPs with allele frequency differences > 20% between the study sample and the 1000 Genomes South Asian reference sample. After these QC steps, we retained 456 samples and 398,118 autosomal markers. After performing the QC steps described earlier, the samples were phased using the program SHAPEIT2 (and imputed at the Sanger Imputation Service, using the PBWT algorithm; Durbin 2014), and the samples of the 1000 Genomes as reference haplotypes.
Statistical Analyses
South Asian Sample from Canada
As a first step of the statistical analyses, we carried out a linear regression with M-index values as the dependent variable, and sex and the first four Principal Component Axis as independent variables and saved the standardized residuals. A similar process was carried out for the L*, a*, b*, and delta iris values, although in this case, given that two different camera bodies were used for taking the pictures, camera body was also used as independent variable. The unstandardized residuals were transformed using the rank-based inverse normal transformation. The M value residuals and the L*, a*, b*, and delta transformed residuals were used as input for the association tests with the program SNPTEST v2 (Marchini and Howie 2010), using an additive model and the expected test (e.g., using genotype dosages) in order to control for genotype uncertainty. For the L*, a*, b* coordinates that define iris color, we also run a Bayesian Multiple Phenotype test implemented in the program SNPTEST (-mpheno option). This test evaluates the three coordinates jointly and provides a log10 Bayes Factor reporting the ratio of two probabilities: the probability of the data under an unconstrained model (M1), and the probability of the data under a null model (M0) in which there is no effect. For example, a log10 Bayes Factor of 3 indicates that the probability of the data under the model M1 is 1,000-fold higher than the probability of the data under the null model with no genotype effects. Of the 333 samples that were retained after the postgenotyping QC step, some samples had missing phenotype data. The final number of samples with valid skin pigmentation data was 264, and the final number of samples with valid iris color data was 329. Our associations identified a strong effect of the well-known loci SLC245A5 on skin pigmentation. For this reason, additional statistical analyses were done conditioning for rs1426654 (skin pigmentation).
South Asian Sample from West Maharashtra, India
For the samples from the India, we carried out a linear regression with M-index values as the dependent variable and sex, age and the first ten Principal Component Axis as independent variables and saved the unstandardized residuals. As the residuals showed deviations from normality, they were transformed using a rank-based inverse normal transformation. These transformed residuals were used as input for the association tests using the program SNPTEST v2 (Marchini and Howie 2010), using an additive model and the expected test (e.g., using genotype dosages) in order to control for genotype uncertainty. Our association tests identified a very strong effect of the well-known SLC24A5 region on skin pigmentation. For this reason, we carried out a second statistical analysis conditioning for the rs1426654 genotypes.
Meta-Analysis of Association Results
A meta-analysis was conducted as genotyping of samples from Canada and West Maharashtra were performed on 2 different chips, namely the Multi-Ethnic Global Array (MEGA) and the Precision Medicine Research Array (PMRA), respectively. The summary statistics from the two GWAS were used to run a meta-analysis using the program METASOFT (Han and Eskin 2011, 2012). This program implements a fixed effects model based on inverse-variance-weighted effect size and also Han and Eskin’s Random effects model (RE2), which has been shown to have more statistical power to detect associations under heterogeneity than the conventional random effects model based on inverse-variance-weighted effect size (Han and Eskin 2011). Additionally, METASOFT provides estimates of the posterior probability that an effect exists in each study (M values; Han and Eskin 2012). Small M values indicate that the study is predicted to not have an effect. Large M values indicate that the study is predicted to have an effect. Intermediate M values indicate ambiguous results.
Annotation of Genome-Wide Association Signals Identified in the Meta-Analysis
The genome-wide association signals identified in the meta-analysis were annotated using the SNPnexus website (SNP Annotation Tool. http://snp-nexus.org/, last accessed 2019 Feb 19). This site provides numerous annotations, including potential effects on protein function (SIFT and PolyPhen), conservation scores (phastCons, GERP), and a range of scores for noncoding variants (CADD, fitCons, EIGEN, FATHMM, GWAVA, DeepSEA, FunSeq2, and ReMM). We also explored potential regulatory effects in HaploReg v4.1. (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php/), last accessed 2018 Jun 05), and Regulome DB (http://www.regulomedb.org/, last accessed June 5, 2018).
Results
Distribution of Pigmentary Traits
Supplementary figure S1 a–f, Supplementary Material online, shows the distribution of M-index values for the South Asian sample from Canada and from West Maharashtra, as well as the L*, a*, b*, and delta values for iris color in the Canadian South Asian sample, respectively. In the sample of West Maharashtra, there are significant differences in skin pigmentation measures between the six populations included in the study (P value < 0.001). The mean M-index values range from 43.02 in the Deshastha Brahmin caste population to 58.83 in the Warli tribal population. A detailed description of the distribution of pigmentation in these samples is available in (Jonnalagadda, Ozarkar, et al. 2016).
