Abstract
Codon usage patterns are affected by both mutational biases and translational selection. The frequency at which each codon is used in the genome is directly linked to the cellular concentrations of their corresponding tRNAs. Transfer RNA abundances—as well as the abundances of other potentially relevant factors, such as RNA-binding proteins—may vary across different tissues, making it possible that genes expressed in different tissues are subject to different translational selection regimes, and thus differ in their patterns of codon usage. These differences, however, are poorly understood, having been studied only in Arabidopsis, rice and human, with controversial results in human. Drosophila melanogaster is a suitable model organism to study tissue-specific codon adaptation given its large effective population size. Here, we compare 2,046 genes, each expressed specifically in one tissue of D. melanogaster. We show that genes expressed in different tissues exhibit significant differences in their patterns of codon usage, and that these differences are only partially due to differences in GC content, expression levels, or protein lengths. Remarkably, these differences are stronger when analyses are restricted to highly expressed genes. Our results strongly suggest that genes expressed in different tissues are subject to different regimes of translational selection.
Keywords: codon usage, tissue specificity, expression, GC content, multivariate analysis
Introduction
Groups (or families) of synonymous codons encode the same amino acid, but are used at largely different frequencies in any genome, a phenomenon known as codon usage bias. Codon bias is affected by both genome nucleotide composition (mutational biases) and translational selection (Sharp et al. 1993). The frequency at which each codon is used by a given genome positively correlates with the cellular concentrations of the corresponding tRNAs, and genes expressed at high levels tend to exhibit increased frequencies of preferred codons (Ikemura 1981, 1982). High tRNA abundances for these codons result in faster and more accurate translation, which makes these codons preferred by natural selection (Ikemura 1982; Andersson and Kurland 1990; Dong et al. 1996; Rocha 2004). The patterns of codon usage vary among species (Kanaya et al. 2001; Duret 2002; Basak and Ghosh 2006; Vicario et al. 2007; Hassan et al. 2009; Du et al. 2014), as expected from the fact that different species exhibit different relative tRNA abundances and nucleotide compositions (Muto and Osawa 1987; Kanaya et al. 2001; Rocha 2004; Goodenbour and Pan 2006). Transfer RNA abundances—in addition to the abundances of other potentially relevant factors, such as RNA-binding proteins—can also differ among the different tissues of an organism (Dittmar et al. 2006), raising the possibility that different patterns of codon usage may be selected in different tissues. However, very few studies, restricted to human and plants, have explored this possibility, producing controversial results.
Using a limited data set (n < 200 genes), Plotkin et al. (2004) found significant differences in codon usage patterns among genes expressed in six human tissues, which they attributed to genes being adapted to the tRNA pools of the tissue in which they are expressed. In line with this interpretation, 1) Dittmar et al. (2006) observed significant differences in the relative abundances of tRNAs among different human tissues, with codons preferred by natural selection usually corresponding to the most abundant tRNAs; 2) the tRNA and codon pools are strongly correlated during development (Schmitt et al. 2014); and 3) proliferation-induced and differentiation-induced genes use different codons that correspond to the tRNA abundances in these two states (Gingold et al. 2014).
In contrast, using internal correspondence analysis and a larger data set of human genes (n = 2,126), Sémon et al. (2006) found that the fraction of the variability of codon usage attributed to tissue specificity was very small (∼2.3%), and mostly due to differences in the GC content of genes expressed in the different tissues, rather than translational selection. Additional analyses by Pouyet et al. (2017) also indicate that heterogeneity in codon usage among genes expressed in different human tissues is largely due to differences in GC content resulting from GC-biased gene conversion. In agreement with this notion, comparison of tRNA and codon abundances of different human cell types suggests that a given tRNA pool translates equally fast different mRNA sets, irrespective of their tissue specificity (Rudolph et al. 2016).
Camiolo et al. (2012) found that genes expressed in different tissues of Arabidopsis thaliana significantly differed in their patterns of codon usage, even after controlling for differences in GC content and expression levels. Similar observations were made in rice (Liu 2012).
The relative importance of translational selection versus nucleotide composition in shaping codon usage is expected to depend on the effective population size (Ne). In organisms with large Ne, natural selection is more effective at driving slightly advantageous mutations to fixation and at removing slightly deleterious mutations, such as synonymous mutations (Kimura et al. 1963; Kimura 1968, 1983). Ne has been estimated to be significantly higher for D. melanogaster (1,000,000–5,000,000 individuals; Shapiro et al. 2007; Du et al. 2013), than for A. thaliana (250,000–400,000 individuals; Yue et al. 2010; Cao et al. 2011) or humans (∼10,000 individuals; Yu et al. 2004). This, together with the fact that D. melanogaster is the best characterized multicellular organism in terms of codon bias (Vicario et al. 2007), makes it suitable to characterize the differences in codon usage among tissues.
Here, we describe significant differences in the patterns of codon usage of genes expressed in 16 D. melanogaster adult tissues. Multivariate analyses indicate that the differences are small but significant and only partially due to differences in GC content. The differences were stronger when analyses were restricted to highly expressed genes. Our results indicate different patterns of translational selection among genes expressed in different tissues of Drosophila, potentially due to adaptation to different tRNA abundances.
Materials and Methods
Genomic Data
We downloaded all D. melanogaster coding sequences (CDSs) from Ensembl BioMart, version 83 (Flicek et al. 2012; Kersey et al. 2016). If a gene had multiple CDSs, then the longest one was chosen for analysis. After filtering, we retained 13,905 D. melanogaster CDSs.
Gene Expression Data
For each D. melanogaster protein-coding gene, the mRNA abundances in the whole adult body and in 16 adult nonredundant tissues/organs (adult carcass, brain, crop, eyes, fat body, head, heart, hindgut, male accessory glands, midgut, ovaries, salivary glands, testes, thoracoabdominal ganglia, tubules, and virgin spermatheca) were obtained from the FlyAtlas database (Chintapalli et al. 2007). Probes were mapped to genes using the Affymetrix annotation file “Drosophila 2,” version 35. We discarded from our analysis those probes that matched multiple genes. If a gene mapped to multiple probes, we used the probe with the highest mRNA signal in the whole fly. After filtering, a total of 13,088 D. melanogaster genes with available mRNA abundance data were retained for our study. Messenger RNA abundances were averaged across 4 biological replicates.
