Abstract
Purine homeostasis is ensured through a metabolic network widely conserved from prokaryotes to humans. Purines can either be synthesized de novo, reused, or produced by interconversion of extant metabolites using the so-called recycling pathway. Although thoroughly characterized in microorganisms, such as yeast or bacteria, little is known about regulation of the purine biosynthesis network in metazoans. In humans, several diseases are linked to purine metabolism through as yet poorly understood etiologies. Particularly, the deficiency in adenylosuccinate lyase (ADSL)—an enzyme involved both in the purine de novo and recycling pathways—causes severe muscular and neuronal symptoms. In order to address the mechanisms underlying this deficiency, we established Caenorhabditis elegans as a metazoan model organism to study purine metabolism, while focusing on ADSL. We show that the purine biosynthesis network is functionally conserved in C. elegans. Moreover, adsl-1 (the gene encoding ADSL in C. elegans) is required for developmental timing, germline stem cell maintenance and muscle integrity. Importantly, these traits are not affected when solely the de novo pathway is abolished, and we present evidence that germline maintenance is linked specifically to ADSL activity in the recycling pathway. Hence, our results allow developmental and tissue specific phenotypes to be ascribed to separable steps of the purine metabolic network in an animal model.
Keywords: ADSL deficiency, AICAR, metabolism, purine salvage pathway, SAICAR, SZMP, ZMP
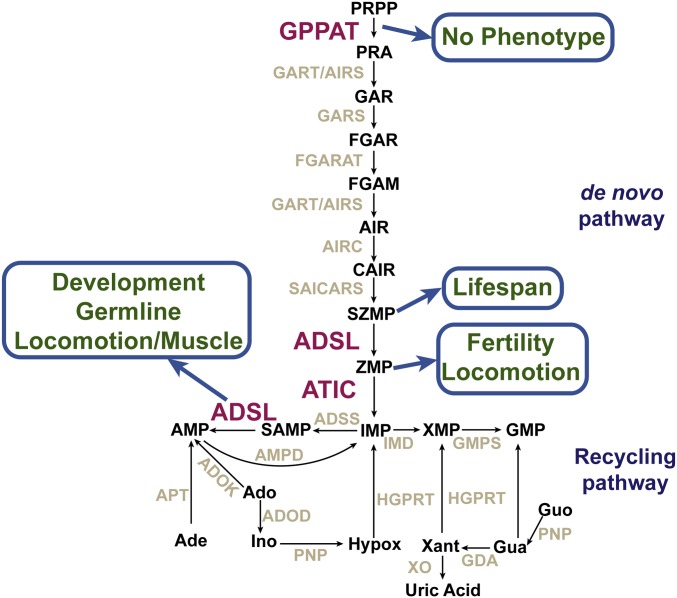
PURINE biosynthesis pathways ensure the production and homeostasis of AMP and GMP in the cell, and are widely conserved throughout evolution. The de novo pathway leads to the synthesis of inosine monophosphate (IMP) using 5-phosphoribosyl-1-pyrophosphate (PRPP) as a precursor. The recycling (salvage) pathway can then transform extant purines, including IMP, synthesized through the de novo pathway, to produce AMP and GMP (Figure 1A; reviewed in Ljungdahl and Daignan-Fornier 2012). Importantly, if available in the extracellular environment, purines can be recovered by specific transporters and subsequently metabolized through the recycling pathway, which requires much less energy expenditure. Indeed, purine synthesis through the de novo pathway requires the hydrolysis of six more molecules of ATP than purine synthesis through the recycling pathway.
Figure 1.
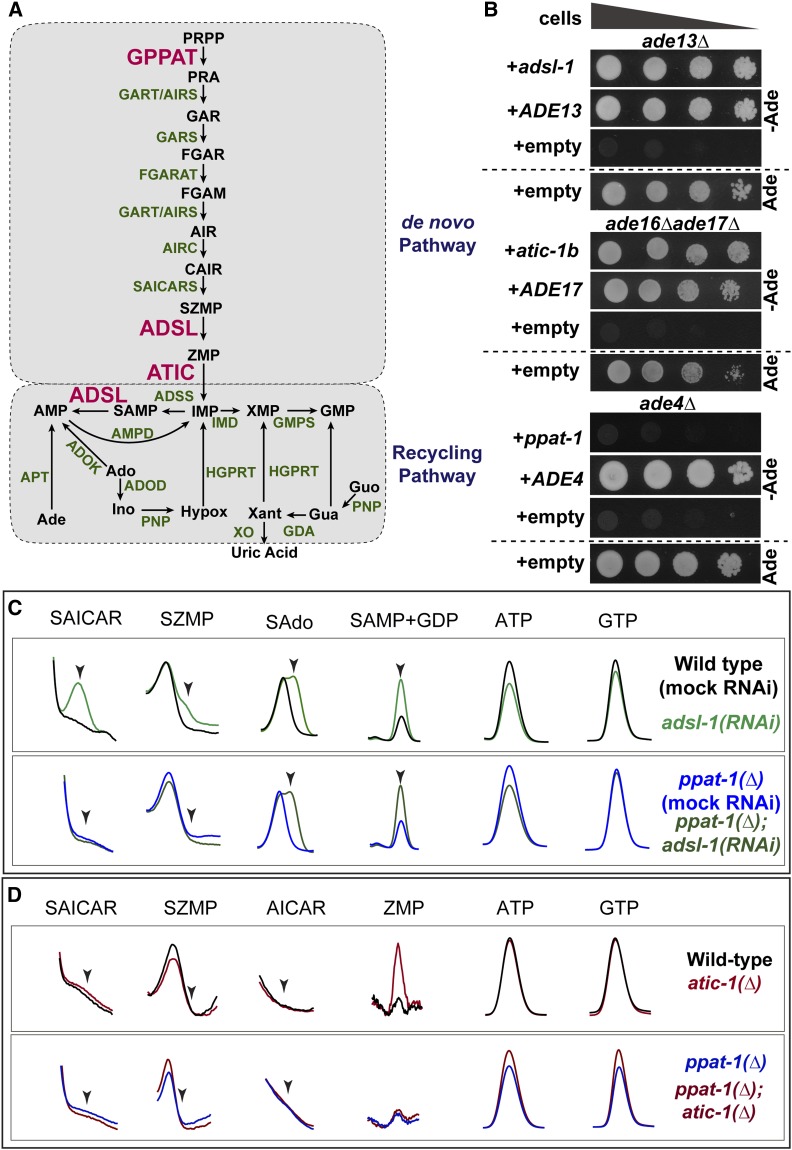
Purine metabolism in C. elegans. (A) Schematics of the purine biosynthesis pathways in C. elegans based on sequence homology. Enzymes subjected to functional analysis are represented in red, other enzymes in green, and metabolites in black. (B) Drop test to assess adenine auxotrophy in yeast S. cerevisiae mutants deficient for enzymes ADSL (ade13∆), ATIC (ade16∆ ade17∆), or GPPAT (ade4∆) expressing C. elegans genes adsl-1, atic-1b and ppat-1, respectively. Expression of S. cerevisiae genes ADE13, ADE17, and ADE4 was used as positive control, and expression vector without insert as negative control. A culture with adenine is shown as auxotrophy control. In all drop tests presented, four drops are shown per condition, corresponding to serial dilutions (1:10) of cellular suspensions, with deceasing cell concentrations from left to right. (C) Zoom in on HPLC chromatogram peaks of specific metabolites SAICAR (SZMP riboside form), SZMP, sAdo (succinyl-adenosine, SAMP riboside form), SAMP, ATP, and GTP, upon adsl-1(RNAi) and ppat-1(∆); adsl-1(RNAi). (D) Zoom in on HPLC chromatogram peaks of specific metabolites SAICAR (SZMP riboside form), SZMP, AICAR (ZMP riboside form), ZMP, ATP, and GTP, in atic-1(∆) and ppat-1(∆); atic-1(∆). Ade, adenine; Ado, adenosine; SAdo, succinyl-adenosine; Ino, Inosine; Hypox, hypoxantine; Xant, Xantine; Gua, guanine; Guo, guanosine. In (C) and (D), scales are adjusted differently among metabolites, in order to highlight differences between genotypes.
Imbalance in purine metabolism is involved in several diseases that have been extensively characterized, most of them associated with mutations in genes encoding enzymes involved in purine biosynthesis (reviewed in Nyhan 2005; Kelley and Andersson 2014). The genetics and biochemistry of purine biosynthesis are well established in unicellular organisms like bacteria or yeast, and are highly conserved. Nonetheless, how the disease-causing enzymatic deficiencies lead to the symptoms observed in patients remains elusive. It is particularly challenging to untangle the contribution of shortage in the final products—AMP and GMP—from mutation-caused accumulation of intermediate metabolites in the pathway. For instance, ZMP (5-amino-4-imidazole carboxamide ribonucleotide monophosphate; Bochner and Ames 1982) and SZMP (succinyl-ZMP)—two intermediates of the purine de novo pathway (Figure 1)—are known to act as signal metabolites regulating gene expression transcriptionally in yeast (Pinson et al. 2009). Moreover, ZMP is known to activate AMPK (AMP activated kinase) by mimicking AMP (Jin et al. 2007), and SZMP has been shown to directly bind and interfere with pyruvate kinase activity in cancer cells (Keller et al. 2012). In addition, SAMP (succinyl-AMP)—an intermediate metabolite in the recycling pathway—has been shown to stimulate insulin secretion in pancreatic cells (Gooding et al. 2015). Hence SZMP, ZMP, or SAMP buildup could potentially disrupt homeostasis upon deficiencies in the enzymes using them as substrates, i.e., ADSL (SZMP and SAMP) and ATIC (ZMP, Figure 1A). Accordingly, mutations in the human genes encoding for these two enzymes are associated with ADSL deficiency and ATIC deficiency (a.k.a. AICAribosiduria), syndromes whose etiology remains unknown (Race et al. 2000; Marie et al. 2004; Spiegel et al. 2006; Jurecka et al. 2008, 2015; Zikanova et al. 2010). Adding to the complexity of the issue, ADSL catalyzes two different reactions, conversion of SZMP in ZMP in the de novo pathway, and conversion of SAMP in AMP—a step required for AMP synthesis whether coming from the de novo or from the recycling pathway (Figure 1A). This raises the question of which of the two steps is associated with ADSL deficiency phenotypes.
In comparison with microorganisms, the study of purine metabolic deficiencies in patients faces yet other challenges. One such challenge is tissue specificity, as different tissues may require different levels of purines depending on their specialized function. For instance, in ADSL-deficiency patients, the most severe symptoms affect the muscles and the nervous system, suggesting these tissues have a larger requirement for purine metabolism, either for their function or during development. Another challenge is purine transport across tissues; the tissue affected by the deficiency not being necessarily the tissue where the enzyme is active. Moreover, purines can be available exogenously from the food source; therefore, the consequences of purine metabolism deficiencies are not necessarily caused by limiting amounts of purines. Finally, purine metabolism might be subject to different regulations throughout development, in which case the impairments observed could be the consequence of a requirement for purine regulation at a specific developmental stage.