Correlation of Skin Pigmentation and Iris Color Measures in Canadian South Asian Sample
Supplementary figure S2 a–d, Supplementary Material online, depict the correlations observed between M-index values capturing constitutive pigmentation variation, and L*, a*, b*, and delta values for iris color in the Canadian South Asian sample. Significant correlations were observed between M-index and all the iris color measures. The correlation coefficients range from 0.4 (M-index and b*) to 0.218 (M-index and delta).
Population Structure
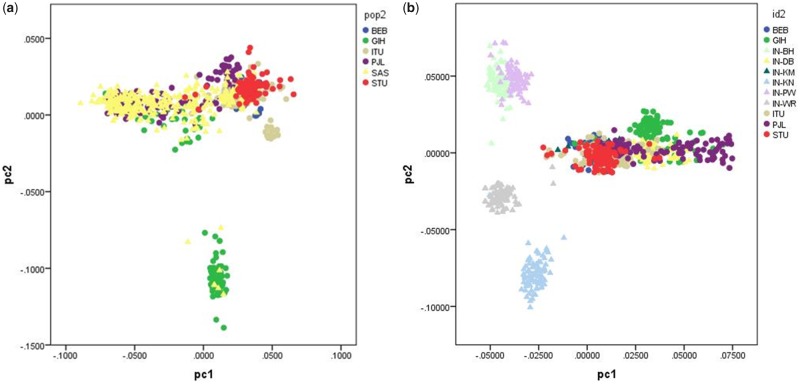
In order to evaluate population structure, we merged the genotype data of each sample with the genotype data of the South Asian 1000 Genome Project samples. The South Asian sample from Canada and the West Maharashtra sample were genotyped with different arrays (Illumina’s MEGA array vs. Affy’s Axiom Precision Medicine Array), and the overlap of markers between these two arrays is very limited, so we carried out these analyses independently. We then carried out a Principal Component Analysis (PCA) with the program PLINK, after pruning markers based on LD patterns (r2>0.1). The resulting PCA plots (axes 1 and 2) for the South Asian sample from Canada and the West Maharashtra sample are presented in figure 1a and b, respectively. As expected based on the broad ancestral origins of the South Asian participants recruited in Canada, there is a large degree of overlap with the South Asian 1000 Genomes samples. However, in the West Maharashtra sample, the two caste groups Deshastha Brahmin and Kunbi Maratha overlap with the 1000 Genomes samples, but the four tribal groups occupy different positions in the PCA space. The PCA plot indicates close genetic affinities between the Pawara and Bhil tribal groups, as per previous reports (Jonnalagadda et al. 2013).
Fig. 1.
—PCA Plots for the South Asian sample from Canada and the West Maharashtra sample.
GWAS of Skin Pigmentation in the South Asian Sample from Canada
Supplementary figure S3 a, Supplementary Material online, shows the Manhattan plot depicting the results of the GWAS of skin pigmentation in the South Asian sample from Canada. The QQ plot is depicted in supplementary figure S3b, Supplementary Material online. Supplementary table S1, Supplementary Material online, shows the genome-wide significant (P < 5×10−8) and suggestive signals (P < 10−5) identified in this analysis. The only genome-wide significant signal was observed in the SLC24A5 region, and the lead SNP is the nonsynonymous variant rs1426654 (P = 4.64×10−14). We repeated the association tests conditioning on this variant. The Manhattan plots and QQ plots corresponding to this analysis are depicted in supplementary figure S3c and d, Supplementary Material online, and the markers with suggestive P values are listed in supplementary table S2, Supplementary Material online. No genome-wide significant signals were identified after conditioning for rs1426654.
GWAS of Skin Pigmentation in the South Asian Sample from West Maharashtra
Supplementary figure S3 e, Supplementary Material online, shows the Manhattan plot depicting the results of the GWAS of skin pigmentation in the sample from India. The QQ plot is depicted in supplementary figure S3f, Supplementary Material online. Supplementary table S3, Supplementary Material online, reports the genome-wide significant (P < 5×10−8) and suggestive signals (P < 10−5) identified in this analysis. A genome-wide significant result was observed in the SLC24A5 region, again with rs1426654 identified as the lead SNP (P = 1.25×10−23). No other genome-wide significant signals were identified in this study, as well as the tests conditioning for rs1426654 (supplementary fig. S3g and h and table S4, Supplementary Material online).
