Abstract
Hypertension is strongly influenced by genetic factors. Although hypertension prevalence in some Hispanic sub-populations is greater than in non-Hispanic whites, genetic studies on hypertension have focused primarily on samples of European descent. A recent meta-analysis of 200,000 individuals of European descent identified 29 common genetic variants that influence blood pressure, and a genetic risk score derived from the 29 variants has been proposed. We sought to evaluate the utility of this genetic risk score in Hispanics. The sample set consists of 1994 Hispanics from two cohorts: the Northern Manhattan Study (primarily Dominican/Puerto Rican) and the Miami Cardiovascular Registry (primarily Cuban/South American). Risk scores for systolic and diastolic blood pressure were computed as a weighted sum of the risk alleles, with the regression coefficients reported in the European meta-analysis used as weights. Association of risk score with blood pressure was tested within each cohort, adjusting for age, age squared, sex, and BMI. Results were combined using an inverse-variance meta-analysis. The risk score was significantly associated with blood pressure in our combined sample (p = 5.65 × 10−4 for systolic and p = 1.65 × 10−3 for diastolic) but the magnitude of the regression coefficients varied by degree of European, African, and Native American admixture. Further studies among other Hispanic sub-populations are needed to elucidate the role of these 29 variants and identify additional genetic and environmental factors contributing to blood pressure variability in Hispanics.
Keywords: blood pressure, genetics, genetic risk score, racial disparities
Introduction
Hypertension is an important risk factor for cardiovascular diseases including heart failure, stroke, and myocardial infarction. In the United States, the prevalence of hypertension is estimated at 40%, 27%, and 26% among non-Hispanic blacks, non-Hispanic whites, and Hispanics, respectively [1]. However, the prevalence in Hispanics varies across sub-populations, with estimates in men/women as follows: 25/24% Cuban, 33/26% Dominican, 21/20% Mexican, 27/29% Puerto Rican, 25/26% Central American, and 20/16% South American [2]. Although hypertension is affected by environmental and behavioral factors (i.e., smoking, physical activity, diet, and BMI) [1], it is also under genetic influence, with heritability estimates greater than 0.50 for both systolic blood pressure (SBP) and diastolic blood pressure (DBP) [3]. Genetic studies of blood pressure (BP) phenotypes have focused primarily on sample sets from European ancestry populations. Through an international collaboration, a recent meta-analysis of data from genome-wide association studies identified 29 single nucleotide polymorphisms (SNPs) from 28 genetic loci that influence BP. Each of these polymorphisms met a genome-wide significance threshold in a sample of 200 000 individuals of European ancestry [4]. A genetic risk score computed using these loci has been replicated in independent samples of European ancestry as well as in samples of East Asian, South Asian, and African ancestry [4, 5]. However, the relationship between the score and BP phenotypes differs drastically across populations; with effect size estimates of ~0.9 in Europeans, ~0.6 in South Asians, ~1.1 in East Asians, and ~0.5 in Africans for both SBP and DBP. Given the disparity in prevalence and the inconsistency of the risk score association across different race-ethnic backgrounds, we investigated the effect of these variants on BP in a genetically admixed sample of Hispanics from two cohorts to determine the utility of the risk score in Hispanic populations.
Subjects and Methods
Subjects and Data collection
This study consisted of Hispanic samples from two independent cohorts: the Northern Manhattan Study (NOMAS) and Miami Cardiovascular Registry (MCR).
NOMAS:
The detailed ascertainment scheme of NOMAS has been reported previously [6]. Briefly, participants were eligible for inclusion if they had never been diagnosed with a stroke, were at least 40 years of age, and resided for at least three months in a household with a telephone in northern Manhattan. At enrollment, demographic characteristics and risk factors were collected through standardized questionnaires and laboratory tests. Body mass index (BMI, kg/m2) was calculated based on measured weight and height. SBP and DBP were measured in a sitting position by a trained research assistant, after a five minute rest. BP measurements were performed twice, 15 minutes apart using a calibrated standard aneroid sphygmomanometer (Omron; Vernon Hills, Illinois). In participants with BP recordings discrepant by more than 10 mm Hg, a third measurement was obtained by the study physician. All BP measurements were then averaged. Data on use of BP medication were collected through a questionnaire. Approximately 65% of NOMAS Hispanics report their country of origin as the Dominican Republic. An additional 12% report Puerto Rico, 8% Cuba, and 15% other. All subjects provided informed consent to participate, and the study was approved by the Institutional Review Boards of Columbia University in New York and the University of Miami.