We used this gene expression data to obtain a list of tissue-specific genes. A gene was considered to be expressed in a certain tissue/organ if it was detectable in at least 3 out of the 4 biological replicates (as in Chakraborty and Alvarez-Ponce 2016). Genes expressed only in one out of the 16 tissues/organs were considered as tissue-specific genes. Using these criteria, we identified a total of 2,046 D. melanogaster tissue-specific genes. RNA-Seq data were retrieved from FlyAtlas 2 (Leader et al. 2018). A total of 833 genes were determined to be tissue-specific, belonging to 8 tissues (central nervous system, virgin spermatheca, mated spermatheca, accessory glands, testes, ovaries, trachea, and fat body). Genes were determined to be tissue-specific if they were expressed in at least 2 out of 3 biological replicates.
Data Analysis
We processed our data using several in-house PERL scripts. Data analysis, including generation of plots and statistical tests, were conducted using R (R Core Team 2013). Codon frequencies and relative synonymous codon usage (RSCU) values for each gene were calculated using the “Bio:: Tools:: CodonOptTable” module of the BioPerl package. We used the seqinr (Charif and Lobry 2007) and ade4 (Dray and Dufour 2007) packages to perform correspondence analysis in R. Additionally, we used the pipeline of Sémon et al. (2006) to perform the internal correspondence analysis (Cazes et al. 1988). We also used the vegan package (https://cran.r-project.org/web/packages/vegan/; last accessed October 3, 2017) to perform PERMANOVA and PERMANCOVA analyses in R. Expression levels were log-transformed for our correspondence and PERMANCOVA analyses to improve normality. Protein lengths were log-transformed for our PERMANCOVA analyses.
Results
Patterns of Codon Usage in D. melanogaster
We first conducted a codon usage analysis based on 13,088 D. melanogaster nucleus-encoded protein-coding genes whose expression level is available in the FlyAtlas database (Chintapalli et al. 2007). We first counted how many times each codon is used. The most frequent codon within each synonymous family were: GCC (Ala), CGC (Arg), AAC (Asn), GAU (Asp), UGC (Cys), CAG (Gln), GAG (Glu), GGC (Gly), CAC (His), AUC (Ile), CUG (Leu), AAG (Lys), UUC (Phe), CCC (Pro), AGC (Ser), ACC (Thr), UAC (Tyr), GUG (Val), and UAA (Stop). AUG and UGG are the only codons coding for Met and Trp, respectively (supplementary table S1, Supplementary Material online).
The most frequently used codons are not necessarily the preferred ones (favored by natural selection). To identify the preferred codon in each of the 18 multicodon synonymous families, we compared the patterns of codon usage of highly and lowly expressed genes. First, we identified the most highly expressed (10% top expression), and the least expressed (10% bottom expression). Second, we compared the relative synonymous codon usage (RSCU) of each codon among highly and lowly expressed genes. We considered a codon as preferred if its RSCU value was significantly higher in the highly expressed gene set (Mann–Whitney U test) after controlling for the false discovery rate associated with multiple testing using the Benjamini and Hochberg approach (Benjamini and Hochberg 1995) (q < 0.05). We identified a total of 22 preferred codons (excluding the three termination codons, the one coding for Met, and the one coding for Trp): UUC (Phe), CUG (Leu), AUC (Ile), GUC and GUG (Val), UAC (Tyr), CAC (His), CAG (Gln), AAC (Asn), AAG (Lys), GAC (Asp), GAG (Glu), UCC and UCG (Ser), CCC (Pro), Thr (ACC), GCC (Ala), UGC (Cys), CGU and CGC (Arg), and GGU and GGC (Gly) (table 1). Of note, most of these codons end in G or C, with the exception of GGU and CGU.
Table 1.
Preferred and Unpreferred Codons in Drosophila melanogaster
| Amino Acid | Codon | High Expression (average RSCU) | Low Expression (average RSCU) | P Value (RSCU) | q Value | Amino Acid | Codon | High Expression (average RSCU) | Low Expression (average RSCU) | P Value (RSCU) | q Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 0.54 | 0.80 | 3.7 × 10−60 | 2.6 × 10−59 | Ser | UCU | 0.58 | 0.50 | 3.9 × 10−1 | 4.1 ×10−1 |
| UUC* | 1.46 | 1.20 | 2.6 × 10−60 | 1.7 × 10−59 | UCC* | 1.76 | 1.43 | 8.6 × 10−22 | 1.3 × 10−21 | ||
| Leu | UUA | 0.23 | 0.35 | 1.0 × 10−47 | 4.0 × 10−47 | UCA | 0.43 | 0.57 | 1.7 × 10−29 | 3.3 × 10−29 | |
| UUG | 1.00 | 1.17 | 6.0 × 10−16 | 8.6 × 10−16 | UCG* | 1.28 | 1.10 | 1.4 × 10−9 | 1.274 × 10−9 | ||
| Leu | CUU | 0.56 | 0.65 | 1.7 × 10−12 | 1.5 × 10−12 | Pro | CCU | 0.49 | 0.55 | 3.1 × 10−9 | 3.8 × 10−9 |
| CUC | 0.95 | 0.92 | 9.0 × 10−1 | 9.0 × 10−1 | CCC* | 1.75 | 1.34 | 7.0 × 10−42 | 1.9 × 10−41 | ||
| CUA | 0.39 | 0.56 | 1.8 × 10−36 | 4.5 × 10−36 | CCA | 0.80 | 1.06 | 7.5 × 10−30 | 1.5 × 10−29 | ||
| CUG* | 2.87 | 2.36 | 1.0 × 10−38 | 2.6 × 10−38 | CCG | 0.96 | 1.06 | 1.2 × 10−6 | 1.4 × 10−6 | ||
| Ile | AUU | 0.96 | 1.02 | 8.1 × 10−4 | 1.0 × 10−3 | Thr | ACU | 0.67 | 0.73 | 1.8 × 10−5 | 2.1 × 10−5 |
| AUC* | 1.70 | 1.37 | 1.1 × 10−47 | 4.0 × 10−47 | ACC* | 1.95 | 1.55 | 1.4 × 10−35 | 2.9 × 10−35 | ||
