Genetic screens in the nematode Caenorhabditis elegans have identified EGF and Notch pathways as key for vulval precursor cell fate patterning. Here, Vargas-Velazquez, Besnard, and Félix report on the molecular identification of...
Keywords: C. elegans, O. tipulae, lin-3/EGF, Wnt, plexin, semaphorin, evolution of development, genetic screens
Abstract
Genetic screens in the nematode Caenorhabditis elegans identified the EGF/Ras and Notch pathways as central for vulval precursor cell fate patterning. Schematically, the anchor cell secretes EGF, inducing the P6.p cell to a primary (1°) vulval fate; P6.p in turn induces its neighbors to a secondary (2°) fate through Delta-Notch signaling and represses Ras signaling. In the nematode Oscheius tipulae, the anchor cell successively induces 2° then 1° vulval fates. Here, we report on the molecular identification of mutations affecting vulval induction in O. tipulae. A single Induction Vulvaless mutation was found, which we identify as a cis-regulatory deletion in a tissue-specific enhancer of the O. tipulae lin-3 homolog, confirmed by clustered regularly interspaced short palindromic repeats/Cas9 mutation. In contrast to this predictable Vulvaless mutation, mutations resulting in an excess of 2° fates unexpectedly correspond to the plexin/semaphorin pathway. Hyperinduction of P4.p and P8.p in these mutants likely results from mispositioning of these cells due to a lack of contact inhibition. The third signaling pathway found by forward genetics in O. tipulae is the Wnt pathway; a decrease in Wnt pathway activity results in loss of vulval precursor competence and induction, and 1° fate miscentering on P5.p. Our results suggest that the EGF and Wnt pathways have qualitatively similar activities in vulval induction in C. elegans and O. tipulae, albeit with quantitative differences in the effects of mutation. Thus, the derived induction process in C. elegans with an early induction of the 1° fate appeared during evolution, after the recruitment of the EGF pathway for vulval induction.
HOW multicellular organisms arise from single cells is a question that has intrigued scientists over the ages. In the 1960s, Sydney Brenner selected Caenorhabditis elegans as a new model organism to study animal development using genetics (Brenner 1974). Vulva precursor cell (VPC) fate patterning rapidly became one of the most studied developmental processes in C. elegans, due to the easy isolation of mutants with a defective vulva (Sternberg 2005).
The C. elegans vulva is an epidermal specialization that develops from a row of six VPCs in the ventral epidermis, called P3.p–P8.p from anterior to posterior. In most animals, the central vulval fate, or primary (1°) fate, is adopted by P6.p, while the outer vulval fate, or secondary (2°) fate, is adopted by its neighbors P5.p and P7.p (Sulston and Horvitz 1977; Sternberg 2005). Finally, P3.p, P4.p, and P8.p are able to replace the central cells (for example if they are destroyed with a laser), but normally adopt an epidermal fate with one division and fusion of the daughters to the large epidermal syncytium hyp7 (Sulston and White 1980). Laser ablation of the anchor cell in the gonad primordium results in all precursor cells adopting a tertiary (3°) fate, showing that the vulval fates are induced by the anchor cell (Kimble 1981).
Upon random chemical mutagenesis, some recurrent phenotypes were isolated with pronounced defects in vulval development, such as the Vul (Vulvaless) and Muv (Multivulva) phenotypes (Horvitz and Sulston 1980; Ferguson and Horvitz 1985). The Vulvaless mutants can be easily seen in the dissecting microscope by the internal hatching of the progeny in their mother (bag of worms). The Vulvaless mutants can be further classified in two classes: (i) those that mimick an anchor cell ablation (cells adopting a 3° fate) or Induction Vulvaless, and (ii) those that prevent the development of competent VPCs or Generation Vulvaless (Ferguson et al. 1987). The Multivulva mutants are recognized by the additional protrusions on the ventral cuticle (pseudovulvae).
The C. elegans Induction Vulvaless and Multivulva mutants allowed the identification of the EGF/Ras/MAP kinase pathway, the former class corresponding to a loss of activity in the pathway and the latter to a gain of activity (Sternberg 2005). In addition, mutations at the lin-12 locus specifically affect 2° fates: reduction-of-function lin-12 alleles transform 2° fates to 1° or 3°, while gain-of-function alleles transformed 1° and 3° fates to the 2° fate (Greenwald et al. 1983). lin-12 was shown to encode a Notch receptor, receiving Delta signals mostly produced by P6.p. Studies of the interplay between the EGF and Delta/Notch pathways in the patterning vulval cell fates have established this system as a textbook example of intercellular signaling and organogenesis (Sternberg and Han 1998).