GWAS of Iris Color in the South Asian Sample from Canada
Supplementary figure S4 a–d, Supplementary Material online, show the Manhattan plots corresponding to the GWAS of the L*, a*, and b* dimensions of the CIELab color space, as well as the delta value that captures the difference in pigmentation between the pupillary and ciliary regions of the iris. Supplementary figure S4e–h, Supplementary Material online, depicts the respective QQ plots. Table 1 reports the markers that reached genome-wide significance in these analyses. Supplementary tables S7–S10, Supplementary Material online, list both the genome-wide significant and suggestive regions identified in these analyses. In each table, we provide the P values observed for all the iris pigmentary measures, as well as the Bayesian multiple phenotype tests implemented in SNPTEST. Genome-wide significant signals were identified in the HERC2 region for L* and delta. For both measures, the top SNPs are rs12898729 (P value L* = 2.54×10−14; P value delta = 4.25×10−11) and rs12913832 (P value L* = 5.52×10−14; P value delta= 5.08×10−11). For b*, a genome-wide significant signal was observed in the gene SLC24A5, and the lead SNP is the nonsynonymous rs1426654 SNP (P value = 8.49×10−9). For a*, a genome-wide significant signal was identified in an intergenic region on chromosome 10 (lead SNP rs28634972, P = 3×10−8). Additionally, a region near the ZNF804A gene on chromosome 2 was very close to genome-wide significance in the original analysis, and it reached genome-wide significance after conditioning for rs12913832 and rs1426654. The lead SNP in this region is rs359899 (P value = 5.7×10−8, P value after conditioning = 1.77×10−8). All these regions are also supported by very low P values for other iris pigmentation measures, as well as the multiple phenotype tests, which in all cases have log10 Bayes factors >3. The regional plots for all these regions are depicted in supplementary figure S5a–e, Supplementary Material online.
Table 1.
Genome-Wide Significant Signals Observed in GWAS of Iris Color in the Canadian South Asian Sample
|
GWAS-L-iris | ||||||||||||||
| Rsid | chr | Pos | NEA | EA | EAF | info | P value | Beta | SE | Gene | a-iris | b-iris | Delta-iris | Iris-mpheno |
| rs12898729 | 15 | 28392261 | G | A | 0.128 | 0.867 | 2.54E-14 | 0.900 | 0.113 | HERC2 | 0.704 | 5.74E-07 | 4.25E-11 | 11.721 |
| rs12913832 | 15 | 28365618 | A | G | 0.115 | 0.939 | 5.52E-14 | 0.904 | 0.115 | HERC2 | 0.678 | 4.66E-07 | 5.08E-11 | 11.328 |
| rs12916300 | 15 | 28410491 | C | T | 0.144 | 0.832 | 5.69E-14 | 0.880 | 0.112 | HERC2 | 0.423 | 1.52E-07 | 1.91E-10 | 10.932 |
| GWAS-a-iris | ||||||||||||||
| Rsid | chr | Pos | NEA | EA | EAF | info | P value | Beta | SE | Gene | L-iris | b-iris | Delta-iris | Iris-mpheno |
| rs28634972 | 10 | 126569229 | C | G | 0.250 | 0.940 | 3.11E-08 | −0.513 | 0.090 | 0.282 | 9.21E-05 | 0.211 | 4.841 | |
| rs359899 | 2 | 185448231 | A | C | 0.155 | 0.973 | 5.70E-08 | 0.620 | 0.111 | 2.87E-04 | 3.45E-06 | 0.180 | 3.293 | |
| GWAS-b-iris | ||||||||||||||
| Rsid | chr | Pos | NEA | EA | EAF | info | P value | Beta | SE | Gene | L-iris | a-iris | Delta-iris | Iris-mpheno |
| rs1426654 | 15 | 48426484 | A | G | 0.150 | 0.956 | 8.49E-09 | −0.594 | 0.100 | SLC24A5 | 9.28E-04 | 2.16E-06 | 0.016 | 4.050 |
| GWAS-delta-iris | ||||||||||||||
| Rsid | chr | Pos | NEA | EA | EAF | info | P value | Beta | SE | Gene | L-iris | a-iris | b-iris | Iris-mpheno |
| rs12898729 | 15 | 28392261 | G | A | 0.128 | 0.867 | 4.25E-11 | 0.788 | 0.115 | HERC2 | 2.54E-14 | 0.704 | 5.74E-07 | 11.721 |
| rs12913832 | 15 | 28365618 | A | G | 0.115 | 0.939 | 5.08E-11 | 0.798 | 0.117 | HERC2 | 5.52E-14 | 0.678 | 4.66E-07 | 11.328 |
| rs1129038 | 15 | 28356859 | C | T | 0.110 | 0.996 | 5.92E-11 | 0.794 | 0.117 | GALNT12 | 6.37E-14 | 0.733 | 2.02E-06 | 10.951 |
aGenome-wide significant after conditioning for rs12913832 and rs1426654 (P = 1.77E-8).