| AUA | 0.34 | 0.61 | 1.9 × 10−80 | 8.1 × 10−79 | ACA | 0.58 | 0.77 | 6.8 × 10−28 | 1.2 × 10−27 | ||
| Met | AUG | — | — | — | — | ACG | 0.80 | 0.95 | 1.6 × 10−13 | 2.1 × 10−13 | |
| Val | GUU | 0.73 | 0.81 | 3.8 × 10−8 | 2.7 × 10−8 | Ala | GCU | 0.80 | 0.82 | 5.7 × 10−1 | 5.9 × 10−1 |
| GUC* | 1.08 | 0.96 | 3.6 × 10−10 | 4.6 × 10−10 | GCC* | 2.12 | 1.74 | 8.6 × 10−49 | 4.0 × 10−48 | ||
| GUA | 0.34 | 0.44 | 2.8 × 10−19 | 4.1 × 10−19 | GCA | 0.52 | 0.73 | 1.9 × 10−43 | 6.1 × 10−43 | ||
| GUG* | 1.85 | 1.80 | 1.4 × 10−2 | 1.7 × 10−2 | GCG | 0.56 | 0.72 | 3.1 × 10−26 | 5.3 × 10−26 | ||
| Tyr | UAU | 0.58 | 0.81 | 3.6 × 10−47 | 1.3 × 10−46 | Cys | UGU | 0.46 | 0.64 | 3.5 × 10−25 | 5.8 × 10−25 |
| UAC* | 1.42 | 1.19 | 5.7 × 10−47 | 1.9 × 10−46 | UGC* | 1.54 | 1.37 | 5.3 × 10−25 | 8.6 × 10−25 | ||
| STOP | UAA | — | — | — | — | STOP | UGA | — | — | — | — |
| STOP | UAG | — | — | — | — | Trp | UGG | — | — | — | — |
| His | CAU | 0.71 | 0.84 | 2.0 × 10−14 | 2.8 × 10−14 | Arg | CGU* | 1.33 | 0.86 | 4.0 × 10−31 | 8.2 × 10−31 |
| CAC* | 1.29 | 1.16 | 3.5 × 10−14 | 4.8 × 10−14 | CGC* | 2.47 | 1.62 | 2.3 × 10−76 | 2.5 × 10−76 | ||
| Gln | CAA | 0.51 | 0.67 | 9.1 × 10−36 | 2.1 × 10−35 | CGA | 0.62 | 0.98 | 1.8 × 10−57 | 1.0 × 10−58 | |
| CAG* | 1.49 | 1.33 | 1.2 × 10−35 | 2.7 × 10−35 | CGG | 0.60 | 0.89 | 5.8 × 10−41 | 1.5 × 10−40 | ||
| Asn | AAU | 0.71 | 0.97 | 2.8 × 10−61 | 2.8 × 10−60 | Ser | AGU | 0.58 | 0.97 | 2.7 × 10−80 | 8.1 × 10−79 |
| AAC* | 1.329 | 1.03 | 2.5 × 10−61 | 2.8 × 10−60 | AGC | 1.37 | 1.42 | 1.1 × 10−2 | 1.4 × 10−2 | ||
| Lys | AAA | 0.45 | 0.69 | 4.5 × 10−61 | 3.5 × 10−60 | Arg | AGA | 0.41 | 0.75 | 3.1 × 10−69 | 4.6 × 10−69 |
| AAG* | 1.55 | 1.31 | 4.7 × 10−61 | 3.5 × 10−60 | AGG | 0.57 | 0.89 | 2.3 × 10−54 | 1.1 × 10−53 | ||
| Asp | GAU | 0.97 | 1.14 | 4.3 × 10−29 | 8.02 × 10−29 | Gly | GGU* | 0.90 | 0.84 | 1.1 × 10−3 | 1.2 × 10−3 |
| GAC* | 1.03 | 0.86 | 4.9 × 10−29 | 8.84 × 10−29 | GGC* | 1.81 | 1.54 | 9.6 × 10−25 | 1.5 × 10−24 | ||
| Glu | GAA | 0.54 | 0.73 | 3.1 × 10−43 | 8.84 × 10−42 | GGA | 1.10 | 1.29 | 1.7 × 10−20 | 2.5 × 10−20 | |
| GAG* | 1.46 | 1.27 | 3.0 × 10−43 | 8.84 × 10−42 | GGG | 0.19 | 0.34 | 3.7 × 10−48 | 1.8 × 10−47 |
Note.—Preferred codons (those for which RSCU is significantly higher for highly expressed genes) are marked with an asterisk and in bold face. P values correspond to the Mann–Whitney U test and q values indicate FDR correction using the Benjamini and Hochberg approach.
Codon Usage Differences among Genes Expressed in Different Tissues of D. melanogaster
For each of the 13,088 D. melanogaster protein-coding genes, we retrieved their levels of expression (mRNA abundances) in the whole adult body, and in 16 individual adult tissues, from the FlyAtlas database (Chintapalli et al. 2007). This information was used to identify a total of 2,046 genes that are expressed in only one tissue (19 in the adult carcass, 77 in the brain, 22 in the crop, 44 in the eyes, 23 in the fat body, 47 in the head, 15 in the heart, 30 in the hindgut, 116 in the male accessory glands, 133 in the midgut, 84 in the ovaries, 10 in the salivary glands, 1,364 in the testes, 10 in the thoracoabdominal ganglia, 28 in the tubules, and 24 in the virgin spermatheca).
For each of these 16 gene sets, we computed the frequencies at which the different codons were used. In the majority of cases, the most frequent codon was the same as that for the entire gene set. However, a number of differences existed. In the hindgut, male accessory glands, testes, thoracoabdominal ganglia, and virgin spermatheca, AAU is the most frequently used codon to code for Asn, instead of AAC (the most commonly used codon genome-wide to code for Asn). Similarly, the most frequent codon for Cys is UGC, except for genes expressed in the salivary glands, which tend to use UGU. Glu is often encoded by GAG, except for genes expressed in the male accessory glands and the virgin spermatheca, which tend to use GAA. In general, Gly is most frequently encoded by GGC; however, genes expressed in the carcass, head, male accessory glands, salivary glands, and virgin spermatheca tend to use GGA. Genes expressed in all tissues prefer CAC to encode His, except those expressed in the male accessory glands, which use CAU more often. Ile is often encoded by AUC, except among genes expressed in the male accessory glands, which tend to use AUU. Phe is generally encoded by UUC, but genes expressed in the male accessory glands use more frequently UUU. The most commonly used codon to encode Pro is CCC, but genes expressed in the crop, male accessory glands, salivary glands, and virgin spermatheca prefer CCA, and those expressed in the brain prefer CCG. The most used codon to encode Ser is AGC; however, genes expressed in the carcass, crop, head, hindgut, midgut, salivary glands, testes, and tubules prefer UCC, and genes expressed in the virgin spermatheca prefer AGU. Finally, Tyr is generally encoded by UAC, but genes expressed in the salivary glands use more frequently UAU (supplementary table S1, Supplementary Material online).