Since the 1990s, studies of vulval development in different Caenorhabditis species and other nematode genera have made vulval development an emblematic example of developmental system drift (True and Haag 2001): while the vulval cell fate pattern remains overall invariant (2°1°2° for P5.p, P6.p, and P7.p), evolution occurs in the manner in which it forms. First, the size of the competence group varies (Sternberg and Horvitz 1982; Sommer and Sternberg 1996; Félix et al. 2000a; Delattre and Félix 2001; Pénigault and Félix 2011a). Second, vulval cell fate patterning does not always require the anchor cell (Sommer and Sternberg 1996; Félix et al. 2000a). Third, when it requires the gonad, ablating the anchor cell at intermediate timepoints has widely different effects depending on the species (Sommer and Sternberg 1994; Félix and Sternberg 1997; Sommer 2005; Kiontke et al. 2007; Félix 2012; Félix and Barkoulas 2012). In particular, in many genera of rhabditids and diplogastrids (Félix and Sternberg 1997, 1998; Sigrist and Sommer 1999; Félix et al. 2000a; Félix 2007; Kiontke et al. 2007), gonad ablation at an intermediate timepoint results in P(5–7).p adopting a 2° fate (vs. a 3° fate for the outer cells), with no apparent differences among these three cells. This contrasts with anchor cell ablation results in C. elegans, where no such intermediate state exists and P6.p adopts a 1° fate earlier, thereby activating lateral induction and inhibition (Félix 2007) (Figure 1). The mode of induction where an intermediate fate is found for all cells has been called a two-step induction (Félix and Sternberg 1997). In this case, the second step of induction of the 1° fate occurs after one division round on P6.p daughters. Even though two steps can be distinguished (induction of P6.p to a 2° fate then after P6.p division to a 1° fate), signaling from the gonad may be continuous (Félix and Sternberg 1997; Sigrist and Sommer 1999; Kiontke et al. 2007).
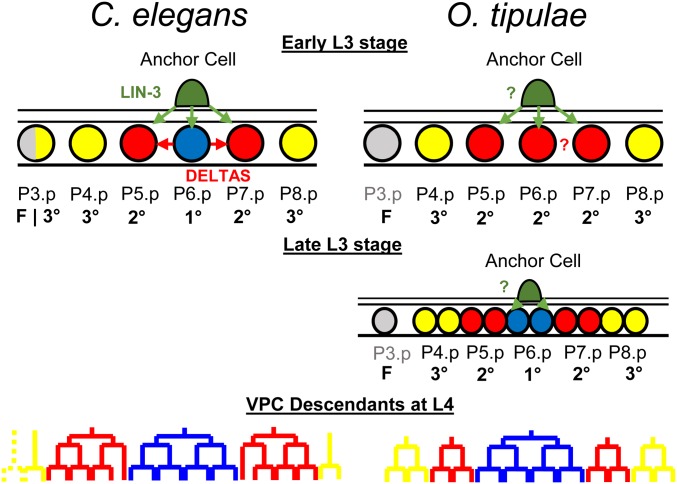
Figure 1.
Vulval cell fate patterning in C. elegans and O. tipulae. In the third larval stage (L3) of C. elegans, a cell from the somatic gonad known as the anchor cell (AC) produces an EGF-like inductive signal (LIN-3, green arrows) that activates the Ras pathway in the central vulval precursor cells (VPCs). High Ras signaling promotes the primary (1°) fate (blue circle) in P6.p, which, in turn, produces Deltas (red arrows) that induce a secondary (2°) fate (red circle), and represses the 1° fate in P5.p and P7.p. Both fates prevent the formation of nonspecialized epidermis [tertiary (3°) fate, yellow circles]. Only VPCs with 1° or 2° fates will give rise to the cells that will form the vulva (bottom). P3.p is not competent to acquire a vulval cell fate (gray) in O. tipulae. Unlike in C. elegans, the AC of O. tipulae has been shown to be required to induce the 1° fate in P6.p descendants after VPC division. While a similar vulval cell fate pattern is conserved between the two nematodes, the cell division patterns of the 2° and 3° fates are different.
Among species with a two-step induction (Figure 1), Oscheius tipulae is a rhabditid nematode found in the same habitat as C. elegans (Félix and Duveau 2012), which can be cultured in the same laboratory conditions (Félix et al. 2000b). In this species, the anchor cell first induces P(5–7).p to a 2° fate (first induction) then induces P6.p daughters to a 1° fate (second induction) (Figure 1). A genetic screen was conducted 20 years ago to isolate vulva development mutants in O. tipulae (Dichtel et al. 2001; Louvet-Vallée et al. 2003; Dichtel-Danjoy and Félix 2004a,b). This genetic screen led to a distinct spectrum of vulval cell fate and lineage phenotypes compared to those found in C. elegans. This result suggested a different sensitivity of the developmental system to mutation in the two species and therefore a different evolutionary potential, as the phenotypic response to mutation may bias phenotypic evolution (Dichtel-Danjoy and Félix 2004a). The distinct spectrum of mutant phenotypes also reflected the difference in vulval development between O. tipulae and C. elegans (Dichtel-Danjoy and Félix 2004a). We further identified a null mutant in the Hox gene lin-39 with the same phenotype as in C. elegans, namely a loss of competence of the VPCs (Louvet-Vallée et al. 2003).
