Abstract
CRISPR-based genome-editing methods in model organisms are evolving at an extraordinary speed. Whereas the generation of deletion or missense mutants is quite straightforward, the production of endogenous fluorescent reporters is more challenging. We have developed Nested CRISPR, a cloning-free ribonucleoprotein-driven method that robustly produces endogenous fluorescent reporters with EGFP, mCherry or wrmScarlet in Caenorhabditis elegans. This method is based on the division of the fluorescent protein (FP) sequence in three fragments. In the first step, single-stranded DNA (ssDNA) donors (≤200 bp) are used to insert the 5′ and 3′ fragments of the FP in the locus of interest. In the second step, these sequences act as homology regions for homology-directed repair using a double-stranded DNA (dsDNA) donor (PCR product) containing the middle fragment, thus completing the FP sequence. In Nested CRISPR, the first step involving ssDNA donors is a well-established method that yields high editing efficiencies, and the second step is reliable because it uses universal CRISPR RNAs (crRNAs) and PCR products. We have also used Nested CRISPR in a nonessential gene to produce a deletion mutant in the first step and a transcriptional reporter in the second step. In the search for modifications to optimize the method, we tested synthetic single guide RNAs (sgRNAs), but did not observe a significant increase in efficiency. To streamline the approach, we combined all step 1 and step 2 reagents in a single injection and were successful in three of five loci tested with editing efficiencies of up to 20%. Finally, we discuss the prospects of this method in the future.
Keywords: CRISPR, Cas9, Caenorhabditis elegans, fluorescent proteins, genome editing
THE advent of CRISPR/Cas9 has allowed genetic engineering to progress at an unprecedented level. Naturally employed by bacteria as a defense mechanism, the Cas9 nuclease has been engineered to introduce blunt double-strand breaks (DSBs) in target DNA when guided by an RNA duplex comprised of a generic trans-activating CRISPR RNA (tracrRNA) and a sequence-specific CRISPR RNA (crRNA) (Mojica et al. 2005; Jinek et al. 2012). This cut is only made at sites complementary to the 20-nucleotide guide sequence within the crRNA in the presence of a downstream protospacer adjacent motif (PAM) comprised of the bases 5′-NGG-3′ (in the case of Streptococcus pyogenes Cas9). Thus, the ease of use and specificity of the technique has made it an attractive tool for genome editing in cellular systems and model organisms.
In the nematode Caenorhabditis elegans, gene editing is achieved via injection of a mix containing crRNA, tracrRNA, and Cas9 into the gonads. These components can be expressed from messenger RNA (mRNA) (Chiu et al. 2013; Katic and Grosshans 2013; Lo et al. 2013), plasmids [expressing Cas9 and single guide RNA (sgRNA) in place of crRNA and tracrRNA] (Dickinson et al. 2013; Friedland et al. 2013; Frøkjær-Jensen 2013; Waaijers et al. 2013) or added as independent molecules (commercially available) that form ribonucleoprotein complexes (RNPs) (Cho et al. 2013; Paix et al. 2015). In the C. elegans germline, the polymerase theta-mediated end-joining repair mechanism is initiated in the absence of a repair template, leaving behind small deletions and/or insertions that are useful for generating nonspecific mutations (Chen et al. 2013; van Schendel et al. 2015). However, when a repair template in the form of a single-stranded oligonucleotide (ssODN) or double-stranded DNA with homology arms is added into the mix, a homology-directed repair (HDR) pathway is initiated, allowing precise changes such as point mutations and defined deletions or insertions to be introduced into the genome (Zhao et al. 2014; Paix et al. 2015; Ward 2015).
Simultaneous editing of other genes is commonly used as positive control of Cas9 activity. Several co-CRISPR strategies are being followed to produce visible phenotypes or to repair lethal mutations (Kim et al. 2014; Ward 2015; Farboud et al. 2019). The editing of dpy-10 is widely used for generating dominant phenotypes and for being suitable for any genetic background (Arribere et al. 2014; Kim et al. 2014). A subset of dpy-10 coedited F1s would then be heterozygous for the desired edit, and these animals, by self-fertilization, will produce 25% of homozygous worms for the edit of interest. Its hermaphroditism, coupled with the fast life cycle of this nematode (∼3 days at 25°), allows the production of homozygous lines in 8 days at 25° (Ward 2015).
Despite the rate at which the optimization of the technique is progressing, it is not without limitations. These include variable efficiency of the sgRNA (Briner et al. 2014; Farboud and Meyer 2015; Liu et al. 2018) and potential off-target mutagenesis (Ran et al. 2013). In the case of insertions, another limiting factor is the length of commercially available ssODNs, which are commonly synthesized with a maximum length of 200 bp. In C. elegans, homology arms of 35–45 bp are required for homologous recombination, and, therefore, the maximum length of a fragment that can be inserted by CRISPR with a single ssODN is ∼110–130 bp (Paix et al. 2014).
Insertions of small size are relatively easy to perform. However, the challenge currently lies in inserting larger fragments of DNA, such as the integration of fluorescent reporters into endogenous loci or gene replacements. Even though it has been demonstrated that this can be achieved by targeting vectors with homology arms (Dickinson et al. 2013; Schwartz and Jorgensen 2016; Farboud et al. 2019; McDiarmid et al. 2018), this method requires additional time to construct the correct plasmid and, in some cases, remove the markers from the genome. An alternative option is to use a PCR product with 35-bp flanking sites that are homologous to the insertion site as a repair template (Paix et al. 2015). However, in our hands, and in the experience of other colleagues (Dokshin et al. 2018), the efficiency of this method is very low.
To overcome this challenge, we developed a Nested CRISPR protocol that can consistently generate protein::FP fusions without the need for cloning. It involves the insertion of the FP of interest in two steps. The first step involves a ∼120-bp in-frame insertion at the N-terminus or C-terminus of the target gene, consisting of the joint 5′ and 3′ ends of EGFP, mCherry, or wrmScarlet using an ssODN as a repair template. This fragment contains a new PAM site and a protospacer sequence that, in a second step, allows the in-frame insertion of the remaining sequence of ∼700 bp, depending on the number and length of introns included in the donor, using a universal PCR product as a repair template. We found, as expected, that the first step is very efficient when using ssODNs of ≤200 bp as a repair template; and we demonstrate that the second step for inserting the larger fragment is reliable. Moreover, we have observed that in three of five attempts, both steps can be achieved with a single injection. In summary, we present data for the integration of three distinct FPs across eight genes, including protein fusions and one transcriptional reporter, demonstrating the efficiency of the Nested CRISPR method.