In order to address these issues, we sought to take advantage of C. elegans as a model organism. Through a genetic approach, we characterized deficiencies in the purine biosynthesis pathways, focusing our analysis on an ADSL-deficient mutant. Our goal was to establish the phenotypes associated with this enzymatic deficiency in order to uncover what functions, tissues, and developmental stages are affected. Given ADSL is required both in the de novo and recycling pathways, we performed a comparative analysis with ATIC and GPPAT deficiencies to dissect the contribution of the de novo vs. recycling pathways to the phenotypes observed. We established that ADSL deficiency strongly disrupts muscle integrity, postembryonic developmental timing and germline stem cell (GSC) maintenance. Importantly, neither abolishing the de novo purine synthesis pathway per se, nor the accumulation of de novo pathway intermediate metabolites ZMP and SZMP, has an impact on these traits. Furthermore, we found no essential requirement for the de novo pathway in C. elegans under standard laboratory conditions.
Materials and Methods
C. elegans strains and culture
Nematodes were maintained under standard culture conditions (Brenner 1974; Stiernagle 2006), on nematode growth medium (NGM), with Escherichia coli (strain OP50) as food source, kept at 20° (except strains carrying the transgene ruIs32 [pie-1p::GFP::H2B + unc-119(+)], which were kept at 25°).
The following C. elegans strains were used in the study: N2 (Bristol, wild-type reference strain) obtained from the Caenorhabditis Genetics Center (CGC), AZ212 ruIs32 [pie-1p::GFP::H2B + unc-119(+)] III, CF1903 glp-1(e2141) III, PE254 feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V, VC2999 R151.2(gk3067) III/hT2 [bli-4(e937) let-?(q782) qIs48] (I,III) - parental strain for the hT2 balancer, WBX52 adsl-1(tm3328) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I,III), WBX53 adsl-1(tm3328) I; ppat-1(tm6344) III/hT2 [bli-4(e937) let-?(q782) qIs48] ppat-1(tm6344) (I,III) (from WBX119 and WBX52), WBX119 ppat-1(tm6344) III, WBX117 atic-1(tm6374) IV, WBX116 ppat-1(tm6344) III, atic-1(tm6374) IV (from WBX119 and WBX117); WBX124 atic-1(tm6374) IV; feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V (from PE254 and WBX116), WBX127 ppat-1(tm6344) I; adsl-1(tm3328) III/hT2 [bli-4(e937) let-?(q782) qIs48] ppat-1(tm6344) (I,III); feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V (from PE254 and WBX53), WBX125 ppat-1(tm6344) III; atic-1(tm6374) IV; feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V (from PE254 and WBX116), WBX130 adsl-1(tm3328) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I,III); feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V (PE254 and WBX53), WBX131 ppat-1(tm6344) III; feIs4 [sur-5p::luciferase::GFP + rol-6(su1006)] V (from PE254 and WBX116), WBX147 ppat-1(tm6344) ruIs32 [pie-1p::GFP::H2B + unc-119(+)] III (from AZ212 and WBX119), WBX149 adsl-1(tm3328) I; ruIs32 [pie-1p::GFP::H2B + unc-119(+)] III/hT2 [bli-4(e937) let-?(q782) qIs48] (I,III) (from AZ212 and WBX52) and WBX152 adsl-1(tm3328) I; ppat-1(tm6344) ruIs32 [pie-1p::GFP::H2B + unc-119(+)] III/hT2 [bli-4(e937) let-?(q782) qIs48] (I,III) (from WBX147 and WBX149). The adsl-1(tm3328), ppat-1(tm6344), and atic-1(tm6374) deletion alleles—referred to as adsl-1(∆), ppat-1(∆), and atic-1(∆) throughout the manuscript—were generated by the laboratory of Shohei Mitani, National Bioresource Project (NBRP), Japan; all strains carrying these alleles used in this study were outcrossed a minimum of four times.
Nematode synchronization:
Adult worms were collected with M9 buffer in 15 ml conical tubes, centrifuged 1 min at 1000 rpm, and the supernatant was discarded. Worm bleach solution (1 M NaOH, 0.25 M NaOCl) was added until the carcasses dissolved and eggs were released (∼8 min). Eggs were washed three times with M9 buffer then transferred onto NGM seeded plates.
RNAi:
adsl-1 PCR fragment was amplified with oligomers 5′-CTTCTTCCGGCAGAATCATCCAGTG-3′ and 5′-TGACTGTCGGACGTACTCATTACC-3′, and adss-1 PCR fragment was amplified with oligomers 5′-GACGTCTTCCAACAAGACTGTGTCC-3′ and 5′-ACGAGACGGCGATACTTCTCCGAG-3′. Both fragments were inserted onto L4440 plasmid (EcoRV site) (Timmons and Fire 1998). For RNAi experiments, HT115(DE3), bacteria transformed with either the adsl-1(RNAi) or adss-1(RNAi) feeding vectors, or L4440 plasmid (Control), were cultured overnight at 37° in LB 150 mg/liter Ampicillin and induced with 2 mM IPTG 1–2 hr prior seeding onto NGM 2 mM IPTG 150 mg/liter Ampicillin, in which synchronous populations of worms were cultured from hatching to adulthood (unless specified).
Embryonic lethality in adsl-1(tm3328)/+ progeny: adsl-1(tm338)/hT2 [bli-4(e937) let-?(q782) qIs48] hermaphrodites—expressing pharyngeal GFP—were crossed to N2 males. Non-GFP hermaphrodites in the progeny were selected as adsl-1(tm338)/+ and allowed to lay eggs, one worm per plate, for ∼24 hr. Upon removal of the adults plates were kept in culture for another 24 hr, upon which unhatched eggs were scored as dead embryos.
Yeast heterologous rescue assay
Yeast expression vectors were constructed by inserting full-length cDNAs of ppat-1 (amplified using oligomers 5′-CACTAAATTACCGGATCAATTCGGGGGATCCATGTGTGGTATATTTGGGATTG-3′ and 5′-CATAACTAATTACATGATGCGGCCCTCCTGCAGTTATAAATCGATAGCAACTGG-3′), adsl-1 (5′-CGGGATCCGTATCATGGCCTCCGAAGACAAGT-3′ and 5′-CCAATGCATGAATCAAACATCCAGCTGAACATTTCC-3′), and atic-1b (5′-GAAGATCTCGACGAATTGTGTGAAATGACCGAC-3′ and 5′-ATAAGAATGCGGCCGCATCTAATGGTGGAAGAGACGAAGTC-3′) onto pCM189 plasmid (Garí et al. 1997). ade4∆ yeast strain, genotype ade4::KanMX4; his3∆1; leu2∆0; lys2∆0; ura3∆0, was used to test heterologous rescue by ppat-1, ade13∆ yeast strain, genotype ade13::kanMX4; his3∆; leu2∆; ura3∆, was used to test heterologous rescue by adsl-1 and ade16∆ ade17∆ double mutant, genotype ade16::KanMX4; ade17::KanMX4; his3∆1; leu2∆0; ura3∆0; lys2∆0 was used to test heterologous rescue with atic-1b.
adsl-1(∆) rescue
A genomic fragment containing the adsl-1 reference allele, ∼1 kb upstream of the start codon and ∼1 kb downstream of the stop codon, was amplified by PCR using oligonucleotides 5′-GCTCCCATGATATTCTACCG-3′ and 5′-GAGGATTCAACGGTGCTTGCA-3′, and as template the cosmid R06C7 (obtained from the Sanger Center). The PCR-amplified fragment was cloned onto pBluescript on the EcoRV site, we named this plasmid pRM01. The insert sequence was verified upon cloning. Transgenic lines were obtained by standard microinjection (Mello et al. 1991), adsl-1/+ animals were cotransformed with pRM01 at 1 or 5 ng/µl, pDD04 at 10 ng/µl as transformation marker—pDD04 expresses GFP in the pharynx, under the control of the myo-2 promotor (Giordano-Santini et al. 2010)—and pJM001 as carrier DNA at 30–90 ng/µl [a kind gift from Denis Dupuy, Institut Européen de Chimie et Biologie (IECB), Bordeaux, France].
Imaging
For adult morphology, differential interference contrast (DIC) (also known as Nomarski) images were taken with a Leitz Diaplan microscope. Adult worms were mounted in M9 buffer, on a 2% agarose cushion. Images were acquired with a VisiCam 5.0 camera, analyzed, and processed with Fiji software. For imaging of the pie-1p::GFP::H2B germline reporter, synchronized adult worms were mounted in M9 Buffer 5 mM Levamisole onto a 2% agarose cushion. Images were acquired with an Olympus IX81 microscope under a 40× oil-immersion objective, with a Hamamatsu Orca R2 camera. Images were analyzed with Fiji software. For size measurements, synchronized populations of young adult worms were used; each worm was measured three times on three different pictures. Images were acquired on a Nikon SMZ18 stereo microscope with VisiCam 5.0 camera, and analyzed and processed with Fiji software.
Immunostaining
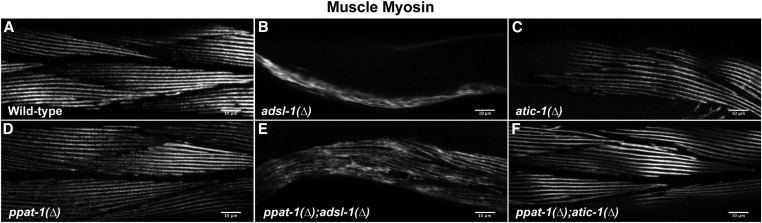
Immunostaining was performed on synchronized populations of worms. N2 and de novo pathway mutants were fixed 72 hr after synchronization; adsl-1(∆) and ppat-1(∆); adsl-1(∆) were fixed 96 hr after synchronization. The fixation protocol was adapted from Benian et al. (1996). Worms were collected in ice-cold PBS buffer in 1.5-ml tubes and centrifuged. The supernatant was discarded, and fixation solution was added (made fresh, 1 ml of MRBW buffer, 320 mM KCl, 80 mM NaCl, 40 mM EGTA, 20 mM spermidine, 60 mM Pipes was added to 2.8 ml of methanol and 200 μl of formaldehyde 20%), followed by five cycles of freezing in liquid N2 and thawing, followed by 1 hr incubation at 4° with gentle agitation, fixation solution was replaced by a permeabilization solution (PBS, Triton X-100 2%, β-mercaptoethanol 1%) and incubated 1 hr at room temperature, permeabilization solution was renewed and incubated 1 hr at room temperature, followed by three washes in PBSt (PBS, 0.5% Triton X-100), after which worms were incubated with primary antibody (diluted 1:1000 in PBSt 5% fetal calf serum) overnight at 4° with gentle agitation, washed three times in PBSt and incubated with secondary antibody (diluted 1:500 in PBSt 5%FCS). Monoclonal antibody 5–6 (Developmental Studies Hybridoma Bank) directed against muscle myosin was used as primary antibody, anti-mouse Alexa555 (Molecular Probes) was used as secondary antibody, worms were mounted on Fluoroshield (Sigma). Images were acquired with a Leica DM6000 TCS SP5 confocal microscope using LAS AF (Leica) software.