A draft of the O. tipulae genome has recently been published, along with a strategy to map the genomic location of loci whose mutation produces a visible phenotype (Besnard et al. 2017). As a proof-of-principle for the mutant identification technique, we described alleles of the Oti-mig-13 locus with an unexpected vulva phenotype (Besnard et al. 2017). Here, we take advantage of the mapping approach to molecularly identify the collection of O. tipulae mutations affecting vulval cell fate patterning. We had found a single Induction Vulvaless locus with a single allele and this turned out to be a cis-regulatory deletion in a tissue-specific enhancer of the O. tipulae lin-3 homolog, which we confirmed by targeted clustered regularly interspaced short palindromic repeat (CRISPR) mutation of the element. We then identified mutations in Wnt pathway components (mom-5/frizzled, mig-14/wingless, and egl-20/Wnt) affecting fates of the O. tipulae VPCs, and discuss similarities and differences with C. elegans and Pristionchus pacificus, another nematode species where similar screens have been conducted (Sommer 2006). Finally, the last class of vulval cell fate mutations caused an excess of 2°-fated cells. Unexpectedly, these mutations corresponded to lesions in Oti-plx-1 and Oti-smp-1, encoding plexin and semaphorin, a cell signaling system that was not found in C. elegans vulva mutagenesis screens.
Materials and Methods
Nematode culture
C. elegans and O. tipulae were handled according to the usual procedures, on standard NGM plates with Escherichia coli strain OP50 as a food source (Brenner 1974; Félix et al. 2000b). C. elegans and O. tipulae strains were maintained, respectively, at 20 and 23°, unless otherwise indicated. N2 was used as a reference strain for C. elegans and CEW1, a wild isolate from Brazil, as a reference strain for O. tipulae. The O. tipulae mutants were all described previously (Dichtel et al. 2001; Louvet-Vallée et al. 2003; Dichtel-Danjoy and Félix 2004a,b). Except for the Vulvaless Oti-iov-1(mf86) mutant, the mutations were all backcrossed one to five times. A list of strains used in this study is presented in Supplemental Material, Table S1.
Mapping-by-sequencing and identification of molecular lesions
The mapping-by-sequencing strategy using the partial genome assembly of O. tipulae includes genetic mapping and whole-genome sequencing in a single step, and has been comprehensively described before (Besnard et al. 2017). In brief, each mutant O. tipulae line previously obtained in the CEW1 genetic background was crossed to males of the molecularly divergent wild isolate JU170. In the case of the fully Vulvaless iov-1(mf86) mutant, males of strain JU432 of genotype iov-1(mf86); him(sy527) were crossed to JU170 hermaphrodites. In all cases, individual F2 progeny with a recessive mutant phenotype were isolated and the mutant phenotype verified on the F3 brood. The pooled DNA of the progeny of mutant F2s was extracted using the Puregene Core Kit A (QIAGEN, Valencia, CA) and whole-genome sequenced at the Beijing Genomics Institute facilities. Pools from 37 to 152 individual F2s were used, depending on the ease of scoring of the mutant phenotype.
Sequencing reads from each mutant pool were mapped to the CEW1 genome using bwa (Li and Durbin 2009) and the resulting alignment converted to bam format using samtools (Li et al. 2009). Each mapping was further processed with the GATK suite (Van der Auwera et al. 2013) and allelic variants were called using HaplotypeCaller on a restricted list of JU170 sites for faster computation. Scaffolds having a mean JU170 allele frequency of < 10% were selected as candidates for possible linkage with a causative locus and processed for homozygous variant calling in an unrestrictive manner. JU170 variants were filtered out from the candidate scaffolds and the remaining variants were analyzed for any functional impact on the O. tipulae gene annotations (CEW1_nOt2) using snpEff (Cingolani et al. 2012). Scripts used for this processing pipeline can be found at: https://github.com/fabfabBesnard/Andalusian_Mapping. The candidate scaffolds were also analyzed using Pindel (Ye et al. 2009) to identify large deletions or insertions, which were confirmed later by visual inspection with the Tablet software (Milne et al. 2013).
