Abstract
Inner nuclear membrane (INM) protein composition regulates nuclear function, affecting processes such as gene expression, chromosome organization, nuclear shape, and stability. Mechanisms that drive changes in the INM proteome are poorly understood, in part because it is difficult to definitively assay INM composition rigorously and systematically. Using a split-GFP complementation system to detect INM access, we examined the distribution of all C-terminally tagged Saccharomyces cerevisiae membrane proteins in wild-type cells and in mutants affecting protein quality control pathways, such as INM-associated degradation (INMAD), ER-associated degradation, and vacuolar proteolysis. Deletion of the E3 ligase Asi1 had the most specific effect on the INM compared to mutants in vacuolar or ER-associated degradation pathways, consistent with a role for Asi1 in the INMAD pathway. Our data suggest that Asi1 not only removes mistargeted proteins at the INM, but also controls the levels and distribution of native INM components, such as the membrane nucleoporin Pom33. Interestingly, loss of Asi1 does not affect Pom33 protein levels but instead alters Pom33 distribution in the nuclear envelope through Pom33 ubiquitination, which drives INM redistribution. Taken together, our data demonstrate that the Asi1 E3 ligase has a novel function in INM protein regulation in addition to protein turnover.
Keywords: inner nuclear membrane, ERAD, split-GFP, Asi1, Pom33, NPC
THE nucleus is the defining feature of eukaryotic cells. Successful propagation of nuclei, and the genome contained within their walls, is vital for an organism’s survival. Although the double lipid bilayer that forms the nuclear envelope (NE) is often viewed as a fortress, the inner and outer nuclear membranes (INM and ONM) are highly dynamic structures that undergo changes in structure and composition throughout development and differentiation, in mitosis and meiosis, and in diseased and dying cells (Dauer and Worman 2009; Chen et al. 2014; Gordon et al. 2014). Lobulated nuclei with aberrant membranes and abnormal chromosome configurations are used to grade many tumors; however, similar changes in nuclear shape and genome organization also occur during the maturation of normal cell types, most notably during hematopoiesis, suggesting a complexity at the NE that we are just beginning to understand (Skinner and Johnson 2017). As many of the unique properties of the NE, such as its mechanical stiffness, distinctive lipid composition, and chromosome organization, are attributed to the INM, understanding the composition, function, and regulation of the INM is a key problem in cell biology.
In all eukaryotes, the INM and ONM are joined together at many spots where nuclear pore complexes (NPCs) reside. NPCs form the first regulator of INM composition by controlling the passage of proteins and other macromolecules into and out of the nucleus. INM proteins travel through central or lateral channels of the NPC to reach the INM [reviewed in Katta et al. (2014), Ungricht and Kutay (2017)]. Most do not have any targeting sequence and reach the INM by diffusion; their retention at the INM occurs through binding to nuclear or NE-associated proteins such as lamins, NPCs, or chromatin (Furukawa et al. 1998; Wu et al. 2002; Antonin et al. 2011; Ungricht and Kutay 2015; Ungricht et al. 2015). A small subset of proteins is targeted to the INM by a specific sequence motif, which is recognized by the nuclear translocation machinery (King et al. 2006; Lusk et al. 2007; Turgay et al. 2010; Gardner et al. 2011; Tapley et al. 2011). Additional mechanisms of INM transport have also been proposed that bypass the NPC, and rely on the budding and fusion of vesicles from the ONM to the INM (Speese et al. 2012; Mettenleiter 2016). While many mechanistic details regarding INM transport are still poorly understood, it seems clear that the INM has a distinct composition from the ONM, which is contiguous with the ER.
In the ER, misfolded or damaged proteins are targeted for degradation by the ER-associated degradation pathway (ERAD) as part of the cell’s surveillance system, to prevent the formation of nonfunctional complexes or aggregates of defective protein (Zattas and Hochstrasser 2015). The conserved E3 ligases Doa10/MARCH6/TEB4 and Hrd1/SYVN1 recognize lesions in the cytosolic or luminal/membrane regions of ER proteins, respectively, resulting in ubiquitination and retro-translocation of defective proteins back into the cytoplasm for destruction by the 26S proteasome (Zattas and Hochstrasser 2015). Although ERAD likely ensures that ONM proteins are functional and present in the correct stoichiometry, it is unclear if this pathway operates at the INM due to its separation from the ONM/ER by NPCs (Boban and Foisner 2016). Instead, an INM-associated degradation (INMAD) pathway has been proposed to remove mistargeted proteins from the INM through ubiquitin-mediated proteolysis (Foresti et al. 2014; Khmelinskii et al. 2014).
Three putative RING finger E3 ligases have been implicated in INMAD in yeast: Doa10, Asi1, and Asi3. A broad range of Doa10 substrates have been described in the nucleus and ER, including the MATα2 transcriptional repressor, a soluble version of the kinetochore protein Ndc10, and a mutant form of the spindle pole body (SPB) protein Mps2 (Swanson et al. 2001; Ravid et al. 2006). Asi1 and Asi3 are thought to form a multimeric E3 ligase complex together with the adapter protein Asi2 (Foresti et al. 2014). The best-characterized substrates of this Asi complex are the transcription factors Stp1 and Stp2, which are ubiquitinated in the nucleus as part of the SPS (Ssy1-Ptr3-Ssy5) sensor pathway that monitors the extracellular amino acid environment (Boban et al. 2006; Zargari et al. 2007; Omnus and Ljungdahl 2014). More recently, roles for Doa10, Asi1, and Asi3 were suggested in the turnover of INM proteins, including Erg11, Nsg1, and Asi2. These and other substrates were identified based on increased whole-cell protein levels in cells lacking the ligases (Boban et al. 2014; Foresti et al. 2014; Khmelinskii et al. 2014). Interestingly, many of the INMAD substrates identified were not INM components but proteins mistargeted to the INM, such as a mutant version of the Sec61 translocon, vacuolar transport complex subunits such as Vtc1, and multiple plasma membrane transporters (Foresti et al. 2014; Khmelinskii et al. 2014), leading to the idea that the primary role of INMAD is to rid the INM of non-INM proteins. The mechanisms by which INMAD distinguishes between “foreign” and “resident” membrane proteins, and whether INMAD targets damaged or misfolded INM components through a pathway similar to ERAD, remains unknown.
Examination of protein stability in rats using isotope labeling suggested that NPCs and INM proteins such as lamins are extremely long-lived, leading to the general idea that the INM is stable, with little protein turnover (Savas et al. 2012). One important exception occurs under nutrient deprivation when nonessential sections of the entire nucleus, including the INM, are pinched off into the vacuole (the yeast lysosome) and degraded, a process known as piecemeal nuclear autophagy (PMNA) (Adnyana et al. 2000; Do et al. 2003; Roberts et al. 2003; Millen et al. 2009). PMNA has not yet been identified in higher eukaryotes, but autophagy of NE proteins in mammalian cells occurs (Park et al. 2009; Dou et al. 2015), and is linked to both cancer and aging (Martinez-Lopez et al. 2015; White et al. 2015). However, a role for INMAD outside of yeast has not been reported.
We previously described a method using split-GFP that allowed us to systematically and unequivocally assay the ability of budding yeast membrane proteins to access the INM (Smoyer et al. 2016). Unlike biochemical methods for studying INM composition that depend on in silico subtraction, or comparative analysis of nuclear and microsomal membrane samples (an ER-derived fraction formed in vitro), our assay is specific for the INM pool of protein, allowing for analysis of proteins that have dual functions. The assay also discriminates the INM from the ONM, using endogenously expressed proteins that serve as the sole copy in the cell and can be used in live cells. To test the contribution of INMAD to INM composition, we compared the distribution of proteins using split-GFP in wild-type cells to that of cells lacking ASI1. Comparison of INM composition in cells lacking other protein quality control components, such as the INMAD/ERAD E3 ligase Doa10, the ERAD E3 ligase Hrd1, the E2 ubiquitin-conjugating enzyme Ubc7, and the vacuolar peptidase Pep4, revealed 64 proteins whose INM levels increased in cells lacking ASI1, suggesting direct or indirect regulation by Asi1. In asi1∆, we also observed an increase in the size and frequency of INM puncta containing Pom33/Tts1/TMEM33, a conserved NPC-localized protein that plays a role in NPC distribution, biogenesis, and/or stability (Chadrin et al. 2010; Urade et al. 2014; Zhang and Oliferenko 2014). We provide evidence that Pom33 is directly ubiquitinated by Asi1; however, ubiquitination of Pom33 does not lead to turnover, but instead contributes to proper Pom33 INM distribution.