Most of these differences represent significant departures from the frequencies at which codons are used in the entire genome (χ2 test, P < 0.05; supplementary table S2, Supplementary Material online). For each tissue (16 tissues) and for each family of synonymous codons with more than one codon (18 codons after excluding those encoding Met and Trp) (i.e., 16 tissues × 18 amino acids = 288 contrasts), we used a χ2 test to compare the frequencies at which the different codons are used in that tissue with the frequencies at which the codons are used in the overall genome. For instance, the D. melanogaster proteome contains a total of 338,998 asparagines, of which 156,904 are encoded by AAU and 182,094 are encoded by AAC; the male accessory glands proteome contains a total of 2,376 asparagines, of which 1,392 are encoded by AAU and 984 are encoded by AAC; the frequencies at which codons are used are significantly different in both gene sets (χ2 = 145.14, 1 degree of freedom, P = 1.88 × 10−33). Out of the 288 contrasts, 168 were significant (χ2 test, P < 0.05; supplementary table S2, Supplementary Material online). Genes expressed in the testes and the male accessory glands were the ones with the highest number of significant differences: contrasts were significant for all 18 amino acids (supplementary table S2, Supplementary Material online). Among genes expressed in other tissues, significant differences were observed in the heart (in 3 amino acids), crop (in 5 amino acids), fatbody (in 5 amino acids), thoracicoabdominal ganglia (in 4 amino acids), hindgut (in 7 amino acids), adult carcass (in 9 amino acids), head (in 9 amino acids), tubules (in 10 amino acids), ovaries (in 11 amino acids), salivary glands (in 11 amino acids), brain (in 13 amino acids), eyes (in 14 amino acids), midgut (in 15 amino acids), and virgin spermatheca (in 15 amino acids). The number of amino acids with significant differences positively correlates with the number of genes expressed in each tissue (Spearman ρ = 0.689, P = 0.003), suggesting that our contrasts are to some extent limited by statistical power.
For the three tissues in which a larger number of genes are expressed (testes, midgut, and male accessory glands) we determined the set of preferred codons by comparing the most highly expressed (top 20%) and the least expressed (bottom 20%) genes. A total of 14 preferred codons were identified among genes expressed in the midgut (supplementary table S3, Supplementary Material online). Four codons (CAG, AAG, CCC, and CGU) were identified as preferred among genes expressed in testes (supplementary table S4, Supplementary Material online). Our analysis of genes expressed in male accessory glands did not identify any preferred codon (supplementary table S5, Supplementary Material online).
Codon usage is strongly correlated with GC content at the third codon positions (GC3) (Sueoka and Kawanishi 2000; Wan et al. 2004) and GC3 content varies among genes expressed in different tissues (ranging from 51.03% in the salivary glands to 66.48% in the eyes; Kruskal–Wallis test, P = 3.13 × 10−23; table 2), in agreement with observations in other species (Vinogradov 2003). Together, these observations raise the possibility that the observed differences in codon usage among genes expressed in different tissues may be due to differences in GC content. In order to discard this possibility, for each tissue, we generated a list of genes with a distribution of GC3 virtually identical to that of the genes expressed in the tissue. For that purpose, for each of the genes expressed in the tissue of interest, we randomly selected a gene not expressed specifically in the tissue with a very similar GC3 content (± 1%). Two lines of evidence indicate that our observations are not explained (at least entirely) by GC content. First, many of the tissue-specific deviations from the codon preferences of the entire genome (i.e., many of the cases in which one codon is preferred in general, but another codon is preferred among genes expressed in a certain tissue) are not observed in the randomized data set (supplementary table S6, Supplementary Material online). For instance, as mentioned earlier, in the original data set AAC is the most commonly used codon to encode Asn, except for genes expressed in the hindgut, male accessory glands, testes, thoracoabdominal ganglia, and virgin spermatheca, in which AAU is preferred (supplementary table S1, Supplementary Material online). In the randomized data set, AAC is the most commonly used codon, except for the gene sets matching the GC3 content of genes expressed in the heart, male accessory glands, and virgin spermatheca (supplementary table S6, Supplementary Material online). Second, in 210 out of the 288 cases the frequencies at which codons are used in each tissue significantly differ (χ2 test, P < 0.05) from the frequencies at which codons are used in the randomized data sets corresponding to the same tissue (supplementary table S7, Supplementary Material online); we would not expect this to be the case if codon preference differences were only dictated by GC3.
Table 2.
GC3, Expression Levels, and Protein Lengths in Different Tissues
| Tissue | GC3 |
Tissue-Specific Expression Level |
Protein Length |
|||
|---|---|---|---|---|---|---|
| Median | Mean | Median | Mean | Median | Mean | |
| All genes | 0.66 | 0.65 | 69.2 | 196.8 | 409.00 | 554.59 |
| Adult carcass | 0.54 | 0.57 | 38.1 | 235.1 | 199.00 | 279.32 |
| Brain | 0.66 | 0.67 | 18.5 | 23.6 | 652.00 | 886.91 |
| Crop | 0.65 | 0.63 | 29.2 | 51.1 | 472.00 | 482.73 |
| Eyes | 0.67 | 0.67 | 23.4 | 54.6 | 229.50 | 360.91 |
| Fat body | 0.60 | 0.59 | 11.1 | 12.3 | 352.00 | 492.48 |
| Head | 0.62 | 0.63 | 19.1 | 178.4 | 398.00 | 552.83 |
| Heart | 0.63 | 0.65 | 40.2 | 46.2 | 362.00 | 448.67 |
| Hingut | 0.62 | 0.63 | 59.9 | 366.4 | 388.00 | 421.27 |
| Male accessory glands | 0.52 | 0.54 | 1,996.7 | 2,741.4 | 344.50 | 399.33 |
| Midgut | 0.66 | 0.66 | 464.9 | 1,593.6 | 335.00 | 429.83 |
| Ovaries | 0.61 | 0.61 | 131.7 | 304.9 | 415.00 | 537.60 |
| Salivary glands | 0.51 | 0.52 | 164.5 | 373.7 | 229.00 | 302.40 |
| Testes | 0.60 | 0.51 | 411.0 | 590.6 | 295.50 | 404.57 |
| Thoracicoabdominal ganglia | 0.67 | 0.66 | 26.4 | 99.3 | 429.00 | 457.60 |
| Tubules | 0.60 | 0.62 | 1,227.3 | 1,849.7 | 414.00 | 435.14 |
| Virgin spermatheca | 0.53 | 0.5 | 15.5 | 1,343.3 | 250.00 | 280.83 |
Note.—Variation of GC3, expression level, and protein length among genes expressed in different tissue is significant (Kruskal–Wallis test, P < 0.05).
