Abstract
Cell size is a complex trait that responds to developmental and environmental cues. Quantitative size analysis of mutant strain collections disrupted for protein kinases and transcriptional regulators in the pathogenic yeast Candida albicans uncovered 66 genes that altered cell size, few of which overlapped with known size genes in the budding yeast Saccharomyces cerevisiae. A potent size regulator specific to C. albicans was the conserved p38/HOG MAPK module that mediates the osmostress response. Basal HOG activity inhibited the SBF G1/S transcription factor complex in a stress-independent fashion to delay the G1/S transition. The HOG network also governed ribosome biogenesis through the master transcriptional regulator Sfp1. Hog1 bound to the promoters and cognate transcription factors for ribosome biogenesis regulons and interacted genetically with the SBF G1/S machinery, and thereby directly linked cell growth and division. These results illuminate the evolutionary plasticity of size control and identify the HOG module as a nexus of cell cycle and growth regulation.
Author summary
The basis for commitment to cell division in late G1 phase, called Start in yeast and the Restriction Point in metazoans, is a critical but still poorly understood aspect of eukaryotic cell proliferation. All eukaryotic cells must grow to a critical cell size before commitment to division occurs. This size threshold couples cell growth to division and thereby establishes long-term size homeostasis. Here, to address the problem of cell size control across different species, we performed the first quantitative survey of the size phenome in the pathogenic yeast Candida albicans by focusing on mutants disrupted for protein kinases and transcriptional regulators. We investigate one of the divergent size regulatory pathways in C. albicans, the p38/HOG MAPK module, to uncover a novel stress-independent function for the HOG module in size control. We show that Hog1 inhibits G1/S transcription to delay the G1/S transition and that Hog1 also controls ribosome biogenesis gene expression. These results suggest that Hog1 represents a critical and previously unsuspected nexus between the growth and division machineries. Our study delineates the overall genetic basis for size control in C. albicans and identifies, to our knowledge, the first specific conduit that couples cell growth to cell division.
Introduction
A central and longstanding problem in cell biology is how cells maintain a uniform cell size, whether in single-celled organisms or in the multitude of tissues of metazoans [1, 2]. In most eukaryotes, attainment of a critical cell size is necessary for commitment to cell division in late G1 phase, called Start in yeast and the Restriction Point in metazoans. This critical cell size threshold coordinates cell growth with cell division to establish a homeostatic cell size [1]. The dynamic control of cell size facilitates adaptation to changing environmental conditions in microorganisms and therefore is essential to maximize fitness [3, 4]. In the budding yeast Saccharomyces cerevisiae, the size threshold is dynamically modulated by nutrients. Pre-Start G1 phase cells grown in the optimal carbon source glucose pass Start at a smaller size if shifted to glycerol, whereas cells shifted from a poor to rich nutrient source pass Start at a larger size [1, 5]. Nutrient conditions similarly dictate cell size in the fission yeast Schizosaccharomyces pombe, although control is primarily exerted at the G2/M transition [5]. In metazoans, cell size control is important for tissue, organ and organism size [6], and is dynamically regulated through changes in growth rate and cell cycle length [7]. Cell size is often perturbed in human disease, for example in diabetes, tuberous sclerosis, mitochondrial disorders, aneuploid syndromes, cancer, and aging [1, 8]. Notably, a loss of cell size homeostasis, termed pleomorphism, correlates with poor cancer prognosis [9].
Cell size is fundamentally dictated by the balance between cell growth and division. The analysis of small-sized mutants in yeast led to key insights into the cell division machinery [10–14]. In all eukaryotes, cell division is controlled by the cyclin dependent kinases (CDKs), which serve to coordinate the replication and segregation of the genome [15]. In S. cerevisiae, the G1 cyclins Cln1, Cln2 and Cln3 trigger Start, whereas the B-type cyclins Clb1-Clb6 catalyze replication and mitosis, all via activation of the same Cdc28 kinase catalytic subunit. The expression of ~200 genes at the end of G1 phase, most vitally CLN1/2, is controlled by transcription factor complexes composed of Swi4 and Swi6 (SBF), and Mbp1 and Swi6 (MBF). Activation of SBF/MBF depends primarily on the Cln3-Cdc28 kinase, the key target of which is Whi5, an inhibitor of SBF/MBF-dependent transcription [16, 17]. Another transcriptional inhibitor called Nrm1 specifically inhibits MBF after Start but does not cause a marked size phenotype under conditions of nutrient sufficiency [17]. Size control in S. pombe is exerted through inhibition of G2/M phase CDK activity by the Wee1 kinase, which is encoded by the first size control gene discovered [10, 12]. Size is also partially regulated at Start in S. pombe through an SBF/MBF- like G1/S transcription factor complex and the Nrm1 inhibitor [18]. The CDK-dependent control of G1/S transcription in metazoans is analogously mediated by the cyclin D-Rb-E2F axis [16, 19, 20].
Cell growth depends on the coordinated synthesis of protein, RNA, DNA and other macromolecules [1, 21, 22]. The production of ribosomes consumes a large fraction of cellular resources and depends on an elaborate ribosome biogenesis machinery [1] that is controlled in part by the conserved TOR (Target Of Rapamycin) nutrient sensing network [6]. In budding yeast, the deletion of ribosome biogenesis (Ribi) factors causes a small cell size, and loss of two master regulators of Ribi gene expression, the transcription factor Sfp1 and the AGC kinase Sch9, causes cell to become extremely small [23]. These observations lead to the hypothesis that the rate of ribosome biogenesis is one metric that dictates cell size [24]. Sfp1 and Sch9 are critical effectors of the TOR pathway and form part of a dynamic, nutrient-responsive network that controls the expression of Ribi and ribosomal protein (RP) genes [24]. Sfp1 activity is controlled through its TOR-dependent nuclear localization [23–26] and is physically linked to the secretory system by its interaction with the Rab escort factor Mrs6 [25, 27]. Sch9 is phosphorylated and activated by TOR, and in turn inactivates a cohort of repressors of RP genes called Dot6, Tod6 and Stb3 [28]. The TOR network also controls size in S. pombe and metazoans [29].
Systematic genetic analyses in various species have uncovered hundreds of genes that directly or indirectly affect cell size. Direct size analysis of all strains in the S. cerevisiae gene deletion collection uncovered a number of potent size regulators, including Whi5, Sfp1 and Sch9 [23, 30, 31], and revealed inputs into size control from ribosome biogenesis, mitochondrial function and the secretory system [24–27]. Subsequent analyses of many of these size mutants at a single cell level have suggested that the critical cell size at Start may depend on growth rate in G1 phase and/or on cell size at birth [32, 33]. Visual screens of S. pombe haploid and heterozygous deletion collections for size phenotypes also revealed dozens of novel size regulators, many of which altered size in a genetically additive fashion [34, 35]. Many genes also influence size in metazoan species. A large-scale RNAi screen in Drosophila melanogaster tissue culture cells revealed hundreds of genes as candidate size regulators, including known cell cycle regulatory proteins [36]. Despite overall conservation of the central processes that control cell growth and division, functionally equivalent size regulators are often not conserved at the sequence level. For example, the G1/S transcriptional regulators SBF/MBF and Whi5 bear no similarly to the metazoan counterparts E2F and Rb, respectively [1]. A number of TOR effectors are also poorly conserved at the sequence level, including the ribosome biogenesis transcription factors Sfp1 in yeast and Myc in metazoans [1].
Candida albicans is a diploid ascomycete yeast that is a prevalent commensal and opportunistic pathogen in humans. C. albicans is a component of the normal human flora, colonizing primarily mucosal surfaces, gastrointestinal and genitourinary tracts, and skin [37]. Although most C. albicans infections entail non-life-threatening colonization of surface mucosal membranes, immunosuppressed patients can fall prey to serious infections, such as oropharyngeal candidiasis in HIV patients and newborns, and lethal systemic infections known as candidemia [38]. Interest in C. albicans is not limited to understanding its function as a disease-causing organism, as it has an ecological niche that is obviously distinct from the classic model ascomycete S. cerevisiae. In this regard, C. albicans has served as an important evolutionary milepost with which to assess conservation of biological mechanisms, and recent evidence suggests a surprising extent of rewiring of central signalling and transcriptional networks as compared to S. cerevisiae [39–43].
In this study, we performed a quantitative analysis of gene deletion mutants from different collections of protein kinases and transcriptional regulators in C. albicans. Our results revealed a noticeable degree of divergence between genes that affect size in C. albicans versus S. cerevisiae and uncovered previously undocumented regulatory circuits that govern critical cell size at Start in C. albicans. In particular, we delineate a novel stress-independent function of the p38/HOG MAPK network in coupling cell growth to cell division. Our genetic and biochemical analysis suggests that the HOG module directly interacts with central components of both the cell growth and cell division machineries in C. albicans.
Results
Analysis of the cell size phenome in C. albicans
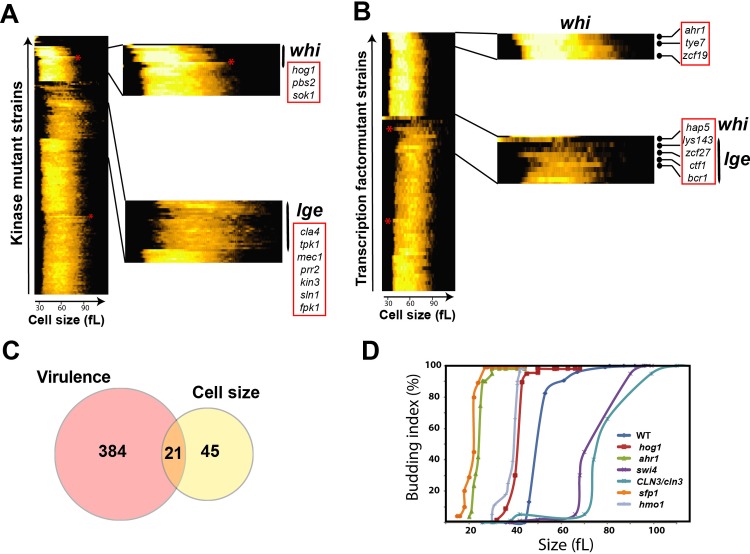
The diploid asexual lifestyle of C. albicans complicates loss-of-function screens because both alleles must be inactivated to reveal a phenotype unless gene function is haploinsufficient [44]. To identify genes required for cell size homeostasis in C. albicans, we directly screened three collections of homozygous diploid gene deletion strains that encompassed 202 transcriptional regulators [42, 45] and 77 protein kinases [41]. We expected transcription factors and kinases to be enriched in cell size regulatory genes based on previous studies in budding yeast and fission yeast. We additionally examined selected homozygous deletion strains of C. albicans orthologs of known size genes in S. cerevisiae (sch9, pop2, ccr4 and nrm1; note that lower case gene names are used to indicate a homozygous mutant) that were not present in these deletion collections (S1 Table). In total, 363 viable mutant strains (279 unique mutants) were individually assessed for their size distribution under conditions of exponential growth in rich medium. Clustering of size distributions across the cumulative datasets revealed distinct subsets of both large and small mutants, relative to the majority of mutants that exhibited size distributions comparable to those of wild-type (wt) control strains (Fig 1A and 1B). Mean, median and mode cell size were estimated for each mutant strain, and mutants were classified as large or small on the basis of a stringent cut-off of a 20% increase or decrease in median size as compared to the parental strain background. This empirical cut-off value was determined based on a benchmark set of conserved small (sch9, sfp1) and large (swi4, pop2, ccr4) sized mutants for which median size was reduced or increased at least 20% as compared to parental strains. Based on this criterion, we identified 66 mutants that exhibited a size defect compared to their parental strain, comprised of 32 small-sized mutants (which we refer to as Whi phenotype, after the "Whiskey" designation used for the first known S. cerevisiae size mutants, [46]) and 34 large-sized mutants (referred to as a Lge phenotype [22]) (S2 and S3 Tables).