The genetic loci with names such as cov-3 and cov-4 were defined by genetic complementation tests (Dichtel et al. 2001; Louvet-Vallée et al. 2003; Dichtel-Danjoy and Félix 2004a,b). When the locus was defined by more than one mutant allele, as described in Besnard et al. (2017) for cov-3, we subjected two alleles to the genetic mapping-by-sequencing approach and determined the intersection between the resulting lists of candidate genes with lesions in the genetic interval, thus pointing to a single common gene. As indicated in the legend to Table 1, for dov-4(sy451) and iov-3(mf78), a candidate gene found by the genetic mapping-by-sequencing approach using one or two other allele(s) was tested by PCR for the presence of a lesion and in all cases we found a lesion in the same gene. Finding two noncomplementing alleles with a lesion in the same gene makes it highly likely that we have identified the correct causal locus. The mutations identified by the mapping-by-sequencing approach were also verified by Sanger sequencing of a PCR product. A list of primers used for sequencing can be found in Table S2.
Table 1 . O.tipulae vulval loci identified by the mapping-by-sequencing approach.
| Locusa | Allele (method) | Phenotypes | Mutagen | Positionb | Mutation | Oti gene | Cel homolog | Type of lesion | Reported before in |
|---|---|---|---|---|---|---|---|---|---|
| cov-4 | sy465 | Competence loss, P5.p centering | EMS | 10:202,548 | G/A | g06014 | mom-5/frizzled | Premature stop | Louvet-Vallée et al. (2003) |
| cov-4 | sy493 | Competence loss, P5.p centering | EMS | 10:201,110 | C/T | g06014 | mom-5/frizzled | Splice acceptor | Louvet-Vallée et al. (2003) |
| cov-5 | mf34 | Competence loss, P5.p centering | EMS | 3:189,504 | T/C | g01986 | mig-14/Wntless | Missense variant | Louvet-Vallée et al. (2003) |
| dov-4 | sy464 | P4.p/P8.p do not divide, some P5.p centering | EMS | 4:882,344 | G/A | g02936 | egl-20/Wnt | Premature stop | Dichtel et al. (2001) |
| dov-4 | sy451 | P4.p/P8.p do not divide, some P5.p centering | EMS | 4:882,874 | T/C | g02936 | egl-20/Wnt | Missense variant | Dichtel et al. (2001) |
| iov-1 | mf86 | Hypoinduction | TMP-UV | 39:154,942–155,133 | 191-bp deletion | g12432 | lin-3 | Cis-regulatory deletion | Dichtel-Danjoy and Félix (2004b) |
| iov-2 | mf76 | Hyperinduction 2° | TMP-UV | 10:74,243–74,877 | 634-bp deletion | g05993 | smp-1/semaphorin | Putative null | Dichtel-Danjoy and Félix (2004b) |
| iov-3 | sy447 | Hyperinduction 2° | EMS | 86:24,647 | A/T | g14741 | plx-1/plexin | Missense variant | Dichtel-Danjoy and Félix (2004b) |
| iov-3 | mf52 | Hyperinduction 2° | EMS | 86:24,066 | A/T | g14741 | plx-1/plexin | Missense variant | Dichtel-Danjoy and Félix (2004b) |
| iov-3 | mf78 | Hyperinduction 2° | EMS | 86:27,580–27,583 | 4-bp deletion | g14741 | plx-1/plexin | Premature stop | Dichtel-Danjoy and Félix (2004b) |
TMP-UV, trimethylpsoralene-ultraviolet; 2°, secondary.
The locus was defined by complementation tests in the article cited in the last column.
The localization corresponds to the genomic position (scaffold: base pair). All molecular lesions in the table were identified by the mapping-by-sequencing approach, except the additional alleles mf78 and sy451, which were identified by PCR and Sanger sequencing of the gene.
All molecular loci in Table 1 were identified using two or more alleles (the identification of Oti-lin-3 was confirmed by CRISPR alleles), except for Oti-mig-14/Wntless and Oti-smp-1/semaphorin, where we had a single mutant allele. The indication that lesions in these two genes are causal relies on the fact that other genes in the same molecular pathway were found with similar mutant phenotypes, respectively: Oti-mom-5/Wnt receptor and Oti-plx-1/plexin.
Identification of homologous genes
The predicted protein sequences of O. tipulae genes were obtained through the genome annotation (Besnard et al. 2017), now available from the Blaxter laboratory website (http://caenorhabditis.org). The sequence of their closest C. elegans homolog was identified using the Basic Local Alignment Search Tool Protein (BLASTP) algorithm (Gish and States 1993), conditioning for highly similar alignments (> 80% identity) and low e-values. Manual curation and reannotation of the O. tipulae gene sequences were then performed using their closest C. elegans homolog as a reference. We aligned the amino acid sequences of the reannotated genes with their respective C. elegans homologs and outgroups using the Muscle algorithm implemented in MEGA X (Kumar et al. 2018) with default parameters. The phylogenetic relationship between the protein sequences was inferred using the neighbor-joining method (Saitou and Nei 1987) and tested for bootstrapping with 1000 replicates.