Metabolic profiling by HPLC
For an extraction, ∼7500 young adult worms, 72 hr after synchronization, were collected with ice-cold water into 15 ml conical tubes, and centrifuged at 1000 rpm for 1 min at 4°; the supernatant was discarded, eliminating most bacteria. The nematode pellet was washed with 10 ml of cold 10 mM Hepes and centrifuged. The worm pellet was resuspended in 10 mM Hepes, 1 ml of worm suspension was transferred into 4 ml of absolute ethanol at 80°, incubated for 5 min and cooled on ice for 10 min. Samples were desiccated on a rotovapor apparatus; dry residue was resuspended in 350 μl of HyClone Molecular Biology-Grade Water (GE Healthcare) and centrifuged at 21,000 × g, 5 min at 4°. The supernatant was transferred and centrifuged again at 21,000 × g, 1 hr at 4°; this final supernatant was used for HPLC. Metabolites were separated by ionic chromatography on an ICS3000 chromatography station (Dionex, Sunnyvale, CA), and measured by an UV diode array (Ultimate 3000 RS; Dionex). Metabolites were separated on a carbopacPA1 column (250 × 2 mm; Dionex) using a sodium acetate gradient (Ceballos-Picot et al. 2015). Metabolite levels were normalized relative to dry weight; the same amount of worms used in the extractions were collected in ice-cold water, 72 hr after synchronization, centrifuged, washed five times with water, allowed to evaporate for 4 days at 70° and weighed.
Lifespan
Upon synchronization (see above), worms were grown on standard conditions until adulthood, whereupon they were transferred onto a new plate every 24 hr, and scored. Immobile worms unresponsive to touch were considered dead. Three replicate experiments were performed on different days, typically with three different plates per genotype, per replicate and 10–40 worms per plate.
Locomotion and behavioral assays
Locomotion speed measurement was adapted from Zheng et al. (1999), Hart (2006); images were acquired on a Leica Z16 APO Macroscope and processed with Fiji software. Worm displacement was measured by tracking the anterior pharynx position, once per second. For each recording, one single worm was placed on one intermediate plate without food, and then transferred onto the recorded plate ∼0.5 cm away from the bacterial food source, with the pharynx toward the bacteria lawn. Locomotion was recorded for 1 min, or until the worm reached the bacteria if it did so in <1 min.
Aldicarb (Sigma) and levamisole (Sigma) assays were performed as described in Gendrel et al. (2009). Synchronized adult worms were placed on 1 mM Aldicarb or 0.4 mM Levamisole plates for 2 hr, after which plates were tapped 10 times and worm movement was scored.
Mechanosensory response assay was performed as described (Hobert et al. 1999; Hart 2006). Adult worms (∼72 hr after synchronization) were isolated on plates without food for 3 min, and stimulated with an eyelash on the posterior pharynx, and scored for avoidance behavior; each worm was stimulated and scored 10 times 3 min apart.
Postembryonic development
Postembryonic development was monitored as described by Olmedo et al. (2015). In brief, upon synchronization, 20 embryos per microliter were incubated in M9 buffer overnight with agitation, allowing L1 larvae to hatch. A single L1 larva was added per well to 96-well plates with S-Basal media, 10 g/liter E. coli OP50 (wet weight) and 100 μM d-Luciferin. The resulting luminescence was measured with a Berthold Centro LB960 XS3 during 1 sec at 5-min intervals, within a temperature controlled Panasonic MIR-154 incubator.
Statistics
Statistical analysis was performed using Graphpad, Excel, Numbers, and R softwares. We considered statistical tests to be significant at P < 0.01.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7642871.
Results
The purine biosynthesis pathway is functionally conserved in C. elegans
In order to establish C. elegans as a model to study purine metabolism, we first sought to investigate conservation of the purine biosynthesis network. We identified, through sequence homology, genes encoding the putative nematode enzymes involved in purine biosynthesis (Supplemental Material, Figure S1A). We found that both de novo and recycling pathways are conserved in C. elegans, as judged by sequence similarity. Specifically, for each enzyme, we identified one single ortholog in all but one case (three genes putatively encoding xanthine oxidase). Notably, in Saccharomyces cerevisiae, the histidine pathway contributes to the purine de novo pathway, ZMP being a byproduct of histidine synthesis (reviewed in Ljungdahl and Daignan-Fornier 2012). Consistent with the histidine pathway loss in worms previously reported (Payne and Loomis 2006), we found no C. elegans orthologs for yeast HIS1, HIS4, HIS6, and HIS7—required for ZMP synthesis through the histidine pathway; thus, ZMP is most likely synthesized exclusively through the purine de novo pathway.
Given the sequence conservation, we undertook a functional analysis focusing on the ADSL enzyme, including ATIC and GPPAT for comparison (see below). Importantly, sequence alignment showed that ADSL, ATIC, and GPPAT are highly conserved (Figure S2). The genes encoding ADSL, ATIC, and GPPAT in C. elegans were named adsl-1, atic-1, and ppat-1 (wormbase.org), respectively. We first tested whether those three C. elegans genes were functionally conserved using an heterologous rescue assay. We transformed yeast S. cerevisiae ade13∆, ade16∆ ade17∆, and ade4∆ mutants with plasmids containing the wild-type sequences of the predicted C. elegans homologous genes adsl-1, atic-1, and ppat-1, respectively. Expression of adsl-1 restored adenine prototrophy of ade13∆ yeast (Figure 1B); likewise, expression of splicing isoform atic-1b restored adenine prototrophy of the ade16∆ade17∆ double mutant (Figure 1B). These results confirm that adsl-1 encodes the ADSL enzyme and that atic-1 encodes the ATIC enzyme. By contrast, expression of ppat-1 alone was not sufficient to restore adenine prototrophy of yeast ade4∆ (Figure 1B). Thus, in our heterologous rescue assay, we could neither confirm nor rule out whether ppat-1 encodes the GPPAT enzyme.
We next tested functional conservation of ADSL, ATIC, and GPPAT activities in vivo. For our functional analysis, we used deletion mutant alleles, putative genetic nulls, henceforth referred to as adsl-1(∆), atic-1(∆), and ppat-1(∆) (see Materials and Methods). One would predict accumulation of ADSL substrates SZMP and SAMP upon adsl-1 loss-of-function as well as ZMP buildup in atic-1(∆), and no detectable de novo pathway intermediate metabolites in ppat-1(∆) (Figure S1B). To test these hypotheses, we performed worm metabolic profiling by HPLC (Figure S1C). We observed a consistent accumulation of SZMP and its riboside form SAICAR upon adsl-1(RNAi), as well as a robust accumulation of SAMP, and its riboside form SAdo (Figure 1C), while neither metabolite was detected in the wild-type (N2) reference strain. Hence, both de novo and recycling pathways are functionally conserved in C. elegans, and adsl-1 is required for ADSL activity in vivo. Furthermore, we observe a modest but consistent accumulation of ZMP both in atic-1(∆) mutants (Figure 1D), as well as atic-1(RNAi) animals (not shown), demonstrating that atic-1 encodes ATIC enzyme. The accumulation of SZMP and ZMP in adsl-1(RNAi) and atic-1(∆) was rather modest. By contrast, we observed a robust accumulation of SAMP in adsl-1(RNAi). These results suggest that C. elegans purine biosynthesis relies mostly on the recycling pathway, at least in standard culture conditions. Notably, ATP levels are slightly lower in adsl-1(RNAi) compared to wild-type (Figure 1C). Otherwise the abundance of ATP and GTP were not severely affected in the deficiencies analyzed.
As to the conservation of GPPAT, the HPLC analysis provided crucial functional evidence: SZMP was not detected in ppat-1(∆); adsl-1(RNAi) (Figure 1C), nor was ZMP detected in ppat-1(∆); atic-1(∆) (Figure 1D), or ppat-1(∆); atic-1(RNAi) (not shown). This demonstrates that the ppat-1 gene is indeed required for purine synthesis through the de novo pathway. Noteworthy, SAMP accumulation in ppat-1(∆); adsl-1(RNAi) is similar to adsl-1(RNAi) alone, providing an internal control for RNAi efficiency and further confirming that SZMP accumulation in adsl-1(RNAi) is dependent on ppat-1. In summary, our metabolic profiling validates functional conservation of ADSL and ATIC, and demonstrates that ppat-1 is required for the de novo pathway, consistent with the prediction that it encodes the GPPAT enzyme in C. elegans.
Functional analysis of adsl-1
Because SZMP and SAMP are known to function as regulatory metabolites in other organisms (Rebora et al. 2001; Pinson et al. 2009; Keller et al. 2014; Gooding et al. 2015), and because they may play an important role in the etiology of the ADSL deficiency, we focused our functional analysis on adsl-1. In order to distinguish the effects of SZMP buildup from those due to the loss of de novo purine synthesis, we further include ppat-1 in our analysis. Moreover, in order to distinguish the effect of ADSL activity strictly in the de novo pathway, converting SZMP to ZMP, from the activity in converting SAMP to AMP, for comparison we used ATIC-deficient worms. ATIC participates only in the de novo pathway accumulating ZMP, which shares some targets with SZMP, at least in yeast (Rebora et al. 2001; Pinson et al. 2009). Finally, in order to test whether phenotypes observed in adsl-1(∆) were due to SZMP buildup, we performed a systematic epistasis analysis of the ppat-1(∆); adsl-1(∆) double mutant. Similarly, to test whether phenotypes in the atic-1(∆) mutant were caused by ZMP accumulation, we performed epistasis in the ppat-1(∆); atic-1(∆) double mutant. Throughout the manuscript, we refer collectively to all three single mutant animals plus the ppat-1(∆); adsl-1(∆) and ppat-1(∆); atic-1(∆) double mutants as “purine mutants”, and to ppat-1(∆), atic-1(∆) single mutants and ppat-1(∆); atic-1(∆) double mutant animals as “de novo” pathway mutants.