Materials and Methods
Yeast strains and plasmids
All strains are derivatives of BY (can1Δ::STE2pr-Sp-HIS5 lyp1Δ his3Δ1 leu2ΔO ura3ΔO met15ΔO LYS2). The split-GFP library was made by first integrating the pRS315-NOP1pr-GFP11-mCherry-PUS1 (pSJ1321) reporter into a MATα derivative to create SLJ7859 (MATα can1Δ::STE2pr-Sp-HIS5 lyp1Δ his3Δ1 leu2ΔO ura3ΔO met15ΔO LYS2 pLEU2-NOP1pr-GFP11-mCherry-PUS1).
Construction of a C-terminally tagged library of membrane proteins with GFP1–10-URA3MX (pSJ1256) by PCR was previously described (Smoyer et al. 2016). Briefly, genes associated with the gene ontology annotation of integral component of membrane or transmembrane were compiled using the Saccharomyces Genome Database (SGD), and TMHMM v2 software (Krogh et al. 2001) was used to predict additional genes containing hydrophobic stretches of > 16 amino acids using a version of the genome downloaded on 6/10/2012. These genes were then tagged by PCR, resulting in a library of 1010 strains. Although 169 of the clones appeared to be soluble, these were kept in the library because it was unknown if their membrane association/integration was regulated, was disrupted by C-terminal tagging, or if they lacked a bona fide membrane domain (Smoyer et al. 2016). The library was crossed to deletion mutants taken from the yeast deletion collection (MATa yfg∆::KANMX LYP1CAN1his3ura3leu2) using the Singer RoToR robot as previously described (Tong and Boone 2006). Following diploid selection on synthetic defined media lacking uracil with monosodium glutamate and G418 (SD/MSG-Ura+G418), cells were sporulated for 3–4 weeks, then haploids containing the deletion and the tags were selected twice on SD/MSG-Ura-Leu-His-Lys-Arg+thialysine+canavanine+G418. All plates were incubated at 23°. We did not recover certain combinations of deletion mutants/tagged genes/selection markers due to linkage and slow growth of certain strains. Therefore, mutant libraries contained fewer genes than the original library. A complete list of genes screened can be found in Supplemental Material, Tables S1 and S2.
For colocalization experiments, we used the pRS315-NOP1pr-GFP11-PUS1 (pSJ1679) reporter so that genes could be C-terminally tagged with mCherry using pFA6-mCherry-KANMX by PCR. Strains used for each experiment are listed in Table S4, with the exception of strains taken directly from our split-GFP libraries.
INM mutant screen
Cells were grown overnight at 23° in SC-Leu in a 96-well plate format, using deep-well dishes (Thermo-Scientific). Imaging plates (Ibidi) were pretreated with 100 μl of polylysine (Sigma [Sigma Chemical], St. Louis, MO) for 1 hr, and then rinsed twice with water and allowed to dry before use. Cells were plated using 100 μl of culture. Each plate was imaged on a Nikon (Garden City, NY) Eclipse TI equipped with a Yokogawa CSU W1 spinning disk head and Andor EMCCD using a Nikon Apo TIRF 100× 1.49NA Oil objective. mCherry was imaged using a 561-nm laser at 100% power and ET605/70m emission filter, with an exposure time of 100 msec. GFP was imaged using a 488-nm laser at 100% power and ET535/30m emission filter, with an exposure time of 200 msec. Four points were automatically selected for each well. An automation script moved to positions, found focus using Nikon perfect focus system (PFS), and imaged each channel with a z-stack of 13 slices and spacing of 0.5 µm. Image processing was performed in ImageJ using custom macros and plugins.
In brief, images were background-subtracted and sum-projected in z, and for each cell a mask of the nucleus was generated based on the mCherry channel, as well as a ring mask surrounding the nucleus to measure the cytoplasmic signal. mCherry and GFP intensity were measured in the nuclear and ring masks for each cell. Cells with a high cytoplasmic signal were discarded as dead cells. For each live cell, the nuclear GFP/mCherry intensity ratio was calculated; from these data the average ratio per sample was calculated, and for each protein the mutant/wild-type ratio was calculated from these averages (given in Table S1). Proteins were counted as hits if the mutant/wild-type ratio was > 1.4.
Wild-type and asi1∆ images were manually inspected for signal in both the 561- and 488-nm channels. If no signal was present upon visual inspection, the sample was categorized as a false positive and was removed from subsequent analysis with all mutants. These genes are listed in Table S2; most correspond to negatives or nonabundant INM hits in wild-type cells (Smoyer et al. 2016). If signal was detected in the nucleus rather than the INM, the sample was categorized as nuclear. This could occur because the protein is not a bona fide transmembrane protein, because its binding partner at the INM is absent, or because C-terminal tagging interfered with membrane association (Smoyer et al. 2016).
Confocal imaging
Cells were grown overnight in SC-Leu in 2-ml cultures, and then harvested for imaging and immobilized on polylysine slides (Polysciences, Warrington, PA). Imaging was conducted on a Zeiss ([Carl Zeiss], Thornwood, NY) LSM780. All images were taken using a 40× water objective, fluorophores were excited using a 488- and 561-nm Argon laser line, GFP emission was collected through a BP 505–540-nm filter, and mCherry was collected through a LP 580-nm filter, with the pinhole set to 1 airy units. Images taken for puncta quantification, puncta measurement, and colocalization experiments had a zoom of 6, and z-stack of 10–12 slices. Puncta widths were measured from manually drawn line profiles over puncta averaged over a thickness of eight pixels. Profiles were then aligned to their peak maxima and averaged to determine average peak profiles. They were then fitted to Gaussian functions using nonlinear least squares to determine their width. Error bars were determined from Monte Carlo analysis with 100 random simulations, as previously described (Bevington and Robinson 2003).
Ubiquitination of Pom33
Liquid nitrogen ground lysates were prepared as previously described in Jaspersen et al. (2006), Bupp et al. (2007), and Friederichs et al. (2012). Cells of the indicated genotypes were grown overnight, then diluted into 1-liter cultures in the morning to an OD600 of ∼0.2. Cells were allowed to grow until midlog phase and then harvested. Extraction buffer was adjusted for each sample so that an OD600 of 1.0 would have 5 ml of extraction buffer added. Cells were then dropped into liquid nitrogen to make yeast dots and frozen at −80°. Yeast powder was made by grinding the frozen yeast dots using a Retsch ball mill 4 × 3 min, 30 Hz. Powder was then collected and weighed out to equal amounts for each sample (∼5 mg). Ground cell powder was thawed on ice, then resuspended in 9 ml of extraction buffer (20 mM HEPES-NaOH, pH 7.5; 300 mM NaCl; 1 mM EDTA; 5 mM EGTA; 50 mM NaF; 50 mM β-glycerophosphate; 0.5% TritonX-100; 1 mM DTT; 1 mM PMSF; and 1 mg/ml each pepstatin A, aprotinin, and leupeptin). Next, samples were homogenized with a Polytron 10/35 for 30 sec, lysates were centrifuged at 3000 × g for 10 min at 4°, and the resulting supernatant was used for immunoprecipitations. In addition, 100 µl of each lysate with 200 µl of 2× SDS (sodium dodecyl sulfate) sample buffer was saved for western blot analysis. Lysates were added to protein G Sepharose beads (GE Healthcare) that had been previously incubated with P4D1 anti-ubiquitin antibody (1:500 µl; Cell Signaling Technology) and rotated for ∼3 hr at 4°. Beads were washed 5× with extraction buffer before western blot analysis, and then 200 µl of SDS sample buffer was added before western blot analysis. Proteins were detected using (1:1000) anti-GFP antibody (Cell Signaling Technology).
To compare the amount of ubiquitinated Pom33 between samples, we first determined the relative abundance of Pom33-GFP1–10 in each whole-cell lysate using histone H4 as a reference [1:10,000 anti-histone H4 antibody (Abcam)]. The amount of Pom33-GFP1–10 protein recovered in each ubiquitin pull-down was then adjusted accordingly. Western blots were quantitated as previously described (Jaspersen et al. 2006; Bupp et al. 2007; Friederichs et al. 2012); because Pom33-GFP1–10 ran as a doublet, the entire region containing both bands was included in the analysis.
Cycloheximide chase experiments
Cells were grown overnight and diluted back the next morning, then allowed to recover to midlog phase before the addition of cycloheximide (125 µg/ml). Samples were removed at given time points and quick-frozen in liquid nitrogen for further analysis by western blotting, using (1:1000) anti-MYC antibody (Cell Signaling Technology) or (1:1000) anti-GFP antibody (Cell Signaling Technology), (1:10,000) anti-histone H4 antibody (Abcam), and (1:1000) anti-Clb2 (Santa Cruz Biotechnology). For the cim3-1 experiments, cells were shifted to the nonpermissive temperature of 37° for 45 min before the addition of cycloheximide.