Nomenclature
We followed C. elegans nomenclature and recommendations for other nematode species in Tuli et al. (2018). Briefly, mutant class names had been given at the time of our screen: iov for induction of the vulva, dov for division of VPCs, and cov for competence and/or centering of VPCs. Once the molecular lesion has been identified, we used the name of the C. elegans homolog preceded by the species prefix for Oscheius tipulae “Oti-”; for example the iov-1(mf86) allele is thus renamed Oti-lin-3(mf86).
Single-molecule fluorescence in situ hybridization
Single-molecule FISM (smFISH) in O. tipulae and in C. elegans was performed as previously described (Barkoulas et al. 2016). Mixed-stage populations were used for an mRNA localization experiment, while bleach-synchronized populations at the L3 larval stage were used for mRNA quantification. Only L3-stage nematodes with a gonad longer than 300 pixels (38.66 μm) were considered for mRNA quantification. The short fluorescently labeled oligos used in this study were acquired from LGC Biosearch Technologies and were used at a concentration of 100–200 mM. A list containing the sequences of the smFISH oligonucleotides is provided in Table S3.
Phenotypic characterization and measurements of distances between nuclei
The cell fates acquired by the O. tipulae VPCs were scored as previously described (Dichtel et al. 2001). In summary, early L4 larvae were mounted with M9 solution on 4% agar pads containing 10 mM sodium azide and analyzed under Nomarski optics. Standard criteria were used to infer cell fates based on the topology and number of cells at different stages. The 1° fate is defined as three rounds of division with a final transverse division of each granddaughter (“TTTT”) and an attachment of the central cells to the anchor cell. Unlike in C. elegans, the cells with 2° and 3° fates undergo two rounds of division; as in C. elegans (Katz et al. 1995), the 2° fate is characterized by the absence of cell fusion to the epidermal syncytium and the detachment from the cuticle (“UUUU” for undivided), whereas the 3° fate is characterized by cell fusion to the epidermal syncytium, which stays attached to the cuticle (“ssss” for syncytial). The attachment to the anchor cell and to the cuticle for the 1° and 3° fates, respectively, are important features of cell fate definitions, since in Dichtel et al. (2001) we isolated mutants with defective divisions but no cell fate alterations in this respect; we do not study these cell division mutants here but only those where cell fates appear homeotically transformed. Half fates were assigned when two daughters of the Pn.p cells acquired distinct fates after the first cell division.
Measurements of distances between the nuclei of Pn.p cells were performed on mounted larvae at three different developmental stages: L2 molt, early L3 (before the division of dorsal uterine DU cells), and mid L3 (after DU cell division and before Pn.p divisions). The distance between the center of the Pn.p and anchor cell nuclei was measured in pixels using a Photometrics CoolSNAP ES camera and Nikon (Garden City, NY) NIS-Elements software (version 3.0.1). To avoid measurement errors due to the animal curvature, the distance between each Pn.p cell (except P6.p) and the anchor cell was calculated via a Pythagorean formula. For example, the distance between P4.p and the anchor cell is equal to:
where is the distance between P6.p and the anchor cell, is the distance between P5.p and P4.p, and is the distance between P6.p and P5.p. Nonnormalized measurements can be found in Table S4.
Genome editing
We followed the CRISPR-Cas9 target design in Paix et al. (2015). We targeted the following sequence at the O. tipulae lin-3 cis-regulatory region 5′-cCACCTGcatgtcctttttgcgc-3′ (E-box site in uppercase, within an underlined NGGNGG protospacer adjacent motif in the negative strand). The mf113 allele was produced with the synthetic Oti_lin-3_A-2 -GCGCAAAAAGGACAUGCAGG- crRNA (CRISPR RNA) manufactured by Dharmacon (GE Healthcare), while mf114 was produced with the same crRNA sequence synthesized by IDT (Integrated DNA Technologies, Inc.). Each crRNA was mixed with tracrRNA (trans-activating CRISPR RNA) (Paix et al. 2015) at an equimolar concentration of 200 μmol/μl. The tracrRNA:crRNA mix was incubated in a thermal ramp between 95 and 25°, decreasing by 5° every 2 min, and then mixed with purified CRISPR-Cas9 protein in HEPES buffer (pH 7.4), reaching a final concentration of 30 μM of the tracrRNA:crRNA duplex and ∼18 μM of purified protein. The final mix was incubated for 15 min at 37° and then injected into the gonad of O. tipulae gravid adults. The F1 progeny of the injected nematodes were placed into new plates and, after letting them lay eggs for 1 day, screened for deletions by PCR with the mf86-EboxA-F and mf86-R primers. Heterozygous F1 animals were identified by band-size separation on 3% agarose gels, and homozygous F2 mutants were easily spotted by their bag phenotype. Only a single mutation per injection session (> 10 P0s and > 200 F1s) was obtained.