adsl-1 is required for the maintenance of GSCs
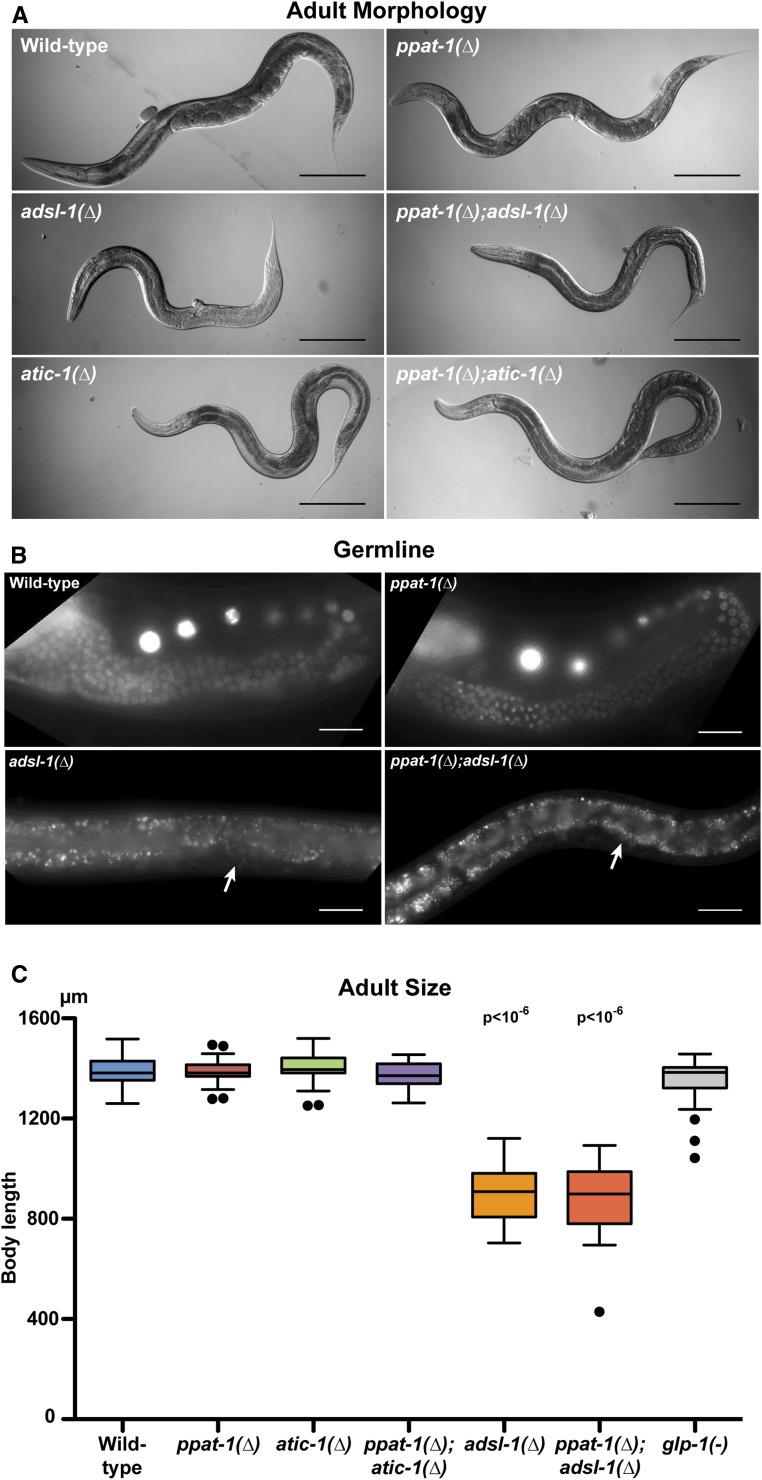
The adsl-1(∆) homozygous animals display striking morphological defects. They are noticeably smaller than wild type, sterile, and often present a protruding vulva (Figure 2A). We observe a complete absence of oocytes and sperm cells in adsl-1(∆) and ppat-1(∆); adsl-1(∆) worms. Only a reduced number of nuclei morphologically resembling GSCs are detectable near the vulva, where the germline distal tip cell is found in wild-type animals (Figure S3A). By contrast, de novo pathway mutants are viable and fertile, and the overall morphology is similar to wild-type worms. To better characterize the GSCs phenotype, we used a germline-specific reporter transgene expressing H2B::GFP fusion in the GSCs, under the control of the pie-1 promoter (Praitis et al. 2001). While in ppat-1(∆) single mutants, the germline is indistinguishable from wild type, in adsl-1(∆) and ppat-1(∆); adsl-1(∆) animals, sterility is fully penetrant and the germline is barely detectable, if at all (Figure 2B). Indeed we observed that 52.2% of adsl-1(∆) (n = 23) and 52% of ppat-1(∆); adsl-1(∆) animals (n = 25) had no detectable H2B::GFP, 43.5% of adsl-1(∆) and 44% of ppat-1(∆); adsl-1(∆) had ≤ 10 H2B::GFP positive nuclei, and 4.3% of adsl-1(∆) and 4% of ppat-1(∆); adsl-1(∆) had barely > 10 H2B::GFP positive nuclei. Hence, adsl-1 is required for proliferation and/or maintenance of GSCs independently of ppat-1.
Figure 2.
Developmental defects in purine mutants. (A) DIC whole body images of representative young adults for each of the genotypes studied (Bar, 200 μm). (B) Fluorescence micrographs of worms expressing H2B::GFP in the germline under the control of the pie-1 promoter. Arrows point at rare germ nuclei expressing H2B::GFP; spotty pattern throughout adsl-1(∆) and ppat-1(∆); adsl-1(∆) corresponds to intestinal auto-fluorescence (Bar, 25 μm). (C) Tukey box plot depicting body length of all purine mutants and glp-1(−) for comparison (means statistically different—one-way ANOVA—indicated P values refer to comparison with wild-type—Bonferroni multiple comparison t-test—otherwise comparison to wild type bears no significant difference; n = 30 for all genotypes).
It has been shown that sterility associated with a deficiency in the pyrimidine recycling pathway could be rescued by supplementation of the culture medium with pyrimidine nucleosides (Chi et al. 2016). Thus, we tested whether supplementation with purine nucleosides could rescue adsl-1(∆) sterility. When cultured with 20 mM adenosine (Ado in Figure 1A) in the medium from hatching until adulthood, both adsl-1(∆) (n = 55) and ppat-1(∆); adsl-1(∆) (n = 54) were sterile, much like the nonsupplemented controls (n = 80 and n = 88, respectively). As expected, 100 mM inosine (Ino in Figure 1A) supplementation had no effect on adsl-1(∆) (n = 28) or ppat-1; adsl-1(∆) (n = 64) sterility. Under the same conditions, wild-type (20 mM adenosine n = 86; 100 mM inosine n = 99) and ppat-1(∆) (20 mM adenosine n = 76; 100 mM inosine n = 100) worms were fertile, to the same extent as nontreated controls [wild type n = 116; ppat-1(∆) n = 114]. We further tested whether adenosine supplementation could rescue adsl-1 partial loss of function. When we performed adsl-1(RNAi) with 20 mM adenosine in the culture medium, we observed 30.8% sterility (n = 182) compared to 17.0% in absence of adenosine supplementation (n = 141), while mock RNAi produced 0% sterility, in both 20 mM adenosine or no adenosine supplementation (n = 101 and n = 103 respectively). In summary, we found no evidence that purine supplementation in the culture medium allows to recover the GSCs defects of adsl-1(∆).
As mentioned above, the de novo pathway mutants present a germline undistinguishable from wild type, in contrast to the complete sterility of adsl-1(∆). In C. elegans, adult hermaphrodites reproduce through self-fertilization, with the number of progeny being modulated by food source and metabolic status (Houthoofd et al. 2002; Watson et al. 2014). In order to investigate whether the de novo pathway could impact the level of fertility, we measured total number of eggs laid by ppat-1(∆), atic-1(∆), and ppat-1(∆); atic-1(∆) hermaphrodites throughout their reproductive life. We observed that ppat-1(∆) fertility is in the wild-type range, while atic-1(∆) fertility is significantly lower (Figure S3B). Importantly, ppat-1(∆) is epistatic to atic-1(∆) regarding fertility, suggesting that ZMP accumulation, rather than de novo pathway impairment, might reduce fertility. To test this hypothesis, we cultured ppat-1(∆); atic-1(∆) in the presence of ZMP precursor AICAR (0.1 mM); under these conditions internal ZMP builds up (see below). Consistent with our hypothesis, we observed reduced fertility in ppat-1(∆); atic-1(∆) treated with AICAR compared to the control (Figure S3B). Furthermore, wild-type worms cultured with 0.5 mM AICAR also displayed reduced fertility. We conclude that ZMP buildup in atic-1(∆) causes reduced fertility, while the de novo pathway per se has no impact on this phenotype, as ppat-1(∆) is similar to wild type. Taken together, these data show that adsl-1 is essential for germline integrity, while de novo pathway genes ppat-1 and atic-1 are dispensable.
The strong germline defects in adsl-1(∆) prompted us to investigate whether adsl-1 is required during early embryonic development. We therefore examined the viability of ADSL deficient embryos. adsl-1(∆) sterility precludes the analysis of homozygous embryos devoid of maternal contribution. We first analyzed zygotic requirement by examining the progeny of adsl-1(∆)/+ heterozygotes (see Materials and Methods); if adsl-1 zygotic expression were strictly essential in the embryo, one would expect no adsl-1(∆) homozygous progeny, and a Mendelian ∼25% proportion of embryonic lethality; instead, we observed only 7.7% of dead embryos. Therefore, either adsl-1 is not essential for embryo development or maternal contribution is, at least in part, sufficient. To further investigate adsl-1 function in the early embryo, we used RNAi, which depletes maternally expressed mRNAs. Because adsl-1 full penetrant loss-of-function leads to sterility, we performed adsl-1(RNAi) knockdown, starting at the L4 stage—resulting in a partial loss-of-function. Under these conditions, we observed 32.2% embryonic lethality in adsl-1(RNAi) (Figure S3C), suggesting that adsl-1 is required for embryonic development, and that maternal expression contributes to adsl-1 function at this stage. To test whether the de novo pathway is necessary for embryo development, we performed adsl-1(RNAi) in the ppat-1(∆), resulting in 31.5% embryonic lethality. The observation that ppat-1(∆) does not enhance the adsl-1(RNAi) hypomorphic phenotype suggests that the de novo pathway does not contribute to embryonic development (see below). Consistent with this hypothesis, de novo pathway mutants display an embryonic development indistinguishable from that of wild type; just as in wild-type controls, ∼100% of the eggs laid hatched in all de novo pathway mutants. Thus, adsl-1 function in the recycling pathway is required for proper embryonic development.
adsl-1(∆) leads to small body size phenotype
The overall morphology observed in adsl-1(∆) and ppat-1(∆); adsl-1(∆) [morphologically indistinguishable from adsl-1(∆) single mutants] suggests that cell lineage specification and morphogenesis are not affected. No defects were observed in somatic tissues of adsl-1(∆) other than the vulva, including pharynx, intestine, or hypodermis, and animals were viable and mobile. Hence, the smaller size does not seem to result from defects in fate specification. To better characterize this phenotype, we measured body length in adult worms. adsl-1(∆) are significantly smaller than wild type and similar to ppat-1(∆); adsl-1(∆), while de novo mutants are indistinguishable from wild type (Figure 2C). Given the volume occupied by the germline in adult C. elegans, we wondered whether sterility could account for the small size in adsl-1(∆). We therefore compared adsl-1(∆) to glp-1(e2141) mutants, which are known to lack a germline when cultured at 25° (Priess et al. 1987). Given that adult glp-1(e2141) length is in the wild-type range (Figure 2C), sterility cannot account for the small size of adsl-1(∆). Notably, newly hatched adsl-1(∆) animals are the same size as wild-type, and morphologically indistinguishable (not shown); hence, size defects are likely to manifest during postembryonic development. We conclude that adsl-1 is required for adult body size independently of the sterility phenotype. By contrast, deficiencies in GPPAT or ATIC in the de novo pathway do not affect this trait.