Meta-Analysis of Skin Pigmentation GWAS
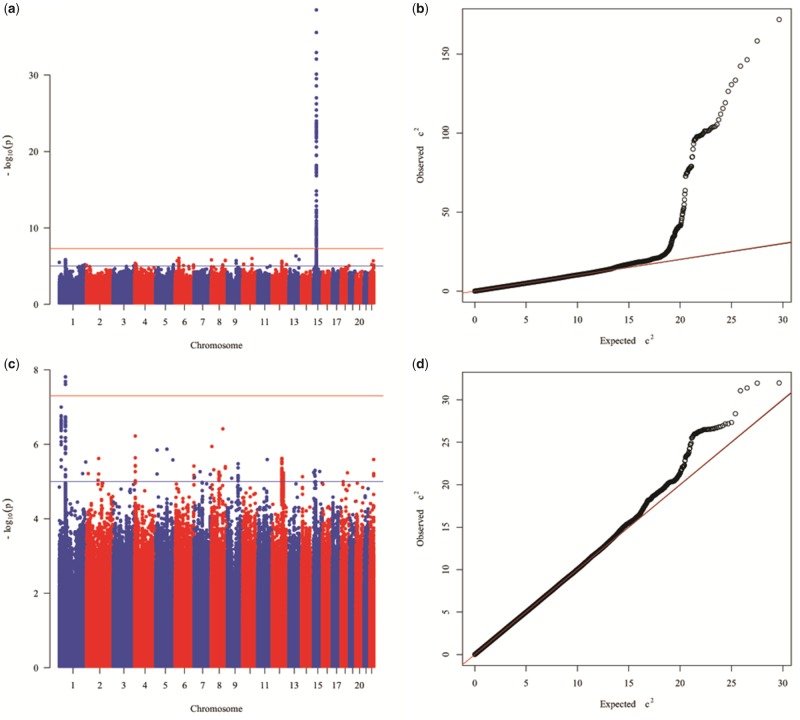
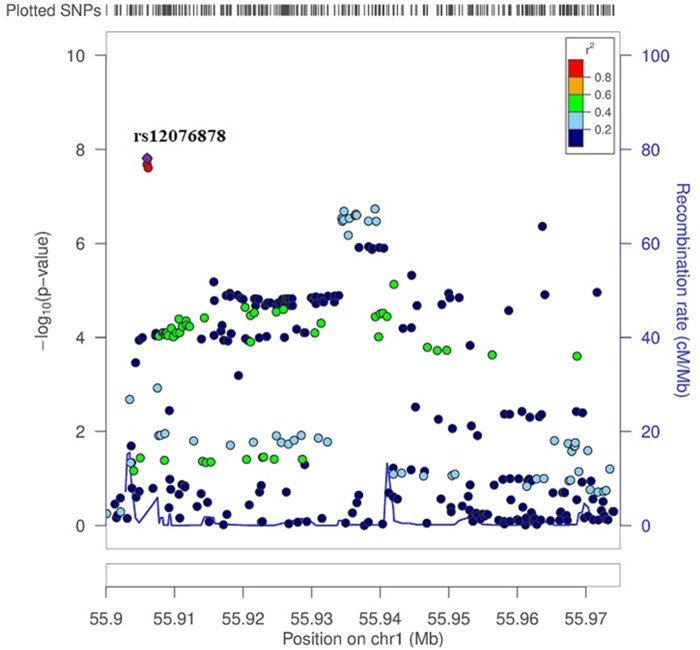
We carried out a meta-analysis of the association results of the South Asian sample from Canada and from West Maharashtra. The Manhattan plot showing the results of the meta-analysis is shown in figure 2a and the QQ plot in figure 2b. Supplementary table S5, Supplementary Material online, provides information about the genome-wide significant and suggestive signals identified in the meta-analysis, including P values, estimates of between-study heterogeneity (Cochran’s Q and corresponding P value and I2 value), P values and M-scores of the individual studies. A genome-wide significant signal was observed on chromosome 15, the lead SNP being the nonsynonymous SNP rs1426654 located within the SLC24A5 gene (P value = 2.94×10−39). Given the very strong effect of this variant, we repeated the meta-analysis after running again the association tests in the two South Asian samples conditioning for rs1426654. The Manhattan plot showing the results of the meta-analysis after conditioning for SNP rs1426654 is shown in figure 2c and the QQ plot in figure 2d. Supplementary table S6, Supplementary Material online, provides information about the genome-wide significant and suggestive signals identified in the meta-analysis after conditioning for SNP rs1426654. After conditioning for rs1426654, several variants on chromosome 1 reached genome-wide significance. The lead SNP is rs12076878 (P value = 1.54×10−8). Figure 3 shows the regional-plot corresponding to this genome-wide signal.