Materials and Methods
Strains
We used the Bristol N2 strain as wild-type background and worms were maintained on Nematode Growth Medium (NGM) plates seeded with Escherichia coli OP50 bacteria (Stiernagle 2006). All strains generated in this study are listed in Supplemental Material, Table S7.
crRNA and ssODN design
The 20-nucleotide guide sequences were selected with the help of CCTop (Stemmer et al. 2015) or Benchling (www.benchling.com), which contain CRISPR/Cas9 target predictors. The crRNAs were ordered as 2 nmol products from Integrated DNA Technologies (IDT) (www.idtdna.com) and were resuspended in 20 µl of nuclease-free duplex buffer to yield a stock concentration of 100 µM. Once the cut site had been determined, ssODN donors for C-terminal fusions were designed in such a way that the FP 1–3 sequences were inserted in-frame immediately before the stop codon of the gene of interest (pgl-1 and prpf-4), or within a few amino acids before the stop codon (gtbp-1 and ubh-4), depending on the availability of a PAM sequence. In contrast, N-terminal fusions rely on ssODN donors that facilitate in-frame insertions immediately after the start codon (sftb-1 and nfki-1). The canonical design is as follows: a 35- to 45-bp left homology arm extending from the region upstream of the start codon (N-terminal fusions) or stop codon (C-terminal fusions), followed by the FP 1–3 sequence, followed by a 35- to 45-bp right homology arm in the coding sequence (N-terminal fusions) or in the 3′ UTR (C-terminal fusions). The exact lengths of the homology arms depend on the distance of the insertion from the cut site and must account for the adjustment of nucleotides to ensure that the FP 1–3 fragment is inserted in-frame. In the case of transcriptional reporters (K12C11.3), two crRNAs were designed to cut within the gene of interest, leaving behind a few amino acids after the start codon and before the stop codon. However, nucleotides encoding for these excess amino acids were not included in the ssODN repair template, thus generating mCherry 1–3 insertions that are immediately flanked by the start and stop codons. ssODNs were ordered as 4 nmol ultramers from IDT and were resuspended in 40 µl of nuclease-free duplex buffer to yield a stock concentration of 100 µM. The sequences of crRNAs and ssODN donors used in all FP 1–3 injections are shown in Tables S4 and S5.
Preparation of FP PCR product repair templates
The plasmids pJJR82 and pJJR83 were gifts from Mike Boxem (plasmids #75027 and #75028, respectively; Addgene). These plasmids contain sequences for the EGFP and mCherry fluorophores, respectively, which are codon-optimized for C. elegans. The pJV003 plasmid was a gift from Denis Dupuy from which the wrmScarlet sequence was amplified. Finally, the fosmid WRM0625C_F10 containing the 2xTY1::EGFP::3xFLAG sequence was obtained from the TransgeneOme resource (https://transgeneome.mpi-cbg.de). Primers were designed to amplify the complete sequences of the FPs and additional motifs using the Phusion High-Fidelity DNA Polymerase (Cat. No. F530S; Thermo Fisher Scientific). The primers and PCR conditions for these universal steps are specified in Figure S2. Five microliters of PCR product were run on a 2% agarose gel to verify correct amplification of the fragments. The amplicon lengths for EGFP, mCherry, wrmScarlet, and 2xTY1::EGFP::3xFLAG are 865, 855, 693, and 1035 bp, respectively. The eight 50-μl reactions were purified with the MinElute PCR purification kit (Cat. No. 28004; QIAGEN, Hilden, Germany) with a yield of ∼800–1200 ng/μl.
Preparation of RNP injection mix
The individual components were combined in the following order: target gene crRNA (or in the case of step 2, the FP 1–3 crRNA), dpy-10 crRNA, tracrRNA (Cat. No. 1072532; IDT), and Cas9 (Cat. No. 1081058; IDT); and were incubated at 37° for 10 min. Cas9 was added to the mix at distinct concentrations, ranging from 0.25 to 1.64 ng/µl (Table 1). The dpy-10 repair template and target gene ssODN (in the case of step 1) or EGFP/mCherry PCR product (in the case of step 2) were added, and the volume brought up to the required amount with nuclease-free H2O. In our experience, it is possible to prepare 5-μl injection mixes to avoid wasting reagents in excess. The mixture was then centrifuged at 13,000 rpm for 2 min to settle particulate matter and was kept on ice prior to loading the capillary needles. Fresh injection mixes were normally prepared in this study. However, we observed that excess mixes stored at −20° for 4 months still work effectively. The recommended concentrations for each component of the injection mix are specified in Figure S3.
Table 1. Summary of two-step Nested CRISPR experiments.
| Experiment | Plates with dpy-10 edits/injected worms | Positives/worms screened (%)a | In-frame sequencesb | Cas9 concentration (ng/μl) |
|---|---|---|---|---|
| prpf-4::EGFP Step 1 | 1/13 | 6/23 (26.09) | 2 of 2 | 1500 |
| prpf-4::EGFP Step 1 | 5/24 | 28/50 (56.00) | 3 of 3 | 250 |
| prpf-4::EGFP Step 2 | 7/23 | 13/32 (40.63) | All | 1640 |
| prpf-4::EGFP Step 2 synthetic sgRNA | 4/20 | 2/24 (8.33)c | N/A | 250 |
| prpf-4::mCherry Step 1 | 6/12 | 3/23 (13.04) | 2 of 3 | 250 |
| prpf-4::mCherry Step 2 | 11/28 | 16/43 (37.21) | All | 250 |
| prpf-4::mCherry Step 2 synthetic sgRNA | 7/28 | 26/108 (24.07) | All | 250 |
| prpf-4::2xTy1::EGFP::3xFLAG Step 1 | 13/21 | 29/32 (90.63) | 3 of 3 | 250 |
| prpf-4::2xTy1::EGFP::3xFLAG Step 2 | 13/36 | 1/164 (0.61) | All | 250 |
| gtbp-1::EGFP Step 1 | 9/14 | 3/60 (5.00)d | 1 of 3 | 1000 |
| gtbp-1::EGFP Step 1 | 3/7 | 37/52 (71.15) | 2 of 3 | 250 |
| gtbp-1::EGFP Step 2 | 9/24 | 10/46 (21.74) | All | 250 |
| gtbp-1::EGFP Step 2 synthetic sgRNA | 4/13 | 3/34 (8.82) | All | 250 |
| gtbp-1::mCherry Step 1 | 6/15 | 22/32 (68.75) | 1 of 3 | 250 |
| gtbp-1::mCherry Step 2 | 7/18 | 7/62 (11.29) | All | 250 |
| pgl-1::EGFP Step 1 | 5/24 | 7/50 (14.00)e | 1 of 1 | 1000 |
| pgl-1::EGFP Step 1 | 4/13 | 25/40 (62.50) | 2 of 5 | 250 |
| pgl-1::EGFP Step 2 | 5/16 | 1/13 (7.69) | All | 250 |
| pgl-1::mCherry Step 1 | 3/13 | 24/45 (53.33) | 2 of 5 | 250 |
| pgl-1::mCherry Step 2 | 4/18 | 14/39 (35.89) | All | 250 |
| K12C11.3p::mCherry Step 1 | 7/37 | 21/56 (37.50) | 3 of 3 | 250 |
| K12C11.3p::mCherry Step 2 | 4/14 | 3/35 (8.57) | All | 250 |
| ubh-4::EGFP Step 1 | 2/8 | 8/30 (26.67)e | 1 of 1 | 500 |
| ubh-4::EGFP Step 2 | 9/27 | 4/39 (10.26)c | All | 250 |
| mCherry::sftb-1 Step 1 | 2/4 | 25/29 (86.21) | 3 of 3 | 250 |
| mCherry::sftb-1 Step 2 | 2/23 | 1/3 (33.33) | All | 250 |
| EGFP::nfki-1 Step 2 | 3/12 | 2/26 (7.69) | All | 250 |
Based on PCR genotyping (amplicons of the correct size are considered positives regardless of whether or not the insertion is in-frame).
In-frame sequences represent the fraction of worms with correct inserts over the PCR positives that were confirmed by Sanger sequencing (step 1). In step 2, all the worms that had PCR products of the correct size exhibited fluorescence, indicating in-frame insertions.
Identification of positives via visual screening only.
Pools of two to three worms.
Pools of two worms. The percentage of positive events in worms screened in pools may be underestimated if two or more animals in the same pool are positive.
