Biological, technical, and environmental confounders are ubiquitous in the high-dimensional, high-throughput functional genomic measurements being used to understand cellular biology and disease processes, and many approaches have been developed to estimate and correct for unmeasured confounders...
Keywords: confounder, molecular trait, quantitative trait loci, eigenvector perturbation
Abstract
High-throughput measurements of molecular phenotypes provide an unprecedented opportunity to model cellular processes and their impact on disease. These highly structured datasets are usually strongly confounded, creating false positives and reducing power. This has motivated many approaches based on principal components analysis (PCA) to estimate and correct for confounders, which have become indispensable elements of association tests between molecular phenotypes and both genetic and nongenetic factors. Here, we show that these correction approaches induce a bias, and that it persists for large sample sizes and replicates out-of-sample. We prove this theoretically for PCA by deriving an analytic, deterministic, and intuitive bias approximation. We assess other methods with realistic simulations, which show that perturbing any of several basic parameters can cause false positive rate (FPR) inflation. Our experiments show the bias depends on covariate and confounder sparsity, effect sizes, and their correlation. Surprisingly, when the covariate and confounder have , standard two-step methods all have -fold FPR inflation. Our analysis informs best practices for confounder correction in genomic studies, and suggests many false discoveries have been made and replicated in some differential expression analyses.
ASSOCIATION studies of molecular phenotypes have helped characterize basic biological processes, including transcription, methylation, chromatin accessibility, translation, ribosomal occupancy, and expression response to stimuli. These tests can be performed on cis and trans genetic variants to search for functional quantitative trait loci [*QTL: eQTL (Montgomery et al. 2010; Pickrell et al. 2010), mQTL (Rakyan et al. 2011), caQTL (Degner et al. 2012), pQTL (Albert et al. 2014), rQTL (Battle et al. 2015), sQTL (Rivas et al. 2015; Li et al. 2016), reQTL (Fairfax et al. 2014; Lee et al. 2014), and iQTL (Barry et al. 2017)]. Functional measurements can also be tested against nongenetic covariates with broad genomic effects, including cell type composition (Houseman et al. 2012; Jaffe and Irizarry 2014; Rahmani et al. 2017; Yao et al. 2017), disease status [e.g., cancer (van’t Veer et al. 2002), autism (Parikshak et al. 2016), and obesity (Horvath et al. 2014)], fetal developmental stage (Colantuoni et al. 2011), and ancestry (Galanter et al. 2017). We mostly refer to gene expression for simplicity, but our arguments apply to any highly structured, high-dimensional measurements.
Unfortunately, unmeasured and unknown factors are common and often have large effects in functional genomic data, reducing power and skewing null transcriptome-wide P-values (Leek and Storey 2007; Gibson 2008). Conditioning on known confounders—like technical batch—is invaluable but incomplete. Because genetic effects are typically small, even modest confounders can induce spurious genetic associations that dwarf real signal (Leek and Storey 2007; Kang et al. 2008).
Fortunately, strong confounders induce large, low-dimensional structure in the transcriptome, which is exactly what principal components (PCs) aim to capture (as do their variants, which we collectively call CCs, for confounding components). This blessing of dimensionality motivates a two-step approach where CCs are first estimated and then conditioned on downstream as surrogates for the confounders (Alter et al. 2000; Leek and Storey 2008). Domain-specific CC methods, like surrogate variable analysis (SVA) (Leek and Storey 2007) and PEER (Stegle et al. 2010), make different assumptions about the structure of the confounders, and often outperform PCA. Two-step CC correction is an essential element of thousands of functional genomics analysis pipelines (Leek et al. 2010; Rakyan et al. 2011; Stegle et al. 2012, 2015; Albert and Kruglyak 2015).
Acknowledging the substantial benefits of CCs in many settings, in this work, we explore their adverse impact on the false positive rate (FPR). Theoretically, we derive an unappreciated source of bias created by conditioning on two types of PCs. Our results suggest the two step approach is biased whenever CCs imperfectly partition the phenotype-covariate correlation. We formalize this in a unifying, unidentified likelihood (1) that the CC methods each resolve with distinct assumptions.
We also study the bias with a range of CCs and simulations using real expression data from the GEUVADIS consortium (Lappalainen et al. 2013). First, we find no nontrivial method that avoids bias even in the simple scenario where the covariate has a small effect on a single gene and is added to white noise. In more complex data, this bias can be negligible compared to confounder-induced miscalibration. Next, we perform a series of simulations varying the number and strength of covariate effects and see substantial inflation in all CC methods when their assumptions fail. Finally, we allow confounders to be correlated with the covariate—the ordinary meaning of a confounder (Leek et al. 2010)—and find that all CCs can be severely miscalibrated; further, we show these false positives replicate out-of-sample.
Confounder Estimation and Correction
We write P molecular phenotypes measured on N samples as and let be the p-th phenotype. The primary covariate of interest is . We assume x and the are standardized to mean 0 and variance 1.
We stylize the standard two-step confounder correction as:
- Estimate a rank-K confounder U by solving
(1)
where is the Frobenius norm and is some penalty representing (potentially implicit) priors on the causal and confounding patterns. Here, α and V are dummy variables.
- 2. Estimate the by regressing each gene on x given :
(2)
where OLS indicates the regression coefficient on x from ordinary linear regression of on x, , and an intercept.
We call approaches “unsupervised” when constrains and “supervised” otherwise.
Solving (1) with amounts to maximum likelihood (ML) estimation under an i.i.d. Gaussian noise model for errors in Y (Leek and Storey 2007, 2008; Stegle et al. 2010). Standard ML inference for would then (pseudo)invert the information matrix for asymptotic standard errors that account for uncertainty in U and V. This exposes one problem with two-step confounder correction: step two conditions on a fixed as if it were known without error. Theoretically, this difficulty can be resolved by appealing to assumptions that ensure perfectly estimates U so that there is no uncertainty to propagate (Leek and Storey 2008; Wang et al. 2017).