Fig. 2.
—Manhattan and QQ plots of the meta-analysis of the association results for skin pigmentation of the South Asian sample from Canada and West Maharashtra before (a and b) and after (c and d) conditioning for the effects of SNP rs1426654 on chromosome 15.
Fig. 3.
—Regional-plot corresponding to the genome-wide signal for the lead SNP rs12076878 (P value = 1.54×10−8) on chromosome 1 identified in the meta-analysis after conditioning for SNP rs1426654.
Follow-up of Genome-Wide Significant Signals Reported in Previous Studies
We evaluated in our South Asian data sets the association of genome-wide significant markers described in previous skin pigmentation GWAS (Stokowski et al. 2007; Liu et al. 2015; Crawford et al. 2017). Table 2 reports the P values of these markers in our meta-analysis, as well as the P values in the individual studies, and the allele frequencies in both samples.
Table 2.
Follow-Up of Genome-Wide Significant Signals Described in Previous Studies
| Marker | Gene | Chr | Pos | NEA | EA | P-meta | Beta | P-WM | P-SAS | EAF-WM | EAF-SAS | Studya | Concordant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs16891982 | SLC45A2 | 5 | 33951693 | C | G | 1.63E-03 | −0.301 | 8.78E-04 | 2.63E-01 | 0.063 | 0.137 | 1 | Yes |
| rs35412 | SLC45A2 | 5 | 33967145 | C | G | 8.35E-03 | −0.140 | 1.54E-02 | 2.50E-01 | 0.366 | 0.392 | 2 | Yes |
| rs12203592 | IRF4 | 6 | 396321 | C | T | 3.33E-01 | −0.266 | 2.77E-01 | 7.26E-01 | 0.011 | 0.016 | 2 | Yes |
| rs11230664 | DDB1 | 11 | 61076372 | T | C | 7.50E-04 | 0.259 | 3.51E-03 | 8.52E-02 | 0.144 | 0.146 | 3 | Yes |
| rs148172827 | TKFC | 11 | 61115821 | C | CATCAA | 2.34E-04 | −0.287 | 1.82E-03 | 4.79E-02 | 0.864 | 0.853 | 3 | Yes |
| rs7948623 | TMEM138 | 11 | 61137147 | T | A | 2.78E-04 | −0.307 | 4.29E-04 | 2.00E-01 | 0.886 | 0.899 | 3 | Yes |
| rs1377457 | TMEM138 | 11 | 61144652 | C | A | 5.56E-04 | −0.271 | 2.71E-03 | 7.89E-02 | 0.870 | 0.862 | 3 | Yes |
| rs1042602 | TYR | 11 | 88911696 | C | A | 5.80E-04 | −0.319 | 1.44E-02 | 1.56E-02 | 0.069 | 0.122 | 1 | Yes |
| rs1800404 | OCA2 | 15 | 28235773 | C | T | 8.17E-03 | −0.148 | 6.71E-03 | 4.04E-01 | 0.343 | 0.360 | 3 | Yes |
| rs12913832 | HERC2 | 15 | 28365618 | A | G | 6.76E-03 | −0.238 | 1.89E-01 | 9.79E-03 | 0.082 | 0.122 | 2 | Yes |
| rs4932620 | HERC2 | 15 | 28514281 | T | C | 6.76E-02 | −0.130 | 1.14E-01 | 2.80E-01 | 0.810 | 0.850 | 3 | Yes |
| rs4268748 | MC1R/DEF8 | 16 | 90026512 | T | C | 9.65E-01 | −0.003 | 3.86E-01 | 2.99E-01 | 0.228 | 0.225 | 2 | Yes |
| rs6510760 | Upstream of MFSD12 | 19 | 3565253 | G | A | 7.06E-02 | 0.128 | 2.95E-01 | 1.28E-01 | 0.322 | 0.297 | 3 | Yes |
| rs112332856 | Upstream of MFSD12 | 19 | 3565599 | T | C | 9.85E-01 | −0.002 | 7.63E-01 | 7.90E-01 | 0.172 | 0.139 | 3 | No |
Discussion
Here, we present genome-wide association analyses of skin pigmentation and iris color in South Asian populations. The skin pigmentation results are based on a meta-analysis of a South Asian sample from Canada and a sample from West Maharashtra in India. Although the distribution of melanin values and the PCA plots indicate the presence of population structure in the samples, particularly in the West Maharashtra sample, the statistical analyses were done incorporating the PCA scores as covariates, and there is little evidence of genomic inflation in the association results. Prior to performing the meta-analysis, the standard errors of the beta coefficients were corrected based on the estimated lambda values (Winkler et al. 2014). No quantitative iris color data were available in the Indian sample, so the iris color analysis is based only on the South Asian sample from Canada.