adsl-1 is required for normal developmental timing and lifespan
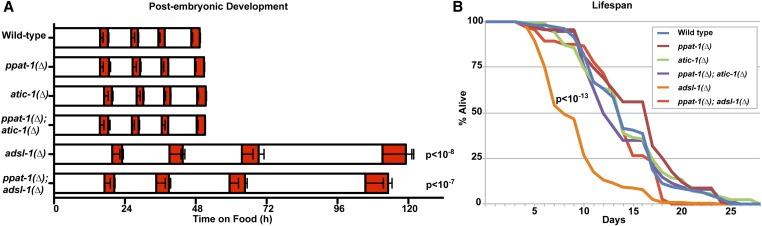
The small size of adsl-1(∆) prompted us to investigate whether postembryonic development was impaired. Given that newly hatched adsl-1(∆) are indistinguishable from wild type, we hypothesized the growth defects were linked to abnormal larval development. Upon hatching, and before adulthood, C. elegans goes through four larval stages, each followed by an inactive state known as lethargus during which a molt takes place (Cassada and Russell 1975). Importantly, this sequence follows a highly reproducible timing under standard culture conditions (Byerly et al. 1976). We therefore measured timing of postembryonic development in purine mutants, using the method described by Olmedo et al. (2015). Briefly, newly hatched L1 larvae carrying a transgenic luciferase are cultured in the presence of luciferin, emitting luminescence when active (Lagido et al. 2008). Since larvae cease feeding during lethargus, they stop taking the substrate and a drastic drop in luminescence is observed. Therefore, monitoring the luminescence of individual larvae allows to precisely time the occurrence of each larval (high luminescence) and lethargus (low luminescence) phase (Figure S4A; Olmedo et al. 2015). We observed that both adsl-1(∆) and ppat-1(∆); adsl-1(∆) present strongly delayed postembryonic development (Figure 3A and Figure S4B). Although all larval and lethargus phases are affected in adsl-1(∆) mutants, the delay is more pronounced as postembryonic development progresses. By contrast, the overall timing of postembryonic development is not affected in de novo pathway mutants. We therefore conclude that adsl-1 is required for normal postembryonic developmental timing, whereas de novo pathway genes ppat-1 and atic-1 are not.
Figure 3.
Life traits in purine mutants. (A) Graph depicting duration of postembryonic developmental stages, white bars represent larval stages, from L1 to L4 (left to right), orange bars represent lethargus phases consecutive to each larval stage (mean durations of larval development, from t0 until exit from fourth lethargus phase, are statistically different—one-way ANOVA—indicated P values refer to comparison with wild type—Bonferroni multiple comparison t-test—otherwise comparison to wild type bears no significant difference; see detailed representation of the data in Figure S4). (B) Graph depicting fraction of live worms over time for all genotypes studied (P value indicated for the adsl-1(∆) vs. wild type comparison, all other genotypes are not significantly different from wild type are significantly different from adsl-1(∆)—log rank test with Bonferroni correction; total combined of three independent experiments for each genotype [wild type n = 106, ppat-1(∆) n = 91, atic-1(∆) n = 126, ppat-1(∆); atic-1(∆) n = 149, adsl-1(∆) n = 227, ppat-1(∆); adsl-1(∆) n = 113].
Postembryonic development, metabolism, and the germline are all known to impact lifespan in C. elegans (reviewed in Antebi 2013). Because of adsl-1(∆) sterility and developmental delay we measured the lifespan of the purine mutants. adsl-1(∆) mutant has a shorter lifespan than wild type (Figure 3B). By contrast, the de novo pathway mutants’ lifespan is indistinguishable from wild type (Figure 3B). Importantly, ppat-1(∆) suppresses the adsl-1(∆) short-lifespan phenotype as ppat-1(∆); adsl-1(∆) double mutant survival profile is in the wild-type range (Figure 3B). These results strongly suggest that SZMP buildup in adsl-1(∆), rather than ADSL activity per se, is responsible for the reduced lifespan. Moreover, we conclude that the developmental delay and sterility of adsl-1(∆) are not linked to lifespan.
Ascribing adsl-1(∆) phenotypes to the de novo vs. recycling pathway
Most of defects we observed in adsl-1(∆)—germline, vulva, adult size, embryonic and postembryonic development—are not observed in ppat-1(∆) or atic-1(∆) mutants, suggesting that the ADSL activity of converting SAMP to AMP is the cause of these phenotypes. However, the question of the relative contributions of the de novo and recycling pathways remains open as SAMP to AMP conversion by ADSL has the peculiarity of being required for the synthesis of AMP both through the de novo and recycling pathways (as is the IMP to SAMP conversion by ADSS, Figure 1). Thus, two hypotheses can explain the adsl-1(∆) defects regarding SAMP to AMP conversion: either these defects are linked to the recycling but not the de novo pathway, or both the recycling and the de novo pathways contribute redundantly to the phenotypes, such that, in ppat-1(∆) mutants, the recycling pathway can compensate for the absence of the de novo pathway. Given that ADSL functions both in the de novo and recycling pathways, the putative null adsl-1(∆) does not allow us to discriminate between de novo and recycling pathway contributions; however, analysis of adsl-1 or adss-1 partial loss-of-function does. In such conditions, if the first hypothesis were true, abolishing the de novo pathway would have no impact on adsl-1 or adss-1 partial loss-of-function phenotypes, while, if the second hypothesis were true, the penetrance of the phenotypes should increase when adsl-1 or adss-1 partial loss-of-function is combined with ppat-1(∆).
We performed adsl-1(RNAi) and adss-1(RNAi) treatment—producing a partial loss-of-function—in wild-type and ppat-1(∆) worms, and analyzed the germline and vulval defects. We observe no increase in penetrance of either phenotype when comparing wild type and ppat-1(∆), in both adsl-1(RNAi) and adss-1(RNAi) (Table 1). Similarly, as described above, adsl-1(RNAi) displays the same penetrance of embryonic lethality in wild-type and ppat-1(∆) backgrounds (Figure S3C). We conclude that adsl-1(−) defects, at least in the germline, vulva, and embryonic development, are linked to the purine recycling pathway, while the de novo pathway makes no detectable contribution.
Table 1. Germline and vulval phenotypes in adsl-1(RNAi) and adss-1(RNAi).
| Genotype (+pie-1p::H2B::GFP) | Sterile no germline (%) | Sterile <10 germ cells (%) | Sterile >10 germ cells (%) | WT-like germline (%) | N = (germline) | Protruding vulva (%) | WT-like vulva (%) | N = (vulva) |
|---|---|---|---|---|---|---|---|---|
| WT | 0 | 0 | 2.3 | 97.7 | 43 | 0 | 100 | 35 |
| ppat-1(Δ) | 0 | 0 | 0 | 100 | 57% | 0 | 100 | 57 |
| adsl-1(RNAi) | 8.1 | 0 | 51.4 | 40.5 | 37 | 26.1 | 73.9 | 23 |
| ppat-1(Δ); adsl-1(RNAi) | 3.4 | 0 | 27.6 | 69.0 | 29 | 0 | 100 | 28 |
| adss-1(RNAi) | 1.7 | 1.7 | 57.9 | 38.8 | 121 | 8.0 | 92.0 | 50 |
| ppat-1(Δ); adss-1(RNAi) | 0 | 0 | 7.7 | 92.3 | 52 | 0 | 100 | 39 |
All strains carry the germline specific pie-1p::H2B:GFP. Three classes of sterile phenotypes were defined based on the number of GFP positive germline nuclei. Vulval phenotypes were scored on DIC (Nomarski) microscopy.
adsl-1(WT) transgene rescues adsl-1(∆) phenotypes
Given that the adsl-1(∆) allele was generated by random mutagenesis, it is possible that the phenotypes observed are caused by other mutations in the genetic background. In order to verify whether adsl-1 loss-of-function was indeed the cause of the observed phenotypes, we performed a transgene rescue assay. We generated a plasmid containing an adsl-1(WT) allele genomic fragment including ∼1 kb upstream of the start codon and ∼1 kb downstream of the stop codon (see Materials and methods) and transformed adsl-1(∆)/+ animals through microinjection. We obtained transgenic animals carrying the adsl-1(∆) allele (either heterozygous or homozygous) and the adsl-1(WT) transgene. In seven independent transgenic clones, we were able to observe rescue of adsl-1 function in adsl-1(∆) homozygous animals. Rescued adsl-1(∆) homozygous laid broods bearing viable embryos; hence, transgenic adsl-1(WT) rescued the germline phenotype. Furthermore, rescued adsl-1(∆) homozygotes presented an overall wild-type-like adult morphology, including wild-type-like vulva; therefore, transgenic adsl-1(WT) rescued body size and vulval defects as well. Given that the extrachromosomal arrays carrying the adsl-1(WT) transgene were poorly transmitted to the progeny, we could not characterize these lines further. We conclude that adsl-1(∆) phenotypes—at least sterility, vulval defects, and body size—are caused by adsl-1 loss-of-function. Further supporting this conclusion, adsl-1(RNAi) leads to sterility and vulval defects (see above, Table 1), matching what would be expected for a partial loss-of-function compared to adsl-1(∆), and adss-1(RNAi), depleting ADSS—the enzyme immediately upstream of ADSL in the recycling pathway—phenocopies adsl-1(RNAi), as would be expected.
Imbalance in purine metabolism impacts locomotion
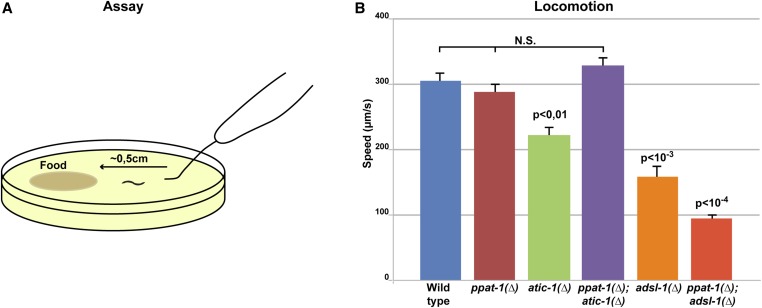
Given the severe muscular and neuronal symptoms of ADSL deficiency patients (Jaeken et al. 1988; Spiegel et al. 2006; Jurecka et al. 2015), we sought to investigate muscle and neural defects in adsl-1(∆) worms. In order to address muscle function, we performed a locomotion assay (Figure 4A; Zheng et al. 1999; Hart 2006), and observed that adsl-1(∆) mutants were slower than wild type (Figure 4B). atic-1(∆) mutants were also slower than wild type, albeit to a lesser extent (Figure 4B), suggesting that the de novo pathway could play a role in locomotion. However, ppat-1(∆) animals were indistinguishable from wild type; thus, the de novo pathway per se does not affect locomotion. Moreover, the locomotion defect in adsl-1(∆) was not suppressed by ppat-1(∆); ppat-1(∆); adsl-1(∆) was actually slower than the adsl-1(∆) single mutant. Thus, ADSL activity, independently of SZMP buildup, accounts for the adsl-1(∆) locomotion defect. By contrast, the atic-1(∆) locomotion defect was suppressed by ppat-1(∆) as ppat-1(∆); atic-1(∆) speed was in the wild type range. Hence, slower locomotion in atic-1(∆) is linked to ZMP build up, rather than the de novo pathway per se.
Figure 4.