MCR:
Samples were ascertained and assessed from the patient population in the cardiac catheterization labs of the University of Miami and Jackson Hospitals. BP was assessed in the catheterization lab or through the medical records; medication use was reported during the assessment. Race/ethnicity was determined by a combination of self-report and principal-components based clustering using genome-wide genotyping data, as some individuals did not report a known race/ethnicity (e.g., reported as “unknown/other”, despite clear clustering with white, non-Hispanics. While country of origin data was not complete for all MCR Hispanics (56% self-reporting a country of origin), of those that did report, the majority were from Cuba (54%) or the United States (14%); the rest were split between the Island countries (10%), South America (10%), Central America (7%), or other (5%). All subjects provided informed consent to participate, and the study was approved by the Institutional Review Board of the University of Miami.
Genotyping and Imputation
Genome-wide genotyping was performed in both cohorts using the Affymetrix (Santa Clara, CA, USA) SNP array 6.0 by the Hussman Institute of Human Genomics (HIHG) at University of Miami, Miller School of Medicine. Extensive quality control was performed in both samples and SNPs to ensure the integrity of the data. Samples were removed if they were determined to be related, had a low genotyping call rate (< 95%), or the self-reported and genetically determined sex were discrepant. After sample quality control, we had data available on 931 and 1063 Hispanic participants from NOMAS and MCR respectively.
SNPs were removed if they were out of Hardy-Weinberg equilibrium (p < 1.0 × 10−06), had a low minor allele frequency (MAF < 0.05), or had a low genotyping call rate (< 95%). Ten of the 29 previously reported SNPs were successfully genotyped in both NOMAS and MCR. Imputation to 1000 Genomes was then performed with IMPUTE2 [7] in each cohort separately, using the phase 1 version 3 reference panel consisting of 1092 individuals from Africa, Asia, Europe, and the Americas. The remaining 19 SNPs were imputed with good quality (INFO > 0.4) in NOMAS; however, only 18 of the remaining 19 were imputed successfully in MCR (rs805303 had low INFO score). After SNP quality control, data were available on all 29 SNPs in NOMAS and 28 SNPs in MCR.
Statistical Analysis
To account for the modifying effect of medication, 10 mm Hg was added to SBP and 5 mm Hg was added to DBP for all participants who were on antihypertensive medication, as was done by the International Consortium for Blood Pressure [4]. This method has been shown to preserve power and reduce bias as compared to including medication as a covariate or analyzing observed blood pressure [8]. In order to study the effect of each of the 29 SNPs on SBP and DBP in Hispanics, a linear regression analysis, assuming an additive genetic model, was performed in each cohort separately using the PLINK software package [9]. To be consistent with the International Consortium for Blood Pressure; age, age squared, sex, and BMI were included as covariates in NOMAS. BMI was not available in MCR and therefore was not included in the MCR analysis. Eigenstrat [10] was run separately in each cohort. After observing the scree plots (see Supplemental Figure 1) [11], the first two principal components were included as covariates in the regression model for each cohort in order to control for population substructure. Risk scores of the 29 SNPs for each of SBP and DBP were computed, using the regression coefficients as identified in 200 000 individuals of European descent and as described by the International Consortium for Blood Pressure [4]. Briefly, risk scores were calculated as a weighted sum of the risk allele counts. Both the risk allele and the weights for each SNP are derived from the published European study, where the risk allele is identified as the allele demonstrating risk in the published study and the weights as the corresponding beta coefficients. Association of the risk scores with SBP and DBP were tested in each cohort, adjusting for the same covariates as in the single SNP analyses. For both the single SNP and risk scores analyses, we then performed an inverse-variance meta-analysis of the two cohorts under a fixed effects model, as implemented in METAL [12]. Variance explained by the risk score was calculated using linear modeling in R.
To help define genetic ancestry we used Eigenstrat to project the samples from NOMAS and MCR onto the principal component (PC) space of HapMap phase II and III samples (PC1 and PC2). HapMap samples used include CEU (Utah residents with Northern and Western European ancestry) YRI (Yoruba in Ibadan, Nigeria), and MEX (Mexican ancestry in Los Angeles, California) populations [13]. These projection-based PCs were used to split a joint dataset into quartiles of both PC1 and PC2 based on ancestry. To look for trends across ancestry groups, the risk score association analyses were performed within each of the quartiles, adjusting for age, age squared, sex, cohort, and the projection-based PC1 and PC2 to account for residual admixture within quartile.
Post-hoc power calculations were performed using G*Power, version 3.1.9.2 [14], using the “linear bivariate regression” approach, with effect size taken from the previous reports [4,5], and sample size and standard deviations taken from our data.