To obtain C. elegans plx-1 null mutants, hermaphrodites bearing the plx-1(ev724) allele were injected as above but with a crRNA sequence (5′-GGAATGCACTCAGAATGGTG-3′) that targeted a plx-1 coding sequence located upstream of the ev724 deletion in the second exon downstream of the second ATG, and a crRNA that targeted dpy-10 and a short DNA oligo that introduces the dpy-10(cn64) as a co-CRISPR marker (Arribere et al. 2014). Roller F1s were isolated onto new plates, and screened for mutations in the plx-1 gene other than the ev724 deletion by PCR using the forward and reverse Cel-plx-1_111- primers.
Immunofluorescence staining
Bleach-synchronized larvae and mixed-stage populations were fixed, and permeabilized for immunostaining using previously described methods (Louvet-Vallée et al. 2003; Kolotuev and Podbilewicz 2004, 2008). In brief, OP50-grown populations were washed three times in distilled water and placed onto poly-l-lysine-coated (P0425-72EA; Sigma [Sigma Chemical], St. Louis, MO) slides prior to freeze-cracking. Worms with an open cuticle were incubated in antibody buffer with the mouse MH27 antibody against the epithelial cell adherent junctions (Francis and Waterston 1991). This antibody was obtained from the Developmental Studies Hybridoma Bank and used at a concentration of 1 mg/ml. As secondary antibody, we used the goat anti-mouse antibody from Abcam labeled fluorescently with Alexa Fluor 488 (reference #ab150113). The slides containing immunofluorescently labeled worms were mounted with GLOX buffer (Ji and van Oudenaarden 2012) containing DAPI, covered with a cover slip, and imaged with a PIXIS camera (Princeton Instruments).
Data availability
Supplemental Tables are available through the Figshare portal (https://doi.org/10.25386/genetics.7624559):
Table S1. List of strains used in this study.
Table S2. Sequences of DNA primers used in this study. Sequencing primers to verify by Sanger sequencing the mutations identified by the mapping-by-sequencing approach, and to identify the molecular lesions in additional alleles.
Table S3. Sequences of smFISH probes used in this study. The fluorophore coupled to each probe is noted at the end of the set name.
Table S4. smFISH quantifications, distance measurements, and vulval cell fates used in this study.
Code for mutant identification is available at https://github.com/fabfabBesnard/Andalusian_Mapping. Sequencing reads used for mapping-by-sequencing can be find at the Sequence Read Archive portal under the bioproject number PRJNA514975.
Results
The sole hypoinduction mutation is due to a cis-regulatory change in Oti-lin-3
Our prior mutagenesis screens had yielded a single mutant with an Induction Vulvaless phenotype, i.e., the 1° and 2° fates are transformed to a 3° fate (two rounds of division and fusion to the hyp7 syncytium, represented in yellow in the figures) but rarely to a noncompetent state (fusion to hyp7 without division, prior to the L3 stage, represented in gray in the figures) (Dichtel-Danjoy and Félix 2004b). This allele, iov-1(mf86), was obtained after TMP-UV (trimethylpsoralene-ultraviolet) mutagenesis. The mapping-by-sequencing approach identified a 191-bp deletion upstream of the coding sequence (second ATG) of O. tipulae lin-3 (Oti-lin-3) (Figure 2B). We hypothesized that this deletion may cause a reduced level of expression in Oti-lin-3 and thus performed smFISH experiments to quantify Oti-lin-3 mRNA number in the anchor cell (Raj et al. 2008; Barkoulas et al. 2013, 2016). Indeed, the Oti-lin-3 mRNA level in the anchor cell was much decreased in animals bearing the Oti-lin-3(mf86) deletion compared to animals of the CEW1 reference strain (Figure 2C, Kolmogorov–Smirnov test, P < 10−14), while Oti-lin-3 expression elsewhere in the pharynx and in the germline was retained (Figure S7B), as in the C. elegans cis-regulatory mutant lin-3(e1417) (Saffer et al. 2011). The deleted region in Oti-lin-3(mf86) contains an E-box motif known to be conserved in Caenorhabditis species (Barkoulas et al. 2016), as well as a second less-characteristic putative E-box motif (Figure 2B and Figure S7A).