Locomotion of purine mutants. (A) Cartoon schematics of the locomotion behavior assay. (B) Graph depicting the average locomotion speed in all purine mutants [error bar, SEM; n = 30 in all genotypes except wild type and ppat-1(∆); atic-1(∆) n = 40; means statistically different—one-way ANOVA—indicated P values refer to comparison with wild–type—Bonferroni multiple comparison t-test—wild type, ppat-1(∆), and ppat-1(∆) adsl-1(∆) are not statistically different from one another].
AICAR treatment
The ADSL product ZMP is known to activate AMPK by mimicking AMP (Jin et al. 2007), and AICAR treatment is known to impact muscle function in mice, presumably through ZMP induced AMPK activation (Narkar et al. 2008). To test whether ZMP is responsible for the locomotion defects in atic-1(∆), we tested whether providing the worms with external AICAR would impact locomotion. We treated late L4 stage wild-type and de novo pathway mutants with AICAR for 24 hr. Somewhat surprisingly, AICAR treatment did not significantly affect locomotion in any genotype tested (Figure S5A). Given this surprising result, we performed metabolic profile analysis of AICAR-treated worms. As expected, we observed higher levels of AICAR upon treatment, and, noticeably, a significant buildup of both ZMP and ZTP in all treated genotypes (Figure S5B), showing that AICAR was efficiently internalized and phosphorylated. As expected, ZMP levels were higher in atic-1(∆) than in other genotypes due to the cumulative buildup of ZMP from the de novo pathway and from AICAR treatment (Figure S5C). Therefore, buildup of ZMP in adult worms per se does not explain the reduced locomotion observed in atic-1(∆). Importantly, while AICAR treatment did not significantly affect ATP levels across genotypes, it led to an increase in uric acid, particularly in atic-1(∆) and ppat-1(∆); atic-1(∆). Moreover, we also observed, in all treated genotypes, accumulation of the ADSL substrate SZMP and its riboside form SAICAR. Therefore, AICAR treatment leads to increased levels of ZMP as expected, but also of uric acid, ZTP, and SZMP (Figure S5B). Accumulation of SAICAR and SZMP indicates not only that SZMP to ZMP conversion by ADSL is inhibited by AICAR treatment, but also that this reaction can be reversed, given that SZMP was detected in ppat-1(∆) and ppat-1(∆); atic-1(∆) mutants (Figure S5B, see Figure S1B). Accordingly, in yeast, ADSL enzyme has been shown to convert ZMP in SZMP when ZMP is in excess (Rebora et al. 2005), which seems also to be the case for C. elegans ADSL.
Muscle structure, but not the neuromuscular junction, is disrupted in adsl-1(∆)
One possible explanation for the locomotion defects observed in atic-1(∆) and adsl-1(∆) is that the neuromuscular junction is compromised. We therefore tested whether purine mutants were sensitive to Levamisole and Aldicarb. Both drugs activate acetylcholine receptors at the neuromuscular junction in C. elegans, at either the presynapse (Aldicarb) or postsynapse (Levamisole) leading to paralysis of wild-type worms (Lewis et al. 1980), while mutants with defective cholinergic neuromuscular junction are not affected (Fleming et al. 1997; Gendrel et al. 2009). We found no significant differences among purine mutants and wild-type animals (Figure S6, A and B), demonstrating that the neuromuscular junction remains functional upon purine metabolism deficiencies. We therefore tested whether other neuronal functions might be impaired in purine mutants. We performed a standard touch assay (Hobert et al. 1999; Hart 2006). We observed a similar response to touch in all purine mutants as in the wild type; thus, in our experiments, the mechanosensory function as well as the behavioral response are intact in purine mutants (Figure S6C). In summary, we found no evidence of neuronal dysfunction in the purine metabolism mutants tested.
Given these results, we hypothesized that the locomotion defects we observed could be explained by muscle malfunction. We performed an analysis of muscle cell structure through immunohistochemistry and confocal microscopy. As expected, we observed a regular parallel arrangement of myosin fibers in the wild type (Figure 5A), while in adsl-1(∆) mutants (n = 13), muscle myosin fibers formed an irregular meshwork and muscle mass seemed reduced (Figure 5B). In atic-1(∆) (n = 14), by contrast, the structure of muscle myosin fibers was indistinguishable from wild-type controls (Figure 5C, n = 10), as was also the case in ppat-1(∆) (Figure 5D, n = 13). Furthermore ppat-1(∆); adsl-1(∆) (Figure 5E, n = 10) displayed the same muscle fiber defects as adsl-1(∆), while muscle fibers in ppat-1(∆); atic-1(∆) were indistinguishable from those in ppat-1(∆) and atic-1(∆) single mutants (Figure 5F, n = 15), showing that de novo pathway deficiency does not affect muscle structure (Figure 5). These results show that ADSL is required for muscle integrity, while deficiencies in the de novo pathway have no discernible effect on this trait. Muscle structure defects could account for the locomotion defects of adsl-1(∆).
Figure 5.
Muscle structure in purine mutants. Confocal micrographs of muscle myosin immunostaining in young adult body wall muscle cells of all studied purine mutants (Bar, 10 µm). (A) Wild-type. (B) adsl-1(∆). (C) atic-1(∆). (D) ppat-1(∆). (E) ppat-1(∆); adsl-1(∆). (F) ppat-1(∆); atic-1(∆).
Discussion
A nematode model to study ADSL deficiency and, more broadly, purine metabolism
We describe here the phenotypes and metabolic profiles associated with three deficiencies in the purine biosynthesis pathway in the nematode C. elegans, focusing on ADSL deficiency. This study provides a reference for future investigation of the underlying molecular mechanisms. The phenotypes observed are, to some extent, genetically separable. In particular, the most severe defects in the adsl-1(∆) mutant—muscle, germline, and development—are not shared with ppat-1(∆) and atic-1(∆) mutants. The model we put forward links these defects to the ADSL activity in converting SAMP in AMP as the critical enzymatic step in ADSL deficiency (Figure 6). For instance, there is a long-standing debate concerning the etiology of the pathology associated with ADSL deficiency. It has been proposed that the SAICAR/S-Ado ratio correlated with specific symptoms (Jaeken et al. 1988; Race et al. 2000), while more recent studies found no evidence for such a correlation (Jurecka et al. 2008; Zikanova et al. 2010). Studies in human subjects are of limited reach, while a genetically amenable model organism will allow investigations of the mechanisms underlying specific phenotypes. In our model, SAMP to AMP conversion is crucial, hence SAMP buildup could play a significant role in the ADSL deficiency. Notwithstanding the evolutionary distance between nematodes and humans, testing this hypothesis may be informative in human pathology. However, at this point, caution is advised when comparing our results to human pathologies; more research will be required to address this issue. Importantly, the reported mutations causing ADSL deficiency in humans are associated with a residual enzymatic activity (reviewed in Nyhan 2005; Kelley and Andersson 2014), contrasting with our analysis of putative null mutants. The use of null mutations allows us to carefully establish the phenotypes associated with ADSL deficiency in the nematode model, and will allow us to further establish tissue specific contributions and to perform epistasis analysis in order to ascribe phenotypes to different steps in the pathway. Only once this characterization is achieved can mutations comparable to human ADSL deficiency be modeled with C. elegans to provide further insight. In summary, although further investigation is required, C. elegans, as metazoan model organism, provides the means to investigate the metabolic regulation of developmental progression, muscle defects or cell proliferation in the context of ADSL deficiency, as well as in other deficiencies in the purine biosynthesis pathway.
Figure 6.
Summary of the functional analysis of the purine biosynthesis pathway in C. elegans reported in this study.
Purine recycling and GSCs maintenance
One striking phenotype of adsl-1(∆) is the severe reduction in GSCs, leading to sterility. In adult C. elegans, the germline is the sole proliferating tissue. More precisely, GSCs in the distal gonad proliferate under the control of Notch signaling from neighboring somatic cells, while, in the proximal region, germline nuclei enter meiotic differentiation (reviewed in Kimble and Crittenden 2005; Hansen and Schedl 2013). Interestingly, deficiencies in both pyrimidine (Chi et al. 2016) and pyridine nucleotides (also known as NAD+) recycling pathways (Vrablik et al. 2009) have also been linked to germline defects in C. elegans, suggesting that nucleotide metabolism plays a critical role in maintaining the GSCs pool. Importantly, however, the adsl-1(∆) phenotype is more severe than in either of the other cases. In pnc-1(−) mutants, affecting the pyridine salvage pathway, animals are fertile despite germline defects. In cdd-1/2(−) mutants, which are deficient in cytidine deaminase in the pyrimidine recycling pathway, GSC proliferation could be restored with pyrimidine supplementation. By contrast, purine supplementation had no effect on either adsl-1(∆) or adsl-1(RNAi)—partial loss-of-function—suggesting that it is not a shortage of the final products of the purine pathway that is the main factor causing the GSC defect. While one cannot exclude the possibility that the defects in pnc-1(−), cdd-1/2(−) and adsl-1(∆) are unrelated, the three metabolic pathways are closely connected. It is therefore plausible that germline defects in these three mutants share a common mechanism through crosstalk among these metabolic pathways. In this context, the severity of the adsl-1(∆) phenotype remains to be explained. One tempting explanation is that the primary metabolic defect lies on the purine pathway, and that imbalance in the other nucleotide biosynthetic pathways impacts purine metabolism as an indirect consequence. It is known that GSC proliferation is influenced by environmental factors, particularly nutrient availability, and that it is likely regulated by metabolic signals (reviewed in Hubbard et al. 2013). Our results are consistent with the purine recycling pathway serving as sensor integrating nucleotide pathway status, and providing a metabolic signal regulating GSC proliferation.
Interestingly, the purine biosynthetic pathway has been shown to play a role in the balance between proliferation and differentiation in both stem cells and cancer cells, with inhibition of purine synthesis leading to increased differentiation at the expense of proliferation (Oburoglu et al. 2014; Tardito et al. 2015). A similar mechanism could explain the adsl-1(∆) GSCs defect; in that scenario, ADSL deficiency would lead to germline nuclei entering meiosis at the expense of proliferation.
Purine metabolism and development
Metazoans often control their development in response to environmental conditions, namely nutrient availability. This control involves metabolic regulation, for instance through insulin/insulin-like or mTOR pathways (reviewed in Danielsen et al. 2013). In C. elegans, adult worm size varies depending on the bacterial food source (Samuel et al. 2016), suggesting that size is one of the developmental phenotypes subjected to metabolic regulation. This regulation could, in theory, be due to metabolic regulation of cell fate decisions, or, alternatively, be a consequence of metabolic regulation of cell size. Based on our analysis, in adsl-1(∆), only GSCs and vulval cells, which have no impact in overall adult size, present cell fate abnormalities in the adult. Hence, the small size we observe is most likely due to an effect of purine metabolism on cell size. In addition to size, adsl-1(∆) also displays a strong delay during postembryonic development. In C. elegans, progression through larval stage and molting cycles is highly regulated, involving oscillations of LIN-42—the ortholog the circadian protein PERIOD (Monsalve et al. 2011; Edelman et al. 2016; reviewed in Monsalve and Frand 2012). The developmental changes associated with this progression involve tight coordination of different cell lineages across the worm body, and are very well characterized, as are the genetic pathways involved (reviewed in Rougvie and Moss 2013). By contrast, the upstream mechanisms and signals triggering or modulating these developmental transitions are poorly understood. The quality and amount of food available certainly play a role, as they have been shown to impact the timing of postembryonic development (Houthoofd et al. 2002; MacNeil et al. 2013). The dramatic developmental delay in adsl-1(∆) points at a metabolic regulation of these developmental transitions involving the purine recycling pathway.