Microinjection
Approximately 1 μl of the injection mix was loaded on Eppendorf Femtotips Microinjection Capillary Tips (Cat. No. 930000035; Eppendorf) using Eppendorf Microloader Pipette Tips (Cat. No. 5242956003; Eppendorf). Approximately 15–20 young adult hermaphrodites were immobilized in 2% agar pads with halocarbon oil and were injected with the corresponding transformation mix using the XenoWorks Microinjection System (Sutter Instrument) and the Nikon Eclipse Ti-S inverted microscope with Nomarski optics. Injected worms were recovered in M9 buffer and were individually separated onto NGM plates. The plates were incubated at 20° for 4 days or at 25° for 3 days.
Screening
dpy-10-edited animals (F1 rollers and dumpies) were, individually or in pools, transferred onto NGM plates and were left to lay F2 progeny. Single-worm or pooled (two to three individuals) PCR was then performed on F1 worms. Primers were designed for each target gene and amplicon size shifts on 2% agarose gel were indicative of insertion events. If the PCR product was of the correct size, eight wild-type-appearing F2 progeny were individually transferred onto NGM plates to isolate homozygous individuals. PCR products for homozygous animals were then purified using the QIAquick PCR purification kit (Catalog No. 28104; QIAGEN) and underwent Sanger sequencing to verify the correctness of the insertion. In step 2 insertions, visual screening was performed through fluorescence microscopy, in addition to genotyping by single-worm PCR. Green (EGFP) or red (mCherry, wrmScarlet) fluorescence were indicative of complete, in-frame insertion events. A list of external primers used for genotyping are shown in Table S8.
Data availability
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions are present within the article, figures, and tables. Table S1 contains a summary of experiments involving megamers. Table S2 shows the sequences of the ssODN bridges used in experiments coupled with megamers. Table S3 shows the sequences of the GFP and mCherry megamers. Table S4 contains a list of the crRNAs used in Nested CRISPR Step 1. Table S5 contains a list of ssODNs used in Nested CRISPR Step 1. Table S6 contains a list of Nested CRISPR universal sequences. Table S7 contains a list of all strains generated in this study. Table S8 contains a list of external primers for genotyping. Figure S1 shows the correlation between the number of dpy-10 edits and successful edits in the target locus. Figure S2 shows the reagents and conditions for the generation of universal PCR product repair templates for Nested CRISPR Step 2. Figure S3 shows the standard composition of Nested CRISPR injection mixes. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7629641.
Results
Endogenous translational fluorescent reporters by Nested CRISPR
Nested CRISPR was conceived after multiple failed attempts to generate an endogenous fluorescent reporter of the locus prpf-4. The most straightforward method existing at the time was the use of Cas9-guide RNA ribonucleoprotein (RNP) complexes and a PCR product with 35-bp homology arms as an HDR donor (Paix et al. 2015). After four experiments consisting of 72 injected P0s and 366 screened F1s, only a single positive event was achieved (0.3% efficiency). The same method was applied to five additional loci, but all attempts to endogenously tag these genes with FPs failed. Therefore, we investigated alternative cloning-free methods to generate endogenous fluorescent reporters in C. elegans.
First, we explored the possibility of using single-stranded DNA (ssDNA) megamers as repair templates. Since the synthesis of megamers is expensive and often presents difficulties, we thought of a strategy that uses universal megamers. We tested the use of two short ssODNs (≤200 bp) that bridge the target sequence to long universal ssODNs (GFP or mCherry megamers, 872 and 702 nt, respectively), taking advantage of C. elegans recombineering (Paix et al. 2016). We combined these three overlapping ssODNs (two short and one long) with concordant polarity as a repair template to tag two different loci, but we only obtained partial insertions of GFP or mCherry ranging from 100 to 200 bp (Tables S1–S3).
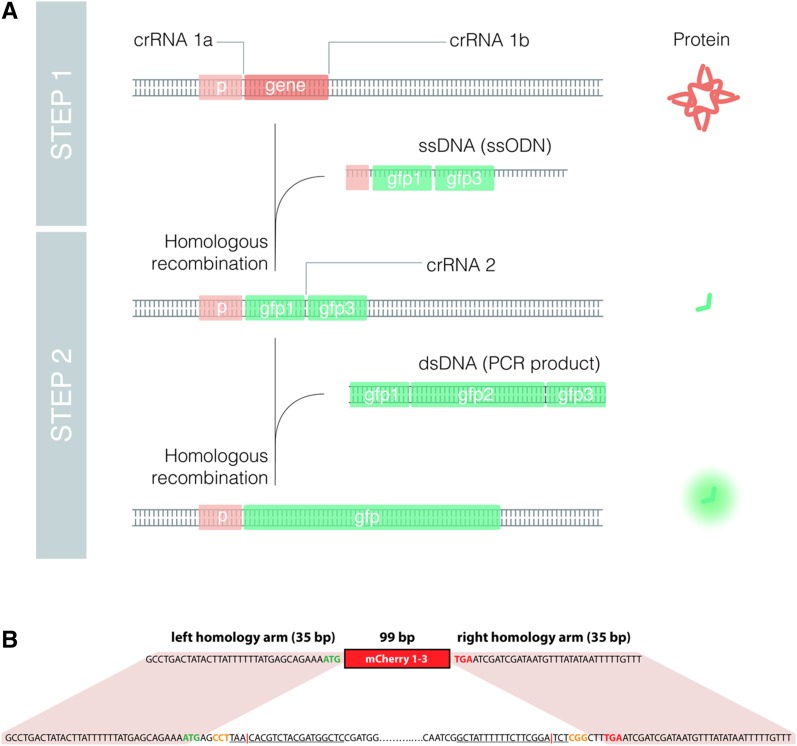
Since the targeted insertion of long double-stranded DNA (dsDNA) templates was much more difficult than genome editing with small (≤200 nt) single-stranded DNA, we reasoned that a nested approach that always uses the same, previously validated dsDNA and crRNA to insert the longer fragment would be an efficient method to produce endogenous fluorescent reporters. Thus, we designed a pipeline to produce homozygous translational reporters in five generations (∼3 weeks in C. elegans) (Figure 1). The pipeline works as follows: in the case of Enhanced Green Fluorescent Protein (EGFP), we divided the 866 bp of the EGFP sequence, including three introns, into three sequences of 58 + 752 + 56 bp that are designated as fragments 1, 2, and 3, respectively (Figure 2). Using a crRNA specific for the target gene, a block containing sequences 1 and 3 can be inserted in-frame in the place of interest by using single-stranded oligodeoxynucleotides (ssODNs) as donors (Tables S4 and S5). To enrich for successfully edited animals, we used the co-CRISPR strategy (Arribere et al. 2014; Kim et al. 2014). Thus, only plates with the presence of dpy-10-edited worms, either dumpies or rollers, were screened for insertions by PCR (Figure 1). Once homozygotes for the 1–3 block were obtained, the insertion was sequenced to ensure that the 1–3 fragment was in-frame. We observed that most of the sequences were correct, with the most common error being the lack of a single nucleotide or the incorporation of an additional nucleotide. Correct in-frame sequences were observed in 68% of the cases (26 out of 38), and, therefore, we recommend sequencing at least three positives (Table 1).