In the meta-analysis of skin pigmentation, we identified a very strong effect of the well-known SLC24A5 region, driven by the nonsynonymous rs1426654 SNP. In a regression model including only this polymorphism, it explains 34.0% and 32.1% of the variation in M-index values observed in the SAS Canadian sample and the West Maharashtra sample, respectively. When using a model including other covariates (sex, age and PCA scores), adding rs1426654 to the model increases substantially the amount of pigmentation variation explained by the model, from 30.2% to 47.9% in the SAS Canadian sample, and from 40.1% to 54.9% in the West Maharashtra sample. Under an additive model of inheritance, each copy of the derived A allele decreases melanin index by ∼5 units in both the SAS Canadian sample and the West Maharashtra sample. The variant rs1426654 has been associated with skin pigmentation in numerous studies (Lamason et al. 2005; Norton et al. 2006; Basu Mallick et al. 2013; Jonnalagadda, Norton, et al. 2016), including the only GWAS carried out in South Asian populations (Stokowski et al. 2007).
In the association tests conditioning for rs1426654 we identified several genome-wide significant SNPs on chromosome 1 (lead SNP rs12076878, P value = 1.54×10−8), in an intergenic region between the RNA genes RNU6-830P and Y_RNA. This marker has good imputation info scores (info > 0.85 in both samples), and the P values are nominally significant in the Canadian South Asian sample and the West Maharashtra sample. There is no evidence of heterogeneity in effect sizes in the two South Asian samples.
We also followed-up in our data set genome-wide significant markers described in previous GWAS (Stokowski et al. 2007; Liu et al. 2015; Crawford et al. 2017), which are also polymorphic in our South Asian samples. These results are depicted in table 2. Although none of the markers surpassed the genome-wide significance threshold in our meta-analysis, there are multiple nominally significant markers, and the direction of effect is concordant with the effects described in previous studies in all but one polymorphism. The nonsynonymous SNP rs16891982 located in the well-known SLC45A2 gene has a P value of 1.63×10−3. Multiple variants in the DDB1/TMEM138 region are also nominally significant in our South Asian samples (P values range from 2.3×10−4 to 7.5×10−4). SNPs in the OCA2/HERC2 region are also nominally significant in the meta-analysis. These include the OCA2 rs1800404 SNP (P value 8.2×10−3) recently reported by Crawford et al. (2017) and the HERC2 rs1291332 SNP (P value 6.8×10−3) that has been associated with blue eye color in multiple studies, including our iris color GWAS. Two of the markers reported in the MFSD12 region (rs6510760 and rs112332856) are polymorphic in the South Asian sample but are not nominally significant. It is important to note that these markers were not imputed with good confidence in the sample from West Maharashtra (info scores < 0.6). Finally, no nominal associations were observed for the markers rs4268748 (MC1R/DEF8) and rs12203592 (IRF4). The latter is the only SNP that appears in frequencies <5% in our South Asian samples.
In the GWAS of iris color using quantitative measures obtained from high-resolution pictures, we observed three genome-wide significant regions. The first region corresponds to the well-known HERC2 gene. Several variants within this gene are strongly associated to L* and delta. One of the top variants is rs12913832 (P value L*=5.52×10−14; P value delta= 5.08×10−11), an intronic SNP that has known regulatory effects on OCA2 expression by disrupting the interaction between an enhancer located on HERC2 and the OCA2 promoter. This SNP is strongly associated with blue eye color in European populations (Eiberg et al. 2008; Visser et al. 2012). It is interesting to note that, as reported in Edwards et al. (2016), there seem to be differences in the iris color effects of this polymorphism in South Asians and Europeans. The South Asian individuals homozygous for the derived G allele in our sample tend to have intermediate iris colors, instead of blue iris colors, as typically observed in Europe. This suggests that the effect of this polymorphism may be modified by other variants that are differentially distributed in both populations. Our data also show that HERC2 is a major determinant of iris heterochromia (e.g., differences in iris color between the pupillary and ciliary regions if the iris). This has been also reported in European populations by Edwards et al. (2016).