Results
The distribution of blood pressure and associated risk factors for NOMAS and MCR are presented in Table 1. The average age at the time of BP reading was similar in the two cohorts (64 years in NOMAS and 65 years in MCR, p=1.84 × 10−03), but the percentage of women was quite different (62% in NOMAS and 33% in MCR, p=1.84 × 10−38). Although the percentage of hypertension was greater in MCR (92% in MCR and 68% in NOMAS, p=1.71 × 10−41), the raw measurements of SBP and DBP, prior to adjustment for use of anti-hypertensive medications, were significantly higher in NOMAS. This could be due to the increased percentage of individuals using anti-hypertensive medications in MCR (85% in MCR and 42% in NOMAS, p=4.18 × 10−90). Despite only 28 of the 29 SNPs being used to compute the risk score in MCR, we also see a higher average risk score in the MCR cohort for both SBP (p=3.62 × 10−05) and DBP (p=5.72 × 10−04). Given that the regression coefficient for the missing SNP, rs805303, is positive, the risk scores for MCR would only have increased had it been included. The distribution of the risk scores within each population are presented in Supplemental Figure 2.
Table 1:
Blood Pressure Measures and Associated Risk Factors
| NOMAS (N=931) | MCR (N=1063) | ||||
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | P | |||
| Age | 63.98 ± 7.99 | 65.45 ± 12.81 | 1.84E-03 | ||
| Systolic Blood Pressure (SBP) | 140.00 ± 19.62 | 136.09 ± 24.29 | 7.44E-05 | ||
| Diastolic Blood Pressure (DBP) | 83.46 ± 10.35 | 75.81 ± 13.12 | 1.62E-45 | ||
| Body Mass Index (BMI) | 28.46 ± 4.71 | NA | NA | ||
| SBP Risk Score | 0.44 ± 1.70 | 0.76 ± 1.77 | 3.62E-05 | ||
| DBP Risk Score | 0.21 ± 1.08 | 0.38 ± 1.12 | 5.72E-04 | ||
| N | % | N | % | ||
| Sex (Female) | 574 | 61.65 | 348 | 32.74 | 1.84E-38 |
| Hypertension | 636 | 68.31 | 976 | 91.82 | 1.71E-41 |
| On Hypertensive Medication | 387 | 41.57 | 863 | 84.52 | 4.18E-90 |
While one SNP for SBP and four SNPs for DBP reached nominal significance (p < 5.0 × 10−2) in the meta-analysis of NOMAS and MCR, none were significant after controlling the family-wise error rate at 5% over 58 tests (Table 2). The most significant SNP in the analysis of both SBP (p = 1.19 × 10−2) and DBP (p = 3.80 × 10−3) was rs7129220 in adrenomedullin (ADM) on chromosome 11. This was also the only SNP which showed p < 5.0 × 10−2 for both SBP and DBP. We did however see significant correlation between the reported effects sizes and our estimated effects sizes for both NOMAS (r2=0.180 and model p-value=0.022 for SBP; r2=0.148 and model p-value=0.039 for DBP) and for MCR (r2=0.4225, model p-value=1.81×10−4 for SBP; r2=0.3676, model p-value=6.26×10−4 for DBP; see Supplemental Figures 3 and 4).
Table 2:
Association Results for 29 Blood Pressure SNPs
| Alleles | SBP | DBP | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | CHR | POS | CA/NCA | CAF NOMAS | CAF MCR | CAF CHARGE European | Beta | P-value | Q-value | Effect in NOMAS/MCR | Beta | P-value | Q-value | Effect in NOMAS/MCR |