Data availability
All strains are listed in Table S4 and are available upon request. Split-GFP and PCR tagging plasmids have been deposited at AddGene. Custom macros and plugins used for image processing and quantitation with Image J are available at http://research.stowers.org/imagejplugins/index.html. Original data underlying this manuscript can be downloaded from the Stowers Original Data Repository at http://www.stowers.org/pubs/LIBPB-1363_2018. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7646975.
Results
Screen for INM changes in protein quality control mutants
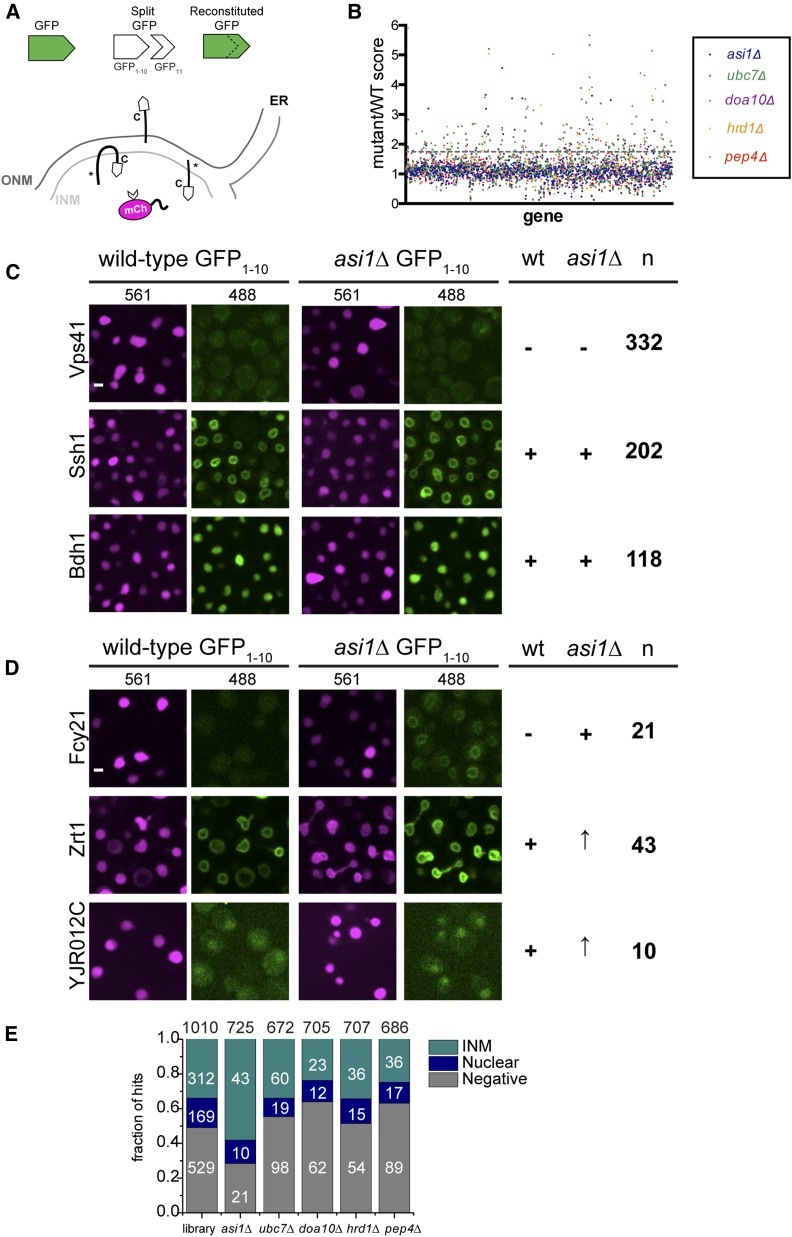
To detect if proteins are able to access the INM, we expressed a soluble nuclear protein at high levels fused to half of split-GFP (GFP11-mCherry-Pus1); this can reconstitute a working GFP when a protein fused to GFP1–10 localizes to the same compartment (Figure 1A). Previously, we showed that 312 of 1010 C-terminally tagged membrane or predicted membrane proteins in yeast have access to the INM using the split-GFP assay (Smoyer et al. 2016). Of the 1010 C-terminally tagged genes in the library, 169 localized throughout the nucleus (nuclear) because they encoded a soluble protein or because C-terminal tagging disrupted membrane association, while 529 did not exhibit any signal (Figure 1E).
Figure 1.
Mutation of ASI1 alters split-GFP signal of a subset of INM proteins. (A) Schematic showing split-GFP and its use to detect proteins that can access the INM (denoted with asterisk). (B) Plot of the split-GFP mutant/WT ratios for each gene reported from 96-well plate imaging analysis of membrane proteins in asi1∆, ubc7∆, doa10∆, hrd1∆, and pep4∆. Dashed line represents the 1.4 cutoff. (C) Representative images of proteins that were categorized previously as negative or positive (INM or soluble) that remained unchanged in asi1∆, based on an asi1∆/WT ratio of 1.39 or lower. (D) Signal of proteins that changed in asi1∆ largely fell into three categories: proteins that were previously negative, and proteins that were positive (INM or soluble) that increased in asi1∆. The number of proteins assigned to each category for (B and C) is on the right, based on an asi1∆/WT ratio of 1.4 or higher. (E) The starting library contained 1010 genes, including 312 that localized to the INM in WT cells, 169 that gave a soluble nuclear signal, and 529 that were negative. Of the 725 genes analyzed in the asi1∆ mutant, 74 were hits using the 1.4 ratio cutoff. The fraction (number) of asi1∆ hits that localized to the INM, nucleus, or were negative in WT cells is shown. Similarly, a summary of starting library localization based on WT results is also plotted for the other mutants, with the total number of genes analyzed listed at the top and the number of hits in each category shown. Bar, 2 μm. INM, inner nuclear membrane; ONM, outer nuclear membrane; WT, wild-type.
To determine how the INM proteome is altered by the removal of quality control systems, we examined INM localization of the same library in cells lacking ASI1 (INMAD), DOA10 (INMAD/ERAD), HRD1 (ERAD), UBC7 (INMAD/ERAD and other pathways), and PEP4 (vacuole). Wild-type and mutant strains were screened using a high-throughput 96-well plate imaging format, and images were quantitatively assessed for INM signal using an automated image analysis pipeline. Although it is unlikely that the amount of nuclear reporter is limiting as it is present in high copy, levels of GFP11-mCherry-Pus1 could affect the amount of 488 fluorescence visualized, so we first normalized to mCherry levels on a cell-by-cell basis. Next, we averaged the split-GFP signal for each protein/mutant combination to eliminate possible cell cycle/cell growth artifacts, and then used this value to determine the final intensity ratio of wild-type and each mutant (Table S1).
Manual inspection of images in cells lacking ASI1 compared to wild-type showed that only 725 of the 833 genes tested in the mutant could be reliably scored. False positives (Table S2) were removed from analysis of all mutants tested, due to a lack of signal in wild-type and asi1∆, or suspected low expression levels or higher amounts of autofluorescence. The mating pheromone receptor Ste2 was also a false positive due to the MATa mating type of strains used for mutant analysis vs. the MATα strain we used for wild-type (Figure S1). If we examined Ste2-GFP1–10 in a wild-type MATa strain, it localized to the INM like the mutants (Figure S1). No other INM components were obviously affected by mating type.
Our analysis showed that the distribution of most proteins was unaffected in the deletions, resulting in ratio values ∼1 (Figure 1, B and C). Using a cutoff of 1.4, which is slightly < 1 SD above the mean for most samples, we found that 10–26% of proteins showed an increase above wild-type. This included increased levels for 74/725 proteins in asi1∆, 177/672 in ubc7∆, 97/705 in doa10∆, 105/707 in hrd1∆, and 141/686 in pep4∆ (Figure 1E). These hits could be further subdivided into three categories based on their localization in wild-type. For example, 21 proteins present at the INM in asi1∆ did not localize to the INM in wild-type, 10 were soluble and nucleoplasmic, and 43 were at the INM but increased in cells lacking ASI1 (Figure 1, D and E). Overrepresentation of INM components in asi1∆ (58%) compared to other mutants (34%) points to the idea that Asi1 affects native INM components. Consistent with this possibility, the well-characterized INM components Pom33, Per33, Asi2, and Heh2 were identified as hits in our asi1∆ screen (Tables S1 and S3), as discussed below.