Codon usage is known to be highly affected by gene expression and by protein length (Duret and Mouchiroud 1999; Powell and Dion 2015), and genes expressed in different tissues differ in their levels of expression (Kruskal–Wallis test, P = 2.93 × 10−83) and in the length of their encoded products (Kruskal–Wallis test, P = 1.56 × 10−18; table 2). Therefore, we repeated our analyses using these variables (instead of GC3) as controlling variables. Similar results were obtained, indicating that our observations are not due to expression levels or protein lengths either (supplementary tables S8 and S9, Supplementary Material online).
Correspondence Analysis and Internal Correspondence Analysis
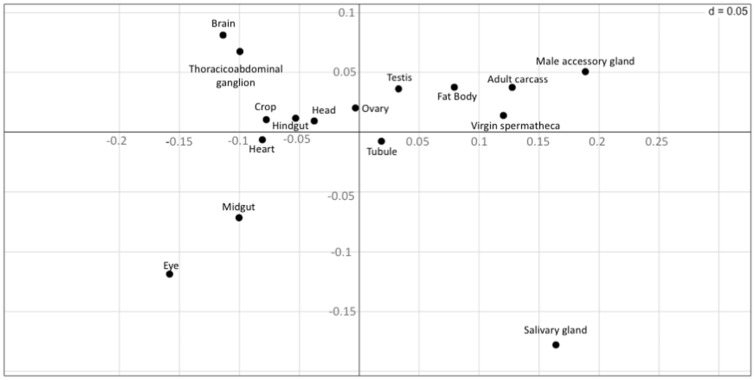
We used correspondence analysis (Grantham et al. 1980) to visualize codon usage differences among genes expressed in the different tissues. Correspondence analysis is a multivariate analysis method that summarizes the information from a high-dimensional space into a low-dimensional space while losing as little information as possible (Lobry and Chessel 2003). In our case, we only considered the two main axes and plotted the centroids of the cluster for each tissue in figure 1. Consistent with the analyses described in the previous section, we found that genes expressed in different tissues exhibit different codon usage patterns (fig. 1).
Fig. 1.
—Position of tissues along the first two major axes of the correspondence analysis based on the centroid of codon usage values. The vertical axis represents principal component 1 and the horizontal axis corresponds to principal component 2.
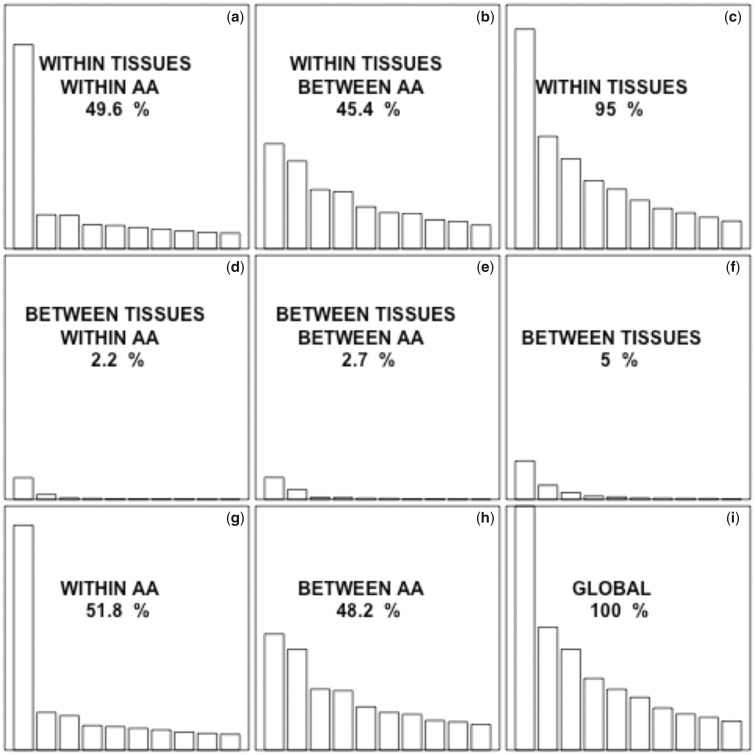
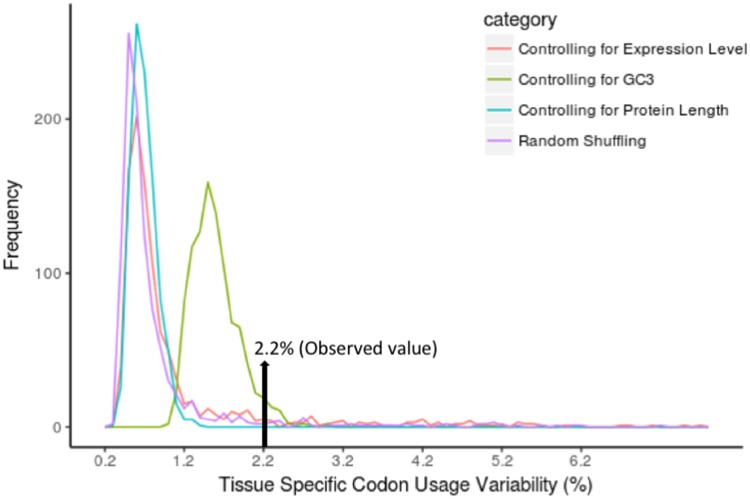
This analysis, however, does not allow us to distinguish between the differences in codon usage due to different amino acid usage (proteins expressed in different tissues tend to use different amino acids) or to differences in the usage of synonymous codons (different codons being preferred to encode a certain amino acid) (Sémon et al. 2006). Therefore, we next used internal correspondence analysis (Cazes et al. 1988; Sémon et al. 2006). This technique is basically a double within-between-correspondence analysis, which allows us to partition the variance of codon usage into different components (Lobry and Chessel 2003). We used the pipeline of Sémon et al. (2006) to partition the codon usage variability into four components: within tissues within amino acids, within tissues between amino acids, between tissues within amino acids, and between tissues between amino acids. Interestingly, we found that 51.8% of the total variability in codon usage is due to variability in synonymous codon usage (fig. 2g), but only 2.2% of the variation in synonymous codon usage is due to tissue specificity. To assess the statistical significance of this value (2.2%), we generated 1,000 randomized data sets and performed internal correspondence analysis in each of them. Each randomized data set was generated by randomly assigning each gene to one of the 16 studied tissues, keeping the same number of genes in each tissue as in the original data set. All of the 1,000 randomized data sets exhibited a lower value compared with the observed one, indicating that the observed value is higher than expected by chance (expected valuemedian = 0.6%, expected valuemean = 0.75%; P < 0.001; fig. 3).