Fig 1. The cell size phenome of C. albicans.
Clustergrams of size profiles of two different systematic mutant collections of C. albicans. (A) a set of 81 kinases [41] and (B) a set of 166 transcription factors [42]. For each strain, the cell volume distribution in femtoliters (fL) was measured over 256 size bins with a Beckman Coulter Z2 Channelizer. Hierarchical clustering was used to self-organize the datasets. Red asterisk in the clustergrams indicates size distribution of wt strains. Sections of the clusters corresponding to small and large size mutants are magnified. (C) Overlap between C. albicans size and virulence phenotypes. Avirulent mutant phenotypes were obtained from CGD based on decreased competitive fitness in mice and/or reduced invasion and damage to host cells. (D) Size regulators in C. albicans act at Start. Early G1-phase cells of different size mutant and wt (SN250 [47]) strains were isolated by centrifugal elutriation, released into fresh YPD medium and monitored for bud emergence and cell size at 10 min intervals until the entire population was composed of budded cells. Budding index was determined as the percentage of budded cells in each sample for at least 200 cells.
Deletion mutants of the HOG MAPK pathway (hog1, pbs2) and the morphogenesis checkpoint kinase (swe1) resulted in a small cell size phenotype. Conversely, mutants defective in functions related to the G1/S phase transition (swi4, ace2), filamentous growth and nitrogen utilization (gat1, gzf3, dal81, rob1) caused a large cell size phenotype (S1B and S1C Fig). As in S. cerevisiae, disruption of the central SBF (Swi4-Swi6) G1/S transcription factor complex increased cell size, whereas mutation of the ribosome biogenesis regulators Sch9 and Sfp1 reduced cell size, as did inactivation of Cbf1, the major transcriptional regulator of ribosomal protein genes in C. albicans and other ascomycetes [48, 49] (S2 Fig).
Interestingly, 21 of the 66 size mutants identified by our screen have been shown previously to be required for pathogenesis (p-value = 1.07e-10; S1 Fig). This set of genes included those with functions in transcriptional control of biofilm and invasive filamentation (cyr1, gcn5, ndt80, ace2, zcf27) as well as known adhesion genes (ahr1, war1). This overlap suggested that cell size homeostasis may contribute to C. albicans fitness inside the host (Fig 1C and S6 Table).
Novel Start regulators in C. albicans
Previous work has shown that disruption of cell growth rate is often accompanied by a small cell size phenotype, for example by mutations in RP or Ribi genes [23, 50]. To identify bona fide negative Start regulators, as opposed to mere growth rate-associated effects, doubling times were determined for the 32 homozygous small size mutants identified in our screens (S3 Table). Mutants that exhibited a greater than 10% increase in doubling time as compared to the wt controls were removed from subsequent consideration for this study. As expected, amongst the 21 remaining candidates predicted to more directly couple growth to division (Table 1), we recovered two known conserved repressors of Start, namely Sfp1 and Sch9. Candidate Start regulators in C. albicans included many conserved genes that do not affect size in S. cerevisiae, including components of the HOG MAPK pathway (Hog1, Pbs2), genes linked to respiration (Hap2, Hap43), invasive filamentous growth (Cph2), adhesion (Ahr1, War1) and metabolism (Ino4, Mig2, Gis2). We also found that inactivation of Nrm1 resulted in whi phenotype, consistent with its role as a repressor of the G1/S transition [51]. Interestingly, loss of the transcription factor Hmo1, a main element in the rewired ribosomal gene regulons in C. albicans [49], caused a small size phenotype. An unexpectedly potent size regulator that emerged from our screens was Dot6, a Myb-like HTH transcription factor that binds to the PAC (Polymerase A and C) motif [52]. The dot6 deletion was among the smallest mutants identified in our screens. C. albicans Dot6 is the ortholog of two redundant transcriptional repressors of rRNA and Ribi gene expression called Dot6 and Tod6 in S. cerevisiae, which cause only a minor large size phenotype when deleted together [28].
Table 1. Candidate Start regulators in C. albicans.
| Size gene | Orf19 ID | Median size | Size reduction /WT (%) | Doubling time (min) | Function | Homolog in S. cerevisiae |
|---|---|---|---|---|---|---|
| Hap2 | orf19.1228 | 38 ± 1.9 | 47 | 132 | Transcription factor that regulates low-iron induction of FRP1 | YGL237C |
| Hap43 | orf19.681 | 38 ± 0.9 | 47 | 130 | Transcription factor required for low iron response | YHL009C |
| Ahr1 | orf19.7381 | 34 ± 2.1 | 46 | 143 | Transcription factor that regulates adhesion | |
| Cph2 | orf19.1187 | 34.1 ± 3.9 | 46 | 115 | Transcription factor that promotes hyphal growth | YOR032C |
| Sch9 | orf19.829 | 34 ± 0.2 | 38 | 139 | Kinase involved in growth control, cell size, filamentation and virulence | YHR205W |
| Nrm1 | orf19.6022 | 35 ± 0.4 | 36 | 139 | Transcriptional regulator of cell cycle genes and DNA replication stress | |
| Gis2 | orf19.3182 | 45 ± 4.3 | 36 | 128 | Translational activator for mRNAs | YNL255C |
| Dot6 | orf19.2545 | 49 ± 0.2 | 32 | 139 | Protein with unknown function | YER088C |
| Pbs2 | orf19.7388 | 49.2 ± 0.7 | 31 | 140 | MAPKK with role in osmotic and oxidative stress responses | YJL128C |
| Hog1 | orf19.895 | 50.2 ± 1.2 | 30 | 136 | MAP kinase that regulates osmotic and general stress responses | YLR113W |
| Hmo1 | orf19.6645 | 51 ± 3.1 | 30 | 116 | Transcription factor that binds upstream of hexose and ergosterol metabolism genes, and cell cycle genes | YDR174W |
| Asg1 | orf19.166 | 47 ± 2.1 | 26 | 103 | Gal4 family transcription factor with similarity to S. cerevisiae Asg1 | YIL130W |
| orf19.1496 | 47 ± 1.9 | 26 | 120 | Putative transcription factor | ||
| Zcf3 | orf19.1168 | 47 ± 1.6 | 26 | 118 | Putative transcription factor | |
| Wor2 | orf19.5992 | 47 ± 3.5 | 26 | 96 | Transcription factor that regulates of white-opaque switching | |
| Mig2 | orf19.5326 | 47 ± 3.9 | 26 | 133 | Putative transcription factor | YGL209W |
| War1 | orf19.1035 | 47 ± 2.3 | 26 | 110 | Transcription factor with a role in resistance to weak organic acids | YML076C |
| Ino4 | orf19.837.1 | 47 ± 2.5 | 26 | 114 | Transcription factor involved in lipid biosynthesis | YOL108C |
| orf19.2821 | 48.4 ± 1.2 | 24 | 136 | Protein of unknown function | ||
| Sfp1 | orf19.5953 | 57 ± 2.1 | 20 | 139 | Transcription factor implicated in regulation of RP and Ribi genes | YLR403W |
| Sok1 | orf19.451 | 57.3 ± 1.1 | 20 | 141 | Protein kinase required for degradation of Nrg1 transcriptional repressor of hyphal growth genes | YDR006C |
| WT strains | ||||||
| DAY286 | 71.7 ± 1.9 | 144 | ||||
| SN250 | 64.1 ± 1.5 | 139 | ||||
| SFY87 | 72.1 ± 0.8 | 138 | ||||
| CAI4 | 55 ± 0.9 | 144 | ||||
We demonstrated the effect of six C. albicans size regulators on the timing of Start by assessing the correlation between size and bud emergence in a synchronous early G1 phase population of cells obtained by centrifugal elutriation. We used this assay to determine the effect of three potent novel size control mutants that conferred a small size phenotype (ahr1, hog1, hmo1) and, as a control, disruption of a conserved known regulator of Start in S. cerevisiae (sfp1). We also characterized two large size mutants, namely swi4 and a heterozygous deletion of CLN3, which is an essential G1 cyclin in C. albicans [53]. The critical cell size of the four small sized mutants ahr1, hog1, hmo1 and sfp1 was markedly reduced as compared to the wt parental strain (Fig 1D), whereas Start was delayed in the CLN3/cln3 and swi4 strains. These results demonstrate that the transcription factors Ahr1 and Hmo1, and the MAPK Hog1 are novel bona fide repressors of Start in C. albicans, and suggested that aspects of the Start machinery have diverged between C. albicans and S. cerevisiae.