Another genome-wide significant signal for iris color was observed in the SLC24A5 gene, and is driven by the nonsynonymous polymorphism rs1426654, which is also the lead SNP in the skin pigmentation meta-analysis. This and other recent studies indicate that SLC24A5 has pleiotropic effects on pigmentation, and determines variation in skin and hair pigmentation, as well as iris color (Valenzuela et al. 2010; Beleza et al. 2013; Edwards et al. 2016).
For the a* dimension of the CIELab color space, we identified two genome-wide significant regions that, to our knowledge, have not been associated with pigmentary phenotypes in previous studies. The first is an intergenic region between the genes FAM175B and ZRANB1 on chromosome 10. The lead SNP is rs28634972 (P = 3×10−8). This SNP has a good imputation score (info = 0.94). However, this is the only genome-significant variant identified in the region, and all the other variants show substantially higher P values (supplementary fig. S28, Supplementary Material online). Based on the 1000 Genomes Project data, no other variants in this region are in linkage disequilibrium with this SNP in South Asian populations. The second region is located near the ZNF804A gene on chromosome 2. Additional studies will be needed to confirm the role of these two regions in iris color variation.
In summary, we report the results of GWAS for skin pigmentation and iris color in South Asian populations. South Asia has a complex history of migrations, and is characterized by substantial pigmentary and genetic diversity. For this reason, it is an ideal region to study the genetic architecture of normal pigmentation variation. Unfortunately, to date, there has been a single GWAS published for skin pigmentation in South Asia (Stokowski et al. 2007), and no studies have been carried out for iris color. Although our sample size was relatively small (742 samples in the meta-analysis of skin pigmentation, and 329 in the GWAS of iris color), we were able to identify genome-wide significant associations of variants within the well-known gene SLC24A5 not only with skin pigmentation but also with iris color. Variants in the HERC2 gene were also associated with iris color and iris heterochromia. Our study highlights the usefulness of quantitative methods to study iris color variation. We also identified novel genome-wide associations with skin pigmentation and iris color, but we could not replicate these associations due to the lack of independent samples.
The strong correlations observed in the Canadian South Asian sample between skin pigmentation (M-index) and iris color measures (L*, a*, b*, and delta) clearly show that these traits share, to some extent, a common genetic architecture. However, as can be observed in supplementary figure S2a–d, Supplementary Material online, these correlations are far from perfect, suggesting the presence of independent or heterogeneous genetic effects in skin pigmentation and iris color. It is important to note that other factors, such as the influence of iris surface features on iris color, may also explain the limited correlations observed between skin pigmentation and iris color. We also observed that for both samples, there was a strong correlation between skin pigmentation and the first PCA axis, indicating that population history is an important determinant of pigmentary phenotypes in these samples (data not shown). In our PCA analyses including the 1000 Genomes project samples (as well as additional plots incorporating the South Asian Simons Genome Diversity Project samples), samples that cluster toward South Asian groups with high ANI ancestry (e.g., Pathan, Sindhi) tend to show lower M-index values (e.g., lighter pigmentation) than samples that cluster toward South Asian samples with low ANI ancestry (e.g., Madiga, Mala) (fig. 2b) Therefore, our data show that the relative ANI/ASI proportions are important determinants of pigmentation in South Asia. It will be critical to expand the number of studies in South Asian populations in order to better understand the genetic architecture of pigmentary traits, as well as the relative role that migration and selection have played in determining the substantial diversity observed in this region. These not only include genetic association studies in contemporary populations but also ancient DNA studies, which could provide insights on the temporal and geographical distribution of relevant pigmentation alleles in South Asian populations. Ancient DNA studies in Europe have been instrumental in exploring the role of selection on pigmentary traits in this continent (Wilde et al. 2014; Allentoft et al. 2015; Mathieson et al. 2015). It is important to note that there have been recent ancient DNA studies that have clarified many previously unknown details of the extremely complex migration history of the South Asian continent (Lazaridis et al. 2016; Narasimhan et al. 2018).
Ethics Approval and Consent to Participate
This study was approved by the University of Toronto Research and Ethics Board (Protocol Reference No. 27015), and all participants were required to provide written informed consent. Participants from West Maharashtra, India were included in the study after obtaining informed written consent. The study was approved by the Institutional Ethics Committee (IEC) at the Savitribai Phule Pune University, Pune (Ethics/2012/16).
Supplementary Material
Supplementary data are available at GenomeBiology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank all the individuals that participated in this study. E.P. was funded by an NSERC Discovery Grant. H.L.N. and E.P. were funded by the US National Institute of Justice (grant 2013-DN-BX-K011). In Canada, computations were performed on the GPC supercomputer at the SciNet HPC Consortium. SciNet is funded by: the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, Ontario Research Fund—Research Excellence, and the University of Toronto.