| MTHFR-NPPB | rs17367504 | 1 | 11,862,778 | G/A | 0.12 | 0.13 | 0.15 | −1.062 | 3.19E-01 | 7.66E-01 | −/+ | −0.539 | 3.49E-01 | 7.66E-01 | −/+ |
| MOV10 | rs2932538 | 1 | 113,216,543 | G/A | 0.80 | 0.80 | 0.75 | 0.434 | 6.20E-01 | 8.24E-01 | +/+ | −0.222 | 6.37E-01 | 8.50E-01 | +/− |
| SLC4A7 | rs13082711 | 3 | 27,537,909 | T/C | 0.85 | 0.82 | 0.78 | −1.194 | 2.14E-01 | 7.30E-01 | −/− | −0.790 | 1.27E-01 | 6.09E-01 | −/− |
| ULK4 | rs3774372 | 3 | 41,877,414 | T/C | 0.80 | 0.81 | 0.83 | −0.377 | 6.63E-01 | 8.50E-01 | +/− | −0.701 | 1.32E-01 | 6.09E-01 | −/− |
| MECOM | rs419076 | 3 | 169,100,886 | T/C | 0.45 | 0.48 | 0.47 | −0.153 | 8.28E-01 | 9.44E-01 | −/+ | 0.396 | 2.96E-01 | 7.66E-01 | −/+ |
| FGF5 | rs1458038 | 4 | 81,164,723 | T/C | 0.22 | 0.24 | 0.29 | 0.728 | 4.11E-01 | 7.89E-01 | +/+ | 0.038 | 9.36E-01 | 9.44E-01 | −/+ |
| SLC39A8 | rs13107325 | 4 | 103,188,709 | T/C | 0.04 | 0.07 | 0.05 | −0.789 | 6.49E-01 | 8.50E-01 | +/− | −1.008 | 2.82E-01 | 7.66E-01 | −/− |
| GUCY1A3-GUCY1B3 | rs13139571 | 4 | 156,645,513 | C/A | 0.76 | 0.74 | 0.76 | 0.654 | 4.23E-01 | 7.89E-01 | −/+ | 0.245 | 5.76E-01 | 8.24E-01 | −/+ |
| NPR3-C5orf23 | rs1173771 | 5 | 32,815,028 | G/A | 0.67 | 0.63 | 0.60 | 0.162 | 8.28E-01 | 9.44E-01 | +/+ | 0.254 | 5.26E-01 | 8.17E-01 | −/+ |
| EBF1 | rs11953630 | 5 | 157,845,402 | T/C | 0.27 | 0.34 | 0.37 | 0.587 | 4.36E-01 | 7.89E-01 | +/+ | 0.075 | 8.52E-01 | 9.44E-01 | −/+ |
| HFE | rs1799945 | 6 | 26,091,179 | G/C | 0.11 | 0.14 | 0.14 | 0.100 | 9.25E-01 | 9.44E-01 | −/+ | 0.473 | 4.13E-01 | 7.89E-01 | −/+ |
| BAT2-BAT5 | rs805303 | 6 | 31,616,366 | G/A | 0.53 | NA | 0.61 | 1.830 | 5.46E-02 | 5.27E-01 | +/? | 1.071 | 3.23E-02 | 3.75E-01 | +/? |
| CACNB2(5') | rs4373814 | 10 | 18,419,972 | G/C | 0.51 | 0.54 | 0.55 | 0.483 | 4.99E-01 | 7.89E-01 | +/− | −0.193 | 6.17E-01 | 8.24E-01 | +/− |
| CACNB2(3') | rs1813353 | 10 | 18,707,448 | T/C | 0.72 | 0.67 | 0.68 | 0.878 | 2.52E-01 | 7.63E-01 | −/+ | 0.666 | 1.07E-01 | 6.09E-01 | +/+ |
| C10orf107 | rs4590817 | 10 | 63,467,553 | G/C | 0.83 | 0.85 | 0.84 | −0.127 | 8.93E-01 | 9.44E-01 | −/+ | −0.098 | 8.48E-01 | 9.44E-01 | −/+ |
| PLCE1 | rs932764 | 10 | 95,895,940 | G/A | 0.35 | 0.42 | 0.44 | −0.771 | 2.80E-01 | 7.66E-01 | −/− | −0.928 | 1.56E-02 | 2.59E-01 | −/− |
| CYP17A1-NT5C2 | rs11191548 | 10 | 104,846,178 | T/C | 0.90 | 0.90 | 0.91 | 2.005 | 8.03E-02 | 6.09E-01 | +/+ | 0.095 | 8.78E-01 | 9.44E-01 | −/+ |
| ADM | rs7129220 | 11 | 10,350,538 | G/A | 0.91 | 0.90 | 0.89 | −2.908 | 1.19E-02 | 2.42E-01 | −/− | −1.800 | 3.80E-03 | 2.33E-01 | −/− |
| PLEKHA7 | rs381815 | 11 | 16,902,268 | T/C | 0.26 | 0.27 | 0.26 | 0.908 | 2.49E-01 | 7.63E-01 | +/+ | 0.328 | 4.38E-01 | 7.89E-01 | +/− |
| FLJ32810-TMEM133 | rs633185 | 11 | 100,593,538 | G/C | 0.28 | 0.31 | 0.28 | −0.595 | 4.38E-01 | 7.89E-01 | −/− | −0.466 | 2.59E-01 | 7.66E-01 | −/− |
| ATP2B1 | rs17249754 | 12 | 90,060,586 | G/A | 0.85 | 0.83 | 0.84 | 1.152 | 2.20E-01 | 7.30E-01 | +/+ | −0.211 | 6.77E-01 | 8.50E-01 | −/+ |