In each mutant analyzed, we identified increased INM access of a unique combination of split-GFPs (Table S3). Cells lacking the E2 ubiquitin-conjugating enzyme Ubc7 partially overlapped with the E3 ligases, consistent with the idea that Ubc7 plays a role in both ERAD and INMAD. The partial overlap between doa10∆ and hrd1∆ or asi1∆ mutants (Figure S2A) is similar to previous reports suggesting that Doa10 also plays a role in both pathways (Foresti et al. 2014; Khmelinskii et al. 2014). We tested 12 of 20 previously reported Asi1 substrates from a whole-cell assay based on the tandem timer (Khmelinskii et al. 2014). From this, we confirmed increased INM localization in 8 of the 12, including the vacuolar transferase complex subunit Vtc1 that is thought to be mistargeted to the INM due to protein tagging (Khmelinskii et al. 2014), the Rab GTPase-interacting protein Yip4, the plasma membrane transporter Zrt2, inositolphosphotransferase Ipt1, and Irc23, a protein of unknown function that is linked to DNA damage (Figure S2B). We also saw similar effects for ubc7∆. Perhaps unsurprisingly, our screen for INM proteins affected by doa10∆ or hrd1∆ showed limited overlap with Doa10 or Hrd1 substrates found by Foresti et al. (2014) and Khmelinskii et al. (2014), who used whole-cell based methods that would detect ER substrates. It is unknown if protein levels and/or INM localization is a direct or indirect consequence of the Asi1 deletion; therefore, we performed more detailed analysis on a subset of mutants and proteins.
Pom33 distribution is altered in cells lacking ASI1
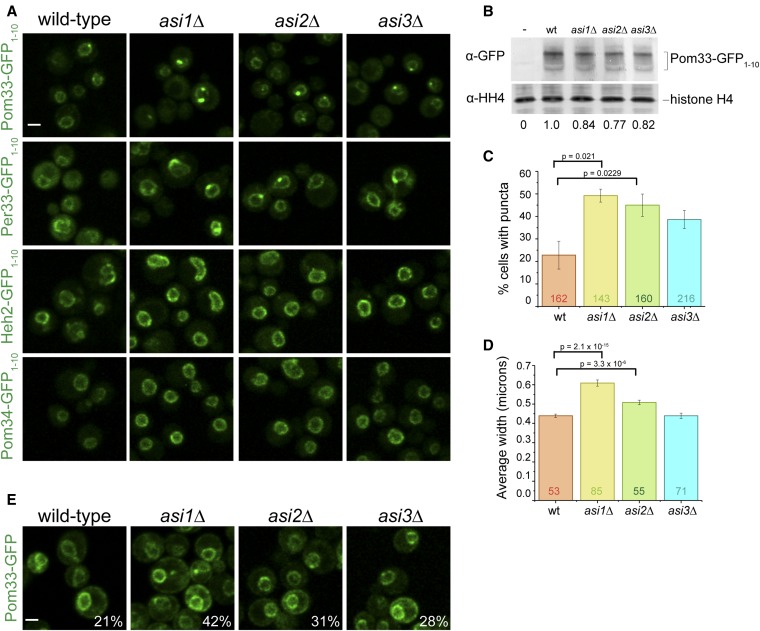
In addition to a change in protein levels, one of the most interesting phenotypes that we observed in INM quality control mutants was the appearance of puncta: increased levels of INM protein at one or more NE locations in the presence of background fluorescence throughout the membrane. A striking example is shown in Figure 2A for the paralogs, Pom33 and Per33, in cells lacking ASI1. The INM component Heh2 and the NPC pore membrane protein Pom34 did not form puncta in any mutant analyzed (Figure 2A and Figure S3), suggesting that puncta formation is specific to Pom33 and Per33, and not a general feature of INM or NPC pore components in asi1∆. Pom34 was identified as a potential Ubc7-dependent, Hrd1- and Doa10-independent target in a previous proteomic screen for Asi1 targets (Foresti et al. 2014), and in our analysis it did exhibit a slight ratio increase for asi1∆ (1.27) and ubc7∆ (1.35). Unlike the puncta formation we previously observed for a number of proteins using split-GFP (Smoyer et al. 2016), the puncta formed by Pom33 were specifically linked to INM quality control pathways, forming in mutant cells lacking ASI1, ASI2, UBC7, and to a lesser extent ASI3 (Figure 2, A and C and Figure S3). Pom33 levels at the INM were reduced in both doa10∆ and hrd1∆ (Figure S3 and Table S1). Per33 puncta also formed in ERAD mutants (Figure S3) and were not studied further.
Figure 2.
Pom33 INM distribution is altered in Asi complex mutants. (A) Comparison of reconstituted GFP signal for Pom33-GFP1–10, Per33-GFP1–10, Heh2-GFP1–10, and Pom34-GFP1–10 with GFP11-mCherry-Pus1 in wt cells and cells lacking ASI1, ASI2, or ASI3. (B) Quantitation of total protein levels for Pom33-GFP1–10 in wt, asi1∆, asi2∆, and asi3∆ cells. An untagged strain was also used (−). Pom33-GFP1–10 levels were first normalized to histone H4 signal and the mutant levels each compared to wt, which was set at 1; ratios set at bottom are an average from four separate blots. (C) The frequency of Pom33-GFP1–10 puncta increases in mutants of ASI1 and ASI2. Total cells counted (n) for each sample depicted on graph. (D) The average puncta width measured by FWHM of fluorescence intensity at the puncta. Total puncta measured (n) depicted on graph. In (C and D), P-values were determined by Students t-test. Error bars equal SEM. (E) Localization of Pom33-GFP in wt, asi1∆, asi2∆, and asi3∆ cells. Percentage of cells with puncta reported in the lower right corner. n > 125 cells. Bar, 2 μm. FWHM, full width at half maximum; INM, inner nuclear membrane; wt, wild-type.
Quantitation of Pom33 at the INM showed that there was little change in intensity or total Pom33 levels in wild-type and mutants (Figure 2, A and B), but that the frequency of puncta more than doubled in asi1∆ and asi2∆ cells (Figure 2C). Importantly, the formation of Pom33 puncta was not simply due to split-GFP as they were also observed in cells expressing Pom33-GFP in asi1∆ and asi2∆ (Figure 2E). Previous work showed that Pom33-GFP not only localizes to the INM but also to the ONM/ER (Chadrin et al. 2010), which can be seen in images with full-length GFP (Figure 2E). As the ONM/ER pool of Pom33 is unlikely to be affected by deletion of ASI1 or ASI2, puncta are not as pronounced as in Pom33-GFP1–10/GFP11-mCherry-Pus1 cells where the nuclear pool is exclusively detected (Figure 2A). The effects of asi3∆ on Pom33 puncta formation were also not as pronounced, similar to previous studies showing that the asi3∆ mutant had a milder effect on INMAD compared to asi1∆ (Khmelinskii et al. 2014). The puncta formed in the mutants, particularly in Pom33-GFP1–10/GFP11-mCherry-Pus1 cells, are considerably larger than in wild-type cells (Figure 2D), suggesting that Asi1 and Asi2 affect Pom33 distribution at the INM.