Fig. 2.
—Contribution to the global codon usage variability of synonymous, nonsynonymous, between-tissues, and within-tissues effects. The eigenvalue for a given factor is proportional to the fraction of the variability in codon usage that is accounted for by that factor. The total contribution to the variance of each component is indicated. All the graphs are on the same scale to allow direct visual comparison. In each graph, only the first 10 eigenvalues are represented. The fraction of the global variability due to synonymous codon usage (a, d, g) is higher than the fraction explained by nonsynonymous codon usage (b, e, h). The fraction explained by the differences in codon usage within tissues (a, b, c) is much higher than the fraction explained by the differences between tissues (d, e, f).
Fig. 3.
—Distribution of tissue-specific codon usage variation in 1,000 randomized data sets.
We repeated this analysis by controlling for GC3. For that purpose, each of the randomized data sets was generated by selecting, for each of the genes in our data set, a gene with a very similar GC3 (± 2%) not expressed specifically in the same tissue. Similar results were obtained (expected valuemedian = 1.5%, expected valuemean = 1.58%; P = 0.037; fig. 3). These results indicate that the variation of tissue-specific codon usage is small but significant, and not due to GC content. In addition, similar results were obtained when using expression level (expected valuemedian = 0.7%, expected valuemean = 1.13%; P < 0.001; fig. 3) or protein length (expected valuemedian = 0.7%, expected valuemean = 0.69%; P < 0.001; fig. 3) as controlling variables, indicating that they are not the cause of the observed differences among tissues either.
We also repeated the internal correspondence analysis restricting our analyses to highly expressed genes. Highly expressed genes are expected to be subject to strong translational selection and thus are expected to exhibit stronger differences if these are due to translational selection. We repeated our analyses on genes whose log-expression levels in their tissue of expression were 25% (n = 1,298), 50% (n = 901), 75% (n = 385), and 100% (n = 105) over the average expression level. Interestingly, we observed that the variation in synonymous codon usage between the genes expressed in different tissues increases as we increase the expression cut-off (table 3), suggesting that the observed trend is due to translational selection.
Table 3.
Internal Correspondence Analysis with Different Cut-Offs
| Cut-Off (percent over mean expression level) | Variation in Synonymous Codon Usages between the Tissues |
|---|---|
| 25% | 2.5% |
| 50% | 3.3% |
| 75% | 5.6% |
| 100% | 13.9% |
Note.—Different cut-offs were used to quantify the variation in synonymous codon usage between genes highly expressed in the different tissues.
The number of genes expressed in certain tissues is very small (supplementary table S1, Supplementary Material online). In order to discard the possibility that this might be inflating the observed differences among tissues, we repeated our analyses after removing all tissues in which <30 genes were expressed. In this case, the fraction of the variability explained by tissue and not by amino acid differences was 2.1%, that is, similar to the fraction estimated from the entire data set.
Permutational Multivariate Analysis of Variance
For further investigation of codon usage differences among genes expressed in different tissues, we used permutational multivariate analysis of variance (PERMANOVA) (Anderson 2001), a permutation-based extension of multivariate analysis of variance. In order to reduce the intrinsic correlation among the RSCU values corresponding to any set of synonymous codons, we generated 1,000 randomized versions of our data set. In each randomization, one codon per amino acid was randomly chosen, and all its RSCU values were removed (18 columns in total). All randomized data sets were analyzed using PERMANOVA (with 999 permutations). In all 1,000 cases, the effect of tissue on codon usage was statistically significant (P < 0.05; average pseudo-F ratio = 3.94; table 4). Given the possibility that the observed results may be affected by the strong variation in the number of genes expressed in the different tissues (Shaw and Mitchell-Olds 1993) we repeated our analyses on a second set of 1,000 randomized versions of our data set, each obtained using a double randomization technique: first, one codon per amino acid was removed (as above), and second, 10 genes from each tissue were selected for analysis. In this case, we observed a significant association between codon usage and tissue of expression in 891 of the randomized data sets (i.e., 89.1% of cases, average pseudo-F ratio = 1.42; table 4).
Table 4.
PERMANOVA Results
| Variable | Internal Single Randomization |
Internal Double Randomization |
||
|---|---|---|---|---|
| Average Pseudo-F | No. of Data Sets with Significance at P < 0.05 | Average Pseudo-F | No. of Data Sets with Significance at P < 0.05 | |
| Tissue | 3.94 | 1,000 (100%) | 1.42 | 891 (89.10%) |
Note.—We generated 1,000 random data sets to calculate the average pseudo-F value, and for each data set, we used 999 permutations to assess the significance of the observed pseudo-F value.
In order to discard the possibility that the observed results might be a by-product of covariation of both codon usage and tissue specificity with GC3, gene expression, or protein length, we repeated our analyses using PERMANCOVA, a nonparametric version of ANCOVA, using the three confounding variables as covariates. Using internal single randomization, the tissue of expression had a significant effect on codon usage in all 1,000 randomized data sets (average pseudo F-ratio = 7.05). Using internal double randomization, tissue had a significant effect in 677 of the randomized data sets (67.7%; average pseudo F-ratio = 1.72). These results strongly indicate that the effect of tissue of expression on codon usage is not due to GC3, expression level, or protein length. Indeed, the effect of tissue was stronger after controlling for these factors (compare tables 4 and 5).
Table 5.
PERMANCOVA Results
| Variable | Internal Single Randomization |
Internal Double Randomization |
||
|---|---|---|---|---|
| Average Pseudo-F | No. of Data Sets with Significance at P < 0.05 | Average Pseudo-F | No. of Data Sets with Significance at P < 0.05 | |
| Tissue | 4.78 | 1,000 (100%) | 1.97 | 1,000 (100%) |
| GC3 | 397.75 | 1,000 (100%) | 36.26 | 994 (99.4%) |
| Expression level* | 7.05 | 1,000 (100%) | 1.97 | 556 (56.6%) |
| Protein length* | 12.41 | 1,000 (100%) | 4.06 | 210 (21.0%) |
Note.—We generated 1,000 random data sets to calculate the average pseudo-F value, and for each data set, we used 999 permutations to assess the significance of the observed pseudo-F value. * Data were normalized using logarithmic transformation using base 10.