| SH2B3 | rs3184504 | 12 | 111,884,608 | T/C | 0.30 | 0.36 | 0.47 | 0.015 | 9.85E-01 | 9.94E-01 | −/+ | −0.281 | 5.01E-01 | 7.89E-01 | −/− |
| TBX5-TBX3 | rs10850411 | 12 | 115,387,796 | T/C | 0.64 | 0.66 | 0.70 | −0.370 | 6.20E-01 | 8.24E-01 | −/− | 0.076 | 8.49E-01 | 9.44E-01 | +/− |
| CYP1A1-ULK3 | rs1378942 | 15 | 75,077,367 | C/A | 0.65 | 0.53 | 0.35 | 1.379 | 6.71E-02 | 5.40E-01 | +/+ | 0.598 | 1.40E-01 | 6.09E-01 | +/+ |
| FURIN-FES | rs2521501 | 15 | 91,437,388 | T/A | 0.26 | 0.30 | 0.31 | 1.049 | 1.92E-01 | 7.30E-01 | +/+ | 0.403 | 3.51E-01 | 7.89E-01 | +/+ |
| GOSR2 | rs17608766 | 17 | 45,013,271 | T/C | 0.92 | 0.90 | 0.86 | −0.903 | 4.99E-01 | 7.89E-01 | −/− | −1.845 | 1.02E-02 | 2.42E-01 | −/− |
| ZNF652 | rs12940887 | 17 | 47,402,807 | T/C | 0.26 | 0.30 | 0.38 | 0.776 | 3.24E-01 | 7.66E-01 | +/+ | 0.653 | 1.23E-01 | 6.09E-01 | +/+ |
| JAG1 | rs1327235 | 20 | 10,969,030 | G/A | 0.41 | 0.43 | 0.46 | −0.563 | 4.20E-01 | 7.89E-01 | −/− | 0.208 | 5.79E-01 | 8.24E-01 | +/− |
| GNAS-EDN3 | rs6015450 | 20 | 57,751,117 | G/A | 0.10 | 0.10 | 0.12 | 1.776 | 1.51E-01 | 7.01E-01 | +/+ | −0.039 | 9.53E-01 | 9.44E-01 | +/− |
Italic SNPs are those that were directly genotyped
Position is based on human genome 19 and dbSNP 137. CA = coding allele, NCA = non-coding allele, CAF = coding allele frequency
Bold P-values indicate two-sided P < 5.0×10−02
Bold β coefficients indicate that the direction of the effect is the same as Europeans from the study conducted by the International Consortium on Blood Pressure
The genetic risk score was replicated in Hispanics through the meta-analysis of NOMAS and MCR for both SBP (p = 5.65 × 10−4) and DBP (p = 1.65 × 10−3) (Table 3). However, the effect size of the risk score in MCR was much larger than in NOMAS (β=1.06 vs 0.50 for DBP, corresponding to a 1.06 and 0.50 mm Hg increase in DBP for each 1 unit increase in risk score). The effect size in MCR was even larger than in the previously reported replication with a European sample (β=0.84), approaching that seen with an East Asian sample (β=1.13) [4]. The NOMAS effect size approaches that in the previously reported replication with an African American sample (β=0.45) [4]. A similar trend was seen for SBP. No significant change in the results was seen after removing the adjustment for BMI in the NOMAS analysis (data not shown). This difference in effect size was also reflected in differences in the blood pressure variation explained by the risk score. In the MCR dataset the risk score explained 0.93% and 0.81% of SBP and DBP respectively (after controlling for relevant covariates), while the same risk score explained 0.30% and 0.24% of the variance of SBP and DBP for the NOMAS dataset. Post-hoc power calculations showed that these datasets were sufficiently powered to detect the reported effect sizes of the risk score [4] from the European population (power of 0.65 to 0.72 for SBP, and 0.76 to 0.85 for DBP).