Basal activity of the HOG MAPK pathway delays Start
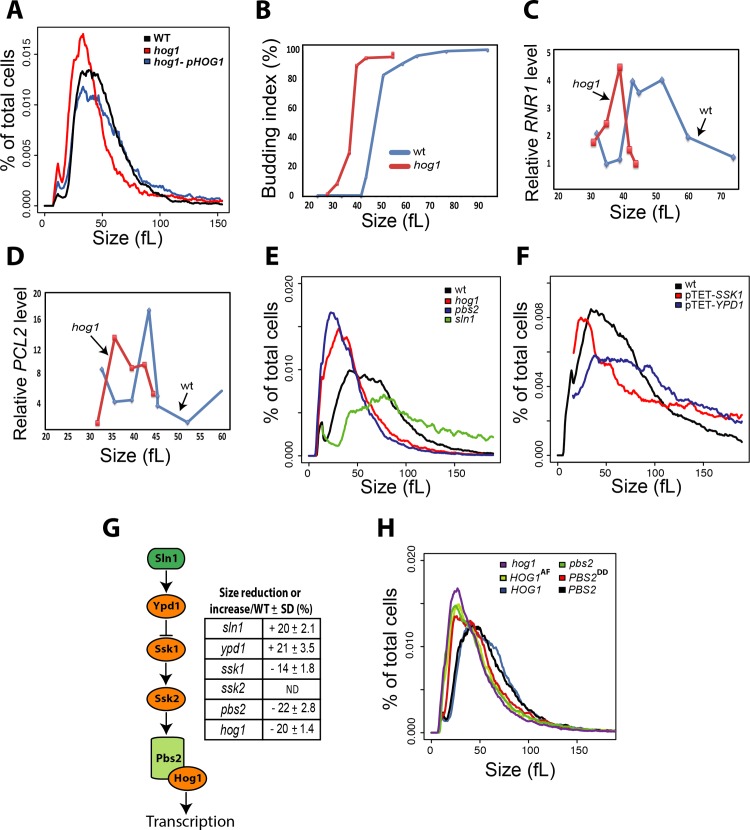
We generated a new hog1 homozygous deletion mutant in C. albicans to confirm the small size phenotype (Fig 2A). The hog1 mutant strain had a median cell volume that was 20% smaller than its congenic parental strain, at 44 and 55 fL respectively. To ascertain that this effect was mediated at Start, we evaluated two hallmarks of Start, namely bud emergence and the onset of SBF-dependent transcription as a function of cell size in synchronous G1 phase cells obtained by elutriation. As assessed by median size of cultures in which 25% of cells had a visible bud, the hog1 mutant passed Start at 41 fL, whereas the parental wt control culture passed Start at 55 fL (Fig 2B). Importantly, in the same experiment, the onset of G1/S transcription was accelerated in the hog1 strain as judged by the peak in expression of the two representative G1 transcripts, RNR1 and PCL2 (Fig 2C and 2D). These results demonstrated that the Hog1 protein kinase normally acts to delay the onset of Start.
Fig 2. Basal activity of HOG pathway is required for normal Start onset and cell size homeostasis.
(A) Confirmation of the Whi phenotype in a newly generated hog1 deletion mutant. Size distributions (i.e., % of cells in each size bin of the Coulter Z2 Channelizer) of a wt (SN148), hog1 and hog1 strain complemented with wild type HOG1 (hog1-pHOG1 are shown. (B-D) Acceleration of Start in a hog1 strain. (B) Elutriated G1 phase daughter cells were released into fresh media and fractions were collected at intervals of 10 min. Bud emergence was assessed in each size fraction. (C-D) Expression of G1/S transcripts. RNR1 and PCL2 transcript levels in elutriated cell fractions relative to pre-elutriated asynchronous cells were assessed by quantitative real-time PCR and normalized to ACT1 levels. (E-F) Size distributions of different mutant strains for the HOG pathway in C. albicans. (G) Schematic of the canonical HOG pathway in C. albicans and summary of size for each mutant strain expressed as mean percentage of reduction or increase of size as compared to the paternal wt strain of each mutant ± standard deviation (four biological replicates). The ssk2 strain exhibited constitutive filamentation that precluded size determination (ND = not determined). (H) Mutation of the two activating phosphorylation sites on Hog1 (T174A and Y176F, termed AF) and Pbs2 (S355D and T359D, termed DD) confers a small size phenotype.
We then tested whether other main elements of the HOG pathway, namely the MAPKK Pbs2, the phosphorelay proteins Ssk1 and Ypd1, and the two-component transducer Sln1, were also required for normal cell size homeostasis (Fig 2E–2G). Disruption of the upstream negative regulators (Ypd1 and Sln1) caused a large size whereas mutation of the core MAPK module (Ssk1, Pbs2 and Hog1) caused a small size phenotype. As the cultures for these experiments were grown in constant normo-osmotic conditions, we inferred that the effect of the HOG module on cell size was unrelated to its canonical role in the osmotic stress response. Consistent with this interpretation, mutation of the known osmotic stress effectors of the HOG pathway in C. albicans, namely the glycerol biosynthetic genes GPD1, GPD2 and RHR2 [54, 55], did not cause a cell size defect (S3 Fig). To address whether basal activity of the HOG MAPK module might be required for size control, we tested the effect of phosphorylation site mutants known to block signal transmission. Mutation of the activating phosphorylation sites on either Hog1 (Thr174 and Tyr176) or Pbs2 (Ser355 and Thr359) to non-phosphorylatable residues phenocopied the small size of hog1 and pbs2 deletion mutants, respectively (Fig 2H). These results demonstrated that a basal level of Hog1 and Pbs2 activity was required for Start repression under non-stress conditions.
To examine the possible role of the HOG pathway in communicating nutrient status to the Start machinery, the effects of different carbon sources on cell size were assessed in hog1 and wt strains. Cell size was reduced on poor carbon sources in the hog1 strain to the same extent as the wt strain, suggesting that the HOG module was not required for carbon source-mediated regulation of cell size (S4 Fig). These results demonstrate that the HOG module relays a stress- and carbon source-independent signal for size control to the Start machinery in C. albicans.
Previous genome-wide screens in S. cerevisiae failed to uncover a role for the HOG pathway in size control [23, 30–32]. To confirm these results, cell size distributions of HOG pathway mutants in S. cerevisiae (hog1, pbs2, ssk1, ssk2, opy2 and sho1 strains) were assessed in rich medium. None of the S. cerevisiae mutants had any discernable size defect as compared to a parental wt strain (S5 Fig).
Hog1 acts upstream of the SBF transcription factor complex
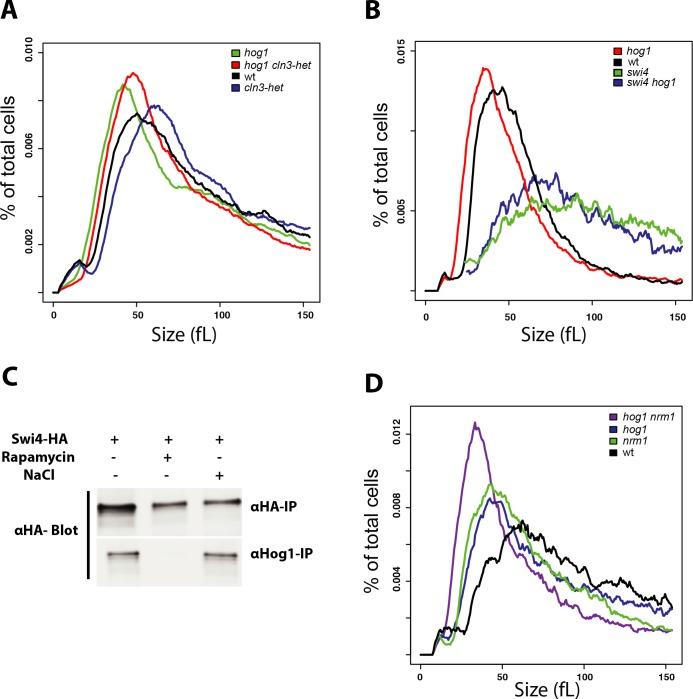
Cln3-dependent activation of the Swi4-Swi6 transcriptional complex drives G1/S progression in both S. cerevisiae and C. albicans [56–59] and we confirmed that CLN3/cln3, swi6 and swi4 mutants all exhibited large size and a G1 phase delay (Figs 1D and S2E). To examine the functional relationship between the HOG pathway and these canonical Start regulators, we characterized their genetic interactions by size epistasis. We observed that the small size of a hog1 mutant strain was partially epistatic to the large size of the heterozygous CLN3/cln3 mutant (Fig 3A), suggesting that the HOG pathway may function in parallel to Cln3. In contrast, a hog1 swi4 double mutant strain had a large size comparable to that of a swi4 mutant, suggesting that Hog1 acts genetically upstream of Swi4 to inhibit Start (Fig 3B). In support of this finding, co-immunoprecipitation assays revealed that Hog1 physically interacted with Swi4 in a rapamycin-sensitive manner and that the Hog1-Swi4 interaction was insensitive to osmotic stress (Fig 3C). In C. albicans, the Nrm1 inhibitor is known to interact with the SBF complex to repress the G1/S transition [51], and consistently a nrm1 mutant exhibited a reduced cell size (Fig 3D). We found that a nrm1 hog1 double mutant had a smaller size than either of the nrm1 or hog1 single mutants, suggesting that Nrm1 and Hog1 act in parallel pathways to inhibit G1/S transcription (Fig 3D). Collectively, these genetic and biochemical results identified Hog1 as a new regulator of SBF in C. albicans, and suggested that Hog1 may transmit signals from the TOR growth control network to the G1/S machinery.
Fig 3. Genetic interactions between the HOG pathway and the G1/S transcriptional machinery.
(A) Additive effect of hog1 and Cln3/cln3 mutations on cell size. The wt strain was in the SN148-Arg+ parental background. (B) A swi4 mutation is epistatic to a hog1 mutation for cell size. The wt strain was in the SN250 background. (C) Co-immunoprecipitation assays for Hog1 and Swi4. Cultures were treated or not as indicated with rapamycin (0.5 μg/ml or NaCl (0.5 M) for 30 min. (D) Additive effect of hog1 and nrm1 mutations on cell size. The wt strain was in the SN250 background.