Ubiquitination of Pom33 regulates INM distribution but not degradation
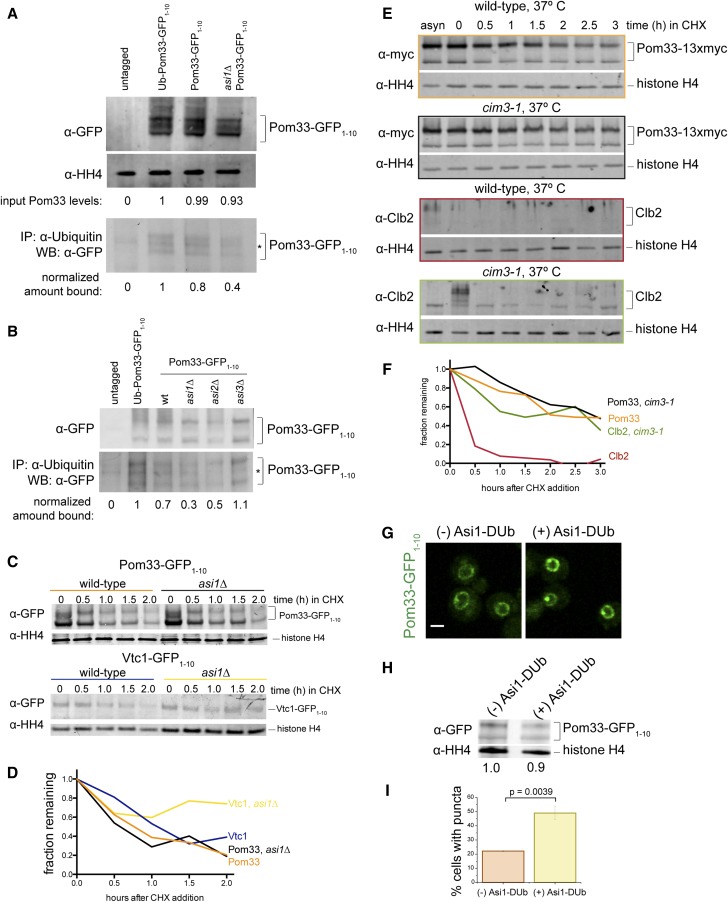
To understand how Asi1 and Asi2 affect Pom33 distribution at the INM, we considered the possibility that Pom33 is a target of the ubiquitin ligase activity of Asi1 and is ubiquitinated in vivo. Ubiquitinated proteins were immunoprecipitated from lysates containing Pom33 and Pom33-GFP1–10, with and without ASI1, with an anti-ubiquitin antibody. These cells also contained the GFP11-mCherry-Pus1 reporter. As a positive control, we immunoprecipitated an N-terminal fusion of ubiquitin to Pom33-GFP1–10 (Ub-Pom33). Pom33 is unlikely to be ubiquitinated at the N-terminus, but this control construct is able to serve as a functional version of POM33, rescuing the NPC clustering phenotype caused by pom33∆ to the same extent as Pom33-GFP1–10 (Figure S4). We analyzed these immunoprecipitates by western blotting with an anti-GFP antibody. Although a ladder of bands was not present to indicate polyubiquitination, we enriched for Pom33-GFP1–10 from wild-type but not asi1∆ cells in these experiments (Figure 3A). Loss of ASI2, but not ASI3, also affected recovery of ubiquitinated Pom33-GFP1–10 (Figure 3B), suggesting that Pom33 is ubiquitinated in an Asi1- and Asi2-dependent manner.
Figure 3.
Asi1-dependent ubiquitination of Pom33 does not lead to its degradation by the proteasome. (A) Lysates from an untagged control strain, Ubiquitin-Pom33-GFP1–10, Pom33-GFP1–10, and asi1∆ Pom33-GFP1–10 were probed with anti-GFP antibodies to determine levels of Pom33 relative to the loading control histone H4. Ubiquitinated proteins were isolated from lysates using an anti-ubiquitin antibody, followed by western blotting using an anti-GFP antibody. The signal in the pull-down was quantitated and adjusted according to input levels, with the level in the untagged and Ub-Pom33-GFP1–10 controls set to 0 and 1, respectively. Note, the lysate is 1% of the protein used for the pull-down. (B) Cells containing asi2∆ and asi3∆ Pom33-GFP1–10 were analyzed alongside strains in (A). The lysate is 0.4% of the pull-down. (C) Time course showing that Vtc1-GFP1–10 but not Pom33-GFP1–10 is stabilized in cells lacking Asi1 after addition of CHX. (D) Quantitation of Vtc1-GFP1–10 and Pom33-GFP1–10 protein levels from WBs in (E) showed the amount of protein remaining at each time point relative to the histone H4 loading control. (E) Time course to determine total levels of Pom33-13xmyc in wild-type and cim3-1 cells at 37°. Cells were shifted to the nonpermissive temperature for 45 min before addition of CHX, which inhibits protein synthesis. Western blotting was done to detect Pom33-13xmyc on the top and Clb2 on the bottom. (F) Quantitation of Pom33-13xmyc in (F) and Clb2 protein levels from WBs in (E) showed the amount of protein remaining at each time point relative to the histone H4 loading control. (G) Pom33-GFP1–10 with and without Asi1-DUb expressed. (H) Total levels of Pom33-GFP1–10 with and without Asi1-DUb expressed. (I) Frequency of Pom33-GFP1–10 puncta, DUb off: n = 95 cells, DUb on: n = 96 cells. Bar, 2 μm. P-values were determined by Students t-test. Note, cells in (A, B, and E–I) also contain GFP11-mCherry-Pus1. CHX, cycloheximide; IP, immunoprecipitation; WB, western blot.
Total Pom33 proteins levels do not change in asi1∆ (Figure 2B) and it does not appear to be polyubiquitinated (Figure 3, A and B), suggesting that Pom33 is not actively degraded. To formally test whether Pom33 stability is affected by loss of ASI1, we performed a cycloheximide chase experiment with cells containing GFP11-mCherry-Pus1 and Pom33-GFP1–10, or Vtc1-GFP1–10, a previously characterized INMAD target whose INM levels are regulated by Asi1 (Khmelinskii et al. 2014). Cycloheximide was added to cells to inhibit protein synthesis so that the stability of proteins could be assayed over time by western blot analysis (Figure 3, C and D). Both Pom33-GFP1–10 and Vtc1-GFP1–10 levels decreased during the cycloheximide chase but only Vtc1-GFP1–10 was stabilized by asi1∆, confirming the idea that Pom33 stability is Asi1-independent. The decrease in Pom33 levels is not the result of 26S proteasome degradation as we observed a similar decrease in the levels of Pom33-13xmyc in wild-type cells and cim3-1, a mutant that disrupts the proteasome lid component Rpt6 (Ghislain et al. 1993; Schork et al. 1995) (Figure 3, E and F). Clb2, a cell cycle protein known to be degraded by the proteasome (Seufert et al. 1995; Deshaies 1997), served as a control that showed stabilization in cim3-1 (Figure 3, E and F), while histone H4 was a loading control since its levels and stability are unaffected by temperature, cycloheximide, or proteasome inhibition. It is unclear why Pom33 protein levels decrease over time during the cycloheximide chase, but the fact that we saw similar decreases with different epitopes indicates that it is unlikely to be related to the epitope tag.
If Pom33 is ubiquitinated but not degraded by the proteasome, what role does Asi1-dependent ubiquitination play in the regulation and function of Pom33? One attractive idea is that Asi1-dependent ubiquitination may modulate Pom33 INM distribution, similar to the way that ubiquitination of proteins is involved in different processes such as nucleocytoplasmic trafficking. Examples of this include p53, in which monoubiquitination is thought to expose a nuclear export signal (Lohrum et al. 2001; Li et al. 2003; Nie et al. 2007), and cytidydyltransferase (CCTα), where monoubiquitination blocks its nuclear localization signal (Chen and Mallampalli 2009). To test this idea, we employed an inducible version of Asi1 fused to the deubiquitinating domain of Herpes Virus UL36 (Asi1-DUb) (MacDonald et al. 2012, 2017). This Asi1-DUb is designed to deubiquitinate Asi1 targets as Asi1 acts on them, allowing us to assay the specific role of ubiquitination without lingering concerns of indirect effects caused by chronic removal of ASI1. Although Asi1-DUb had little effect on total Pom33 protein levels, its induction resulted in increased frequency and extent of Pom33-GFP1–10 puncta formation at the INM (Figure 3, G–I), a phenotype very similar to that of asi1∆. The appearance of the Pom33 puncta upon induction of the Asi1-DUb suggest that their formation is a direct consequence of deubiquitination and supports the idea that Asi1-dependent ubiquitination of Pom33 regulates its INM distribution.