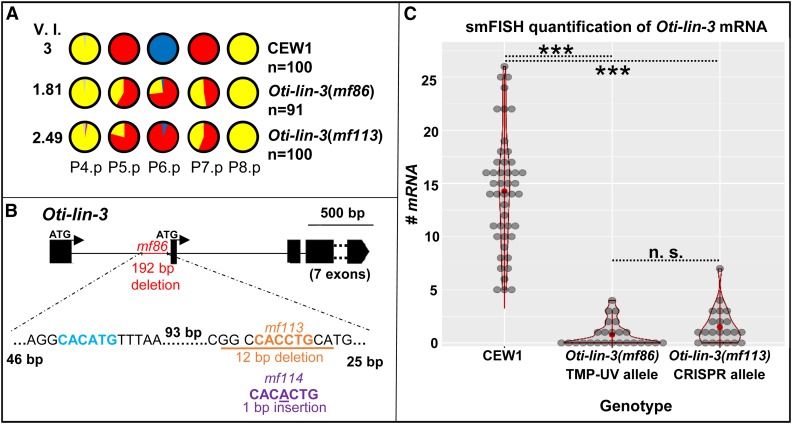
Figure 2.
Cis-regulatory lesions in Oti-lin-3/EGF cause hypoinduction of primary (1°) and secondary (2°) vulval cell fates. (A) P(4–8) cell fates in the wild-type CEW1 O. tipulae reference strain and mutants for lin-3/EGF. The pie diagrams represent the percentage of cell fates over individuals. Yellow, red, and blue are for the tertiary (3°), 2°, and 1° fates, respectively. Gray denotes an undivided cell fused to the hypodermis. The Vulva Index (V.I.) is calculated as the average number of cells acquiring a vulval cell fate in a set of animals. The quantifications of Oti-lin-3(mf86) are from Dichtel-Danjoy and Félix (2004b). (B) Position of the deletions in the trimethylpsoralene-ultraviolet (TMP-UV) and clustered regularly interspaced short palindromic repeat (CRISPR) alleles. As in C. elegans, the lin-3 gene of O. tipulae is predicted to have two alternative ATGs, with the anchor cell cis-regulatory element upstream of the second ATG. However, unlike C. elegans, O. tipulae has a single E-box “CACCTG” binding site and no nuclear hormone receptor (NHR) binding site. Note that the seven exons following the second ATG were excluded from the diagram. (C) Distributions of Oti-lin-3 mRNA number in the anchor cell of wild-type CEW1 and lin-3 cis-regulatory mutants, as quantified by single-molecule FISH. n.s., not significant. ***: p<0.001.
To test whether the conserved E-box motif was required for the expression of Oti-lin-3 and also confirm that the 191-bp deletion was causal for the vulva phenotype, we performed a CRISPR/Cas9 experiment specifically targeting this site. We obtained two new mutations, a smaller 12-bp deletion (mf113) and a 1-bp insertion in the E-box (mf114). Both showed a strong decrease in the level of induction, confirming that the lesion in the Oti-lin-3 gene is causal for the phenotype (Figure 2A and Table S4). Further smFISH experiments in the Oti-lin-3(mf113) mutant revealed a similar level of mRNAs as in the Oti-lin-3(mf86) mutant that was different from that of the O. tipulae CEW1 reference strain (Kolmogorov–Smirnov test, P = 0.94 and P < 10−9, respectively, Figure S7B). We conclude that the conserved E-box site is required in O. tipulae for lin-3 expression, and that LIN-3 secreted from the anchor cell is necessary for the induction of both 2° and 1° fates.
The Wnt pathway plays a role in vulval precursor competence/induction and fate pattern centering
A large class of mutants in our screen displayed a lower number of competent Pn.p cells (transformation to quaternary/gray fate) and a displacement of the 1° fate from P6.p to P5.p. In C. elegans, this phenotype has not been seen at this high level of penetrance. The mapping-by-sequencing approach had already identified one locus in this class as Oti-mig-13 (Besnard et al. 2017). In this class, we further identified mutations in two Wnt pathway components:
1. A Wnt receptor gene, Oti-mom-5 (supported by two alleles, including an early stop before the transmembrane domain) (Figure 3B). Relationships among Wnt receptor paralogs in the different species are shown in Figure S4. Curiously, the Oti-mom-5 putative null allele, sy465, is viable in O. tipulae, while an early stop before the transmembrane domain such as mom-5(ne12) is embryonic lethal in C. elegans (embryonic mesoderm vs. endoderm specification; Rocheleau et al. 1997). We cannot rule out that another mutation suppresses the lethality in the O. tipulae strain.