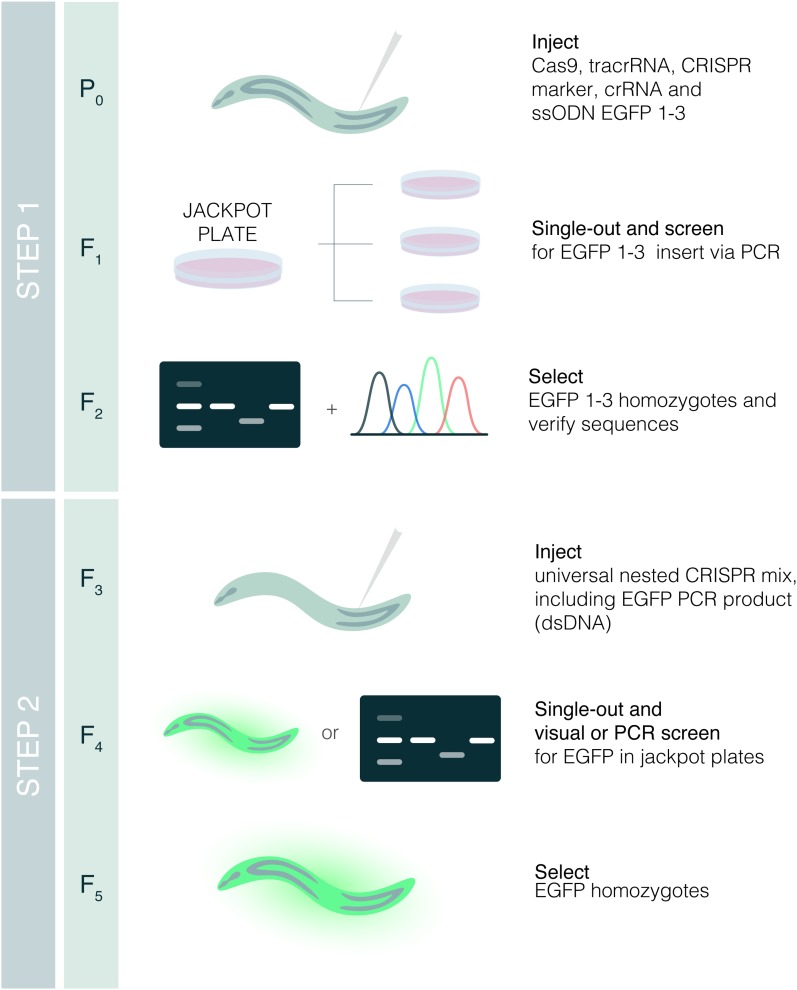
Figure 1.
Nested CRISPR pipeline. Young adult hermaphrodites are injected in the germline with a mix containing Cas9, tracrRNA, crRNA, and ssODN repair template for the fluorescent protein of interest (EGFP in this example), and a marker of CRISPR efficiency [crRNA and ssODN for dpy-10(cn64) in our case as in Arribere et al. (2014)]. Injected animals are singled out onto NGM plates. After 3–4 days at 20°, animals from plates with dumpies and/or rollers (jackpot plates) are singled out onto NGM plates and genotyped for the insertion of interest once they have laid progeny. The progeny of F1 animals positive for the insertion are singled out to obtain F2 homozygous animals, and insertions are verified by Sanger sequencing. Bands in the gel and sequencing peaks are illustrative. In the second step, young F3 hermaphrodites with the in-frame insertions of fragments 1 and 3 are injected with a universal mix containing Cas9, tracrRNA, crRNA for fragment 1–3, dsDNA (PCR product of the FP of interest) as repair template, and CRISPR comarker. Successful edits in the F4 can be detected either through screening visually for fluorescence or through PCR. F4 animals heterozygous for the insertion will give rise to F5 homozygotes for the translational reporter of interest.
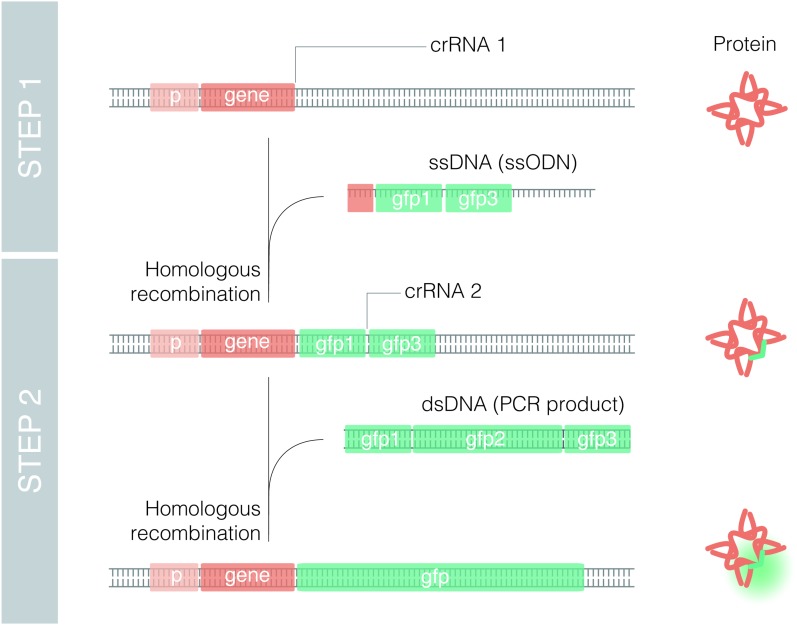
Figure 2.
Translational endogenous reporters by Nested CRISPR. Scheme of molecular events to generate a translational fluorescent reporter (GFP in this case) by Nested CRISPR. A gene-specific crRNA (crRNA 1) is required to assemble Cas9 RNP complexes that cut at the 5′ or 3′ end of the gene. Along with these RNPs, the injection mix contains an ssODN with two homology arms of 35–45 bp (depending on the distance from the cut site) that is inserted in the place of interest by homologous recombination. In the second step, RNPs contain a universal crRNA (crRNA 2) that cuts the gfp 1–3-specific targeted sequence or protospacer. Then, a universal dsDNA molecule, resulting from PCR amplification of EGFP, is used as a repair template to generate a translational reporter.
Next, the EGFP 1 and 3 sequences were used as homology regions to insert the remaining EGFP sequence using an EGFP 1–3 specific crRNA and a PCR product amplified from the pJJR82 plasmid that contains EGFP (Figure 3A). Importantly, the injection mix for this second step is universal as it uses the same crRNA and PCR product, increasing reproducibility and reducing costs (Table S6). Scoring of F1 positives can be performed visually if the fluorescence signal is detectable by microscopy. Otherwise, it is necessary to perform a PCR-based screen among animals, as individuals or in pools, from dpy-10-edited plates.
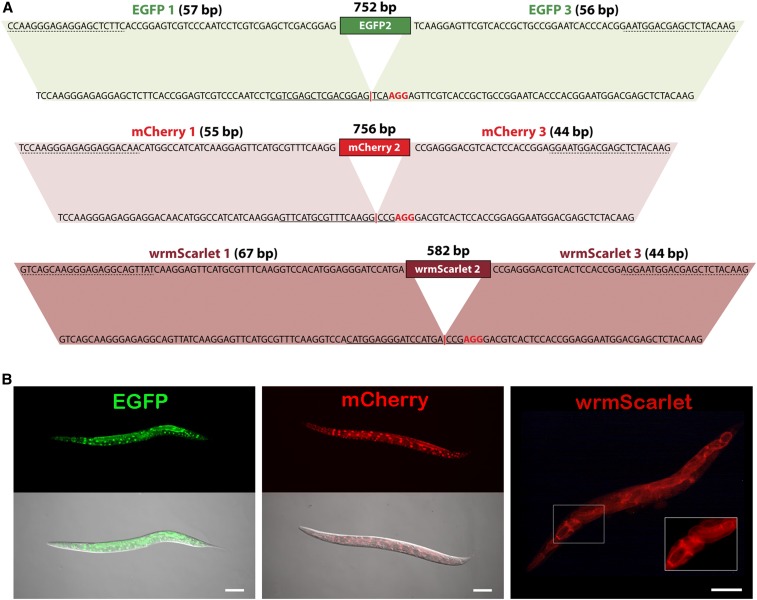
Figure 3.