The Ptc1 and Ptc2 phosphatases control Start via Hog1
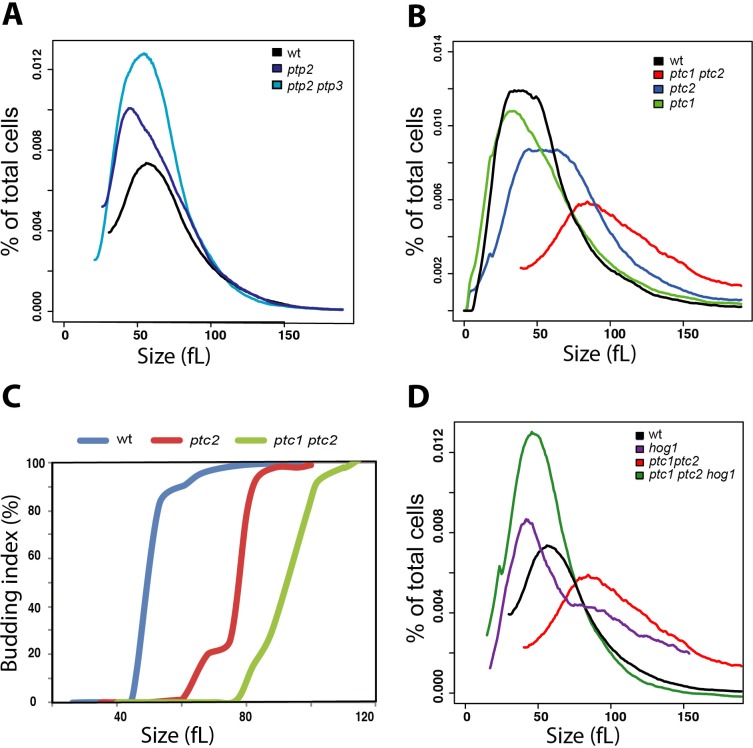
MAPK activity is antagonized by the action of serine/threonine (Ser/Thr) phosphatases, tyrosine (Tyr) phosphatases, and dual specificity phosphatases that are able to dephosphorylate both Ser/Thr and Tyr residues [60]. In S. cerevisiae, after adaptation to osmotic stress, components of the HOG pathway are dephosphorylated by Tyr phosphatases and type 2C Ser/Thr phosphatases [60, 61]. In C. albicans, recent work has identified the two Tyr phosphatases Ptp2 and Ptp3 as modulators of the basal activity of Hog1 [62]. A prediction of the HOG-dependent size control model is that disruption of the phosphatases that modulate Hog1 basal activity should cause a large cell size. However, none of the Tyr-phosphatase single mutants ptp1, ptp2 or ptp3, nor a ptp2 ptp3 double mutant exhibited a noticeable cell size defect (Fig 4A). In contrast, deletion of the type 2C Ser/Thr phosphatase Ptc2 conferred a median size of 84.9 fL, which was 24% larger than the parental wt control size of 68 fL, while a ptc1 ptc2 double mutant strain had an even larger size of 90.5 fL (Fig 4B). To confirm that the large size phenotype of the ptc mutants was mediated directly via effects on Start, we evaluated the critical cell size of both ptc2 and ptc1 ptc2 mutants in elutriated G1 cells. Whereas wt control cells passed Start at 49 fL, the critical cell size of the ptc2 and ptc1 ptc2 mutant strains was increased by 59% to 78 fL and 87% to 92 fL, respectively (Fig 4C). To determine whether Hog1 is an effector of Ptc1 and Ptc2 at Start, we examined the epistatic relationship between the hog1 and ptc1 ptc2 mutations. The size of the hog1 ptc1 ptc2 triple mutant was identical to that of hog1 single mutant, indicating that Hog1 functions downstream of Ptc1 and Ptc2 for the control of cell size (Fig 4D). These data suggest that Ptc1 and Ptc2 phosphatases may modulate the phosphorylation state of Hog1 to govern the timing of Start onset and critical cell size.
Fig 4. Ptc1 and Ptc2 control Start via Hog1.
(A) Size distributions of a wt strain and ptc1, ptc2 and ptc1 ptc2 deletion mutants. (B) Size distributions of a wt strain, a ptp2 single mutant, and a ptp2 ptp3 double mutant. (C) Start is delayed in ptc mutants. Elutriated G1 phase daughter cells were released into fresh media and monitored for bud emergence as a function of size. (D) The small cell size of a hog1 mutant is epistatic to the large size of a ptc1 ptc2 double mutant.
Hog1 activates ribosome biosynthetic gene transcription and inhibits G1/S transcription
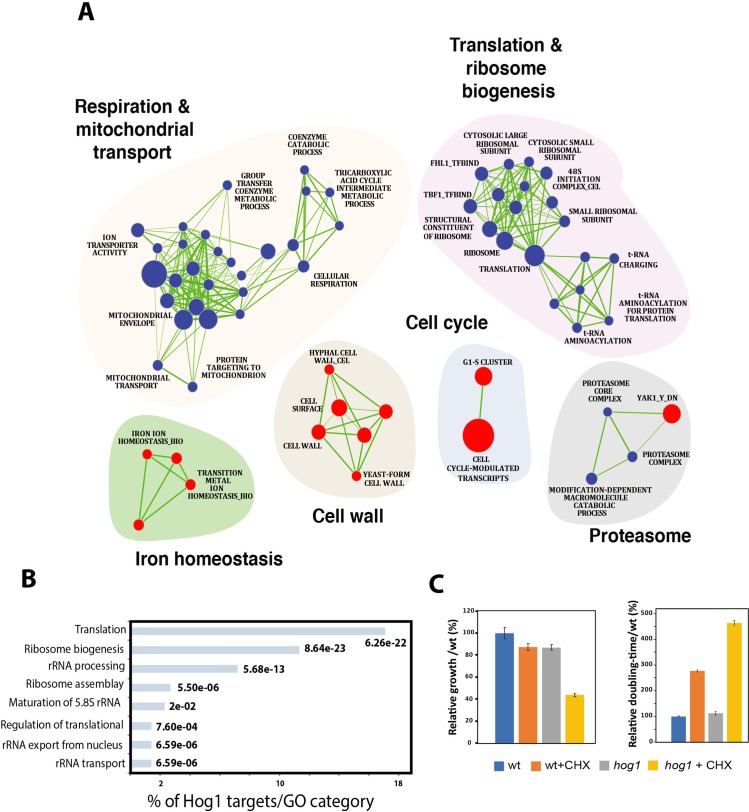
To explore the role of Hog1 at Start, we assessed genome-wide transcriptional profiles using custom microarrays. G1 phase cells for hog1 mutant and wt strains were collected by centrifugal elutriation, followed by microarray analysis of extracted total RNA. Gene set enrichment analysis (GSEA) of transcriptional profiles [63, 64] revealed that the hog1 strain was defective in expression of genes that function in protein translation, including members of the 48S/43S translation initiation complex, structural components of the small and large subunits of the ribosome, and tRNA-charging components (Fig 5A and S4 Table). Transcription of genes that function in mitochondrial transport, the tricarboxylic acid cycle, protein degradation by the 26S proteasome and respiration were also downregulated in a hog1 deletion. Conversely, the G1/S transcriptional program [56] was hyperactivated in a hog1 mutant, consistent with the above results for RNR1 and PCL2. These results suggested that Hog1 activates multiple processes that underpin cellular growth in addition to its role as a negative regulator of the G1/S transcriptional program.
Fig 5. A Hog1-dependent transcriptional program in G1 phase cells.
(A) GSEA analysis of differentially expressed genes in a hog1 mutant relative to a congenic wt strain. Cells were synchronized in G1 phase by centrifugal elutriation and released in fresh YPD medium for 15 min and analyzed for gene expression profiles by DNA microarrays. Up-regulated (red circles) and down-regulated (blue circles) transcripts are shown for the indicated processes. The diameter of the circle reflects the number of modulated gene transcripts in each gene set. Known functional connections between related processes are indicated (green lines). Images were generated in Cytoscape with the Enrichment Map plug-in. (B) Genome-wide promoter occupancy of Hog1 in G1 phase cells. Gene categories bound by Hog1 were determined by GO term enrichment. p-values were calculated using hypergeometric distribution. (C) Growth rate and cycloheximide (CHX; 200 μg/ml) sensitivity of wt and hog1 mutant strains. Relative growth rate was calculated as time to reach half maximal OD600 for each culture normalized to the value for the untreated WT control strain, which was 24 h of growth in SC medium at 30°C. Doubling times were calculated during the exponential phase of each strain treated or not with cycloheximide (200 μg/ml) and represented as a percentage relative to the value of the untreated WT control strain. Results are the mean of three replicates. Bars show the means +/- standard errors of the means.
It has been previously reported that Hog1 in S. cerevisiae and its ortholog p38 in humans directly bind and activate downstream transcriptional target genes [65–70]. In S. cerevisiae, Hog1 thus associates with DNA at stress-responsive genes and is required for recruitment of general transcription factors, chromatin modifying activities and RNA Pol II [66, 69, 71, 72]. However, although mechanisms of Hog1-dependent transcription have been investigated under osmotic stress conditions in C. albicans, the function of this kinase in normal growth conditions in the absence of stress has not been explored. In order to assess whether Hog1 might directly regulate gene expression relevant to cell size control in C. albicans, we profiled the genome-wide localization of Hog1 in G1 phase cells obtained by centrifugal elutriation from TAP-tagged Hog1 and untagged control strains. Hog1 binding sites in the genome were determined in duplicate by chromatin immunoprecipitation and microarray analysis (ChIP-chip). These experiments revealed that Hog1TAP was significantly enriched at 276 intergenic regions and 300 ORFs when compared to the untagged control (S5 Table). The ORF and promoter targets of Hog1 were strongly represented for translation and Ribi genes (Fig 5B), in accord with the above expression profiles. These data suggested that Hog1 may directly activate expression of the Ribi regulon and other translation-associated genes. The strong enrichment for Hog1 at translation and Ribi loci suggested that Hog1 may be required for maximal translational capacity as G1 phase cells approach Start. Consistently, we observed that a hog1 mutant exhibited increased sensitivity to the protein translation inhibitor cycloheximide as compared to a wt strain (Fig 5C). These results suggest that Hog1 may directly activate ribosome biogenesis and protein translation as cells approach Start.
Hog1 is required for Sfp1-dependent gene expression and recruitment to target promoters
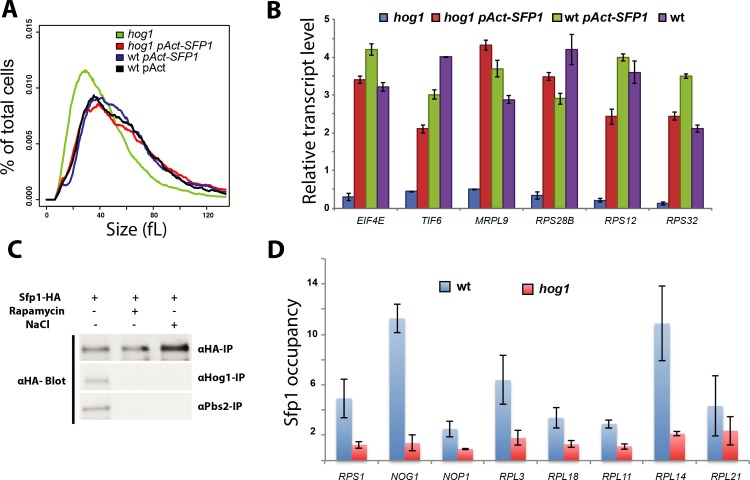
Based on the conserved role of the Sfp1 transcription factor and the kinase Sch9 in ribosome biogenesis and cell size control in C. albicans, we examined genetic interactions between these factors and the HOG pathway. To identify potential epistatic interactions, we overexpressed SCH9 or SFP1 in a hog1 strain. The overexpression of SFP1 but not SCH9 restored the hog1 strain to a near wt cell size distribution (Fig 6A). These results suggested that Sfp1 might act downstream of Hog1. Consistent with this interpretation, we found that the gene expression defects of six Ribi and translation genes (RPS12, RPS28B, RPS32, EIF4E and TIF6) in a hog1 strain were rescued by the overexpression of SFP1 (Fig 6B).