Table 3:
Association Results for Blood Pressure Risk Scores
| SBP | DBP | ||||||
|---|---|---|---|---|---|---|---|
| N | Beta | SE | P | Beta | SE | P | |
| Current Hispanic Data Sets | |||||||
| NOMAS | 931 | 0.65 | 0.39 | 9.41E-02 | 0.5 | 0.33 | 1.31E-01 |
| MCR | 1,063 | 1.3 | 0.4 | 1.30E-03 | 1.06 | 0.35 | 2.80E-03 |
| Combined | 1,994 | 0.97 | 0.28 | 5.65E-04 | 0.76 | 0.24 | 1.65E-03 |
| Previous Validation Data Sets | |||||||
| European (Stage 1–3) | 133,661 | 0.85 | 0.03 | 2.76E-156 | 0.84 | 0.03 | 3.59E-153 |
| African | 19,775 | 0.41 | 0.12 | 9.83E-04 | 0.45 | 0.11 | 5.30E-05 |
| South Asian | 23,977 | 0.55 | 0.08 | 2.92E-13 | 0.57 | 0.07 | 9.53E-15 |
| East Asian | 29,719 | 1.06 | 0.08 | 1.11E-40 | 1.13 | 0.08 | 2.91E-48 |
All validation results are from the study conducted by the International Consortium on Blood Pressure
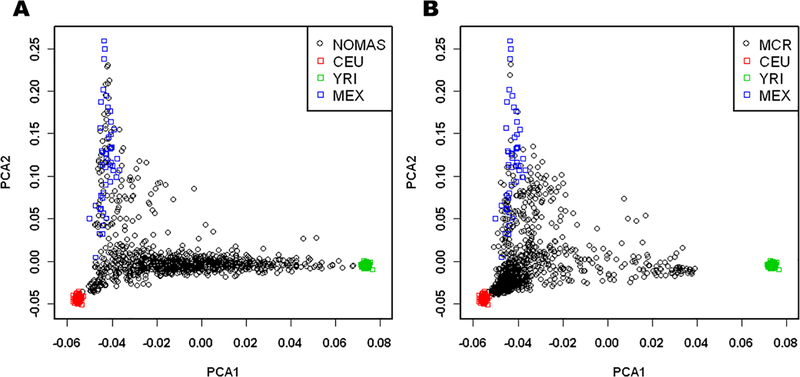
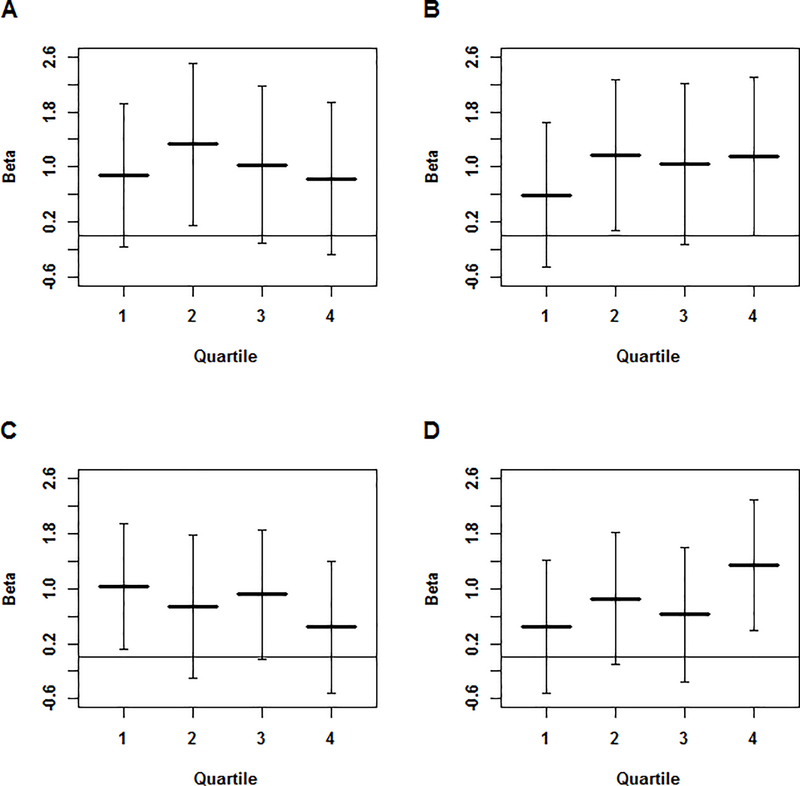
When projecting the samples from NOMAS and MCR onto the HapMap population space, we see that the NOMAS cohort is made up predominantly of samples with both African and European ancestry (Figure 1A), while there is more European ancestry in the MCR cohort (Figure 1B). Both cohorts show a small portion of samples with Native American ancestry, as seen by their clustering with the Mexican samples. For PC1, the quartiles correspond to differing levels of African ancestry, with quartile 4 having the most African ancestry and quartile 1 having the least. For PC2, the quartiles correspond to differing levels of Native American or Asian ancestry, with quartile 4 having the most Native American or Asian ancestry and quartile 1 having the least. For the PC1 quartiles, we noted a downward trend in the magnitude of the regression coefficients from quartile 1 to quartile 4 for DBP (p = 3.83 × 10−01 for the difference in βs between the 1st and 4th quartile in Figure 2C). A downward trend is seen from quartile 2 to quartile 4 with SBP (p = 5.49 × 10−01 for the difference in βs between the 2nd and 4th quartile), though quartile 1 is not consistent with this trend (Figure 2A). For the quartiles of PC2, we see an upward trend in the magnitude of the regression coefficients from quartile 1 to quartile 4 for both SBP and DBP (p = 4.87 × 10−01 and p = 1.91 × 10−01 for the difference in βs between the 1st and 4th quartile in Figure 2B and 2D respectively).