Another difficulty for ML inference in (1) is that its solution is not unique (even when requiring, e.g., ), as
Because satisfies the rank-K constraint, can obtain the same likelihood as α: adding any vector in span(V) to any α admits an equivalent solution. [We ignore the nonidentifiability of from the product because only span is used in (2).]
This nonidentifiability means all (well-defined) CCs must use nontrivial penalty functions ; below, we describe the choices of roughly made by several popular CC methods. Moreover, this nonidentification means CCs depend heavily on their chosen capturing the true, unknown parameter structure. In particular, we can easily design simulations where any particular CC behaves badly. This means that choice of CC method is important in practice and should be dataset-specific.
We note that we do not claim to take significant steps toward solving this identification problem, which has been analyzed from various theoretical perspectives elsewhere, e.g. West 2003; Leek and Storey 2008; Gagnon-Bartsch and Speed 2012; Sun et al. 2012; Gerard and Stephens 2017; Wang et al. 2017.
Studied Confounder Estimation Methods
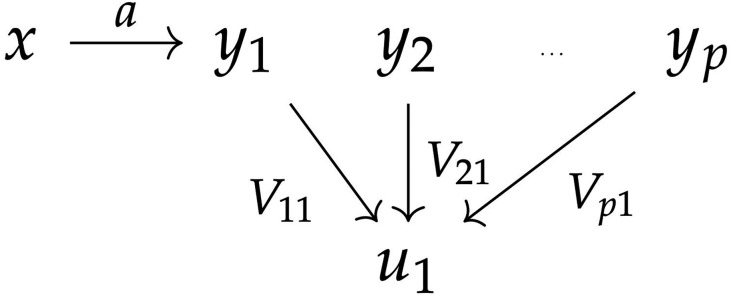
Unsupervised PCA takes as the top eigenvectors of or, equivalently, the top left singular vectors of Y. By definition, PCA solves (1) if solely constrains . The key problem is that the effect of x on Y leads PCs to partially capture x, analogous to unshielded colliders in a directed graphical model (Figure 1). That is, conditioning on genomic PCs can cause, rather than remove, bias. Related concerns arise when conditioning on heritable covariates in other contexts (Aschard et al. 2015, 2017; Day et al. 2016). This bias creates test misspecification even marginally, for each gene; in contrast, previous theory assumed marginal tests were valid and focused on correlations between tests of different genes (Leek and Storey 2008).
Figure 1.
Graphical model suggesting CC conditioning causes bias. is the top PC, which is indirectly captures x. Although x affects only , conditioning on the collider induces spurious correlation with all other .
We approximate the PC conditioning bias for small causal effects with textbook eigenvector perturbation theory. Conditioning on one phenotypic PC, our bias approximation for gene p coincides with the bias that naively derived from Figure 1
where a is the causal effect, q is the causally affected gene, and V are the right singular vectors of Y.
A similar result can be derived for conditioning on genotypic PCs in genome-wide association studies. In Figure 1, this stylistically corresponds to replacing the phenotypes with SNPs, the covariate (x) with the tested phenotype, and reversing the arrow from x to . However, genotype matrices typically contain three orders of magnitude more variables than expression matrices, concomitantly reducing the bias [entries of V are ]. Intuitively, causal SNPs have much lower leverage on genetic PCs than causal genes have on expression PCs.
We also test an approach we call supervised PCA, which aims to protect from x by first residualizing Y on x. This solves (1) when constrains α to its unconditional estimate, . We show this method simply amplifies biases in the unconditional estimate: unsupervised PCs are too correlated with x, but supervised PCs are too uncorrelated.
We study several other approaches through simulation. SVA penalizes (1) by assuming α is sparse, which is often plausible. We used the “two-step” and “irw” algorithms to implement unsupervised and supervised SVA, respectively (“irw” requires supervision, but “two-step” is infrequently used). The “irw” version learns which genes are determined by the signal α vs. the confounder V by testing with q-values. These association strengths are used in turn to weight the relative importance of each gene inside a singular value decomposition, effectively weighting PCA toward more-confounded genes to better estimate confounders. We also tested a recent reimplementation, SmartSVA (Chen et al. 2017); we found that it performs very similar to SVA with iterations, but much faster.
PEER explicitly penalizes U and α through priors on their respective sizes. PEER uses automatic relevance determining priors for the factors U and V and fits parameters with variational Bayes. In simulations, its default hyperparameters perform well when x explains of transcriptome-wide variation but less well for larger α. PANAMA is a closely related approach that greedily adds the most relevant SNPs while learning latent factors. While PANAMA can improve performance over PEER for SNPs with large effects, it is more computationally expensive and is rarely used in practice for human datasets.
RUV and related methods estimate confounders by using only a submatrix of Y that is known, a priori, to be unaffected by the primary signal (Lucas et al. 2006; Gagnon-Bartsch and Speed 2012; Gerard and Stephens 2017; Wang et al. 2017). This prior information breaks the identifiability problem by constraining for some subset of genes S (corresponding to a barrier penalty function for ). Latent factors can be safely identified by restricting to the negative control data, and then their effects on other genes can be extrapolated. We assume control genes or samples are unavailable, which is common, and do not study these approaches further.
We also assess LEAPP (Sun et al. 2012), a recent method that uses sparsity assumptions to disentangle α and V. Unlike other methods we study, LEAPP provably obtains oracle performance, asymptotically and assuming that confounders are strong, signals are sparse, and noise is independent across genes modulo confounders (Wang et al. 2017).