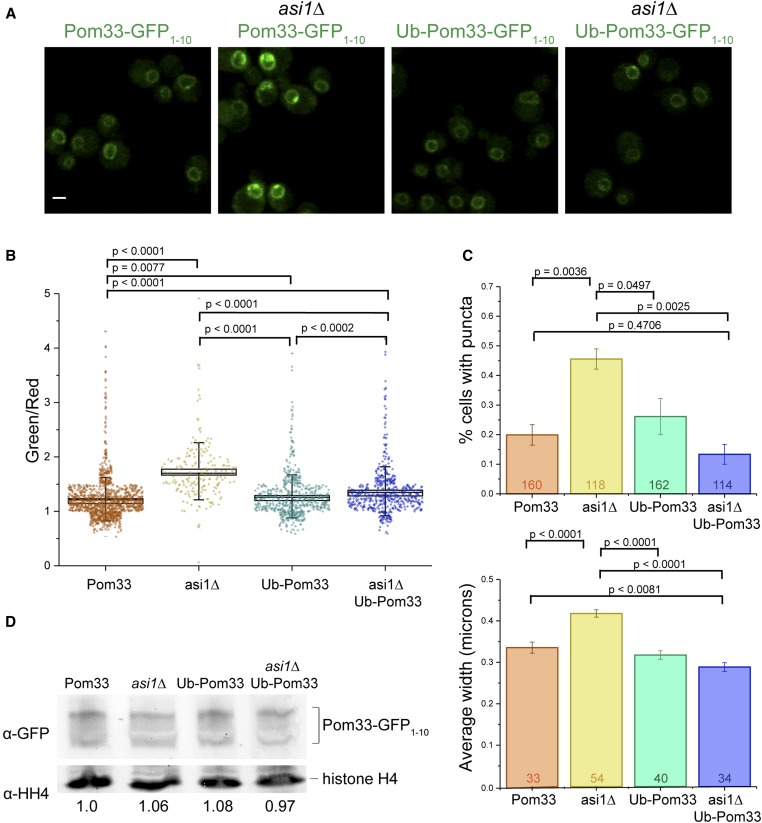
If ubiquitination of Pom33 contributes to its normal INM distribution, then constitutive ubiquitination by fusion of ubiquitin to Pom33 might rescue the puncta phenotype seen in asi1∆ mutants. To test this, we first measured the INM intensity, using a split-GFP signal, of Pom33-GFP1–10 and Ub-Pom33-GFP1–10 in wild-type and asi1∆ cells carrying GFP11-mCherry-Pus1, finding that Ub-Pom33-GFP1–10 partially rescued overall INM intensity in asi1∆ (Figure 4, A and B). Next, we compared the frequency and size of puncta in wild-type and asi1∆ cells, with GFP11-mCherry-Pus1 and Pom33-GFP1–10, or Ub-Pom33-GFP1–10, expressed in single copy at the URA3 locus as the sole copy of POM33 in the cell. As expected, the frequency and size of Pom33-GFP1–10 puncta increased in asi1∆ compared to wild-type (Figure 4, A–C). However, expression of Ub-Pom33-GFP1–10 rescued the frequency and size phenotypes in asi1∆ cells, with fewer puncta (Figure 4, A–C) as well as smaller puncta size (Figure 4C, bottom). Therefore, attaching ubiquitin to Pom33 bypasses the requirement for Asi1 for INM distribution, strongly suggesting that Asi1 directly modifies Pom33. Because modification of N-terminal residues can affect protein stability through the N-end rule pathway (Varshavsky 1992; Tasaki et al. 2012), we examined expression levels by western blotting. This analysis showed that Pom33-GFP1–10 and Ub-Pom33-GFP1–10 were present at similar levels in wild-type cells (Figure 4D).
Figure 4.
Expression of Ub-Pom33-GFP1–10 reduces puncta frequency and size in asi1∆. (A) Example images of Pom33-GFP1–10, asi1∆ Pom33-GFP1–10, Ub-Pom33-GFP1–10, and asi1∆ Ub-Pom33-GFP1–10. Cells are also expressing the nuclear reporter GFP11-mCherry-Pus1. (B) Quantification of strains in (A); intensities measured as a ratio of reconstituted GFP (488) signal over Pus1 (561) signal. (C) For strains depicted in (A), the frequency of Pom33-GFP1–10 puncta was counted (top) and the average puncta width measured by FWHM of fluorescence intensity at the puncta (bottom). Total n depicted on graphs. Error bars equal SEM. (D) Quantitation of total Pom33 protein levels for Pom33-GFP1–10, asi1∆ Pom33-GFP1–10, Ub-Pom33-GFP1–10, and asi1∆ Ub-Pom33-GFP1–10 strains by western blot, normalizing to histone H4 signal. Average ratios from four different experiments are shown below. Bar, 2 μm. FWHM, full width at half maximum.
Pom33 INM puncta contain NPC components
Punctate distribution at the NE has been previously reported for the SPB, nuclear–vacuolar junction, intranuclear quality control, and storage for incomplete NPC (SINC) components: Spc42, Nvj1, Cmr1, and Chm7, respectively (Huh et al. 2003; Webster et al. 2014, 2016; Gallina et al. 2015; Webster and Lusk 2016). After replacing GFP11-mCherry-Pus1 with GFP11-Pus1, we examined colocalization of the split-GFP signal in Pom33-GFP1–10 strains using mCherry-tagged proteins marking each of these NE puncta-associated structures. In most cells, Pom33-GFP1–10/GFP11-Pus1 did not colocalize with any of the tested NE proteins (Figure S5). Moreover, if we deleted key components involved in the formation of nuclear subcomplexes, we did not see an effect on the size or frequency of Pom33 puncta (Figure S5; data not shown). This result was particularly surprising as it suggests that Pom33 puncta represent a novel nuclear subcompartment that is distinct, particularly from the SINC that has been previously described for incompletely assembled NPCs (Webster et al. 2014, 2016).
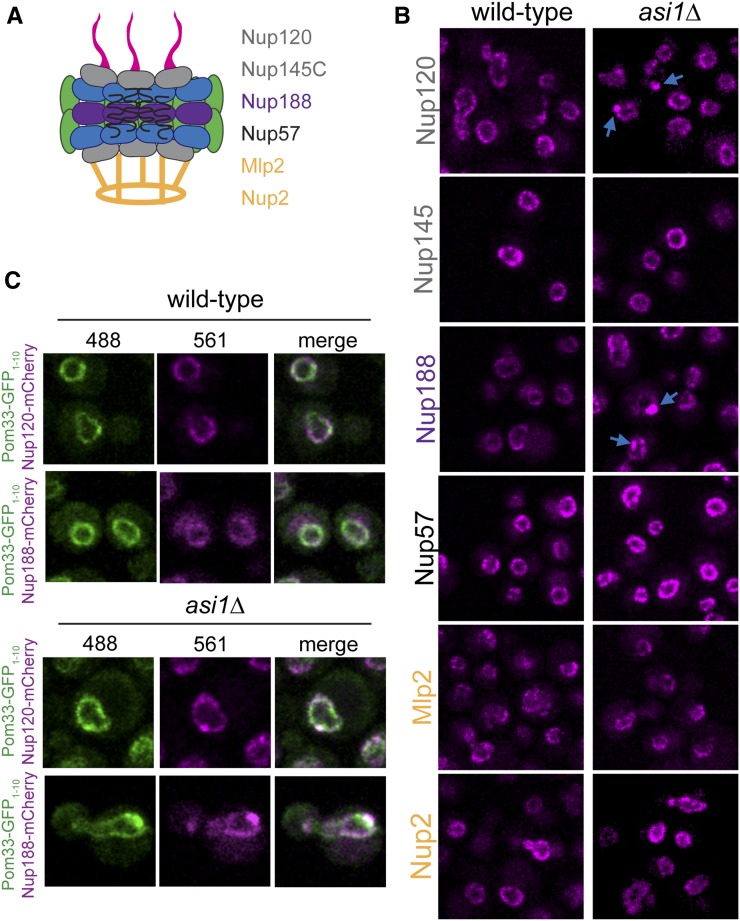
To better understand the nature of the Pom33 puncta, we were interested in determining whether soluble nucleoporins not present in our screen were also present in foci. We localized at least one component from each NPC subcomplex in wild-type and asi1∆ cells, including the outer-ring components Nup120 and Nup145C, the inner-ring protein Nup188, the central channel component Nup57, and the basket proteins Nup2 and Mlp2 (Figure 5A) (Alber et al. 2007; Aitchison and Rout 2012). Nup145C, Nup57, Nup2, and Mlp2 were unaffected by removal of ASI1; however, Nup120 and Nup188 exhibited punctate foci specifically in asi1∆ mutants (Figure 5B). Over half of the Nup120-mCherry or Nup188-mCherry puncta seen in asi1∆ colocalize with Pom33-GFP1–10/GFP11-Pus1 (Figure 5C). It is tempting to speculate that these puncta contain full or complete NPCs; however, it is important to note that we only observe puncta formation with a few nucleoporins. While we detected Pom33-GFP1–10 in asi1∆ by immuno-electron microscopy (EM) at NPCs (data not shown), consistent with previous immuno-EM data in wild-type cells (Chadrin et al. 2010), NPCs do not cluster in cells lacking ASI1 (data not shown). Further, Pom33 did not localize to any recognizable NE landmark other than NPCs (data not shown), pointing to the possibility that Pom33 distributes to a novel subnuclear component in the absence of Asi1.
Figure 5.
Nup188 and Nup120 colocalization with Pom33 puncta in asi1∆. (A) Cartoon of the NPC. Nucleoporins tested included the outer ring components Nup120 and Nup145C, the inner ring Nup188, the central channel nucleoporin Nup57, and the basket-associated Mlp2 and Nup2. (B) Localization of nucleoporins in (A) was assayed in wild-type and asi1∆ cells. Nup120 and Nup188 displayed more puncta in asi1∆, as indicated by blue arrows. (C) Example images of Pom33-GFP1–10/GFP11-Pus1 with either Nup120-mCherry or Nup188-mCherry, in wild-type and asi1∆. Bar, 2 μm. NPC, nuclear pore complex.