Our results also suggest a role for ADSL in embryonic development. Presumably, a more penetrant adsl-1 loss-of-function would lead to higher levels of embryonic lethality; however, the ensuing sterility precludes further study of early development. With the tools currently available, we cannot determine conclusively the extent of adsl-1 requirement in embryonic development. Given most homozygous adsl-1(∆) embryos—where only zygotic adsl-1 expression is affected—are viable, and RNAi—depleting maternal mRNA—leads to embryonic lethality, it is plausible that proper embryo development requires adsl-1 function, ensured, at least in part, by maternal contribution.
One possibility to explain the developmental phenotypes in adsl-1(∆) could be limiting ATP causing slow development and smaller size. We have observed that, in adsl-1(∆), food uptake is reduced compared to wild type (Rodríguez-Palero et al. 2018); thus, not only ATP but also limiting overall bulk nutrients could account for the developmental defects observed. Although limiting nutrients and ATP levels are important factors, and certainly have a sizable impact, other elements suggest that this hypothesis is not sufficient to explain the adsl-1(∆) defects. In eat mutants, known to have feeding impairment, food uptake defects do not always correlate with developmental delay (M.O. and M.A.S. unpublished results). As for ATP levels, our metabolic profiles show them to be reduced only slightly in adsl-1(RNAi) compared with wild type. This is not so surprising given that adenine or adenosine available from the bacterial food source could be used to produce ATP despite ADSL deficiency (see Figure 1A). An alternative hypothesis is that postembryonic developmental and adult size are regulated by the wiring of the purine recycling pathway, rather than the synthesis of its final products. In this view, purine intermediate metabolites could function as metabolic sensors modulating developmental progression.
Purine metabolism and neuromuscular function
Both neuronal and muscle tissues are severely affected in purine metabolism syndromes, suggesting that these two tissues are particularly susceptible upon purine metabolism impairment in patients. In our nematode model, we observed locomotion defects associated with both ATIC and ADSL deficiencies. Somewhat surprisingly, no discernible neuronal dysfunction was observed in any purine mutant. Like wild-type controls, all the genotypes tested displayed sensitivity to Levamisole and Aldicard, showing that the neuromuscular junction was still functional. Moreover, the mechanosensory response in purine mutants was also not affected. Hence, we could not identify any neuronal dysfunction that would account for locomotion defects. The results notwithstanding, given that we did not investigate neuron integrity directly, it remains possible that purine mutants present neuronal impairments that the functional assays reported here cannot detect.
We did, however, find evidence of muscle abnormalities that could explain the locomotion defects. Notably, the locomotion defects we observed in atic-1(∆) and adsl-1(∆) were linked to different mechanisms. Contrary to adsl-1(∆), the locomotion delay in atic-1(∆) was suppressed in the ppat-1(∆); atic-1(∆) double mutant, and hence linked to ZMP buildup. One possible explanation is that ZMP accumulation leads to metabolic changes that, in turn, impact muscle function, possibly through AMPK activation (Jin et al. 2007). Importantly, the structure of muscle myosin fibers in atic-1(∆) are undistinguishable from wild type; thus, muscle integrity seems not to be affected in atic-1(∆). The ZMP effect on locomotion, however, is not straightforward; upon AICAR treatment of L4 larvae/young adults, ZMP builds up as expected; nonetheless, locomotion is not affected. Although the underlying mechanism remains elusive, our results suggest either that the ZMP effect on locomotion is developmental and occurs prior to the L4 stage, or that ZMP accumulation from external AICAR does not affect the same tissues as endogenous ZMP build up in atic-1(∆).
On a side note, because ZMP is an AMPK agonist (Jin et al. 2007), its riboside form AICAR is often added to culture media of different model organisms, including C. elegans (Moreno-Arriola et al. 2016; Ahmad and Ebert 2017), to trigger AMPK activation upon intracellular conversion to ZMP. Our results show that increase of ZMP is not the only metabolic consequence of AICAR treatment; therefore, caution is recommended when attributing phenotypes to AMPK activation.
As to the locomotion defects in adsl-1(∆), our results point to ADSL activity in the purine recycling pathway, given that ppat-1(∆) does not suppress the locomotion phenotype; indeed, quite the contrary. Furthermore, adsl-1(∆) is associated with significant damage to the structure of muscle myosin fibers, suggesting that locomotion defects are caused by muscle defects. While the underlying cause of these defects is yet to be determined, we hypothesize that the developmental delay in adsl-1(∆) may contribute to muscle defects.
Purine metabolic network in a metazoan model
The results we present show the functional conservation of both the de novo and recycling purine biosynthesis pathways in C. elegans. Furthermore, we characterized phenotypes associated with deficiencies in the critical enzymes ADSL and ATIC. While several phenotypes are genetically separable (Figure 6), it stands out that the most severe phenotypes—lack of GSCs, developmental delay, and muscle integrity—are associated with ADSL activity, while deficiencies in the de novo pathway are viable and lead to rather subtle phenotypes suc as reduced fertility and slower locomotion. Notably, deletion of ppat-1, which we show is required for the de novo pathway, produces no detectable phenotype in our culture conditions. The phenotypes observed in atic-1(∆), locomotion defects and reduced fertility, are associated with ZMP accumulation, rather than with shortage in purine synthesis through the de novo pathway, as though the de novo pathway per se is dispensable. It might seem paradoxical that the purine de novo pathway is conserved while having no essential function. Indeed, a number of biosynthetic pathways, namely those of some amino acids, have been lost during metazoan evolution (Payne and Loomis 2006), likely because amino acids are available from food sources, as are nucleotides. However, our culture conditions may not reflect the natural environment in which C. elegans evolved; it is plausible that the de novo pathway is essential in different environments where purines are scarce.
Contrasting with our results in C. elegans, Drosophila mutants in GPPAT-encoding genes are lethal prior to adulthood (Malmanche and Clark 2004; Ji and Clark 2006). These Drosophila mutants are purine auxotroph—lethality is higher in purine deprived food source—and lethality is observed during the pupal phase, i.e., a closed developmental stage during which external nutrients are not available. These different consequences of GPPAT deficiency in C. elegans and Drosophila could be due to substantially different metabolic requirements in the two species for GPPAT, and the whole de novo pathway; they could also reflect different life cycles and different experimental conditions. Further highlighting the contrast between Drosophila and C. elegans, fly mutants deficient for ADSL and ADSS are also lethal, in this case at larval stages prior to pupation (Stenesen et al. 2013). Interestingly, the latter authors show that the lower levels of ADSS and ADSL in heterozygotes lead to increased lifespan through reduced AMP synthesis. Notably, purine pathway mutants in the yeast S. cerevisiae have been reported to increase chronological age (Matecic et al. 2010). In both cases, purine supplementation of the culture media restores the wild-type phenotype, presumably through raising AMP levels to a wild-type range. If the same scenario occurs in C. elegans, our results on the lifespan of purine mutants could be explained by the abundance of purines in the bacterial food source. This possibility emphasizes the contribution of purine availability in the external medium to phenotypic outcomes. Further investigation of the purine pathway in worms will certainly involve controlled manipulation of purine supplementation, which requires a suitable defined culture medium (Flavel et al. 2018).
Actually, if one draws a parallel with yeast, it is the severity of the adsl-1(∆) phenotypes that seems surprising, considering that Ade13 null mutants in S. cerevisiae are viable when cultured with adenine (Dorfman 1969). Given that purines are available from the food source, one would have expected the worm to synthesize AMP and GMP despite ADSL deficiency. Indeed, external adenine and adenosine can be converted to AMP by adenine phosphoribosyltransferase and adenosine kinase, respectively, both of which have orthologs in C. elegans (Figure S1A), and AMP can be converted to GMP (Figure 1A). The severity of adsl-1(∆) phenotypes suggests that ADSL became essential even when external purines are available. Importantly, in humans, purines are also available from the food source, and all but one disease affecting purine metabolism are linked to enzymes in the recycling pathway; with the notable exception being AICA-ribosiduria—a deficiency in ATIC—in which ZMP buildup may be a crucial disease factor (Marie et al. 2004; Nyhan 2005; Kelley and Andersson 2014). The emerging picture in metazoans is that the less energetically costly recycling pathway evolved to become the predominant form of purine metabolism, at least in favorable environmental conditions. Hence, specific functions evolved to rely strongly on the recycling pathway, independently of the abundance of external purine sources. The defects associated with ADSL deficiency, as with other recycling pathway deficiencies, could be the consequence of insufficient synthesis of the final products AMP and GMP as much as the unbalanced wiring of the pathway, namely deregulation caused by intermediate metabolites.
Acknowledgments
We thank Yuji Kohara (N.I.G., Mishima, Japan) for providing cDNAs. We thank the members of the B.D.-F. laboratory for helpful discussions, and particularly M. Moenner for critically reading the manuscript. We thank D. Dupuy and Jonathan Millet (I.E.C.B., Bordeaux, France) for helpful feedback, and for sharing reagents and equipment. Deletion alleles adsl-1(tm3328), ppat-1(tm6344) and atic-1(tm6374) were generated by the National Bioresource Project, Japan, directed by Shohei Mitani. Confocal microscopy, and locomotion assay imaging were done in the Bordeaux Imaging Center—a service unit of the CNRS-INSERM and Bordeaux University, and member of the national infrastructure France BioImaging. Some C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC; https://cgc.umn.edu), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). We thank Wormbase, which is supported by NIH grant U41 HG002223. This work was supported by an AFM-Téléthon Trampoline grant (ref. 18313). R.M. is supported by AFM-Téléthon Ph.D. scholarship (ref. 20450). R.M. stay in M.A.-S. laboratory was supported by a Short Term Scientific Mission financed by GENiE (COST Action BM1408). M.A.-S. and M.O. are supported by the Ramón y Cajal program of the Spanish Ministerio de Economía y Competitividad, RYC-2010-06167 and RYC-2014-15551, respectively.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7642871.