Generation of EGFP, mCherry, and wrmScarlet reporters via Nested CRISPR. (A) Details of sequences and homology regions of EGFP, mCherry, and wrmScarlet for the universal step 2. The nucleotides at the bottom represent the sequences of the 1–3 fragments of EGFP, mCherry, or wrmScarlet that result from step 1 insertions. Solid lines correspond to the 20-nt guide sequence and nucleotides in red represent the PAM sequence. Targeted (protospacer) sequences result from the fusion of the native 1 and 3 fragments, without the need to modify any nucleotides. The red vertical bar represents the DSB site. At the top is the sequence of the dsDNA repair template with homology to the 1–3 fragments. Primer annealing sites for the PCR amplification of the dsDNA repair templates are labeled with dashed lines. Parallelograms mark homology regions. (B) Representative images of prpf-4::EGFP, prpf-4::mCherry, and F27C1.2::wrmScarlet translational reporters generated by Nested CRISPR. Bar, 100 µm.
We first applied the Nested CRISPR pipeline to prpf-4 to produce an EGFP reporter. In the first step, from 13 injected P0s and 23 screened F1s, 26% were positive for the EGFP 1–3 insertion. In the second step, from 23 injected P0s and 32 screened F1s, 40% had full EGFP integration. Whereas in the past we spent months to generate one strain, we obtained 13 independent lines for prpf-4::EGFP in 3 weeks using the Nested CRISPR approach (Figure 3B). Thus, we decided to validate and consolidate this method by applying the same approach using different FPs in various loci. We generated a prpf-4::mCherry reporter with similar efficiencies and also made endogenous EGFP and mCherry reporters for gtbp-1 and pgl-1, genes that were previously used to test CRISPR/Cas9 methods (Paix et al. 2015, 2016). In addition, we also successfully created a translational reporter for ubh-4. Efficiencies for tagging these four loci ranged from 12 to 70% in the first step, and from 7 to 41% in the second step (Figure 4A and Table 1).
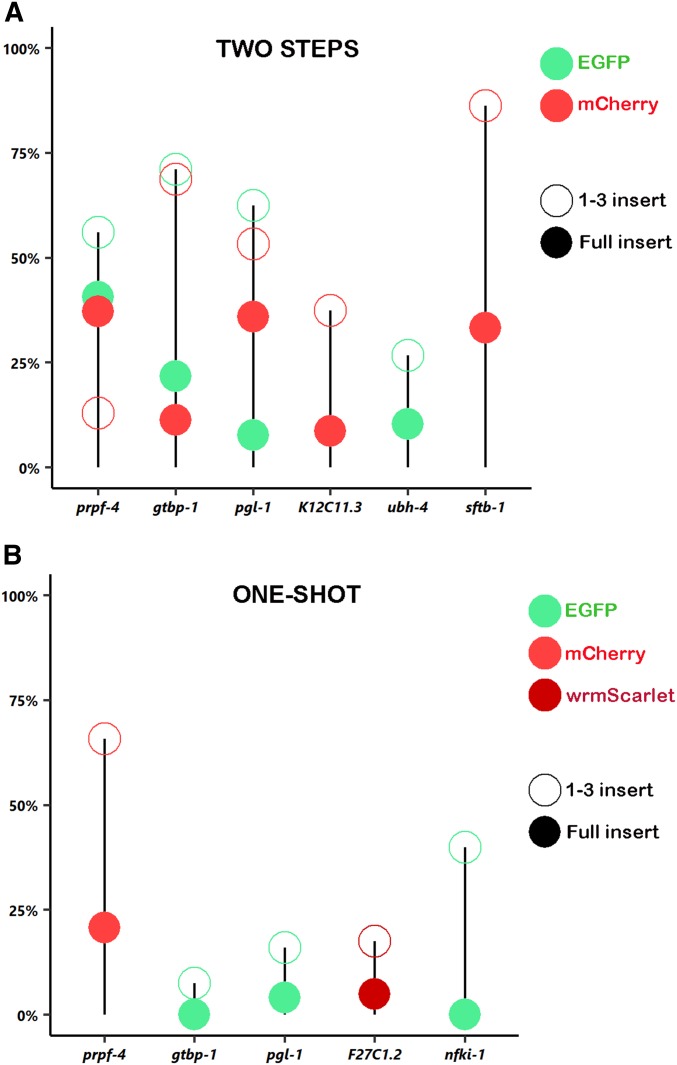
Figure 4.
Efficiency of Nested CRISPR experiments. Percentage of insertion events in two-step (A) and one-shot (B) Nested CRISPR experiments based on the values from Table 1 and Table 2. Open circles represent fragment 1–3 insertions in step 1 experiments, whereas solid circles symbolize in-frame insertions of EGFP (green), mCherry (red), or wrmScarlet (dark red) in step 2. Efficiencies for 1–3 inserts are based on PCR genotyping (amplicons of the correct size are considered positives), whereas efficiencies for full inserts are based on in vivo fluorescence.
As previously mentioned, a universal mix can be used for all step 2 injections to increase the reproducibility of HDR efficiency. However, we observed variability in the efficiency of the second step (Figure 4A), which could be attributed to local chromatin structure or other factors that we do not yet understand. Nevertheless, we believe that a major advantage of this method is its reliability and modularity for obtaining endogenous fluorescent reporters in a cloning-free manner.
Nested CRISPR pipeline to generate a deletion mutant and a transcriptional reporter
Based on our experience, the removal of an entire open reading frame (ORF) of a given gene by CRISPR using two crRNAs at the 5′ and 3′ ends (Chen et al. 2014) is very efficient if an ssODN donor is added to the injection mix, producing mutants with precise deletions. We reasoned that in the case of nonessential genes, which comprise ∼80–85% of the C. elegans genome (Kemphues 2005), the Nested CRISPR approach could be used to produce both a deletion mutant and a transcriptional reporter in the same pipeline (Figure 5A). We tested this approach in the gene K12C11.3, which encodes a nonessential copper transporter. First, a 1339-bp deletion was made, removing most of the K12C11.3 coding sequence. Two crRNAs were used to cut right after the start codon and right before the stop codon, with a donor consisting of an ssODN containing the mCherry 1–3 fragment with two 35-bp homology arms outside of the coding sequence (Figure 5B). In the first step, we observed that 37% of all screened F1s had the deleted ORF replaced with the mCherry 1–3 fragment. Meanwhile, in the second step, three independent transcriptional reporter lines were obtained after injecting 14 P0s and screening 35 F1s (Table 1). This demonstrates that Nested CRISPR is an advantageous option when generating deletion mutants of nonessential genes, as the knockout can be primed for the subsequent generation of a transcriptional reporter.
Figure 5.