Finally, we also test the linear mixed model-based method ICE, which uses a random effect with covariance kernel to capture confounding, and tests the fixed effects of x against each gene individually (Kang et al. 2008). Conceptually, ICE seeks a few genes that are highly correlated with x compared to typical genes. Like in RUV methods, control genes can be used to improve power, which we did not study (Joo et al. 2014).
The bias for one unsupervised PC
In this section we take Y as a *QTL plus some deterministic :
| (3) |
We assume . We allow to be fully general to capture all noise and confounding. This is closely related to the spiked covariance model (Johnstone 2001), though we use a general in place of i.i.d. Gaussian noise. We assume and for , and we call x a local covariate for gene q.
Ideally, the OLS step (2) would condition exactly on the true, unknown confounders. We evaluate conditioning, instead, on the top PCs of Y , which we call U and define as the top left singular vectors of Y . We compare conditioning on U to rather than the true confounders for two reasons. First, is assumed independent of x, hence conditioning on does not cause bias. Second, this allows us not to assume any particular form for confounding, or even its existence.
We aim to quantify the error at gene from conditioning on the top feasible PC, , instead of the top oracle PC, :
where solves (2) given . Since for , any error can only be caused by the effect of x on U.
The bias is the expected error over x, the only randomness:
We study rather than to focus away from the ordinary regression error due to noise and the onto the error caused by x’s perturbation of U. This enables stronger results—particularly, that deterministically. Because of this, we often refer to this perturbed PC conditioning error as the bias, i.e., the randomness in is negligible.
We assume α is small so that we can use a standard approximation to the perturbed eigenvector [e.g., (Allez and Bouchaud 2012), Sec. II]: for any small E, the first eigenvector of is approximately
| (4) |
where is the j-th eigenvalue of .
Under our assumption on α, , giving the perturbation approximation
| (5) |
This uses simplifying definitions based on rotating with U:
Note that is still a spherical Gaussian random variable.
We show in Supplemental Material, Section S1.1 that this approximation can be combined with the standard two-step least squares expression for to give
| (6) |
is a condition number for and are random weights:
The partition the perturbation among PCs and are proportional to the (random) squared correlations between x and the PCs (i.e., ). The are nonnegative and sum to one in expectation. is deterministic—depending only on the spectrum of —and quantifies the susceptibility of the first PC to perturbation.
These properties of mean the error in (6) is a (randomly) weighted correlation between the projections of genes p and q—the tested and the causal genes—onto the eigen-axes, i.e.
| (7) |
where is the correlation weighted by some π. In particular, if is a vector of 1s, is the ordinary correlation between the two genes. In contrast, randomly weights the eigen-axes, but with far greatest weight on axis 1 and successively less expected weight on subsequent axes.
is the only remaining randomness, so the error depends on x only through this (random) notion of correlation. And even this randomness is often negligible. First, for (the former is the sum over the latter), and this gap grows for increasingly confounded data (Figure S1). Second, should be very well approximated by its expectation because it is an average over variables.
Together, this suggests the approximations and for , giving a deterministic approximation to the random error. Because the error is approximately deterministic, it can immediately be recognized a bias approximation, as well:
| (8) |
This uses the approximation (Equation S3). We find (8) is accurate in a realistic simulation (Figure S2).
While is biased conditional on q and p, this conditional bias itself has mean zero on average over q or p ( is mean zero). Nonetheless, our conditional definition of bias conveys the fact that p and q are biologically meaningful and replicable indices. Moreover, even random biases with mean zero introduce overdispersion that still causes false positive inflation.
We have not generalized these calculation to PCs, though we suspect an analogous result will hold after appropriately modifying w. If correct, will move toward as K grows, suggesting the correlation between causal and tested traits is a good intuitive proxy for the bias.
The bias for supervised PCs
An apparent solution is to project out x before computing PCs, which we call supervised PCA. We show this is deeply flawed, even when x has no causal effect, supporting existing simulation results (Leek and Storey 2007).
First, after residualizing x from Y, x is the bottom supervised PC, hence orthogonal to the others. Thus the unconditional OLS estimates for are unchanged by conditioning on supervised PCs. Similarly, the ratio of the conditional and unconditional SEs is just the ratio of the overall regression error estimates, i.e., . Together, the ratio of t-statistics testing is
| (9) |
By definition, U explains large amounts of variance in Y, making smaller than . Formally, the ratio (9) is inflated on average (over genes and x) and, in practice, is usually inflated (Figure S3 and Section S1.4).
Local covariate simulations with white noise
We now demonstrate the CC conditioning bias in a simplistic simulation using (3): the background expression is drawn i.i.d. standard normal; the are (independently) i.i.d. standard normal; and , so that x is a local *QTL for gene q; finally, q is drawn, independently of x and Y, uniformly from , and its effect a is varied over . We chose to match the GEUVADIS data (see below for details).
After simulating Y and x, we test for using either PCA, SVA, PEER, or their supervised versions to estimate . We also test the mixed model implemented in ICE (which failed to converge in a few simulated datasets).
For 1000 independently simulated datasets, we perform one-sided Kolmogorov-Smirnov (KS) tests for deflation in the regression P-values at noncausally affected genes. A two-sided test should be used in the first step when testing for general miscalibration (Leek and Storey 2007, 2008); we, however, are testing for estimator bias, which decreases P-values, and discriminates inflation from deflation.