Fig 6. Hog1-dependent recruitment of Sfp1 to promoter DNA.
(A) Size distributions of wt (wt-pAct), hog1 (hog1/hog1), SFP1-overexpression (wt/pAct-SFP1) and hog1 SFP1-overexpression (hog1/hog1/pAct-SFP1) strains. (B) Increased SFP1 dosage restores expression of representative Ribi and RP transcripts in a hog1 mutant strain. Relative expression levels of the six transcripts were assessed by real-time qPCR as normalized to ACT1. Values are the mean from two independent experiments. (C) Sfp1 interactions with Pbs2 and Hog1. Anti-HA immunoprecipitates from a strain bearing an integrated SFP1HA allele grown in the absence or presence of NaCl (0.5 M) or rapamycin (0.5 μg/ml) were probed with anti-HA, anti-Hog1 or anti-Pbs2 antibodies. (D) Reduced Sfp1 localization to Ribi gene promoters in a hog1 mutant strain. Values are the mean from three independent ChIP-qPCR experiments for each indicated promoter.
Given the apparent genetic relationship between Hog1 and Sfp1, we examined whether the two proteins might physically interact. We evaluated the interaction at endogenous levels using a chromosomal HA-tagged Sfp1 allele and polyclonal antibodies that recognize Pbs2 and Hog1. Capture of Sfp1HA from cell lysates followed by antibody detection revealed that Sfp1 interacted with both Pbs2 and Hog1 (Fig 6C). Notably, the Sfp1 interaction with both Hog1 and Pbs2 was abolished by either osmotic stress or rapamycin (Fig 6C). These results suggested that the timing of Start may be governed in part by modulation of the Hog1-Sfp1 interaction by stress and nutrient signals.
We then examined whether Sfp1 might play an analogous role in Start control in C. albicans as in S. cerevisiae. As described above, an sfp1 deletion strain was extremely small and passed Start at only 42% of wt size (Figs 1D and S2A). Consistently, transcriptional profiles of a strain bearing a tetracycline-regulated allele of SFP1 demonstrated that expression of the Ribi regulon was partially Sfp1-dependent (S6A Fig). We also found that an sfp1 strain was as sensitive to the protein translation inhibitor cycloheximide as a hog1 strain (S6B Fig). These data demonstrated that Sfp1 is a transcriptional activator of Ribi genes and a negative regulator of Start in C. albicans.
The finding that both Hog1 and Sfp1 controlled the expression of Ribi genes, together with the finding that Hog1 acted upstream of Sfp1, led us to hypothesize that Hog1 might be required for the recruitment of Sfp1 to its target genes. To test this hypothesis, we used ChIP-qPCR to measure in vivo promoter occupancy of Sfp1HA at eight representative Ribi and RP genes that were also bound by Hog1. While Sfp1 was detected at each of these promoters in a wt strain the ChIP signals were abrogated in the hog1 mutant strain (Fig 6D). From these data, we concluded that Sfp1 regulates the Ribi regulon in a Hog1-dependent manner, and that the HOG module lies at the interface of the G1/S transcriptional and growth control machineries in C. albicans
Discussion
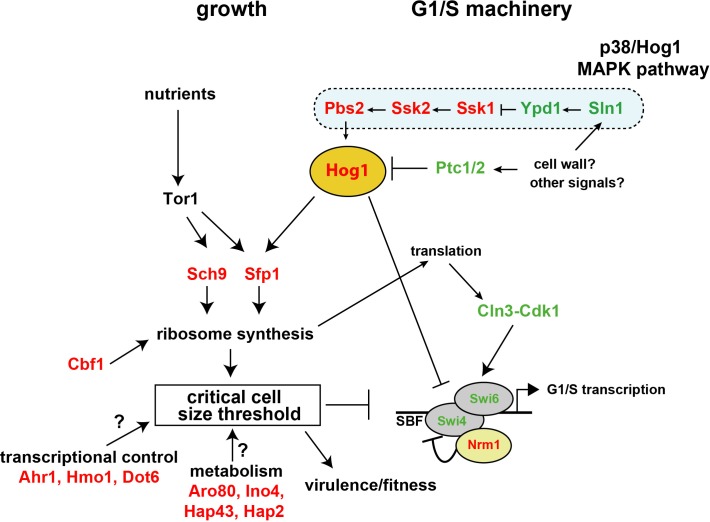
This genetic analysis of size control in C. albicans represents the first detailed characterization of the mechanisms underlying regulation of growth and division in a pathogenic fungus. As is the case for other species that have been examined to date, cell size in C. albicans is a complex trait that depends on diverse biological processes and many genes [23, 30–32, 34, 36]. Of particular note, our screen and subsequent molecular genetic analysis uncovered a novel function for Hog1 as a critical nexus of the growth and division machineries. The HOG module thus represents a direct linkage between cell growth and division (Fig 7).
Fig 7. Architecture of the Start machinery in C. albicans.
Hog1 inhibits the SBF G1/S transcription factor complex and in parallel controls Sfp1 occupancy of Ribi gene promoters, and thereby directly links growth and division. The activity of Hog1 is modulated by the phosphatases Ptc1 and Ptc2 to govern the timing of Start onset. Parallel Start pathways revealed by genetic interactions with Hog1, as well as other prominent size control genes in C. albicans revealed by size screens, are also indicated. Other potential size regulators for which gene inactivation led to small and large size phenotypes are indicated in red and green, respectively.
Conservation and divergence of cell size control mechanisms
Inactivation of genes that control ribosome biogenesis and protein translation in C. albicans resulted in a small cell size, consistent with the notion that the rate of ribosome biogenesis is a component of the critical size threshold [1, 24]. In particular, mutation of the key conserved Ribi regulators Sch9 and Sfp1 dramatically reduced cell size in C. albicans. Previous studies have shown that several RP and Ribi trans-regulatory factors have been evolutionarily rewired in C. albicans compared to S. cerevisiae [48]. Consistently, we found that deletion of CBF1, which encodes a master transcriptional regulator of RP genes in C. albicans but not S. cerevisiae, also caused a small size phenotype. Our analysis also unexpectedly revealed that size regulators may switch between positive and negative functions between the two yeasts. For example, mutation of the conserved transcription factor Dot6 that controls rRNA and Ribi expression caused a strong Whi phenotype in C. albicans, in contrast to the Lge phenotype conferred in S. cerevisiae [28]. These results illustrate the evolutionary plasticity of size control mechanisms at the transcriptional level.
In C. albicans, the G1/S phase cell cycle machinery remains only partially characterized but nevertheless appears to exhibit disparities compared to S. cerevisiae. For instance, despite conservation of SBF and Cln3 function [59, 73], the G1/S repressor Whi5 [16, 19] and the G1/S activator Bck2 [74] appear to have been lost in C. albicans. In S. cerevisiae, cells lacking cln3 are viable and able to pass Start due to the redundant role of Bck2 [74], whereas in C. albicans Cln3 is essential, presumably due to the absence of a Bck2 equivalent [75]. Nrm1 also appears to have replaced Whi5 as it interacts physically with the SBF complex and acts genetically as a repressor of the G1/S transition in C. albicans [51]. Consistently, we observe that nrm1 mutant exhibits a reduced cell size as a consequence of accelerated passage through Start. In addition, the promoters of genes that display a peak of expression during the G1/S transition lack the SCB cis-regulatory element recognized by the SBF complex in S. cerevisiae and are instead enriched in MCB-like motifs [56].
Control of Start by the HOG network
Our systematic size screen uncovered a new stress-independent role of the HOG signaling network in coordinating cell growth and division. Hog1 and its metazoan counterparts, the p38 MAPK family, respond to various stresses in fungi [76] and metazoans [77]. In contrast to these stress-dependent functions, our data suggests that the basal level activity of the module is required to delay the G1/S transition under non-stressed homeostatic growth conditions. This function of the HOG module appears specific to C. albicans as compared to S. cerevisiae. However, the p38 MAPK family is implicated in size control in metazoan species. In the fruit fly D. melanogaster loss of p38β causes small cell and organism size [78], while in mice inactivation of the two Hog1 paralogs p38γ and p38δ alters both cell and organ size, including in the heart and the liver [79, 80]. Recent elegant work in human cells has shown that p38 MAPK activity enforces size homeostasis by controlling the length of G1 phase in proportion to cell size [81]. In S. pombe, there are two critical cell size threshold at both G1/S and G2/M phases [1, 5]. Previous studies have shown that the p38/Hog1 homolog in S. pombe, Sty1, controls mitotic commitment and cell size in a nutrient-dependent manner [34, 82–84]. Deletion of STY1 resulted in a large cell size phenotype which is in accordance with its role as a positive regulator of mitotic onset [83]. These observations suggest the overall role of Hog1 in size control is conserved and that C. albicans may be a suitable yeast model to dissect the mechanisms whereby Hog1 links growth to cell cycle commitment. In particular, we show that the entire HOG module is required for cell size control in C. albicans, and further demonstrate a unique role for the type 2C phosphatases Ptc1 and Ptc2 in size control. In contrast, modulation of the basal activity of Hog1 by the tyrosine phosphatases Ptp2 and Ptp3 in response to a reduction of TOR activity is required for the separate response of hyphal elongation [62]. The mechanisms whereby the same MAPK module can specifically respond to stress, nutrient and cell size remains to be resolved.
A key question raised by our study is the nature of the signal(s) sensed by the HOG network that mediate the coupling of growth to division. Deletion of the upstream negative regulators of the HOG module, Ypd1 and Sln1, caused an increase in cell size, consistent with the negative regulation of Start by the entire HOG network. Previous studies have suggested that Sln1 histidine phosphotransferase activity is required for cell wall biogenesis in both S. cerevisiae and C. albicans [85, 86]. Interestingly, we also found that disruption of the beta-1,3-glucan synthase subunit Gsc1 also caused a reduced size in C. albicans [53]. We speculate that accumulation of cell wall materials, such as glucans, and/or cell wall mechanical proprieties may be sensed through basal activity of the HOG module in order to link growth rate to division. This model is analogous to that postulated in bacteria, whereby the enzymes that synthesize cell wall peptidoglycan help establish cell size control by maintaining cell width [87]. In support of this notion, perturbation of the cell wall leads to a G1 phase cell cycle arrest in S. cerevisiae via the PKC/Slt2 signalling network [32, 88, 89].