Discussion
Mechanisms that control INM protein dynamics are poorly understood. Using split-GFP, we performed a systematic screen of over 700 membrane proteins, comparing INM levels between wild-type cells and mutants in the E3 ubiquitin ligases Asi1, Doa10, and Hrd1, the E2 ubiquitin-conjugating enzyme Ubc7, and the vacuolar peptidase Pep4. Our data overlap to a limited degree with previous studies examining substrates of these enzymes (Foresti et al. 2014; Khmelinskii et al. 2014). However, our ability to specifically and unequivocally assay changes at the INM enabled us to extend our understanding of INM quality control in several important ways. Previous work hypothesized that the primary role of INMAD is to remove foreign substrates from the INM, i.e., proteins that have diffused in via NPCs but failed to diffuse back out, due to associated protein tags or lesions within the polypeptide (Foresti et al. 2014; Khmelinskii et al. 2014). However, we found that deletion of ASI1 preferentially affected INM components compared to other mutants examined. In both our screen and our follow-up studies, we show that loss of ASI1 results in increased levels of bona fide INM proteins, such as Pom33, Per33, Asi2, and Heh2. This suggests that Asi1, and by extension INMAD, plays a role in proteostasis of wild-type INM proteins, not just proteins mistargeted to the INM.
Asi1 and Asi3 are thought to form a heterodimeric E3 ligase, which binds to the accessory factor Asi2 to ubiquitinate substrates in the nucleus and at the INM (Zargari et al. 2007; Foresti et al. 2014; Khmelinskii et al. 2014). Although subtle differences between deletion mutants have been reported (Foresti et al. 2014; Khmelinskii et al. 2014; Pantazopoulou et al. 2016), the prevailing model in the field supports the idea of an Asi complex (Boban and Foisner 2016). Several lines of evidence presented here suggest that Pom33 is a direct Asi1 substrate and, by extension, a substrate of the Asi complex. First, Pom33 is ubiquitinated in an Asi1-dependent manner. Second, expression of Asi1-DUb recapitulated the Pom33 puncta phenotype seen in asi1∆ mutants. Lastly, a constitutively ubiquitinated Pom33 (Ub-Pom33) largely reversed puncta formation in asi1∆. Taken together, these data suggest that Asi1, or the Asi complex, normally acts to ubiquitinate a pool of Pom33, which results in its uniform NE distribution. Given that the Asi complex is nonessential except when cells are placed under stress, this uniform distribution of Pom33 is likely not required for NPC function and immunoprecipitation experiments suggest that only a small fraction of Pom33 is ubiquitinated.
Asi1-dependent Pom33 ubiquitination does not appear to be a signal for degradation. A ladder of polyubiquitinated Pom33 was not observed. In addition, Pom33 levels did not change when it was fused to ubiquitin or expressed in a proteasome mutant. Our data suggest that ubiquitination regulates Pom33 localization in much the same way that monoubiquitination is used throughout the endomembrane system to control subcellular localization or endocytosis (Hicke and Dunn 2003; d’Azzo et al. 2005). Analysis of Pom33 localization determinants in budding yeast suggests that the C-terminal 65 amino acids are important for both stability and NPC localization (Floch et al. 2015), making it tempting to speculate that residues in this region are ubiquitinated. Attempts to map a ubiquitinated lysine residue were unsuccessful, suggesting that multiple lysines are sufficient (data not shown). Examination of our Asi1 substrates, along with previously described targets, did not show a particular motif targeting them to the ligase. However, it has been previously proposed that amphipathic helices play a role in Asi2 degradation and Doa10 substrate recognition (Ravid et al. 2006; Boban et al. 2014). Given that Pom33 has at least two amphipathic helices in its C-terminus that play a role in NPC targeting and membrane binding, one hypothesis is that these domains are also important for Asi1-dependent ubiquitination. More generally, INMAD-dependent control of these common motifs could play a role in the regulation of NPC assembly and NE compartmentalization.
What is the role of INMAD regulation of Pom33 and other resident INM proteins? No obvious changes to nuclear structure, including NPC distribution, have been reported for cells lacking ASI1 by EM and the ligase is not essential under normal growth conditions (Foresti et al. 2014). Colocalization with Nup120 and Nup188 suggests that the Pom33 puncta contain at least some additional NPC components, even though no recognizable NPCs were seen cytologically (data not shown). We do not believe that these are improperly assembled NPCs, as puncta did not colocalize with the SINC. Instead, our data suggest that Pom33 aggregates in a novel INM structure. Since the INMAD has only been investigated for its role in the degradation of targets, it will be of future interest to determine whether the INMAD components mediate ubiquitination of other proteins to alter their localization or if TMEM33, the metazoan ortholog of Pom33, is also ubiquitinated. These types of studies will elucidate the role of INMAD in normal nuclear function.
Acknowledgments
We are grateful to Chris MacDonald and Robert Piper for the Asi1-DUb construct; Richard Alexander, Sean McKinney, Brian Slaughter, and Melainia McClain for help during this project; and to Brian Slaughter and members of the Jaspersen laboratory for comments on the manuscript. S.L.J. is supported by the Stowers Institute for Medical Research. C.J.S. is a predoctoral researcher in the Graduate School of the Stowers Institute.
Author contributions: C.J.S. and S.L.J. conceived of applying split-GFP to study degradation of INM proteins. S.M. made the yeast libraries that were screened by C.J.S. with assistance from S.E.S. Intensity ratios were measured and analyzed by C.J.S. and S.E.S., with assistance from J.M.G., using image analysis tools developed by J.R.U. and S.E.S. C.J.S. and S.L.J. wrote the paper with input from all the authors.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7646975.
Communicating editor: O. Cohen-Fix
Literature Cited
- Adnyana I. K., Tezuka Y., Awale S., Banskota A. H., Tran K. Q., et al. , 2000. Quadranosides VI-XI, six new triterpene glucosides from the seeds of Combretum quadrangulare. Chem. Pharm. Bull. (Tokyo) 48: 1114–1120. 10.1248/cpb.48.1114 [DOI] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., 2012. The yeast nuclear pore complex and transport through it. Genetics 190: 855–883. 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Antonin W., Ungricht R., Kutay U., 2011. Traversing the NPC along the pore membrane: targeting of membrane proteins to the INM. Nucleus 2: 87–91. 10.4161/nucl.2.2.14637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington P., Robinson D. K., 2003. Data Reduction and Error Analysis for the Physical Sciences, pp. 194–218. McGraw-Hill, New York. [Google Scholar]
- Boban M., Foisner R., 2016. Degradation-mediated protein quality control at the inner nuclear membrane. Nucleus 7: 41–49. 10.1080/19491034.2016.1139273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban M., Zargari A., Andréasson C., Heessen S., Thyberg J., et al. , 2006. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Biol. 173: 695–707. 10.1083/jcb.200601011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban M., Pantazopoulou M., Schick A., Ljungdahl P. O., Foisner R., 2014. A nuclear ubiquitin-proteasome pathway targets the inner nuclear membrane protein Asi2 for degradation. J. Cell Sci. 127: 3603–3613. 10.1242/jcs.153163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp J. M., Martin A. E., Stensrud E. S., Jaspersen S. L., 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179: 845–854. 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin A., Hess B., San Roman M., Gatti X., Lombard B., et al. , 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189: 795–811. 10.1083/jcb.200910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. B., Mallampalli R. K., 2009. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol. Cell. Biol. 29: 3062–3075. 10.1128/MCB.01824-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Smoyer C. J., Slaughter B. D., Unruh J. R., Jaspersen S. L., 2014. The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J. Cell Biol. 204: 523–539. 10.1083/jcb.201307043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W. T., Worman H. J., 2009. The nuclear envelope as a signaling node in development and disease. Dev. Cell 17: 626–638. 10.1016/j.devcel.2009.10.016 [DOI] [PubMed] [Google Scholar]
- d’Azzo A., Bongiovanni A., Nastasi T., 2005. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6: 429–441. 10.1111/j.1600-0854.2005.00294.x [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., 1997. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr. Opin. Genet. Dev. 7: 7–16. 10.1016/S0959-437X(97)80103-7 [DOI] [PubMed] [Google Scholar]
- Do L. G., Spencer J. A., Roberts-Thomson K., Ha D. H., Tran T. V., et al. , 2003. Periodontal disease among the middle-aged Vietnamese population. J. Int. Acad. Periodontol. 5: 77–84. [PubMed] [Google Scholar]
- Dou Z., Xu C., Donahue G., Shimi T., Pan J. A., et al. , 2015. Autophagy mediates degradation of nuclear lamina. Nature 527: 105–109. 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch A. G., Tareste D., Fuchs P. F., Chadrin A., Naciri I., et al. , 2015. Nuclear pore targeting of the yeast Pom33 nucleoporin depends on karyopherin and lipid binding. J. Cell Sci. 128: 305–316. 10.1242/jcs.158915 [DOI] [PubMed] [Google Scholar]