Figure 1: Plots of Principal Components 1 and 2, for NOMAS, MCR, and HapMap data.
Principal Components (PC) 1 and 2 of the (A) NOMAS dataset and (B) MCR dataset projected onto a HapMap population-based PC space. The X axis indicates PC1, differentiating African ancestry from non-African ancestry. The Y axis indicates PC2, differentiating Asian/Native American ancestry from non-Asian/non-Native American ancestry.
Figure 2. β Coefficients by PC-based Quartiles.
β coefficients and 95% confidence intervals for the analysis of (A) SBP by quartiles of PC1, (B) SBP by quartiles of PC2, (C) DBP by quartiles of PC1, and (D) DBP by quartiles of PC2. For plots A and C, lower quartiles indicate less African ancestry. For plots B and D, lower quartiles indicate less Asian/Native American ancestry.
Discussion
While our study confirmed the utility of the genetic risk scores for SBP (β = 0.97 and p = 5.65 × 10−04) and DBP (β = 0.76 and p = 1.65 × 10−03) in a sample of 1994 Hispanics, we did observe differences in the magnitude of the regression coefficients and amount of variance explained by the risk score between NOMAS and MCR. The magnitude of the regression coefficients and the amount of variance explained were larger in MCR than NOMAS for both SBP (NOMAS: β = 0.65 and 0.30% variance explained; MCR: β = 1.30 and 0.93% variance explained) and DBP (NOMAS: β = 0.50 and 0.24% variance explained; MCR: β = 1.06 and 0.81% variance explained).
One explanation could be the differences in genetic ancestry between NOMAS and MCR, with NOMAS showing more individuals with African admixture. The countries of origin reported most frequently in NOMAS are the Dominican Republic (65%) and Puerto Rico (12%). The African admixture observed in NOMAS is consistent with previous studies of population structure and admixture among Hispanic and Latino populations, which have shown that both Dominicans (42%) and Puerto Ricans (24%) have high levels of African Ancestry [15]. It is interesting to note that this is also consistent with the higher prevalence of hypertension observed in Dominicans and Puerto Ricans as compared with other Hispanic sub-populations [3]. Country of origin most frequently reported in the MCR are Cuba (54%), United States (14%), and countries from Central and South America (17%); these tend to have less African ancestry than Island countries such as the Dominican Republic and Puerto Rico [16]. The genetic admixture among Hispanic populations from different regions is well understood and likely a result of multiple factors; including the extent and rate at which European settlers displaced native populations and the whether or not slavery was introduced [15].
To determine whether the difference in admixture between cohorts contributed to the observed difference in effect of the risk scores on BP, we first combined the two cohorts and then divided the data into quartiles based on HapMap projection-based PC1 and PC2. The association analysis of risk score within each quartile confirmed a trend of lower risk score effect estimates among quartiles with increased African ancestry, and higher risk score effect estimates among quartiles with increased Asian admixture, even when controlling for cohort. This is consistent with the previous meta-analysis (β coefficients of ~0.9, 1.1, and 0.5 were observed in Europeans, East Asians, and Africans respectively), and shows that the utility and interpretation of the risk scores for SBP and DBP depends at least in part on the underlying genetic ancestry of the populations. This is of particular concern if the risk scores are used in a predictive or risk-stratification context, as the genetic risk of the individual may change based on their underlying ancestry. We also note that the observed difference between cohorts and across quartiles may actually become larger if the local ancestry for the 29 variants in the risk score were used to measure ancestry, rather than the total global (genome-wide) ancestry.