Finally, in addition to its role in size control in C. albicans, other stress-independent functions have been attributed to the HOG pathway in different fungi. In Aspergillus fumigatus and A. nidulans, Hog1 controls growth [90], conidial germination [91] and sexual development [92, 93]. In Cryptococcus neoformans, Hog1 is required for mating and, together with the PKA pathway, contributes to the modulation of cellular response to glucose availability [94, 95]. Future efforts on the mechanisms by which Hog1 control these processes will lend further insights into how this central MAPK conduit transmits multiple different signals.
The HOG network lies at the nexus of growth and cell cycle control
The nature of the linkage between growth to division represents a longstanding general problem in cell biology. The complex genetics of size control, reflected in the 66 genes identified in this study that directly or indirectly affect size, confounds the notion of a simple model of size control [2]. Our analysis of Hog1 interactions with the known growth and division machineries nevertheless suggests that the HOG module may directly link growth and division to establish the size threshold at Start. We demonstrated that the HOG module acts genetically upstream of Sfp1 to activate Ribi and translation-related genes, and specifically that Hog1 is required for the expression of many genes implicated in ribosome biogenesis and the recruitment of Sfp1 to the relevant promoters. We also demonstrated that Hog1 and its upstream kinase Pbs2 both physically interact with Sfp1, and that Hog1 localizes to many ribosome biogenesis promoters, consistent with a direct regulatory mechanism. These data suggest that basal activity of the HOG module help set ribosome biogenesis and protein synthesis rates. The HOG module also exhibits strong genetic interactions with the SBF transcriptional machinery since the loss of SBF function is epistatic to HOG module mutations and Hog1 physically interacts with SBF. The HOG module is therefore ideally positioned to communicate the activity of the growth machinery to the cell cycle machinery. We speculate that under conditions of rapid growth, Hog1 and/or other components of the HOG module may be sequestered away from SBF, thereby delaying the onset of G1/S transcription. In the absence of Hog1 basal activity, this balance is set to a default state, in which SBF is activated prematurely for a given rate of growth. Taken together, these observations suggest a model whereby the HOG module directly links growth to cell cycle commitment (Fig 7). The control of SBF by the HOG module appears to operate in parallel to Cln3, Nrm1 and nutrient conditions, suggesting that multiple signals are integrated at the level of SBF, perhaps to optimize adaptation to different conditions [2]. Further analysis of the functional relationships between the HOG module and the numerous other genes that affect size in C. albicans should provide further insights into the linkage between growth and division.
Plasticity of the global size control network and organism fitness
It has been argued that optimization of organism size is a dominant evolutionary force because fitness depends exquisitely on adaptation to a particular size niche [96]. The strong link between size and fitness has been elegantly demonstrated through the artificial evolution of E. coli strains adapted to different growth rates [3]. Comparison of the size phenomes of the opportunistic pathogen C. albicans and the saprophytic yeasts S. cerevisiae and S. pombe reveals many variations in the growth and cell cycle machineries that presumably reflect the different lifestyles of these yeasts. Intriguingly, one third of the size regulators identified in our focused C. albicans reverse-genetic screens have been previously identified as virulence determinants for this pathogen, similar to our previous study of genes that are haploinsufficient for cell size in C. albicans [53]. We speculate that cell size may be an important virulence trait. Other fungal pathogens such as Histoplasma capsulatum, Paracoccidioides brasiliensis, C. neoformans and Mucor circinelloides also exploit cell size as a virulence determinant [97] to access specific niches in the host and/or to escape from host immune cells. In C. albicans, the recently discovered gray cell type is characterized by a small size, a propensity to cause cutaneous infections, and reduced colonization of internal organs [98, 99]. Conversely, the response of the host immune system appears to sense C. albicans size to mitigate tissue damage at the site of infection [100]. The evident scope and plasticity of the global size control network provides fertile ground for adaptive mechanisms to optimize organism size and fitness.
Methods
Strains, mutant collections and growth conditions
C. albicans strains were cultured at 30°C in yeast-peptone-dextrose (YPD) medium supplemented with uridine (2% Bacto peptone, 1% yeast extract, 2% w/v dextrose, and 50 mg/ml uridine). Alternative carbon sources (glycerol and ethanol) were used at 2% w/v. Wt and mutant strains used in this study together with diagnostic PCR primers are listed in S7 Table. The kinase [41] and the transcriptional factor [42] mutant collections used for cell size screens were acquired from the genetic stock center (http://www.fgsc.net). The transcriptional regulator [45] mutant collection was kindly provided by Dr. Dominique Sanglard (University of Lausanne). Growth assay curves were performed in triplicate in 96-well plate format using a Sunrise plate-reader (Tecan) at 30°C under constant agitation with OD595 readings taken every 10 min for 24h. TAP and HA tags were introduced into genomic loci as previously described [101]. Overexpression constructs were generated with the CIp-Act-cyc plasmid which was linearized with the StuI restriction enzyme for integrative transformation [102].
Cell size determination
Cell volume distributions, referred to as cell size, were analyzed on a Z2-Coulter Counter (Beckman). C. albicans cells were grown overnight in YPD at 30°C, diluted 1000-fold into fresh YPD and grown for 5h at 30°C to an early log phase density of 5x106–107 cells/ml. For the tetracycline repressible mutants, all strains and the wt parental strain CAI-4 were grown overnight in YPD supplemented with the antibiotic doxycycline (40μg/ml) to achieve transcriptional repression. We note that high concentration of doxycycline (100 μg/ml) cause a modest small size phenotype in C. albicans but the screen concentration of 40 μg/ml doxycycline did not cause an alteration in cell size. 100 μl of log phase (or 10 μl of stationary phase) culture was diluted in 10 ml of Isoton II electrolyte solution, sonicated three times for 10s and the distribution measured at least 3 times in 3 different independent experiments on a Z2-Coulter Counter. Size distributions were normalized to cell counts in each of 256 size bins and size reported as the peak mode value for the distribution. Data analysis and clustering of size distributions were performed using custom R scripts (S1 File).
Centrifugal elutriation
The critical cell size at Start was determined by plotting budding index as a function of size in synchronous G1 phase fractions obtained using a JE-5.0 elutriation rotor with 40 ml chamber in a J6-Mi centrifuge (Beckman, Fullerton, CA) as described previously [103]. C. albicans G1 phase cells were released in fresh YPD medium and fractions were harvested at an interval of 10 min to monitor bud index. For the hog1 mutant strain, additional size fractions were collected to assess transcript levels of the RNR1, PCL2 and ACT1 as cells progressed through G1 phase at progressively larger sizes.
Gene expression profiles and quantitative real-time PCR
Overnight cultures of hog1 mutant and wt strains were diluted to an OD595 of 0.1 in 1 L fresh YPD-uridine media, grown at 30°C to an OD595 of 0.8 and separated into size fractions by elutriation at 16°C. A total of 108 G1 phase cells were harvested, released into fresh YPD medium and grown for 15 min prior to harvesting by centrifugation and stored at -80°C. Total RNA was extracted using an RNAeasy purification kit (Qiagen) and glass bead lysis in a Biospec Mini 24 bead-beater. Total RNA was eluted, assessed for integrity on an Agilent 2100 Bioanalyzer prior to cDNA labeling, microarray hybridization and analysis [104]. The GSEA Pre-Ranked tool (http://www.broadinstitute.org/gsea/) was used to determine statistical significance of correlations between the transcriptome of the hog1 mutant with a ranked gene list [105] or GO biological process terms as described by Sellam et al. [105]. Data were visualized using the Cytoscape [106] and EnrichmentMap plugin [107]. Gene expression data are available at GEO with the accession number GSE126732. For quantitative real-time PCR (qPCR), cells were grown as for the microarray experiment. cDNA synthesis and qPCR procedure were performed as previously described [108].
Promoter localization by ChIP-chip and ChIP-qPCR
ChIP analyses were performed as described using a custom Agilent microarray containing 14400 (8300 intergenic and 6100 intragenic) 60-mer oligonucleotides that covered all intergenic regions, ORFs and different categories of non-coding RNAs (tRNAs, snoRNAs, snRNAs and rRNA [101]. A total of 107 G1 phase cells were harvested from log phase cultures by centrifugal elutriation and released into fresh YPD medium for 15 min. Arrays were scanned with a GenePix 4000B Axon scanner, and GenePix Pro software 4.1 was used for quantification of spot intensities and normalization. Hog1 genomic occupancy was determined in duplicate ChIP-chip experiments, which were averaged and thresholded using a cutoff of two standard deviations (SDs) above the mean of log ratios (giving a 2-fold enrichment cutoff). ChIP-chip data are available at GEO with the accession number GSE126732. For ChIP analysis of HA-tagged Sfp1, qPCR was performed using an iQ 96-well PCR system for 40 amplification cycles and QuantiTect SYBR Green PCR master mix (Qiagen) using 1 ng of captured DNA and total genomic DNA extracted from the whole cell extract. The coding sequence of the C. albicans ACT1 gene was used as a reference for background in all experiments. Values were calculated as the mean of triplicate experiments.
Protein immunoprecipitation and immunoblot
Cultures of epitope-tagged strains were grown to OD595 of 1.0–1.5 in YPD and either treated or not with rapamycin (0.2 μg/ml) or NaCl (0.5 M) for 30 min. Cells were harvested by centrifugation and lysed by glass beads in IP150 buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM MgCl2, 0.1% Nonidet P-40) supplemented with Complete Mini protease inhibitor cocktail tablet (Roche Applied Science) and 1 mM phenylmethylsulfonyl fluoride (PMSF). 1 mg of total protein from clarified lysates was incubated with 50 μl of monoclonal mouse anti-HA (12CA5) antibody (Roche Applied Science), or 20 μl anti-Pbs2 rabbit polyclonal antibody or 20μl anti-Hog1 rabbit polyclonal antibody (Santa Cruz) and captured on 40 μl Protein A-Sepharose beads (GE) at 4°C overnight. Beads were washed three times with IP150 buffer, boiled in SDS-PAGE buffer, and resolved by 4–20% gradient SDS-PAGE. Proteins were transferred onto activated polyvinylidene difluoride (PVDF) membrane and detected by rabbit anti-HA (1:1000) antibody (QED Biosciences) and IRDye680 secondary antibody (LI-COR).
Supporting information
(A) GO biological process term enrichment of all 66 size mutants identified in this study. (B) GO term enrichment of small sized mutants. (C) GO term enrichment of large sized mutants. p-values were calculated based on a hypergeometric distribution (see http://go.princeton.edu/cgi-bin/GOTermFinder).
(TIF)
The indicated mutant strains and congenic wt control strains were grown to early log phase in rich YPD medium and sized on a Beckman Coulter Z2 Channelizer. Size distributions were shown for (A) sfp1, (B) sch9, (C) cbf1, (D) hmo1, (E) swi4 and swi6, (F) pop2 and ccr4, (G) ahr1, and, (H) hog1 and pbs2 mutants.