- Foresti O., Rodriguez-Vaello V., Funaya C., Carvalho P., 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science 346: 751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Friederichs J. M., Gardner J. M., Smoyer C. J., Whetstine C. R., Gogol M., et al. , 2012. Genetic analysis of Mps3 SUN domain mutants in Saccharomyces cerevisiae reveals an interaction with the SUN-like protein Slp1. G3 (Bethesda) 2: 1703–1718. 10.1534/g3.112.004614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Fritze C. E., Gerace L., 1998. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J. Biol. Chem. 273: 4213–4219. 10.1074/jbc.273.7.4213 [DOI] [PubMed] [Google Scholar]
- Gallina I., Colding C., Henriksen P., Beli P., Nakamura K., et al. , 2015. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat. Commun. 6: 6533 10.1038/ncomms7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. M., Smoyer C. J., Stensrud E. S., Alexander R., Gogol M., et al. , 2011. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A. Z. J Cell Biol 193: 489–507. 10.1083/jcb.201011017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C., 1993. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362. 10.1038/366358a0 [DOI] [PubMed] [Google Scholar]
- Gordon L. B., Rothman F. G., Lopez-Otin C., Misteli T., 2014. Progeria: a paradigm for translational medicine. Cell 156: 400–407. 10.1016/j.cell.2013.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Dunn R., 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19: 141–172. 10.1146/annurev.cellbio.19.110701.154617 [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Martin A. E., Glazko G., Giddings T. H., Jr., Morgan G., et al. , 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174: 665–675. 10.1083/jcb.200601062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S. S., Smoyer C. J., Jaspersen S. L., 2014. Destination: inner nuclear membrane. Trends Cell Biol. 24: 221–229. 10.1016/j.tcb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Blaszczak E., Pantazopoulou M., Fischer B., Omnus D. J., et al. , 2014. Protein quality control at the inner nuclear membrane. Nature 516: 410–413. 10.1038/nature14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Lusk C. P., Blobel G., 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442: 1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L., 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., et al. , 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975. 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- Lohrum M. A., Woods D. B., Ludwig R. L., Balint E., Vousden K. H., 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21: 8521–8532. 10.1128/MCB.21.24.8521-8532.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Blobel G., King M. C., 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8: 414–420. 10.1038/nrm2165 [DOI] [PubMed] [Google Scholar]
- MacDonald C., Buchkovich N. J., Stringer D. K., Emr S. D., Piper R. C., 2012. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 13: 331–338. 10.1038/embor.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., Winistorfer S., Pope R. M., Wright M. E., Piper R. C., 2017. Enzyme reversal to explore the function of yeast E3 ubiquitin-ligases. Traffic 18: 465–484. 10.1111/tra.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N., Athonvarangkul D., Singh R., 2015. Autophagy and aging. Adv. Exp. Med. Biol. 847: 73–87. 10.1007/978-1-4939-2404-2_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T. C., 2016. Breaching the barrier-the nuclear envelope in virus infection. J. Mol. Biol. 428: 1949–1961. 10.1016/j.jmb.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Millen J. I., Krick R., Prick T., Thumm M., Goldfarb D. S., 2009. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy 5: 75–81. 10.4161/auto.5.1.7181 [DOI] [PubMed] [Google Scholar]
- Nie L., Sasaki M., Maki C. G., 2007. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J. Biol. Chem. 282: 14616–14625. 10.1074/jbc.M610515200 [DOI] [PubMed] [Google Scholar]
- Omnus D. J., Ljungdahl P. O., 2014. Latency of transcription factor Stp1 depends on a modular regulatory motif that functions as cytoplasmic retention determinant and nuclear degron. Mol. Biol. Cell 25: 3823–3833. 10.1091/mbc.e14-06-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou, M., M. Boban, R. Foisner, and P. O. Ljungdahl, 2016 Cdc48 and Ubx1 participate in an inner nuclear membrane associated degradation pathway that governs the turnover of Asi1. J. Cell Sci. 129: 3770–3780. [DOI] [PubMed]
- Park Y. E., Hayashi Y. K., Bonne G., Arimura T., Noguchi S., et al. , 2009. Autophagic degradation of nuclear components in mammalian cells. Autophagy 5: 795–804. 10.4161/auto.8901 [DOI] [PubMed] [Google Scholar]
- Ravid T., Kreft S. G., Hochstrasser M., 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25: 533–543. 10.1038/sj.emboj.7600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., et al. , 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 129–141. 10.1091/mbc.e02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas J. N., Toyama B. H., Xu T., Yates J. R., III, Hetzer M. W., 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science 335: 942 10.1126/science.1217421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork S. M., Thumm M., Wolf D. H., 1995. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 270: 26446–26450. 10.1074/jbc.270.44.26446 [DOI] [PubMed] [Google Scholar]
- Seufert W., Futcher B., Jentsch S., 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81. 10.1038/373078a0 [DOI] [PubMed] [Google Scholar]
- Skinner B. M., Johnson E. E., 2017. Nuclear morphologies: their diversity and functional relevance. Chromosoma 126: 195–212. 10.1007/s00412-016-0614-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoyer C. J., Katta S. S., Gardner J. M., Stoltz L., McCroskey S., et al. , 2016. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215: 575–590. 10.1083/jcb.201607043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S. D., Ashley J., Jokhi V., Nunnari J., Barria R., et al. , 2012. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149: 832–846. 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M., 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15: 2660–2674. 10.1101/gad.933301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley E. C., Ly N., Starr D. A., 2011. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol. Biol. Cell 22: 1739–1752. 10.1091/mbc.e10-08-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T., Sriram S. M., Park K. S., Kwon Y. T., 2012. The N-end rule pathway. Annu. Rev. Biochem. 81: 261–289. 10.1146/annurev-biochem-051710-093308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Turgay Y., Ungricht R., Rothballer A., Kiss A., Csucs G., et al. , 2010. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 29: 2262–2275. 10.1038/emboj.2010.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R., Kutay U., 2015. Establishment of NE asymmetry-targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol. 34: 135–141. 10.1016/j.ceb.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Ungricht R., Kutay U., 2017. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18: 229–245. 10.1038/nrm.2016.153 [DOI] [PubMed] [Google Scholar]
- Ungricht R., Klann M., Horvath P., Kutay U., 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 209: 687–703. 10.1083/jcb.201409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade T., Yamamoto Y., Zhang X., Ku Y., Sakisaka T., 2014. Identification and characterization of TMEM33 as a reticulon-binding protein. Kobe J. Med. Sci. 60: E57–E65. [PubMed] [Google Scholar]
- Varshavsky A., 1992. The N-end rule. Cell 69: 725–735. 10.1016/0092-8674(92)90285-K [DOI] [PubMed] [Google Scholar]
- Webster B. M., Lusk C. P., 2016. Border safety: quality control at the nuclear envelope. Trends Cell Biol. 26: 29–39. 10.1016/j.tcb.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B. M., Colombi P., Jager J., Lusk C. P., 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159: 388–401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B. M., Thaller D. J., Jager J., Ochmann S. E., Borah S., et al. , 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35: 2447–2467. 10.15252/embj.201694574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., Mehnert J. M., Chan C. S., 2015. Autophagy, metabolism, and cancer. Clin. Cancer Res. 21: 5037–5046. 10.1158/1078-0432.CCR-15-0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Lin F., Worman H. J., 2002. Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J. Cell Sci. 115: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Zargari A., Boban M., Heessen S., Andreasson C., Thyberg J., et al. , 2007. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 282: 594–605. 10.1074/jbc.M609201200 [DOI] [PubMed] [Google Scholar]
- Zattas D., Hochstrasser M., 2015. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 50: 1–17. 10.3109/10409238.2014.959889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Oliferenko S., 2014. Tts1, the fission yeast homologue of the TMEM33 family, functions in NE remodeling during mitosis. Mol. Biol. Cell 25: 2970–2983. 10.1091/mbc.e13-12-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are listed in Table S4 and are available upon request. Split-GFP and PCR tagging plasmids have been deposited at AddGene. Custom macros and plugins used for image processing and quantitation with Image J are available at http://research.stowers.org/imagejplugins/index.html. Original data underlying this manuscript can be downloaded from the Stowers Original Data Repository at http://www.stowers.org/pubs/LIBPB-1363_2018. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7646975.