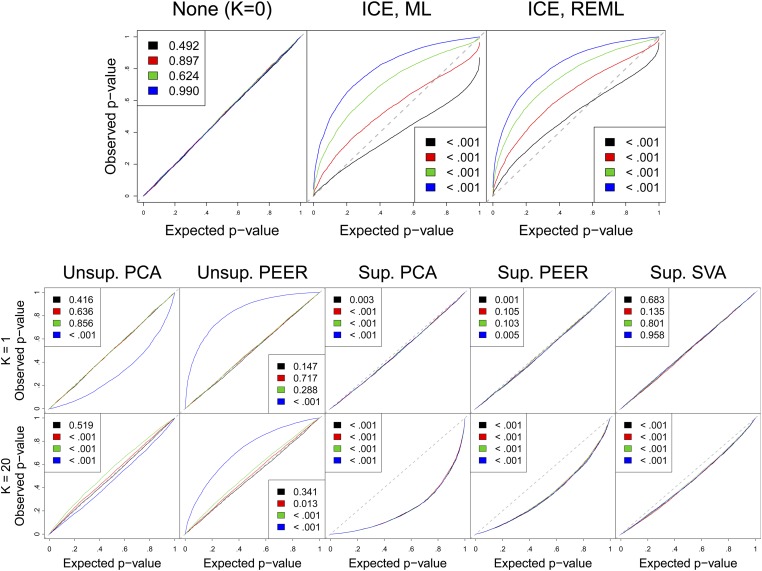
Figure 2 presents the QQ plots for the resulting KS P-values. Unsurprisingly, excluding all CCs (None) delivers well-calibrated P-values because we did not simulate confounders. Confirming our theory, unsupervised PCs cause noticeable bias for large a; however, no bias is detected for small a, emphasizing that the bias can be negligible for local covariates. Unsupervised PEER results are similar for small a, but for large a PEER becomes conservative (such observations require one-sided KS tests for the regression P-values). ICE is mostly conservative, especially when using REML and . Unsupervised SVA correctly declares [with the permutation test from (Buja and Eyuboglu 1992)] and is thus equivalent to “None”; although this is ideal behavior, any CC method could use this (or other) tests to choose K, and analysts in practice often turn to a different method in this situation (e.g., Pierce et al. 2014).
Figure 2.
Confounder correction causes P-value miscalibration in simulations with a local effect and white noise. QQ plots show one-sided KS test P-values for the nominally null regression P-values in 5000 simulations. 2-sided KS tests of the KS P-values are in the legends. Variation explained in the causal gene is 0% (black), 30% (red), 60% (green), or 90% (blue).
The three supervised methods qualitatively share a different type of bias, growing with K and depending little on a. We theoretically characterized this for supervised PCA, but PEER and, especially, SVA seem less biased.
Overall, Figure 2 shows that all tested (nontrivial) CC methods create P-value miscalibration even in the complete absence of confounding, though the problem is small for small a.
Data availability
We used a high-quality RNA-sequencing dataset from the GEUVADIS consortium (Lappalainen et al. 2013) as a realistic simulation baseline. We aligned the raw transcript reads from the European individuals to the reference hg19 transcriptome using RSEM (Li and Dewey 2011). We removed perfectly correlated genes and quantile-normalized the rest to standard normal. The final matrix has samples (rows) and genes (columns) and column-means and -variances equal to 0 and 1. Its spectrum is shown in Figure S10. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7040186.
Global Covariate Simulations with Real Traits
We now simulate a global effect, meaning α is much denser and larger. We set 90% of its entries to 0 and the others to i.i.d. Gaussian with mean zero and variance such that x explains 1% of transcriptome-wide variation. Nongenetic x can easily have these sorts of effects, e.g., even early studies with small sample sizes found broad expression profiles that still inform breast cancer treatment (Sparano et al. 2015; Cardoso et al. 2016). If x were genetic, it would be an extremely strong trans-*QTL.
We now use the GEUVADIS expression for the noise to make the simulation more realistic, and let Binomial(2,20%). Finally, as we do not aim to match the perturbation theory here, we adopt standard practice and normalize x and columns of Y to mean 0, variance 1.
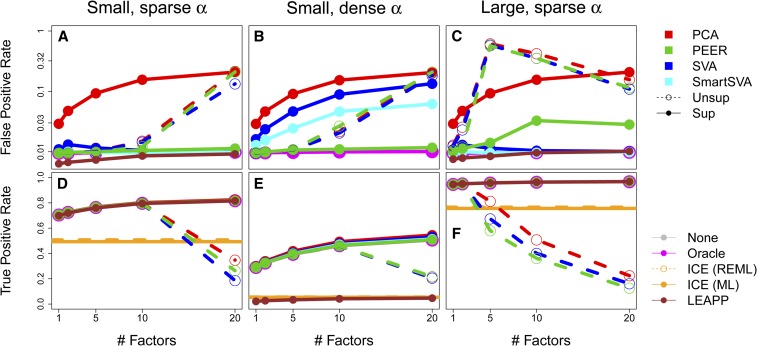
We assess empirical FPR and true positive rate (TPR) at the nominal level and average over 250 independently simulated datasets (averaging before log-transforming, Figure 3, a and d). All unsupervised methods and supervised PCA are badly miscalibrated for , while other supervised CC methods, ICE, and LEAPP were calibrated. These calibrated methods had power similar to Oracle, which uses PCs of the pure noise term , except ICE.
Figure 3 .
Mean FPR (a–c) and TPR (d–f) (on log scale) for a simulated global α added to GEUVADIS expression using three settings. The oracle is always calibrated and often covered by other lines. Two-step CC methods are at top-right; others are bottom-right. ICE has essentially 0 FPR and is omitted from the top plots.
We then decrease the sparsity of α from 90% zeros to 5% zeros, violating the sparsity assumptions of supervised (Smart)SVA and LEAPP. This leads (Smart)SVA to roughly 10-fold FPR inflation at (Figure 3b) and LEAPP to lose essentially all power (Figure 3e). Supervised PEER, however, retains near-oracle power and calibration.