Deletions and transcriptional endogenous reporters by Nested CRISPR. (A) Scheme of molecular events to generate a deletion and a transcriptional fluorescent reporter (GFP in this case) by Nested CRISPR. Two gene-specific crRNAs (crRNA 1a and crRNA 1b) and an ssODN donor are required to produce a deletion of the gene and an in-frame insertion of fragment 1–3. In the second step, RNPs contain a universal crRNA (crRNA 2) that cuts the gfp 1–3-specific targeted sequence. Then, a universal dsDNA molecule, resulting from PCR amplification of GFP, is used as a repair template to generate a transcriptional reporter. (B) Details of sequences and homology regions for simultaneous K12C11.3 knockout and mCherry 1–3 insertion. The nucleotides at the bottom represent the native sequence of the K12C11.3 gene. The entire coding sequence is removed. Nucleotides in blue represent the 5′ and 3′ untranslated regions (UTRs) while nucleotides in green and red represent the start and stop codons, respectively. Solid lines correspond to the 20-nt guide sequence, and nucleotides in orange represent PAM sequences. It can be noted that the 5′ guide sequence is in the antisense orientation. The red vertical bar represents the DSB site. The nucleotides at the top represent the sequence of the 169-bp ssODN repair template consisting of the 99-bp mCherry 1–3 sequence flanked by 35-bp homology arms.
Synthetic sgRNAs are functional
When Cas9 and guide RNAs are injected in the form of RNPs, researchers normally utilize crRNA:tracrRNA duplexes that are synthesized separately and subsequently annealed in a ∼1:1 ratio to form the sgRNA that is later combined with Cas9 to assemble the final RNP complex. However, synthetic sgRNAs consisting of a single RNA oligonucleotide containing both the target-specific crRNA and the universal tracrRNA have recently become commercially available (IDT). Synthetic sgRNAs contain chemical modifications for improved stability and performance, and thus, we investigated if they could increase editing efficiencies in C. elegans. We found that synthetic sgRNAs work well, but we did not observe a clear improvement in editing efficiency (Table 1).
Optimization of the method
Nested CRISPR can be developed and applied in different ways, based on the specific requirements of the reporter desired. Beyond the widely used EGFP and mCherry, there are other FPs that can be of interest for generating endogenous reporters. For instance, wrmScarlet is a codon-optimized version of mScarlet that has recently been described as the brightest red FP (Bindels et al. 2017; El Mouridi et al. 2017). We designed a crRNA and a repair template for wrmScarlet integration via Nested CRISPR in F27C1.2, an essential copper membrane transporter, and found that it was successfully inserted at the C-terminal region of the protein after the first attempt (Figure 3).
We also used Nested CRISPR for a multiple tag that includes EGFP and the epitopes 2xTY1 and 3xFLAG. This tag was amplified from one of the TransgeneOme fosmids that have been widely distributed to the C. elegans community (Sarov et al. 2012). In this case, the efficiency was 91% for the first step and close to 1% for the second step. The reduced efficiency of the second step could be due to the larger size of the insertion (927 bp) or an inefficient crRNA, resulting in insufficient cutting and stimulation of HDR (Paix et al. 2014) (Table S6).
To reduce the number of injections and the number of generations needed to obtain the desired reporters, we attempted to combine both steps in a single injection. We performed this “one-shot” approach in five different loci and obtained full insertions of the desired fluorescent tag in three cases (Figure 4B and Table 2). However, in all cases, several independent lines containing in-frame insertions of the 1–3 fragment were isolated. Thus, it is advisable to first attempt tagging genes with the one-shot approach, followed by a subsequent step 2 injection over a 1–3 insertion background, in cases where full insertions are not achieved with a single injection. Such is the case with the N-terminal EGFP reporter we generated for nfki-1. The one-shot approach failed to yield full EGFP insertion at the locus after screening 70 dpy-10-edited F1s. However, EGFP 1–3 insertion was observed in 40% of the screened worms. After microinjection of these strains with the universal step 2 mix, we successfully obtained an endogenous EGFP::nfki-1 reporter with 7.69% efficiency (Table 1).
Table 2. Summary of one-shot Nested CRISPR experiments.
| Experiment | Plates with dpy-10 edits/injected worms | Step 1 positives/worms screened (%)a | Step 2 positives/worms screened (%)a |
|---|---|---|---|
| prpf-4::mCherry | 11/29 | 25/38 (65.79) | 16/77 (20.78)b |
| gtbp-1::EGFP | 6/28 | 3/40 (7.50) | 0/40 (0) |
| pgl-1::EGFP | 2/22 | 4/25 (16.00) | 1/25 (4.00) |
| F27C1.2::wrmScarlet | 7/22 | 7/40 (17.50) | 7/144 (4.86)b |
| EGFP::nfki-1 | 5/26 | 28/70 (40.00) | 0/70 (0) |
Based on PCR genotyping (amplicons of the correct size are considered positives regardless of whether or not the insertion is in-frame).
Identification of positives via visual screening.
Discussion
CRISPR/Cas9 technology is evolving at such a speed that the standardization of protocols between distinct laboratories is difficult. For instance, a reported efficiency of 16% for inserting two loxP sites for producing conditional KOs in mice (Yang et al. 2013) has recently been proved to be close to 1% by the mice research community (Gurumurthy et al. 2018). Meanwhile, in our hands, the CRISPR-based insertion of fluorescent tags in C. elegans using PCR products with 35-bp homology arms as donors was not as efficient as originally reported (Paix et al. 2015). This discrepancy has been formally mentioned for the first time in a recent publication (Dokshin et al. 2018). These inconsistencies between efficiencies are probably due to factors that are uncontrolled or unrecognized, arising from peculiarities of the methodology in distinct laboratories. For instance, some labs use in-house purified Cas9, whereas other labs use commercial Cas9, and each lab uses different equipment for performing microinjections in the worm germline. Unfortunately, in contrast to the mice research community, there is no coordinated effort to determine the true efficiency of different gene-editing techniques in C. elegans.
CRISPR/Cas9 reagents are now commercially available at a cost that can be afforded by most labs. Thus, the use of common reagents should help in standardizing the outcomes of CRISPR experiments. However, protocols for performing microinjections in C. elegans vary from lab to lab, and from person to person, resulting in variability in the final concentration of Cas9 in the germline. Since high concentrations of Cas9 appear to be toxic (Dokshin et al. 2018), the microinjection process itself could be considered a critical point. However, despite having used high concentrations of Cas9 (1000–1640 ng/μl) in our initial experiments, we were able to obtain positives, albeit at lower efficiencies than when injecting with a lower Cas9 concentration (250 ng/μl) (Table 1).
The Nested CRISPR approach has several advantages (Box 1) and has shortened the time to obtain endogenous reporters in our lab. While we previously spent months performing CRISPR experiments attempting to generate reporters of six distinct genes and being successful only once, by following the Nested CRISPR method we have succeeded in all our attempts to make endogenous fluorescent reporters (Table 1 and Table S7). Other researchers should be able to reproduce our success rate since all the required elements are commercially available, and, in addition, the reagents and conditions for the second step are universal. Thus, this method is feasible for researchers with limited knowledge of molecular cloning and will facilitate the production of endogenous reporters for studying the real expression pattern of a given gene. Therefore, expression patterns inferred from extrachromosomal reporters or molecular constructs inserted randomly in the genome, often as multicopy, should be reviewed and validated.
Box 1. Advantages of the Nested CRISPR method.
It is cloning-free, and the homogeneity of the reagents allows interlaboratory comparative analysis of CRISPR efficiency.
The second step utilizes the same reagents and conditions in all cases. Thus, it is reliable and the difficulty of inserting long dsDNA fragments is bypassed.
Given the universality of the second step, it is scalable to genome-wide projects.
Different fluorescent protein variants can be used, with distinct sequences and introns that may influence expression levels and silencing of exogenous DNA.
A deletion mutant and a transcriptional reporter for the gene of interest can be produced in the same Nested CRISPR pipeline.