(TIF)
(A) gpd2 mutant and wt (SN250) strains were grown to early log phase in rich YPD medium prior to cell size determination. (B) The indicated wt (CAI4), gpd1 and rhr2 strains were grown to early log phase in rich YPD medium in the presence of doxycycline and sized on a Z2 coulter channelizer.
(TIF)
Size distribution of log-phase cultures of the indicated wt (A) and hog1 (B) strains (SN250 background) grown in synthetic glucose (red curve), glycerol (blue) and ethanol (black) medium. Cultures were sized on a Beckman Coulter Z2 Channelizer.
(TIF)
Cultures of the indicated strains were grown to early log phase in rich YPD medium and sized on a Beckman Coulter Z2 Channelizer. Wt (BY4741) and sfp1Δ strains were included as controls.
(TIF)
(A) Network visualization of transcriptional changes in a tet-SFP1/sfp1 conditional mutant strain. Genes expressed at reduced (blue) or elevated (red) levels after Sfp1 repression were organized into functionally connected networks (green lines) based on Gene Ontology biological process terms. Node size indicates the magnitude of change. Data were visualized using Cytoscape and the Enrichment Map plug-in. (B) A pTET-SFP1/sfp1 conditional mutant exhibited increased sensitivity to the protein translation inhibitor cycloheximide (CHX, 200 μg/ml). Cells were grown in YPD at 30°C, and OD595 readings were taken every 10 min on an automated shaker reader.
(TIF)
Mean, median and mode size of each strain are indicated.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data were extracted from CGD database.
(XLSX)
(XLSX)
(RTF)
Acknowledgments
We are grateful to the Fungal Genetics Stock Center (FGSC), Dominique Sanglard (University of Lausanne), Catherine Bachewich (Concordia University), Ana Traven (Monash University), Joachim Ernst (Heinrich-Heine-Universität), Janet Quinn (Newcastle University), Haoping Liu (University of California) and Daniel Kornitzer (Technion) for providing strains.
Data Availability
All relevant data are within the manuscript and its Supporting Information files
Funding Statement
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (#06625) to AS, the Canadian Foundation for Innovation to AS and MT, the Canadian Institutes for Health Research to MT (MOP 366608) and AS (IC118460), the National Institutes of Health (R01RR024031) to MT, and from the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec to MT. JC was supported by a Université Laval Faculty of Medicine and CHUQ foundation PhD scholarships. AS was supported by a start-up award from the Faculty of Medicine, Université Laval and the CHUQ, and by a Fonds de Recherche du Québec-Santé FRQS J1 salary award. MT was supported by a Canada Research Chair Systems and Synthetic Biology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14(23):R1014–27. 10.1016/j.cub.2004.11.027 . [DOI] [PubMed] [Google Scholar]
- 2.Kafri M, Metzl-Raz E, Jonas F, Barkai N. Rethinking cell growth models. FEMS Yeast Res. 2016;16(7). 10.1093/femsyr/fow081 . [DOI] [PubMed] [Google Scholar]
- 3.Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci U S A. 1994;91(15):6808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kafri M, Metzl-Raz E, Jona G, Barkai N. The Cost of Protein Production. Cell Rep. 2016;14(1):22–31. 10.1016/j.celrep.2015.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Curr Biol. 2012;22(9):R350–9. 10.1016/j.cub.2012.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook M, Tyers M. Size control goes global. Curr Opin Biotechnol. 2007;18(4):341–50. 10.1016/j.copbio.2007.07.006 . [DOI] [PubMed] [Google Scholar]
- 7.Ginzberg MB, Chang N, D'Souza H, Patel N, Kafri R, Kirschner MW. Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. Elife. 2018;7 10.7554/eLife.26957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guertin DA, Sabatini DM. Cell Size Control. eLS: John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 9.Zhao ZL, Song N, Huang QY, Liu YP, Zhao HR. [Clinicopathologic features of lung pleomorphic (spindle/giant cell) carcinoma—a report of 17 cases]. Ai Zheng. 2007;26(2):183–8. . [PubMed] [Google Scholar]
- 10.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49(4):559–67. . [DOI] [PubMed] [Google Scholar]
- 11.Sudbery PE, Goodey AR, Carter BL. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature. 1980;288(5789):401–4. . [DOI] [PubMed] [Google Scholar]
- 12.Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256(5518):547–51. Epub 1975/08/14. . [DOI] [PubMed] [Google Scholar]
- 13.Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7(13):4335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(11):4675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyers M. Cell cycle goes global. Curr Opin Cell Biol. 2004;16(6):602–13. 10.1016/j.ceb.2004.09.013 . [DOI] [PubMed] [Google Scholar]
- 16.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117(7):899–913. 10.1016/j.cell.2004.05.024 . [DOI] [PubMed] [Google Scholar]
- 17.de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J 3rd, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23(4):483–96. 10.1016/j.molcel.2006.06.025 . [DOI] [PubMed] [Google Scholar]
- 18.Travesa A, Kalashnikova TI, de Bruin RA, Cass SR, Chahwan C, Lee DE, et al. Repression of G1/S transcription is mediated via interaction of the GTB motifs of Nrm1 and Whi5 with Swi6. Mol Cell Biol. 2013;33(8):1476–86. 10.1128/MCB.01333-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117(7):887–98. 10.1016/j.cell.2004.05.025 . [DOI] [PubMed] [Google Scholar]
- 20.Schaefer JB, Breeden LL. RB from a bud's eye view. Cell. 2004;117(7):849–50. 10.1016/j.cell.2004.06.008 . [DOI] [PubMed] [Google Scholar]
- 21.Ginzberg MB, Kafri R, Kirschner M. Cell biology. On being the right (cell) size. Science. 2015;348(6236):1245075 10.1126/science.1245075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmoller KM, Skotheim JM. The Biosynthetic Basis of Cell Size Control. Trends Cell Biol. 2015;25(12):793–802. Epub 2015/11/18. 10.1016/j.tcb.2015.10.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297(5580):395–400. 10.1126/science.1070850 . [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18(20):2491–505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33(6):704–16. 10.1016/j.molcel.2009.01.034 . [DOI] [PubMed] [Google Scholar]
- 26.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A. 2004;101(40):14315–22. 10.1073/pnas.0405353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh J, Tyers M. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev. 2009;23(16):1944–58. 10.1101/gad.1804409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, et al. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30(15):3052–64. 10.1038/emboj.2011.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davie E, Petersen J. Environmental control of cell size at division. Curr Opin Cell Biol. 2012;24(6):838–44. 10.1016/j.ceb.2012.08.003 . [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Schneider C, Ottmers L, Rodriguez R, Day A, Markwardt J, et al. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr Biol. 2002;12(23):1992–2001. . [DOI] [PubMed] [Google Scholar]
- 31.Dungrawala H, Hua H, Wright J, Abraham L, Kasemsri T, McDowell A, et al. Identification of new cell size control genes in S. cerevisiae. Cell Div. 2012;7(1):24 10.1186/1747-1028-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soifer I, Barkai N. Systematic identification of cell size regulators in budding yeast. Mol Syst Biol. 2014;10:761 10.15252/msb.20145345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrezuelo F, Colomina N, Palmisano A, Gari E, Gallego C, Csikasz-Nagy A, et al. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun. 2012;3:1012 10.1038/ncomms2015 . [DOI] [PubMed] [Google Scholar]
- 34.Navarro FJ, Nurse P. A systematic screen reveals new elements acting at the G2/M cell cycle control. Genome Biol. 2012;13(5):R36 10.1186/gb-2012-13-5-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moris N, Shrivastava J, Jeffery L, Li JJ, Hayles J, Nurse P. A genome-wide screen to identify genes controlling the rate of entry into mitosis in fission yeast. Cell Cycle. 2016;15(22):3121–30. 10.1080/15384101.2016.1242535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439(7079):1009–13. 10.1038/nature04469 . [DOI] [PubMed] [Google Scholar]
- 37.Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3(12):918–30. 10.1038/nrg948 . [DOI] [PubMed] [Google Scholar]
- 38.Odds FC. Candida infections: an overview. Crit Rev Microbiol. 1987;15(1):1–5. 10.3109/10408418709104444 . [DOI] [PubMed] [Google Scholar]
- 39.Lavoie H, Hogues H, Whiteway M. Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr Opin Microbiol. 2009;12(6):655–63. 10.1016/j.mib.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, Nantel A. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol Biol Cell. 2009;20(9):2389–400. Epub 2009/03/13. 10.1091/mbc.E08-11-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6(2):e1000752 Epub 2010/02/09. 10.1371/journal.ppat.1000752 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5(12):e1000783 Epub 2009/12/31. 10.1371/journal.pgen.1000783 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, et al. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. MBio. 2012;3(6). 10.1128/mBio.00495-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noble SM, Johnson AD. Genetics of Candida albicans, a diploid human fungal pathogen. Annu Rev Genet. 2007;41:193–211. 10.1146/annurev.genet.41.042007.170146 . [DOI] [PubMed] [Google Scholar]
- 45.Vandeputte P, Pradervand S, Ischer F, Coste AT, Ferrari S, Harshman K, et al. Identification and functional characterization of Rca1, a transcription factor involved in both antifungal susceptibility and host response in Candida albicans. Eukaryot Cell. 2012;11(7):916–31. Epub 2012/05/15. 10.1128/EC.00134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudbery P. Cell biology. When wee meets whi. Science. 2002;297(5580):351–2. 10.1126/science.1073042 . [DOI] [PubMed] [Google Scholar]
- 47.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42(7):590–8. Epub 2010/06/15. 10.1038/ng.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, et al. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29(5):552–62. Epub 2008/03/18. 10.1016/j.molcel.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8(3):e1000329 10.1371/journal.pbio.1000329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol. 1979;138(1):92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ofir A, Hofmann K, Weindling E, Gildor T, Barker KS, Rogers PD, et al. Role of a Candida albicans Nrm1/Whi5 homologue in cell cycle gene expression and DNA replication stress response. Mol Microbiol. 2012;84(4):778–94. 10.1111/j.1365-2958.2012.08056.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19(4):556–66. Epub 2009/01/23. 10.1101/gr.090233.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaillot J, Cook MA, Corbeil J, Sellam A. Genome-Wide Screen for Haploinsufficient Cell Size Genes in the Opportunistic Yeast Candida albicans. G3 (Bethesda). 2017;7(2):355–60. Epub 2017/01/04. 10.1534/g3.116.037986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kayingo G, Wong B. The MAP kinase Hog1p differentially regulates stress-induced production and accumulation of glycerol and D-arabitol in Candida albicans. Microbiology. 2005;151(Pt 9):2987–99. 10.1099/mic.0.28040-0 . [DOI] [PubMed] [Google Scholar]