Next, we return to 90% sparsity in α but increase its variance explained from 1 to 25%. This apparently violates PEER’s assumptions as it is five-fold inflated at (Figure 3c).
These conclusions qualitatively remain when using a threshold (Storey 2003) or (Figures S4 and S5).
Confounders Correlated with a Global Covariate
The previous simulations only added a causal x effect to some independent . We now add confounders correlated with x, which we feel is common in practice for nongenetic x (genetic x can only be confounded by population structure). For example, even within-tissue, PEER factors had ranging from 10% to 60% with known covariates (GTEx Consortium et al. 2015). Further, experimental procedures are often correlated with biological factors (Gilad and Mizrahi-Man 2015). And correlation between technical confounders and primary biological signal can be pernicious: tissue dissociation in quiescent muscle stem cells can resemble cellular activation from muscle injury (van den Brink et al. 2017).
We now add a confounder, u, that has correlation ρ with x:
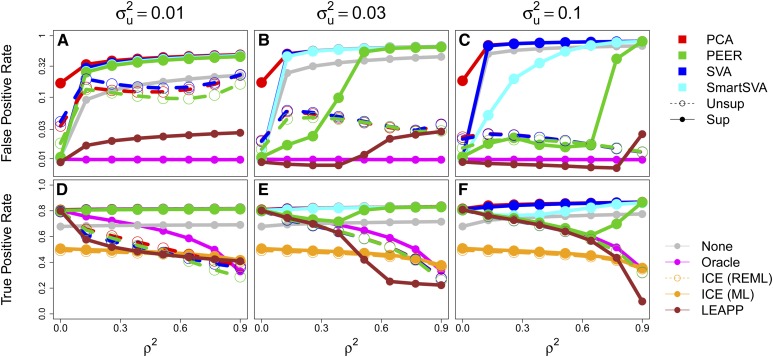
We draw each pair i.i.d. from a bivariate Gaussian with mean zero, variances equal to 1 and correlation ρ. We repeated our above pipeline, simulating 250 independent datasets and plotting the FPR in Figure 4. Here, we always take CCs, because was used originally for (Lappalainen et al. 2013) and we have added two rank-one effects.
Figure 4 .
FPR and TPR at a nominal level for testing a strong covariate x correlated with a confounder u. x and u have squared correlation , and their respective variances explained are and (a and d), 3% (b and e), or 10% (c and f). Two-step CC methods are in the top legend; others are in the bottom legend.
We draw α as in Figure 3a: 10% of its entries are nonzero, drawn i.i.d.Gaussian with mean zero and variance such that is the transcriptome-wide fraction of variance explained. Because u represents a confounder, we draw β i.i.d. Gaussian with variance such that . This is roughly in line with GTEx (Aguet et al. 2017), where the top 15–35 PEER factors collectively explained 59−78% of transcriptome-wide variation. We vary from 0 to 1 (ρ and are equivalent).
The results in Figure 4 show that supervised PCA badly inflates FPR, even at . The other supervised methods are nearly as inflated for , except PEER for larger and modest ρ. Unsupervised CCs also inflate FPR for , though this diminishes as grows and u becomes near-perfectly captured. In particular, the apparently naive approach of simply ignoring confounding (None) can be less inflated than all CC methods, particularly for small or . This shows that, even when confounding exists, CC adjustment can cause more harm than good. Qualitatively similar patterns hold using or thresholds (Figures S6 and S7).
We found that ICE was always calibrated and LEAPP was always close to calibrated, with a maximum of roughly threefold inflation. The LEAPP inflation occurred for smaller and larger , scenarios excluded by assumptions in Wang et al. (2017). LEAPP was often more powerful than ICE, but these were typically settings where CC methods also performed well. For example, LEAPP was similar to supervised PEER/(Smart)SVA when , and LEAPP was similar to unsupervised CCs and supervised PEER when (though better calibrated). In the intermediate range (e.g., and ), however, LEAPP outperformed all competitors. LEAPP was far slower than the other methods, taking 2 hr on average over simulated datasets in Figure 4, compared to 1–5 min for two-step methods and 10 min for ICE (Figure S8) nonetheless, recent re-implementations of LEAPP are faster (Wang et al. 2017).
Similar simulations can be found in Figure S2 of Leek and Storey (2008), but they reached the opposite conclusion, i.e. that SVA is calibrated even when . To test if this discrepancy is due to their smaller tested data dimensions, , we repeated our simulations in Figure 4 after downsampling to 20 samples and 1000 genes (uniformly without replacement, and independently downsampling for each simulation). This reduced FPR for SVA, though SVA can still be inflated (e.g., 10-fold for and , Figure S9). This remaining discrepancy is likely partially because there is, in fact, inflation in Figure S2 of Leek and Storey (2008): e.g., row 2, column 4 visually seems miscalibrated.
False positive replication
Assuming a and q are biologically determined and universal, the PCA bias we derived depends only on the top PCs of Y. As N grows large, the bias remains, precision grows, and the null is rejected for every gene if x affects even one.
Analogously, biases will be similar between datasets with similar top PCs, which occurs in the presence of strong and similar confounders, e.g. batch effects (Leek et al. 2010), population structure (Listgarten et al. 2010), cell type composition (Jaffe and Irizarry 2014), or the signal x itself if it is strong.
We empirically assess replication rates by simulating as in Figure 4 with , , and 10 of α’s entries drawn i.i.d. Gaussian. We then split the data (x and Y) into halves, test each separately, compute the replication rate as the fraction of false positive discoveries from the first half that are deemed positive in the second half, and repeat after transposing the splits. We use a significance level of in each split. We independently repeat the process 250 times, ignoring initial splits without false discoveries. Splitting an existing dataset simulates worst-case confounder sharing between discovery and replication cohorts.