Differences in admixture between the two cohorts may not be the only cause of the observed differences in the magnitude of regression coefficients and amount of variance explained by the risk score between the two cohorts. There is an ascertainment bias that could contribute to the observed differences, with MCR being derived from a high risk population. The observed differences between the two studies with respect to sex ratio and use of anti-hypertensive medications are likely reflective of the difference in ascertainment schemes: NOMAS samples being largely unselected at time of enrollment, while MCR samples were highly selected for cardiovascular risk factors. The lower percentage of women observed in MCR is consistent with the increased prevalence of cardiovascular events in males as compared to females [17]. The higher percentage of individuals on anti-hypertensive medications observed in MCR is expected given their increased risk for cardiovascular disease. In the European sample from which the risk score was derived, the percentage of women is 59%, similar to that of NOMAS, and the percentage of individuals using anti-hypertensives is 14%, lower than both NOMAS and MCR. This could be due in part to the lower average age in the European sample (52 compared to 64 and 65 in NOMAS and MCR respectively) [4]. There may also be other unmeasured or unaccounted for environmental and lifestyle factors which contribute to the observed differences, some of which may be correlated with ancestry. For example, the effect of antihypertensive medicines may not be consistent across subpopulations, and thus the correction (10 SBP and 5 DBP) may not be appropriate for both populations.
While our current Hispanic sample sets may be underpowered to detect weaker associations for the 29 variants individually, we do see a number of variants with nominal significance. Furthermore, we confirm the utility of the risk score in Hispanics. In particular we note that our observed association is almost as strong (as with DBP, Table 3) or slightly stronger (as with SBP, Table 3) than in the previous replication with an African population [4], despite our sample size being much smaller. Although we have shown the risk score to be useful in Hispanics, we acknowledge that admixture plays a large role in the utility and interpretation within specific Hispanic sub-populations. This also shows the need for further genetic and epidemiological studies in Hispanics. Given the small effect size of the risk score, and the high variance in blood pressure, currently the genetic risk score is likely of minimal clinical impact (see Supplemental Figure 5) [18,19]. Sub-population specific studies may reveal important risk loci or environmental factors that are not associated in the particular Hispanic sub-populations, or may show population-specific risk scores or weights that are more readily interpretable.
Supplementary Material
Summary Table.
What is known about topic:
Hypertension is strongly influence by genetic factors and a genetic risk score has been developed in European ancestry datasets
The performance of the risk score is well described in European populations, but is not well-described in Hispanic populations
What this study adds:
This study confirms that the risk score is associated with hypertension in Hispanic datasets
This study shows that underlying ancestry (e.g., European, African) ancestry influences the degree to which the risk score is associated.
Acknowledgements
This research was supported by grants from the National Institute of Neurological Disorders and Stroke grants R37NS029993 (RLS, TR), K24NS06273 (TR), the Evelyn F. McKnight Brain Institute (RLS, SB), the National Heart Lung and Blood Institute grant R01HL102487 (GWB, DS), and a gift from the John P. Hussman Foundation. We thank Dr. William K. Scott at the John P. Hussman Institute for Human Genomics for his helpful comments on the manuscript.
SOURCES OF SUPPORT: This research was supported by grants from the National Institute of Neurological Disorders and Stroke grants R37NS029993 (RLS, TR), K24NS06273 (TR), the Evelyn F. McKnight Brain Institute (RLS, SB), the National Heart Lung and Blood Institute grant R01HL102487 (GWB, DS), and a gift from the John P. Hussman Foundation.
Footnotes
PREVIOUS PUBLICATION: This is original work that has not been previously published.
CONFLICT OF INTEREST: None; there are no conflicts of interest to report
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daviglus ML, Talavera GA, Avilés-Santa ML, Allison M, Cai J, Criqui MH, et al. Prevalence of major cardiovascular risk factors and cardiovascular diseases among Hispanic/Latino individuals of diverse backgrounds in the United States. JAMA. 2012;308:1775–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, et al. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham heart study. Hypertension. 2000;36:477–483. [DOI] [PubMed] [Google Scholar]
- 4.International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013;93: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacco RL, Anand K, Lee HS, Boden-Albala B, Stabler S, Allen R, et al. Homocysteine and the risk of ischemic stroke in a triethnic cohort: the NOrthern MAnhattan Study. Stroke. 2004;35:2263–2269. [DOI] [PubMed] [Google Scholar]
- 7.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007;39:906–913. [DOI] [PubMed] [Google Scholar]
- 8.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24(19):2911–2935. [DOI] [PubMed] [Google Scholar]
- 9.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 11.Cattell RB. The scree test for the number of factors. Multivariate Behavioral Research. 1966;1:245–276. [DOI] [PubMed] [Google Scholar]
- 12.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faul F, Erdfelder E, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behavior Research Methods. 2009;41, 1149–1160. [DOI] [PubMed] [Google Scholar]
- 15.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA. 2010;107 Suppl 2:8954–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. Int J Cardiol. 2013;168(2):934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: Markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5(2):e1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pencina MJ, D’Agostino RB Sr, D’Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med. 2008:27(2):157–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.