An insertion in the first step indicates an accessible chromatin environment for a cut in the second step.
We have used the co-CRISPR system as a marker of injection quality, and, similarly to others, found a positive correlation between successful edits and plates with a high number of dpy-10-edited animals (Figure S1) (Arribere et al. 2014; Paix et al. 2015). Alternatively, other co-injection markers such as extrachromosomal plasmids harboring fluorescent reporters, antibiotic resistance genes, or dominant alleles can be used (Norris et al. 2015; Prior et al. 2017; Dokshin et al. 2018). However, the use of plasmids could be a source of variability (mutations, distinct DNA preparations, etc.) between labs that can be circumvented by using a common crRNA for dpy-10.
Since the structure of the guide RNA can influence the activity of Cas9 (Lim et al. 2016), we tested the use of synthetic sgRNAs, but we did not observe an improvement in editing efficiency compared to using tracrRNA and crRNA. Positive aspects of synthetic sgRNAs are that the volume of the injection mix can be reduced and the potential competition between several crRNAs for the tracrRNA can be avoided, and the fact that they are commercially available or can be rapidly synthesized in vitro (Leonetti et al. 2016).
We also tried using universal EGFP and mCherry megamers (long ssDNA) plus two small ssODNs as homology bridges to induce the in vivo assembly of linear DNAs (Paix et al. 2016), but this only resulted in partial insertions. Once the Nested CRISPR method was established, we also failed in the attempt of using EGFP or mCherry megamers as repair templates during step 2 (Table S1). Although the price of megamers is still high and the synthesis of long ssDNA can present difficulties, the utility of megamers in CRISPR should be explored and evaluated in the future (Quadros et al. 2017). The use of plasmids as repair templates in Nested CRISPR should also be investigated as it has been previously demonstrated that plasmid donors with short homology arms of just 50–60 bp can efficiently direct insertion into the C. elegans genome (Schwartz and Jorgensen 2016).
The modular and flexible nature of the second step of Nested CRISPR is of great value. Once the 1–3 fragment is inserted, the sequence of the dsDNA repair template for the second step can be modified to avoid piRNA-mediated transgene silencing in the germline (Wu et al. 2018) or to alter protein expression levels by modifying codons according to the codon adaptation index (Redemann et al. 2011). The number, length, and sequences of introns can also be modified in the second step. This is relevant because the number and length of introns can influence the transcriptional rate, and the sequence of these introns can influence germline silencing (Frøkjær-Jensen 2013; Heyn et al. 2015).
In the second step of Nested CRISPR, we have efficiently inserted PCR fragments ranging from 582 to 792 bp, depending on the presence or absence of introns. Our attempt to insert a longer fragment of 927 bp consisting of the 2xTY1 and 3xFLAG epitopes flanking EGFP was successful but with much lower efficiency (Table 1). Although Nested CRISPR was initially conceived to produce reporters with standard FPs, we plan to expand the toolkit for Nested CRIPSR and will carefully study the influence of fragment length on editing efficiency and the benefits of inducing two DSBs in the targeted regions to insert longer fragments as recently reported (Farboud et al. 2019). The lower efficiency observed when inserting this larger fragment could also be due to an inefficient crRNA. When we chose the crRNAs for step 2, we prioritized the absence of off-targets in the genome, followed by the in silico predicted activity. However, since all the crRNAs used for EGFP, mCherry, and wrmScarlet were reasonably efficient, we found that it was not necessary to test the in vitro efficiency of different crRNAs candidates. However, searching for the optimal crRNA would be recommendable when working with large-scale projects.
We used Nested CRISPR to generate a deletion mutant and a transcriptional reporter in the same pipeline. This is a strategy that could be considered for large-scale projects because at the time a collection of deletion mutants is made, these strains can be prepared for a universal second step that generates transcriptional reporters.
The need for homozygous animals with the 1–3 fragment in our deletion plus transcriptional reporter pipeline is a handicap for essential genes (∼20% of genes) whose deletions need to be maintained as heterozygous strains. In these cases, a one-shot approach can be considered an option.
Recently, a scalable strategy to create mutants in C. elegans has been suggested. This strategy relies on the insertion of an ssODN with stop codons in the three different reading frames (Wang et al. 2018). This is a smart approach that could be particularly convenient when a full deletion is not desired because of the presence of functional sequences like intronic genes, enhancers, or regulatory RNAs. However, if a null mutant removing the whole gene is of interest, then Nested CRISPR can produce a deletion mutant strain that is primed for tagging with a fluorescent reporter.
The efficiency of Nested CRISPR may be attributed to the use of a universal and reliable step to insert long fragments of DNA. Perhaps, the prior editing of a genomic region (step 1) facilitates the subsequent insertion of a longer piece of DNA in the same location (step 2). It is known that chromatin state influences CRISPR-Cas9 editing efficiencies (Verkuijl and Rots 2019), and it is possible that the first cut makes the chromatin more accessible or sensitive for a subsequent cut. Nevertheless, the cut required for the first step of Nested CRIPSR indicates that the chromatin in that region is accessible for Cas9 to perform the second and more limiting step.
The mechanisms of CRISPR/Cas9 genome editing in distinct organisms seem to be very similar; therefore, any technical advance in C. elegans could be applied to other models. In the case of Nested CRISPR, the need for five generations to obtain homozygous animals could be a handicap in other animals with longer life cycles. In these cases, the single-shot approach should be explored. However, if a deletion mutant for a given gene is planned, it will be advantageous to simultaneously prepare it for a second step that facilitates the insertion of an endogenous fluorescent reporter.
The race to develop more efficient and reliable CRISPR methodologies has not stopped. C. elegans researchers have found several factors that can influence the efficiency of CRISPR gene editing. For instance, the orientation of the ssDNA repair template seems to influence the efficiency of insertion (Katic et al. 2015; Ward 2015; Paix et al. 2016; Farboud et al. 2019), certain nucleotides before the PAM sequence could be more convenient for Cas9 cutting (Farboud and Meyer 2015; Farboud et al. 2019), and new variants of Cas9 protein may exhibit greater specificity and efficiency (Bell et al. 2016; Zhao et al. 2016). Despite these, the insertion of long DNA fragments still has limited efficiency. Recently, two studies have proposed modifications in dsDNA templates to improve the efficiency of genome editing. These studies suggest the use of 5′ modifications in the dsDNA donors and the use of hybrid PCR products with 120 bp of ssDNA overhangs (Dokshin et al. 2018; Ghanta et al. 2018). The generation of hybrid PCR products requires long primers (140 nt) that may need to be optimized, whereas in comparison, the PCR conditions for the second step in Nested CRISPR are already optimized, and the product can be reused for several experiments. These studies and ours are open to the community and will certainly help different labs in finding a convenient method that suits their resources and expertise. In any case, a coordinated effort from the C. elegans community is necessary to compare distinct approaches for inserting long DNA fragments by CRISPR. The onset of different but efficient CRISPR methodologies will facilitate the widespread generation of endogenous reporters that will push forward many research projects.
Acknowledgments
We thank Mike Boxem and Denis Dupuy for sharing the plasmids. We also thank Denis Dupuy, Montserrat Porta-de-la-Riva, and Peter Askjaer for critical reading of the manuscript and members of the Cerón lab for their comments on this study. Thanks to @shookstudio for their assistance in making Figure 1, Figure 2, and Figure 5A. This work has been supported by a grant from the Instituto de Salud Carlos III (ISCIII) to J.C. (PI15-00895), cofunded by FEDER funds/European Regional Development Fund (ERDF) — a way to Build Europe. This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 713673. We thank CERCA (Centres de Recerca de Catalunya) Program/Generalitat de Catalunya for their institutional support. J.V. has an INPhINIT PhD fellowship from “la Caixa” Foundation (LCF/BQ/IN17/11620065), and X.S. has an FPU (Formación de Personal Universitario) Ph.D. fellowship from MINECO.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7629641.