- 55.San Jose C, Monge RA, Perez-Diaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol. 1996;178(19):5850–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cote P, Hogues H, Whiteway M. Transcriptional analysis of the Candida albicans cell cycle. Mol Biol Cell. 2009;20(14):3363–73. 10.1091/mbc.E09-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hussein B, Huang H, Glory A, Osmani A, Kaminskyj S, Nantel A, et al. G1/S transcription factor orthologues Swi4p and Swi6p are important but not essential for cell proliferation and influence hyphal development in the fungal pathogen Candida albicans. Eukaryot Cell. 2011;10(3):384–97. 10.1128/EC.00278-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24(17):2746–55. 10.1038/sj.onc.1208606 . [DOI] [PubMed] [Google Scholar]
- 59.Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4(1):95–102. 10.1128/EC.4.1.95-102.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito H, Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem. 2004;136(3):267–72. 10.1093/jb/mvh135 . [DOI] [PubMed] [Google Scholar]
- 61.Brewster JL, Gustin MC. Hog1: 20 years of discovery and impact. Sci Signal. 2014;7(343):re7 10.1126/scisignal.2005458 . [DOI] [PubMed] [Google Scholar]
- 62.Su C, Lu Y, Liu H. Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol Biol Cell. 2013;24(3):385–97. 10.1091/mbc.E12-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sellam A, Tebbji F, Whiteway M, Nantel A. A novel role for the transcription factor Cwt1p as a negative regulator of nitrosative stress in Candida albicans. PLoS One. 2012;7(8):e43956 10.1371/journal.pone.0043956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23(2):241–50. 10.1016/j.molcel.2006.05.031 . [DOI] [PubMed] [Google Scholar]
- 66.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9(6):1307–17. . [DOI] [PubMed] [Google Scholar]
- 67.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313(5786):533–6. 10.1126/science.1127677 . [DOI] [PubMed] [Google Scholar]
- 68.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7(4):767–77. . [DOI] [PubMed] [Google Scholar]
- 69.Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003;22(10):2433–42. 10.1093/emboj/cdg243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36(7):738–43. 10.1038/ng1378 . [DOI] [PubMed] [Google Scholar]
- 71.Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol. 2007;27(11):3900–10. 10.1128/MCB.00089-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nadal-Ribelles M, Conde N, Flores O, Gonzalez-Vallinas J, Eyras E, Orozco M, et al. Hog1 bypasses stress-mediated down-regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol. 2012;13(11):R106 10.1186/gb-2012-13-11-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chapa y Lazo B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4(1):90–4. 10.1128/EC.4.1.90-94.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wijnen H, Futcher B. Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics. 1999;153(3):1131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun. 2015;6:6741 10.1038/ncomms7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krantz M, Becit E, Hohmann S. Comparative genomics of the HOG-signalling system in fungi. Curr Genet. 2006;49(3):137–51. 10.1007/s00294-005-0038-x . [DOI] [PubMed] [Google Scholar]
- 77.Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192(2):289–318. 10.1534/genetics.112.140863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cully M, Genevet A, Warne P, Treins C, Liu T, Bastien J, et al. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol Cell Biol. 2010;30(2):481–95. 10.1128/MCB.00688-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gonzalez-Teran B, Lopez JA, Rodriguez E, Leiva L, Martinez-Martinez S, Bernal JA, et al. p38gamma and delta promote heart hypertrophy by targeting the mTOR-inhibitory protein DEPTOR for degradation. Nat Commun. 2016;7:10477 10.1038/ncomms10477 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tormos AM, Arduini A, Talens-Visconti R, del Barco Barrantes I, Nebreda AR, Sastre J. Liver-specific p38alpha deficiency causes reduced cell growth and cytokinesis failure during chronic biliary cirrhosis in mice. Hepatology. 2013;57(5):1950–61. 10.1002/hep.26174 . [DOI] [PubMed] [Google Scholar]
- 81.Liu S, Ginzberg MB, Patel N, Hild M, Leung B, Li Z, et al. Size uniformity of animal cells is actively maintained by a p38 MAPK-dependent regulation of G1-length. Elife. 2018;7 10.7554/eLife.26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378(6558):739–43. 10.1038/378739a0 . [DOI] [PubMed] [Google Scholar]
- 83.Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9(11):1263–72. 10.1038/ncb1646 . [DOI] [PubMed] [Google Scholar]
- 84.Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9(17):2117–30. . [DOI] [PubMed] [Google Scholar]
- 85.Kruppa M, Jabra-Rizk MA, Meiller TF, Calderone R. The histidine kinases of Candida albicans: regulation of cell wall mannan biosynthesis. FEMS Yeast Res. 2004;4(4–5):409–16. 10.1016/S1567-1356(03)00201-0 . [DOI] [PubMed] [Google Scholar]
- 86.Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, Smith FJ, et al. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19(7):2741–51. 10.1091/mbc.E08-02-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chien AC, Hill NS, Levin PA. Cell size control in bacteria. Current Biology. 2012;22(9):R340–9. 10.1016/j.cub.2012.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kono K, Al-Zain A, Schroeder L, Nakanishi M, Ikui AE. Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2016;113(25):6910–5. 10.1073/pnas.1523824113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189(4):1145–75. 10.1534/genetics.111.128264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du C, Sarfati J, Latge JP, Calderone R. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol. 2006;44(3):211–8. Epub 2006/05/17. 10.1080/13693780500338886 . [DOI] [PubMed] [Google Scholar]
- 91.Xue T, Nguyen CK, Romans A, May GS. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot Cell. 2004;3(2):557–60. Epub 2004/04/13. 10.1128/EC.3.2.557-560.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawasaki L, Sanchez O, Shiozaki K, Aguirre J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol. 2002;45(4):1153–63. Epub 2002/08/16. . [DOI] [PubMed] [Google Scholar]
- 93.Jaimes-Arroyo R, Lara-Rojas F, Bayram O, Valerius O, Braus GH, Aguirre J. The SrkA Kinase Is Part of the SakA Mitogen-Activated Protein Kinase Interactome and Regulates Stress Responses and Development in Aspergillus nidulans. Eukaryot Cell. 2015;14(5):495–510. Epub 2015/03/31. 10.1128/EC.00277-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16(5):2285–300. Epub 2005/02/25. 10.1091/mbc.E04-11-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Banerjee D, Bloom AL, Panepinto JC. Opposing PKA and Hog1 signals control the post-transcriptional response to glucose availability in Cryptococcus neoformans. Mol Microbiol. 2016;102(2):306–20. Epub 2016/07/09. 10.1111/mmi.13461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bonner J. Why Size Matters: From Bacteria to Blue Whales. Princeton University Press; ed2006. [Google Scholar]
- 97.Wang L, Lin X. Morphogenesis in fungal pathogenicity: shape, size, and surface. PLoS Pathog. 2012;8(12):e1003027 Epub 2012/12/14. 10.1371/journal.ppat.1003027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tao L, Du H, Guan G, Dai Y, Nobile CJ, Liang W, et al. Discovery of a "white-gray-opaque" tristable phenotypic switching system in candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 2014;12(4):e1001830 10.1371/journal.pbio.1001830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sellam A, Whiteway M. Recent advances on Candida albicans biology and virulence [version 1; referees: 2 approved]. 2016;5(2582). 10.12688/f1000research.9617.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15(11):1017–25. 10.1038/ni.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578 Epub 2008/12/06. 10.1186/1471-2164-9-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blackwell C, Russell CL, Argimon S, Brown AJ, Brown JD. Protein A-tagging for purification of native macromolecular complexes from Candida albicans. Yeast. 2003;20(15):1235–41. 10.1002/yea.1036 . [DOI] [PubMed] [Google Scholar]
- 103.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12(5):1955–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sellam A, Tebbji F, Nantel A. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot Cell. 2009;8(8):1174–83. Epub 2009/06/23. doi: EC.00074-09 [pii] 10.1128/EC.00074-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sellam A, van het Hoog M, Tebbji F, Beaurepaire C, Whiteway M, Nantel A. Modeling the transcriptional regulatory network that controls the early hypoxic response in Candida albicans. Eukaryot Cell. 2014;13(5):675–90. 10.1128/EC.00292-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–76. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garcia C, Tebbji F, Daigneault M, Liu NN, Kohler JR, Allen-Vercoe E, et al. The Human Gut Microbial Metabolome Modulates Fungal Growth via the TOR Signaling Pathway. mSphere. 2017;2(6). 10.1128/mSphere.00555-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) GO biological process term enrichment of all 66 size mutants identified in this study. (B) GO term enrichment of small sized mutants. (C) GO term enrichment of large sized mutants. p-values were calculated based on a hypergeometric distribution (see http://go.princeton.edu/cgi-bin/GOTermFinder).
(TIF)
The indicated mutant strains and congenic wt control strains were grown to early log phase in rich YPD medium and sized on a Beckman Coulter Z2 Channelizer. Size distributions were shown for (A) sfp1, (B) sch9, (C) cbf1, (D) hmo1, (E) swi4 and swi6, (F) pop2 and ccr4, (G) ahr1, and, (H) hog1 and pbs2 mutants.
(TIF)
(A) gpd2 mutant and wt (SN250) strains were grown to early log phase in rich YPD medium prior to cell size determination. (B) The indicated wt (CAI4), gpd1 and rhr2 strains were grown to early log phase in rich YPD medium in the presence of doxycycline and sized on a Z2 coulter channelizer.
(TIF)
Size distribution of log-phase cultures of the indicated wt (A) and hog1 (B) strains (SN250 background) grown in synthetic glucose (red curve), glycerol (blue) and ethanol (black) medium. Cultures were sized on a Beckman Coulter Z2 Channelizer.
(TIF)
Cultures of the indicated strains were grown to early log phase in rich YPD medium and sized on a Beckman Coulter Z2 Channelizer. Wt (BY4741) and sfp1Δ strains were included as controls.
(TIF)
(A) Network visualization of transcriptional changes in a tet-SFP1/sfp1 conditional mutant strain. Genes expressed at reduced (blue) or elevated (red) levels after Sfp1 repression were organized into functionally connected networks (green lines) based on Gene Ontology biological process terms. Node size indicates the magnitude of change. Data were visualized using Cytoscape and the Enrichment Map plug-in. (B) A pTET-SFP1/sfp1 conditional mutant exhibited increased sensitivity to the protein translation inhibitor cycloheximide (CHX, 200 μg/ml). Cells were grown in YPD at 30°C, and OD595 readings were taken every 10 min on an automated shaker reader.
(TIF)
Mean, median and mode size of each strain are indicated.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data were extracted from CGD database.
(XLSX)
(XLSX)
(RTF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files