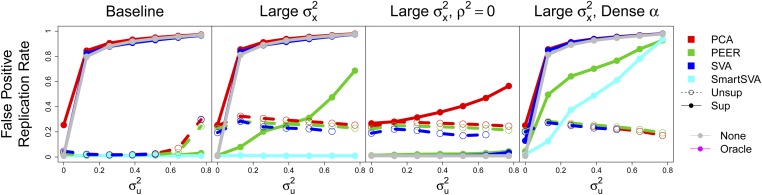
The left panel of Figure 5 shows the average (positive) false replication rate. Unsupervised methods perform reasonably well and supervised SmartSVA and PEER are calibrated. Either performing no confounder correction or using supervised SVA or PCA creates severe spurious replication, with nearly all false positives replicating when is large (e.g., ). In this section, we do not assess ICE as it has low total positive rates.
Figure 5.
False positive replication rate at a nominal level. is the confounder strength. The signal strength is either 1% (baseline) or 10% (others). The squared signal-confounder correlation is 25%, or 0 for the third panel.
We then increased the signal to . Unsupervised methods performed worse, as they more readily capture the larger x effect, and supervised PEER became miscalibrated, especially for larger . Next, we reduced the confounder correlation to 0: unsupervised methods performed slightly better, while supervised methods (except PCA) became roughly calibrated. Finally, we increased the density of α to 90%, causing supervised SmartSVA to suffer similarly to PEER.
Discussion
We have evaluated unappreciated sources of bias induced by conditioning on estimated confounders in functional genomic association tests. We used a combination of theory and simulation to cover different cases of interest. Overall, no two-step CC method we evaluated generally had calibrated FPR; moreover, most studies use PCA, one of the worst-performing methods in our simulations. Although all methods behave well when their assumptions hold, and these assumptions are often reasonable in practice, confounders even modestly correlated with the primary signal can cause substantial bias. We also showed these false positives can replicate at a high rate.
*QTL studies are often performed only within local genomic windows, which are called cis-*QTL studies. In this context, genetic effects are small and restricted to nearby genes, and our results suggest that the bias induced from CC conditioning is minimal. However, unlike in our Figure 2 simulations, cis-windows may contain many highly correlated genes, and cis-*QTL often causally affect genes other than the nearest one (Zhu et al. 2016), both of which serve to inflate FPR.
*QTL studies can also be performed genome-wide, which are called trans-*QTL studies. While such *QTL are biologically central, they are difficult to reliably uncover because the signals tend to be dispersed across the transcriptome and genome. Unlike cis-*QTL, trans-*QTL can have much larger effects on CCs, which has led modern studies to diametrically opposed methodology for testing trans-*QTL. For example, Brynedal et al. (2017) do not use CCs, despite the fact that “all [significant] gene sets were significantly correlated to ... the top 20 PEER factors.” On the other hand, Yao et al. (2017) adjust for 20 PEER factors and cell type composition, and many of the resulting “hotspot” signals are in fact loci known to affect cell type composition1. Similar to (Yao et al. 2017), (Aguet et al. 2017) adjust for PEER factors computed per tissue. We have proposed GBAT to address these and other limitations by testing gene-level trans associations (Liu et al. 2018). GBAT uses supervised SmartSVA, which performs best in our simulations without correlated confounders. This is feasible because we test only thousands of genes rather than millions of SNPs, though SNPs could be prescreened with unsupervised CCs. More generally, we recommend ICE or LEAPP when feasible: ICE was more reliably calibrated but had low power than LEAPP, while two-step CCs could perform very poorly when their assumptions are violated.
Our global covariate simulations are relevant for differential expression studies performed with linear regression and/or CC correction. Such tests have been broadly applied, including to differences between tissues, sexes, or ages. These factors can easily correlate with latent confounders when experiments are not randomized and can substantially affect the expression of many genes. This is the setting in our simulations underlying Figure 4, where no approach (except ICE) generally gives calibrated P-values and CC correction can be worse than a completely uncorrected analysis.
A key limitation of two-step approaches is that step 1 uncertainty is not propagated to the test in step 2. To address this, a multiple imputation-style approach can be used, performing step 2 on several draws from the step 1 CC posterior. We have not evaluated this concept as it is currently developed only within the RUV framework (Gerard and Stephens 2017). Related, the two steps can be integrated, which analytically conveys first-step uncertainty, though this has much greater computational cost when run genome-wide (Stegle et al. 2010; Fusi et al. 2012; Sun et al. 2012; Wang et al. 2017).
In the future, it may be useful to pursue other assumptions on in the bias calculation we derived for unsupervised PCA. For example, we could use a spiked covariance model for , using results from Nadler (2008) to approximate the perturbed eigenvector (e.g., replacing our Equation 5 with their Equation 2.15). An advantage of our approach, however, is that we allow general correlations between traits.
In more complex scenarios, the appropriate covariate and confounding model can be unclear. For example, coexpression studies learn complex and subtle graphical models from (partial) covariance (Horvath 2011; Shin et al. 2014). But uncorrected confounders (or biased corrections) will yield statistically significant, biologically meaningless networks. Latent variable graphical models may suit this problem (Chandrasekaran et al. 2012), and a related two-step approximation was recently proposed for genomics (Parsana et al. 2017). Finally, mixed models that learn specifically-genetic graphical models may be adaptable to adjust for low-dimensional confounding (Dahl et al. 2013).
Acknowledgments
We are grateful to Brunilda Balliu and Antonio Berlanga-Taylor for discussions about CC methods. This work was partially supported by National Institutes of Health grants 1U01HG009080-01, 5K25HL121295-03, and 1R03DE025665-01A1.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7040186.