Communicating editor: O. Hobert
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. T., Fu B. X. H., Fire A. Z., 2016. Cas9 variants expand the target repertoire in Caenorhabditis elegans. Genetics 202: 381–388. 10.1534/genetics.115.185041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels D. S., Haarbosch L., van Weeren L., Postma M., Wiese K. E., et al. , 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14: 53–56. 10.1038/nmeth.4074 [DOI] [PubMed] [Google Scholar]
- Briner A. E., Donohoue P. D., Gomaa A. A., Selle K., Slorach E. M., et al. , 2014. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell 56: 333–339. 10.1016/j.molcel.2014.09.019 [DOI] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193 10.1093/nar/gkt805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu F., Zhu C., Ji J., Zhou X., et al. , 2014. Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci. Rep. 4: 7581 10.1038/srep07581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. 10.1534/genetics.113.155879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. 10.1534/genetics.113.155853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin G. A., Ghanta K. S., Piscopo K. M., Mello C. C., 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210: 781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mouridi S., Lecroisey C., Tardy P., Mercier M., Leclercq-Blondel A., et al. , 2017. Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype. G3 (Bethesda) 7: 1429–1437. 10.1534/g3.117.040824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. 10.1534/genetics.115.175166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Severson A. F., Meyer B. J., 2019. Strategies for efficient genome editing using CRISPR-Cas9. Genetics. 211: 431–457 10.1534/genetics.118.301775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., 2013. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics 195: 635–642. 10.1534/genetics.113.156521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta K. S., Dokshin G. A., Mir A., Krishnamurthy P. M., Gneid H., et al. , 2018. 5′ modifications improve potency and efficacy of DNA donors for precision genome editing. bioRxiv. Preprint. 10.1101/354480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy C., Quadros R., Adams J., Alcaide P., Ayabe S., et al. , 2018. Re-evaluating one-step generation of mice carrying conditional alleles by CRISPR-Cas9-mediated genome editing technology. bioRxiv. Preprint. 10.1101/393231 [DOI] [Google Scholar]
- Heyn P., Kalinka A. T., Tomancak P., Neugebauer K. M., 2015. Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences. BioEssays 37: 148–154. 10.1002/bies.201400138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Grosshans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. 10.1534/genetics.113.155754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Xu L., Ciosk R., 2015. CRISPR/Cas9 genome editing in Caenorhabditis elegans: evaluation of templates for homology-mediated repair and knock-ins by homology-independent DNA repair. G3 (Bethesda) 5: 1649–1656. 10.1534/g3.115.019273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues, K., 2005 Essential genes (December 24, 2005), WormBook, ed. The C. elegans Research Community WormBook. https://www.ncbi.nlm.nih.gov/books/NBK19771/
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D. J., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. 10.1534/genetics.114.166389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti M. D., Sekine S., Kamiyama D., Weissman J. S., Huang B., 2016. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. USA 113: E3501–E3508. 10.1073/pnas.1606731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Bak S. Y., Sung K., Jeong E., Lee S. H., et al. , 2016. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat. Commun. 7: 13350 10.1038/ncomms13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li S., Zhang Y., Li J., Li Y., 2018. Efficient CRISPR-based genome editing using tandem guide RNAs and editable surrogate reporters. FEBS Open Bio 8: 1167–1175. 10.1002/2211-5463.12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. 10.1534/genetics.113.155382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDiarmid T. A., Au V., Loewen A., Liang J. J. H., Mizumoto K., et al. , 2018. CRISPR-Cas9 human gene replacement and phenomic characterization in Caenorhabditis elegans to understand the functional conservation of human genes and decipher variants of uncertain significance. Dis Model Mech. 11: 1–15. 10.1242/dmm.036517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J., Díez-Villaseñor C., García-Martinez J., Soria E., 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60: 174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Norris A. D., Kim H.-M., Colaiacovo M. P., Calarco J. A., 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201: 449–458. 10.1534/genetics.115.180679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-cas9 ribonucleoprotein complexes. Genetics 201: 47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Schmidt H., Seydoux G., 2016. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 44: e128 10.1093/nar/gkw502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior H., Jawad A. K., MacConnachie L., Beg A. A., 2017. Highly efficient, rapid and Co-CRISPR-independent genome editing in Caenorhabditis elegans. G3 (Bethesda) 7: 3693–3698. 10.1534/g3.117.300216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros R. M., Miura H., Harms D. W., Akatsuka H., Sato T., et al. , 2017. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18: 92 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S., Schloissnig S., Ernst S., Pozniakowsky A., Ayloo S., et al. , 2011. Codon adaptation-based control of protein expression in C. elegans. Nat. Methods 8: 250–252. 10.1038/nmeth.1565 [DOI] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. 10.1016/j.cell.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. L., Jorgensen E. M., 2016. SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Genetics 202: 1277–1288. 10.1534/genetics.115.184275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J. L., 2015. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10: e0124633 [corrigenda: PLoS One 12: e0176619 (2017)]. 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006). WormBook, ed. The C. elegans Research Community WormBook. 10.1895/wormbook.1.101.1 [DOI] [Google Scholar]
- van Schendel R., Roerink S. F., Portegijs V., van den Heuvel S., Tijsterman M., 2015. Polymerase Theta is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun. 6: 7394 10.1038/ncomms8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkuijl S. A., Rots M. G., 2019. The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr. Opin. Biotechnol. 55: 68–73. 10.1016/j.copbio.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B. L. G., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. 10.1534/genetics.113.156299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Park H., Liu J., Sternberg P. W., 2018. An efficient genome editing strategy to generate putative null mutants in Caenorhabditis elegans using CRISPR/Cas9. G3 (Bethesda) 8: 3607–3616. 10.1534/g3.118.200662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. 10.1534/genetics.114.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.-S., Huang W.-C., Brown J. S., Zhang D., Song X., et al. , 2018. pirScan: a webserver to predict piRNA targeting sites and to avoid transgene silencing in C. elegans. Nucleic Acids Res. 46: W43–W48. 10.1093/nar/gky277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D.,2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. 10.1038/cr.2014.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Lv X., Zhao X., Suehiro Y., et al. , 2016. One-step homozygosity in precise gene editing by an improved CRISPR/Cas9 system. Cell Res. 26: 633–636. 10.1038/cr.2016.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions are present within the article, figures, and tables. Table S1 contains a summary of experiments involving megamers. Table S2 shows the sequences of the ssODN bridges used in experiments coupled with megamers. Table S3 shows the sequences of the GFP and mCherry megamers. Table S4 contains a list of the crRNAs used in Nested CRISPR Step 1. Table S5 contains a list of ssODNs used in Nested CRISPR Step 1. Table S6 contains a list of Nested CRISPR universal sequences. Table S7 contains a list of all strains generated in this study. Table S8 contains a list of external primers for genotyping. Figure S1 shows the correlation between the number of dpy-10 edits and successful edits in the target locus. Figure S2 shows the reagents and conditions for the generation of universal PCR product repair templates for Nested CRISPR Step 2. Figure S3 shows the standard composition of Nested CRISPR injection mixes. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7629641.