Communicating editor: B. Grant
Literature Cited
- Ahmad W., Ebert P. R., 2017. Metformin attenuates Aβ pathology mediated through levamisole sensitive nicotinic acetylcholine receptors in a C. elegans model of Alzheimer’s disease. Mol. Neurobiol. 54: 5427–5439. 10.1007/s12035-016-0085-y [DOI] [PubMed] [Google Scholar]
- Antebi A., 2013. Steroid regulation of C. elegans diapause, developmental timing, and longevity. Curr. Top. Dev. Biol. 105: 181–212. 10.1016/B978-0-12-396968-2.00007-5 [DOI] [PubMed] [Google Scholar]
- Benian G. M., Tinley T. L., Tang X., Borodovsky M., 1996. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 132: 835–848. 10.1083/jcb.132.5.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Ames B. N., 1982. ZTP (5-amino 4-imidazole carboxamide riboside 5′-triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell 29: 929–937. 10.1016/0092-8674(82)90455-X [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L., Cassada R. C., Russell R. L., 1976. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev. Biol. 51: 23–33. 10.1016/0012-1606(76)90119-6 [DOI] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. 10.1016/0012-1606(75)90109-8 [DOI] [PubMed] [Google Scholar]
- Ceballos-Picot I., Le Dantec A., Brassier A., Jaïs J. P., Ledroit M., et al. , 2015. New biomarkers for early diagnosis of Lesch-Nyhan disease revealed by metabolic analysis on a large cohort of patients. Orphanet J. Rare Dis. 10: 7 10.1186/s13023-014-0219-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C., Ronai D., Than M. T., Walker C. J., Sewell A. K., et al. , 2016. Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes Dev. 30: 307–320. 10.1101/gad.275107.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen E. T., Moeller M. E., Rewitz K. F., 2013. Nutrient signaling and developmental timing of maturation. Curr. Top. Dev. Biol. 105: 37–67. 10.1016/B978-0-12-396968-2.00002-6 [DOI] [PubMed] [Google Scholar]
- Dorfman B. Z., 1969. The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics 61: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman T. L., McCulloch K. A., Barr A., Frøkjær-Jensen C., Jorgensen E. M., et al. , 2016. Analysis of a lin-42/period null allele implicates all three isoforms in regulation of Caenorhabditis elegans molting and developmental timing. G3 (Bethesda) 6: 4077–4086. 10.1534/g3.116.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavel M. R., Mechler A., Shahmiri M., Mathews E. R., Franks A. E., et al. , 2018. Growth of Caenorhabditis elegans in defined media is dependent on presence of particulate matter. G3 (Bethesda) 8: 567–575. 10.1534/g3.117.300325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. T., Squire M. D., Barnes T. M., Tornoe C., Matsuda K., et al. , 1997. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 17: 5843–5857. 10.1523/JNEUROSCI.17-15-05843.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E., Piedrafita L., Aldea M., Herrero E., 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848. [DOI] [PubMed] [Google Scholar]
- Gendrel M., Rapti G., Richmond J. E., Bessereau J. L., 2009. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature 461: 992–996. 10.1038/nature08430 [DOI] [PubMed] [Google Scholar]
- Giordano-Santini R., Milstein S., Svrzikapa N., Tu D., Johnsen R., et al. , 2010. An antibiotic selection marker for nematode transgenesis. Nat. Methods 7: 721–723. 10.1038/nmeth.1494 [DOI] [PubMed] [Google Scholar]
- Gooding J. R., Jensen M. V., Dai X., Wenner B. R., Lu D., et al. , 2015. Adenylosuccinate is an insulin secretagogue derived from glucose-induced purine metabolism. Cell Rep. 13: 157–167. 10.1016/j.celrep.2015.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. 10.1007/978-1-4614-4015-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, A. C., 2006 Behavior (July 3, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.87.1, http://www.wormbook.org.
- Hobert O., Moerman D. G., Clark K. A., Beckerle M. C., Ruvkun G., 1999. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J. Cell Biol. 144: 45–57. 10.1083/jcb.144.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd K., Braeckman B. P., Lenaerts I., Brys K., De Vreese A., et al. , 2002. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp. Gerontol. 37: 1371–1378. 10.1016/S0531-5565(02)00173-0 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Korta D. Z., Dalfó D., 2013. Physiological control of germline development. Adv. Exp. Med. Biol. 757: 101–131. 10.1007/978-1-4614-4015-4_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeken J., Wadman S. K., Duran M., van Sprang F. J., Beemer F. A., et al. , 1988. Adenylosuccinase deficiency: an inborn error of purine nucleotide synthesis. Eur. J. Pediatr. 148: 126–131. 10.1007/BF00445919 [DOI] [PubMed] [Google Scholar]
- Ji Y., Clark D. V., 2006. The purine synthesis gene Prat2 is required for Drosophila metamorphosis, as revealed by inverted-repeat-mediated RNA interference. Genetics 172: 1621–1631. 10.1534/genetics.105.045641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Townley R., Shapiro L., 2007. Structural insight into AMPK regulation: ADP comes into play. Structure 15: 1285–1295. 10.1016/j.str.2007.07.017 [DOI] [PubMed] [Google Scholar]
- Jurecka A., Zikanova M., Tylki-Szymanska A., Krijt J., Bogdanska A., et al. , 2008. Clinical, biochemical and molecular findings in seven Polish patients with adenylosuccinate lyase deficiency. Mol. Genet. Metab. 94: 435–442. 10.1016/j.ymgme.2008.04.013 [DOI] [PubMed] [Google Scholar]
- Jurecka A., Zikanova M., Kmoch S., Tylki-Szymańska A., 2015. Adenylosuccinate lyase deficiency. J. Inherit. Metab. Dis. 38: 231–242. 10.1007/s10545-014-9755-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller K. E., Tan I. S., Lee Y.-S., 2012. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science 338: 1069–1072. 10.1126/science.1224409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller K. E., Doctor Z. M., Dwyer Z. W., Lee Y.-S., 2014. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol. Cell 53: 700–709. 10.1016/j.molcel.2014.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. E., Andersson H. C., 2014. Disorders of purines and pyrimidines. Handb. Clin. Neurol. 120: 827–838. 10.1016/B978-0-7020-4087-0.00055-3 [DOI] [PubMed] [Google Scholar]
- Kimble, J., and S. L. Crittenden, 2005 Germline proliferation and its control (August 15, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.13.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Lagido C., Pettitt J., Flett A., Glover L. A., 2008. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 8: 7 10.1186/1472-6793-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., Wu C. H., Berg H., Levine J. H., 1980. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95: 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl P. O., Daignan-Fornier B., 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190: 885–929. 10.1534/genetics.111.133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil L. T., Watson E., Arda H. E., Zhu L. J., Walhout A. J., 2013. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153: 240–252. 10.1016/j.cell.2013.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanche N., Clark D. V., 2004. Drosophila melanogaster Prat, a purine de novo synthesis gene, has a pleiotropic maternal-effect phenotype. Genetics 168: 2011–2023. 10.1534/genetics.104.033134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie S., Heron B., Bitoun P., Timmerman T., Van den Berghe G., et al. , 2004. AICA-ribosiduria: a novel, neurologically devastating inborn error of purine biosynthesis caused by mutation of ATIC. Am. J. Hum. Genet. 74: 1276–1281. 10.1086/421475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M., Smith D. L., Pan X., Maqani N., Bekiranov S., et al. , 2010. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 6: e1000921 10.1371/journal.pgen.1000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Frand A. R., 2012. Toward a unified model of developmental timing: a “molting” approach. Worm 1: 221–230. 10.4161/worm.20874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Van Buskirk C., Frand A. R., 2011. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21: 2033–2045. 10.1016/j.cub.2011.10.054 [DOI] [PubMed] [Google Scholar]
- Moreno-Arriola E., El Hafidi M., Ortega-Cuéllar D., Carvajal K., 2016. AMP-activated protein kinase regulates oxidative metabolism in Caenorhabditis elegans through the NHR-49 and MDT-15 transcriptional regulators. PLoS One 11: e0148089 10.1371/journal.pone.0148089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar V. A., Downes M., Yu R. T., Embler E., Wang Y.-X., et al. , 2008. AMPK and PPARd agonists are exercise mimetics. Cell 134: 405–415. 10.1016/j.cell.2008.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhan W. L., 2005. Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 86: 25–33. 10.1016/j.ymgme.2005.07.027 [DOI] [PubMed] [Google Scholar]
- Oburoglu L., Tardito S., Fritz V., de Barros S. C., Merida P., et al. , 2014. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 15: 169–184 (erratum: Cell Stem Cell 15: 666–668). 10.1016/j.stem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Olmedo M., Geibel M., Artal-Sanz M., Merrow M., 2015. A high-throughput method for the analysis of larval developmental phenotypes in Caenorhabditis elegans. Genetics 201: 443–448. 10.1534/genetics.115.179242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S. H., Loomis W. F., 2006. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5: 272–276. 10.1128/EC.5.2.272-276.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson B., Vaur S., Sagot I., Coulpier F., Lemoine S., et al. , 2009. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 23: 1399–1407. 10.1101/gad.521809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess J. R., Schnabel H., Schnabel R., 1987. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell 51: 601–611. 10.1016/0092-8674(87)90129-2 [DOI] [PubMed] [Google Scholar]
- Race V., Marie S., Vincent M. F., Van den Berghe G., 2000. Clinical, biochemical and molecular genetic correlations in adenylosuccinate lyase deficiency. Hum. Mol. Genet. 9: 2159–2165. 10.1093/hmg/9.14.2159 [DOI] [PubMed] [Google Scholar]
- Rebora K., Desmoucelles C., Borne F., Pinson B., Daignan-Fornier B., 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol. Cell. Biol. 21: 7901–7912. 10.1128/MCB.21.23.7901-7912.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora K., Laloo B., Daignan-Fornier B., 2005. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170: 61–70. 10.1534/genetics.104.039396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Palero M. J., López-Díaz A., Marsac R., Gomes J. E., Olmedo M., et al. , 2018. An automated method for the analysis of food intake behaviour in Caenorhabditis elegans. Sci. Rep. 8: 3633 10.1038/s41598-018-21964-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Moss E. G., 2013. Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105: 153–180. 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- Samuel B. S., Rowedder H., Braendle C., Félix M. A., Ruvkun G., 2016. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. USA 113: E3941–E3949. 10.1073/pnas.1607183113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel E. K., Colman R. F., Patterson D., 2006. Adenylosuccinate lyase deficiency. Mol. Genet. Metab. 89: 19–31. 10.1016/j.ymgme.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Stenesen D., Suh J. M., Seo J., Yu K., Lee K.-S., et al. , 2013. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab. 17: 101–112. 10.1016/j.cmet.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org.
- Tardito S., Oudin A., Ahmed S. U., Fack F., Keunen O., et al. , 2015. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 17: 1556–1568. 10.1038/ncb3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Vrablik T. L., Huang L., Lange S. E., Hanna-Rose W., 2009. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development 136: 3637–3646. 10.1242/dev.028431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E., MacNeil L. T., Ritter A. D., Yilmaz L. S., Rosebrock A. P., et al. , 2014. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156: 759–770 (erratum: Cell 156: 1336–1337). 10.1016/j.cell.2014.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Brockie P. J., Mellem J. E., Madsen D. M., Maricq A. V., 1999. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron 24: 347–361. 10.1016/S0896-6273(00)80849-1 [DOI] [PubMed] [Google Scholar]
- Zikanova M., Skopova V., Hnizda A., Krijt J., Kmoch S., 2010. Biochemical and structural analysis of 14 mutant ADSL enzyme complexes and correlation to phenotypic heterogeneity of adenylosuccinate lyase deficiency. Hum. Mutat. 31: 445–455. 10.1002/humu.21212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7642871.