Communicating editor: C. Sabatti
Thanks to Alexander Gusev for this observation.
Literature Cited
- Aguet F., Brown A. A., Segre A. V., Strober B. J., Zappala Z., et al. , 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Albert F. W., Treusch S., Shockley A. H., Bloom J. S., Kruglyak L., 2014. Genetics of single-cell protein abundance variation in large yeast populations. Nature 506: 494–497. 10.1038/nature12904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allez R., Bouchaud J. P., 2012. Eigenvector dynamics: general theory and some applications. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 86: 046202 10.1103/PhysRevE.86.046202 [DOI] [PubMed] [Google Scholar]
- Alter O., Brown P. O., Botstein D., 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97: 10101–10106. 10.1073/pnas.97.18.10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Vilhjálmsson B. J., Joshi A. D., Price A. L., Kraft P., 2015. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 96: 329–339. 10.1016/j.ajhg.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Guillemot V., Vilhjalmsson B., Patel C. J., Skurnik D., et al. , 2017. Playing musical chairs in big data to reveal variables associations. bioRxiv. 10.1038/ng.3975 [DOI] [Google Scholar]
- Barry J. D., Fagny M., Paulson J. N., Aerts H., Platig J., et al. , 2017. Histopathological image QTL discovery of immune infiltration variants. bioRxiv. https://www.biorxiv.org/content/10.1101/126730v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A., Khan Z., Wang S. H., Mitrano A., Ford M. J., et al. , 2015. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347: 664–667. 10.1126/science.1260793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynedal B., Choi J., Raj T., Bjornson R., Stranger B. E., et al. , 2017. Large-scale trans -eQTLs affect hundreds of transcripts and mediate patterns of transcriptional Co-regulation. Am. J. Hum. Genet. 100: 581–591. 10.1016/j.ajhg.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja A., Eyuboglu N., 1992. Remarks on parallel analysis. Multivariate Behav. Res. 27: 509–540. 10.1207/s15327906mbr2704_2 [DOI] [PubMed] [Google Scholar]
- Cardoso F., van’t Veer L. J., Bogaerts J., Slaets L., Viale G., et al. , 2016. 70-Gene signature as an aid to treatment decisions in early-stage. Breast Cancer 375: 717–729. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran V., Parrilo P. A., Willsky A. S., 2012. Latent variable graphical model selection via convex optimization. Ann. Stat. 40: 1935–1967. 10.1214/11-AOS949 [DOI] [Google Scholar]
- Chen J., Behnam E., Huang J., Moffatt M. F., Schaid D. J., et al. , 2017. Fast and robust adjustment of cell mixtures in epigenome-wide association studies with SmartSVA. BMC Genomics 18: 413 10.1186/s12864-017-3808-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C., Lipska B. K., Ye T., Hyde T. M., Tao R., et al. , 2011. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478: 519–523. 10.1038/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, A., V. Hore, V. Iotchkova, and J. Marchini, 2013 Network inference in matrix-variate Gaussian models with non-independent noise. arxiv: 1312.1622v1.
- Day F. R., Loh P. R., Scott R. A., Ong K. K., Perry J. R., 2016. A robust example of collider bias in a genetic association study. Am. J. Hum. Genet. 98: 392–393. 10.1016/j.ajhg.2015.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Pai A. A., Pique-Regi R., Veyrieras J.-B., Gaffney D. J., et al. , 2012. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482: 390–394. 10.1038/nature10808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax B. P., Humburg P., Makino S., Naranbhai V., Wong D., et al. , 2014. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. S 343: 1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi N., Stegle O., Lawrence N. D., 2012. Joint modelling of confounding factors and prominent genetic regulators provides increased accuracy in genetical genomics studies. PLoS Comput. Biol. 8: e1002330 10.1371/journal.pcbi.1002330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Bartsch J. A., Speed T. P., 2012. Using control genes to correct for unwanted variation in microarray data. Biostatistics 13: 539–552. 10.1093/biostatistics/kxr034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanter J. M., Gignoux C. R., Oh S. S., Torgerson D., Pino-Yanes M., et al. , 2017. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife 6: e20532. 10.7554/eLife.20532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard D., Stephens M., 2017. Unifying and generalizing methods for removing unwanted variation based on negative controls. arXiv: 1705.08393v1. [DOI] [PMC free article] [PubMed]
- Gibson G., 2008. The environmental contribution to gene expression profiles. Nat. Rev. Genet. 9: 575–581. 10.1038/nrg2383 [DOI] [PubMed] [Google Scholar]
- Gilad Y., Mizrahi-Man O., 2015. A reanalysis of mouse ENCODE comparative gene expression data. F1000 Res. 4: 121 10.12688/f1000research.6536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium , 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., 2011. Weighted Network Analysis. Springer, New York: 10.1007/978-1-4419-8819-5 [DOI] [Google Scholar]
- Horvath S., Erhart W., Brosch M., Ammerpohl O., von Schönfels W., et al. , 2014. Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. USA 111: 15538–15543. 10.1073/pnas.1412759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E. A., Accomando W. P., Koestler D. C., Christensen B. C., Marsit C. J., et al. , 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. E., Irizarry R. A., 2014. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15: R31 10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I., 2001. On the distribution of the largest eigenvalue in principal components analysis. Ann. Stat. 29: 295–327. 10.1214/aos/1009210544 [DOI] [Google Scholar]