Authors’ Contributions
E.P., H.L.N., and M.J. designed the study. M.J., M.A.F., and S.O. collected the data. E.P., M.J., M.A.F., and H.L.N. analyzed the data. E.P., M.J., M.A.F., S.O., and H.L.N. drafted the article. All authors reviewed the article and provided critical comments on the article.
Literature Cited
- Allentoft ME, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522(7555):167–172. [DOI] [PubMed] [Google Scholar]
- Basu Mallick C, et al. 2013. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 9(11):e1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S, et al. 2013. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 9(3):e1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, et al. 2011. Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol. 28(2):1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Weiss G, Saha N, Stoneking M.. 2004. The northeast Indian passageway: a barrier or corridor for human migrations? Mol Biol Evol. 21(8):1525–1533. [DOI] [PubMed] [Google Scholar]
- Crawford NG, et al. 2017. Loci associated with skin pigmentation identified in African populations. Science 358(6365):eaan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Mukherjee DP.. 1963. A spectrophotometric skin colour survey among four Indian castes and tribes. Z Morphol Anthropol. 54:190–200. [Google Scholar]
- Durbin R. 2014. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics 30(9):1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, et al. 2016. Iris pigmentation as a quantitative trait: variation in populations of European, East Asian and South Asian ancestry and association with candidate gene polymorphisms. Pigment Cell Melanoma Res. 29(2):141–162. [DOI] [PubMed] [Google Scholar]
- Eiberg H, et al. 2008. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 123(2):177–187. [DOI] [PubMed] [Google Scholar]
- Han B, Eskin E.. 2011. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 88(5):586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Eskin E.. 2012. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 8(3):e1002555.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaswal IJ. 1979. Skin colour in north Indian populations. J Hum Evol. 8(3):361–366. [Google Scholar]
- Jaswal IJ. 1983. Pigmentary variation in Indian populations. Acta Anthropogenet. 7(1):75–83. [PubMed] [Google Scholar]
- Jonnalagadda M, Nagare T, Chitale A, Ozarkar S.. 2013. Population affinities of select tribal populations of Maharashtra: a study using dental morphology. Indian J Phys Anthropol Hum Genet. 32:97–112. [Google Scholar]
- Jonnalagadda M, Norton H, Ozarkar S, Kulkarni S, Ashma R.. 2016. Association of genetic variants with skin pigmentation phenotype among populations of west Maharashtra, India. Am J Hum Biol. 28(5):610–618. [DOI] [PubMed] [Google Scholar]
- Jonnalagadda M, Ozarkar S, Ashma R, Kulkarni S.. 2016. Skin pigmentation variation among populations of West Maharashtra, India. Am J Hum Biol. 28(1):36–43. [DOI] [PubMed] [Google Scholar]
- Lamason RL, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310(5755):1782–1786. [DOI] [PubMed] [Google Scholar]
- Lazaridis I, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature. 536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, et al. 2015. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet. 134(8):823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B.. 2010. Genotype imputation for genome-wide association studies. Nat Rev Genet. 11(7):499–511. [DOI] [PubMed] [Google Scholar]
- Mathieson I, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528(7583):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra Anshuman, et al. 2017. Genotype-Phenotype Study of the Middle Gangetic Plain in India Shows Association of rs2470102 with Skin Pigmentation. J. Invest. Dermatol. 137:670–677. doi: 10.1016/j.jid.2016.10.043. [DOI] [PubMed] [Google Scholar]
- Narasimhan VM, et al. 2018. The Genomic Formation of South and Central Asia. bioRxiv. 292581. doi: 10.1101/292581. [Google Scholar]
- Norton HL, et al. 2006. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 24(3):710–722. [DOI] [PubMed] [Google Scholar]
- Norton HL, et al. 2016. Quantitative assessment of skin, hair, and iris variation in a diverse sample of individuals and associated genetic variation. Am J Phys Anthropol. 160(4):570–581. [DOI] [PubMed] [Google Scholar]
- Reddy BM, Tripathy V, Kumar V, Alla N.. 2010. Molecular genetic perspectives on the Indian social structure. Am J Hum Biol. 22(3):410–417. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.. 1989. Molecular cloning: a laboratory manual. 2nd ed New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Stokowski RP, et al. 2007. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 81(6):1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela RK, et al. 2010. Predicting phenotype from genotype: normal pigmentation. J Forensic Sci. 55(2):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra R-J.. 2012. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 22(3):446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde S, et al. 2014. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci U S A. 111(13):4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, et al. 2014. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 9(5):1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.