Analyses of RNA-Seq Expression Data
Given the known biases in microarray-based gene expression level data sets with respect to GC content (Swindell et al. 2014), we repeated our main analyses using RNA-Seq data (Leader et al. 2018). Genes were classified according to their tissue of expression as specific to the central nervous system (n = 35), ovaries (n = 30), testes (n = 651), trachea (n = 48), fat body (n = 16), accessory glands (n = 43), mated spermatheca (n = 7), or virgin spermatheca (n = 3) (supplementary table S11, Supplementary Material online). The codons most used by genes specific to each tissue were in general the same as those found in our analysis of microarray data (supplementary table S12, Supplementary Material online), and the ratios of usage of each codon in each set of tissue-specific genes are often significantly different from those for the entire genome (supplementary table S13, Supplementary Material online). Correspondence analysis shows that 2.3% of the variability of codon usage is due to the tissue in which genes are expressed (fig. 4). This percent was higher for highly expressed genes (supplementary table S14, Supplementary Material online). This percent was lower when evaluated in sets of 1,000 randomized data sets with matching expression levels (expected valuemedian = 0.6%, expected valuemean = 1.41%), protein lengths (expected valuemedian = 0.5%, expected valuemean = 0.55%), and GC3 values (expected valuemedian = 0.9%, expected valuemean = 1.00%), or when randomly matching genes to tissues (expected valuemedian = 0.6%, expected valuemean = 0.77%; supplementary table S15, Supplementary Material online).
Fig. 4.
—The eigenvalue for a given factor is proportional to the fraction of the variability in codon usage that is accounted for by that factor. The total contribution to the variance of each component is indicated. All the graphs are on the same scale to allow direct visual comparison. In each graph, only the first 10 eigenvalues are represented. The fraction of the global variability due to synonymous codon usage (a, d, g) is higher than the fraction explained by nonsynonymous codon usage (b, e, h). The fraction explained by the differences in codon usage within tissues (a, b, c) is much higher than the fraction explained by the differences between tissues (d, e, f).
PERMANOVA analyses on 1,000 data sets randomized by removing one possible codon from each group of synonymous codons showed statistically significant effects of tissue in all 1,000 cases (average pseudo-F = 2.62; supplementary table S16, Supplementary Material online). Two-factor randomization was used to generate 1,000 data sets, limiting the imbalanced distribution of genes between tissues, and a statistically significant effect of tissue was found in 900 of the 1,000 (90%) data sets (average pseudo-F = 1.71; supplementary table S16, Supplementary Material online). PERMANCOVA analyses were repeated using the same single and double randomization strategies to determine the independent effects of tissue specificity, GC3, protein length, and expression level. Single randomization analyses showed statistically a significant effect of tissue in all 1,000 cases (average pseudo-F = 4.78; supplementary table S17, Supplementary Material online). Significant effects were also found for GC3, expression level and protein length in all 1,000 data sets (average pseudo-F = 397.75, 7.05, and 12.41, respectively; supplementary table S15, Supplementary Material online). Double randomization analyses showed statistically significant effects of tissue in all 1,000 cases (average pseudo-F = 1.97), GC3 in 994 cases (99.4%), expression level in 556 cases (55.6%), and protein length in 210 cases (21.0%) (average pseudo-F = 36.26, 1.97, and 4.06, respectively; supplementary table S17, Supplementary Material online).
Discussion
We analyzed the patterns of codon usage of 2,046 genes, each expressed in one D. melanogaster tissue/organ. We observed significant differences among the genes expressed in the different tissues. Our multivariate analyses showed that codon usage differences are not due to differences in GC content, expression level, or protein length. This strongly suggests that the observed differences in codon usage reflect, at least in part, different translational selection regimes acting on genes expressed in different tissues.
One possibility is that the codon usage of genes expressed in different tissues is adapted to the different relative tRNA concentrations of the different tissues. Unfortunately, tRNA abundance data for the different tissues of D. melanogaster are not available at the moment. Therefore, our expectation that the codons preferred in each tissue are the ones with more abundant tRNAs in that tissue cannot be tested directly. Methodologies to directly sequence tRNA are under development (Smith et al. 2015), thus it may eventually be possible to directly test our expectation. Consistent with our hypothesis that our observations are due at least in part to translational selection, the differences in codon usage among genes expressed in different tissues are more pronounced among highly expressed genes (which are expected to be subject to stronger translational selection). Another possibility is that the tissue-specific differences in codon usage may be the result of tissue-specific differences in the concentrations of other potentially relevant factors, such as RNA-binding proteins (e.g., splice factors).
Plotkin et al. (2004), Camiolo et al. (2012), and Liu (2012) also found differences in the patterns of codon usage of genes expressed in different tissues of human, A. thaliana and rice, respectively. In A. thaliana and rice, the differences were shown to be independent of GC content and expression level. In contrast, in human the differences appear to be largely due to differences in GC content rather than to translational selection (Sémon et al. 2006; Pouyet et al. 2017).
At least two factors may account for the differences observed between human and D. melanogaster and A. thaliana and rice. First, humans have a much lower Ne than D. melanogaster and plants (humans: ∼10,000 individuals; D. melanogaster: 1,000,000–5,000,000 individuals; A. thaliana: 250,000–400,000 individuals; Yu et al. 2004; Shapiro et al. 2007; Yue et al. 2010; Cao et al. 2011; Du et al. 2013), which is expected to reduce the efficacy of translational selection (Kimura 1983; Charlesworth 2009). Second, mammalian genomes exhibit a strong isochoric structure (presence of large chromosomic regions with uniform GC content), which produces strong regional variation in GC content (Bernardi 1989, 2000), and genes coexpressed in specific tissues tend to cluster next to each other in the genome (Lercher et al. 2002), making them likely to exhibit similar GC contents and thus similar patterns of codon usage. The D. melanogaster and plant genomes, however, do not exhibit an isochoric structure (Thiery et al. 1976; Jabbari and Bernardi 2000; Nekrutenko and Li 2000; Oliver et al. 2001).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors acknowledge Sandip Chakraborty for his assistance with an early version of this work. We are grateful to Tong Zhou for critically reading an early version of the manuscript. This work was supported by a grant from the National Science Foundation (MCB 1818288), by funds from the University of Nevada, Reno and by a Pilot Grant from Nevada INBRE, funded by the National Institute of General Medical Sciences from the National Institutes of Health (grant P20GM103440).