- Joo J. W., Sul J. H., Han B., Ye C., Eskin E., 2014. Effectively identifying regulatory hotspots while capturing expression heterogeneity in gene expression studies. Genome Biol. 15: r61 10.1186/gb-2014-15-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Ye C., Eskin E., 2008. Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics 180: 1909–1925. 10.1534/genetics.108.094201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T., Sammeth M., Friedländer M. R., ’t Hoen P. A., Monlong J., et al. , 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. N., Ye C., Villani A. C., Raj T., Li W., et al. , 2014. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343: 1246980 10.1126/science.1246980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Storey J. D., 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3: e161 10.1371/journal.pgen.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Storey J. D., 2008. A general framework for multiple testing dependence. Proc. Natl. Acad. Sci. USA 105: 18718–18723. 10.1073/pnas.0808709105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Scharpf R. B., Bravo H. C., Simcha D., Langmead B., et al. , 2010. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11: 733–739. 10.1038/nrg2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. I., van de Geijn B., Raj A., Knowles D. A., Petti A. A., et al. , 2016. RNA splicing is a primary link between genetic variation and disease. Science 352: 600–604. 10.1126/science.aad9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listgarten J., Kadie C., Schadt E. E., Heckerman D., 2010. Correction for hidden confounders in the genetic analysis of gene expression. Proc. Natl. Acad. Sci. USA 107: 16465–16470. 10.1073/pnas.1002425107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Mefford J. A., Dahl A., Subramaniam M., Battle A., et al. , 2018. GBAT: a gene-based association method for robust trans-gene regulation detection. bioRxiv. 10.1101/395970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J., Carvalho C., Wang Q., Bild A., Nevins J. R., et al. , 2006. Sparse statistical modelling in gene expression genomics, pp. 155–176 in Bayesian Inference for Gene Expression and Proteomics, edited by K.-A. Do, Muller P. Cambridge University Press, Cambridge: 10.1017/CBO9780511584589.009 [DOI] [Google Scholar]
- Montgomery S. B., Sammeth M., Gutierrez-Arcelus M., Lach R. P., Ingle C., et al. , 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777. 10.1038/nature08903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler B., 2008. Finite sample approximation results for principal component analysis: a matrix perturbation approach. Ann. Stat. 36: 2791–2817. 10.1214/08-AOS618 [DOI] [Google Scholar]
- Parikshak N. N., Swarup V., Belgard T. G., Irimia M., Ramaswami G., et al. , 2016. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540: 423–427. 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsana P., Ruberman C., Jaffe A. E., Schatz M. C., Battle A., et al. , 2017. Addressing confounding artifacts in reconstruction of gene co-expression networks. bioRxiv: 10.1101/202903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Marioni J. C., Pai A. A., Degner J. F., Engelhardt B. E., et al. , 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772. 10.1038/nature08872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce B. L., Tong L., Chen L. S., Rahaman R., Argos M., et al. , 2014. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 10: e1004818 10.1371/journal.pgen.1004818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani E., Zaitlen N., Baran Y., Eng C., Hu D., et al. , 2017. Correcting for cell-type heterogeneity in DNA methylation: a comprehensive evaluation. Nat. Methods 14: 218–219. 10.1038/nmeth.4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V. K., Down T. A., Balding D. J., Beck S., 2011. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12: 529–541. 10.1038/nrg3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M. A., Pirinen M., Conrad D. F., Lek M., Tsang E. K., et al. , 2015. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348: 666–669. 10.1126/science.1261877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.-Y., Fauman E. B., Petersen A.-K., Krumsiek J., Santos R., et al. , 2014. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46: 543–550. 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparano J. A., Gray R. J., Makower D. F., Pritchard K. I., Albain K. S., et al. , 2015. Prospective validation of a 21-gene expression assay in breast cancer. N. Engl. J. Med. 373: 2005–2014. 10.1056/NEJMoa1510764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O., Parts L., Durbin R., Winn J., 2010. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 6: e1000770 10.1371/journal.pcbi.1000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O., Parts L., Piipari M., Winn J., Durbin R., 2012. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 7: 500–507. 10.1038/nprot.2011.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O., Teichmann S. A., Marioni J. C., 2015. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16: 133–145. 10.1038/nrg3833 [DOI] [PubMed] [Google Scholar]
- Storey J. D., 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31: 2013–2035. 10.1214/aos/1074290335 [DOI] [Google Scholar]
- Sun Y., Zhang N. R., Owen A. B., 2012. Multiple hypothesis testing adjusted for latent variables, with an application to the AGEMAP gene expression data. Ann. Appl. Stat. 6: 1664–1688. 10.1214/12-AOAS561 [DOI] [Google Scholar]
- van den Brink S. C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., et al. , 2017. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14: 935–936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- van ’t Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A. M., et al. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao Q., Hastie T., Owen A. B., 2017. Confounder adjustment in multiple hypothesis testing. Ann. Stat. 45: 1863–1894. 10.1214/16-AOS1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., 2003. Bayesian factor regression models in the “large p, small n” paradigm, in Bayesian Statistics, Oxford University Press, Oxford. [Google Scholar]
- Yao C., Joehanes R., Johnson A. D., Huan T., Liu C., et al. , 2017. Dynamic role of trans regulation of gene expression in relation to complex traits. Am. J. Hum. Genet. 100: 571–580. 10.1016/j.ajhg.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M. R., et al. , 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48: 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We used a high-quality RNA-sequencing dataset from the GEUVADIS consortium (Lappalainen et al. 2013) as a realistic simulation baseline. We aligned the raw transcript reads from the European individuals to the reference hg19 transcriptome using RSEM (Li and Dewey 2011). We removed perfectly correlated genes and quantile-normalized the rest to standard normal. The final matrix has samples (rows) and genes (columns) and column-means and -variances equal to 0 and 1. Its spectrum is shown in Figure S10. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7040186.