Literature Cited
- Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32–46. [Google Scholar]
- Andersson SGE, Kurland CG.. 1990. Codon preferences in free-living microorganisms. Microbiol Rev. 54(2):198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Ghosh TC.. 2006. Temperature adaptation of synonymous codon usage in different functional categories of genes: a comparative study between homologous genes of Methanococcus jannaschii and Methanococcus maripaludis. FEBS Lett. 580(16):3895–3899. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57(1):289–300. [Google Scholar]
- Bernardi G. 1989. The isochore organization of the human genome. Annu Rev Genet. 23:637–661. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241(1):3–17. [DOI] [PubMed] [Google Scholar]
- Camiolo S, Farina L, Porceddu A.. 2012. The relation of codon bias to tissue-specific gene expression in Arabidopsis thaliana. Genetics 192(2):641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, et al. 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 43(10):956–963. [DOI] [PubMed] [Google Scholar]
- Cazes P, Chessel D, Doledec S.. 1988. L’analyse des correspondances internes d’un tableau partitionné: son usage en hydrobiologie. Rev Stat Appl. 36:39–54. [Google Scholar]
- Chakraborty S, Alvarez-Ponce D.. 2016. Positive selection and centrality in the yeast and fly protein-protein interaction networks. Biomed Res Int. 2016:4658506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D, Lobry JR. 2007. SeqinR 1.9.2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: Molecules, networks, populations. New York: Springer Verlag. p. 207–232. [Google Scholar]
- Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 10(3):195–205. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow J.. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39(6):715–720. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T.. 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2:2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HJ, Nilsson L, Kurland CG.. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 260(5):649–663. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour AB.. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 22(4):1–20. [Google Scholar]
- Du J, Dungan SZ, Sabouhanian A, Chang BS.. 2014. Selection on synonymous codons in mammalian rhodopsins: a possible role in optimizing translational processes. BMC Evol Biol. 14:96.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Lipman DJ, Cherry JL.. 2013. Why does a protein’s evolutionary rate vary over time? Genome Biol Evol. 5(3):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. 2002. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 12(6):640–649. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D.. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, Arabidopsis. Proc Natl Acad Sci U S A. 96(8):4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. 2012. Ensembl 2013. Nucleic Acids Res. 41(D1):D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158(6):1281–1292. [DOI] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T.. 2006. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 34(21):6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M.. 1980. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 8(9):1893–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Mahalingam V, Kumar V.. 2009. Synonymous codon usage analysis of thirty two mycobacteriophage genomes. Adv Bioinformatics. 2009:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. 1981. Correlation between the abundance of Escherichia coli transfer-RNAs and the occurrence of the respective codons in its protein genes – a proposal for a synonymous codon choice that is optimal for the Escherichia coli translational system. J Mol Biol. 151(3):389–409. [DOI] [PubMed] [Google Scholar]
- Ikemura T. 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 158(4):573–597. [DOI] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G.. 2000. The distribution of genes in the Drosophila genome. Gene 247(1–2):287–292. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T.. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 53(4–5):290–298. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, et al. 2016. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44(D1):D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217(5129):624–626. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kimura M, Maruyama T, Crow JF.. 1963. The mutation load in small populations. Genetics 48:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader DP, Krause SA, Pandit A, Davies SA, Dow J.. 2018. FlyAtlas2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq, and sex-specific data. Nucleic Acids Res. 48:D809–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD.. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 31(2):180–183. [DOI] [PubMed] [Google Scholar]
- Liu Q. 2012. Mutational bias and translational selection shaping the codon usage pattern of tissue-specific genes in rice. PLoS One 7(10):e48295.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR, Chessel D.. 2003. Internal correspondence analysis of codon and amino-acid usage in thermophilic bacteria. J Appl Genet. 44(2):235–261. [PubMed] [Google Scholar]
- Muto A, Osawa S.. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 84(1):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH.. 2000. Assessment of compositional heterogeneity within and between eukaryotic genomes. Genome Res. 10(12):1986–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JL, Bernaola-Galvan P, Carpena P, Roman RR.. 2001. Isochore chromosome maps of eukaryotic genomes. Gene 276(1–2):47–56. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Robins H, Levine AJ.. 2004. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 101(34):12588–12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyet F, Mouchiroud D, Duret L, Sémon M.. 2017. Recombination, meiotic expression and human codon usage. eLife 6:e27344.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J, Dion K.. 2015. Effects of codon usage on gene expression: empirical studies on Drosophila. J Mol Evol. 80(3–4):219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Rocha E. 2004. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 14(11):2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, et al. 2016. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLoS Genet. 12(5):e1006024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt BM, et al. 2014. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA–tRNA interface. Genome Res. 24(11):1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Lobry JR, Duret L.. 2006. No evidence for tissue-specific adaptation of synonymous codon usage in humans. Mol Biol Evol. 23(3):523–529. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, et al. 2007. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 104(7):2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Stenico M, Peden JF, Lloyd AT.. 1993. Codon usage – mutational bias, translational selection, or both. Biochem Soc Trans. 21(4):835–841. [DOI] [PubMed] [Google Scholar]
- Shaw RG, Mitchell-Olds T.. 1993. ANOVA for unbalanced data: an overview. Ecology 74(6):1638–1645. [Google Scholar]
- Smith AM, Abu-Shumays R, Akeson M, Bernick DL.. 2015. Capture, unfolding, and detection of individual tRNA molecules using a nanopore device. Front Bioeng Biotechnol. 3:91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N, Kawanishi Y.. 2000. DNA G+C content of the third codon position and codon usage biases of human genes. Gene 261(1):53–62. [DOI] [PubMed] [Google Scholar]
- Swindell WR, et al. 2014. Integrative RNA-seq and microarray data analysis reveals GC content and gene length biases in the psoriasis transcriptome. Physiol Genomics. 46(15):533. 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Macaya G, Bernardi G.. 1976. Analysis of eukaryotic genomes by density gradient centrifugation. J Mol Biol. 108(1):219–235. [DOI] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR.. 2007. Codon usage in twelve species of Drosophila. BMC Evol Biol. 7:226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. 2003. Isochores and tissue‐specificity. Nucleic Adids Res. 31(17):5212–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XF, Xu D, Kleinhofs A, Zhou JZ.. 2004. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol Biol. 4(1):19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH.. 2004. Nucleotide diversity in gorillas. Genetics 166(3):1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J-X, et al. 2010. Genome-wide investigation reveals high evolutionary rates in annual model plants. BMC Plant Biol. 10:242.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.