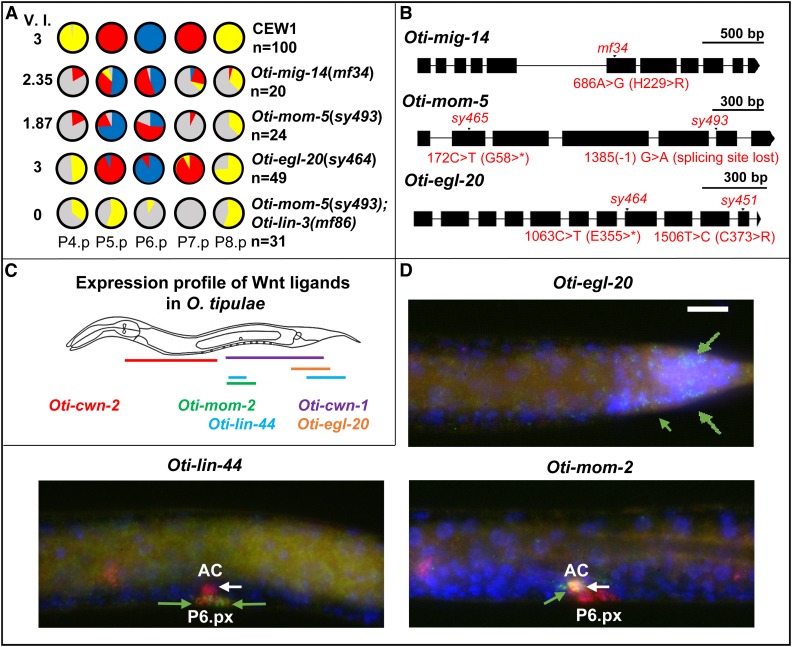
Figure 3 .
O. tipulae mutants in Wnt signaling display defects in competence and centering of the 1° fate on P5.p. (A) Pie diagrams representing the percentage of P(4–8).p cells acquiring one of the four possible cell fates (blue, red, yellow, and gray for the 1°, 2°, 3°, or 4° fates, respectively) for animals of different genotypes. The V.I. is calculated as the average number of cells acquiring a vulval cell fate in a set of animals. The quantifications in Oti-mig-14(mf34) and Oti-mom-5(sy493) animals are from Louvet-Vallée et al. (2003), and that of Oti-egl-20(sy464) from Dichtel et al. (2001). (B) Position of different mutations in genes encoding Wnt pathway components. A star designates a stop codon. (C) Diagram of Wnt ligand expression profiles in O. tipulae at mid-L3 stage. smFISH images of Oti-cwn-1 and Oti-cwn-2 are found in Figure S3. (D) smFISH images of Oti-egl-20, Oti-lin-44, and Oti-mom-2 Wnt ligands after P6.p division at the L3 stage. mRNAs are visible as green dots. The animals were also labeled with DAPI (in blue, labeling nuclei) and fluorophore probes for Oti-lag-2/Δ (in red, labeling the anchor cell, P6.p descendants, and distal tip cells outside the field of view). Oti-egl-20 is visible only in the posterior part of the animal (green arrows). Oti-mom-2 mRNAs (green arrow) are found in the anchor cell (white arrow), while Oti-lin-44 mRNAs (green arrows) appear in P6.p daughters (as well as sex myoblast precursors outside the focal plane). All the images are set to the same scale. The size of the white bar is 10 μm. Anterior is to the left in all images and the ventral side is down. 1°, primary; 2°, secondary; 3°, tertiary; AC, anchor cell; smFISH, small-molecule FISH; V.I., Vulva Index.
2. A Wnt-processing protein, Oti-mig-14 (homolog of Drosophila Wntless) (Bänziger et al. 2006; Yang et al. 2008). The mf34 mutant displays an amino acid substitution in the Oti-mig-14 gene and is thus likely a hypomorph that may negatively affect the activity of all Wnts. Although this gene identification relies on a single available allele at the cov-5 locus and is thus less reliable than the above, this lesion makes Oti-mig-14/Wntless a very likely candidate for the causal mutation for this phenotype, since both cov-4/Wnt receptor and cov-5 loci show similar mutant phenotypes (Louvet-Vallée et al. 2003).
We had somewhat arbitrarily distinguished classes of vulval mutations that affect competence and centering (cov mutants) from those that affect division of VPCs (dov mutants) (Dichtel et al. 2001). Among the latter class, we found that the dov-4 locus encodes a Wnt-type ligand, Oti-egl-20 (supported by two alleles, including a premature stop). The Oti-egl-20 mutation results in a lower competence and division frequency of P4.p and P8.p, but hardly affects P(5–7).p. Centering of the 1° fate on P5.p only occurs at low penetrance. Overall, the Oti-egl-20 phenotype is similar to that of Oti-mig-14/cov-5 or Oti-mom-5, albeit much weaker, suggesting the involvement of other Wnt family ligands.
The O. tipulae genome contains five genes coding for Wnt signaling molecules, which we found to be 1:1 orthologs to the five Wnt genes in C. elegans (Figure S4). By smFISH, the expression pattern of each of these five genes was found to be quite similar in L1–L3 larvae to that of each ortholog in C. elegans, as determined in Song et al. (2010) and Harterink et al. (2011). Specifically, Oti-egl-20 is expressed in the posterior region of the animal from the L1 stage (Figure 3D). Oti-cwn-1 is also expressed quite posteriorly (Figure S3). Oti-cwn-2 is expressed in the anterior region (Figure S3). Oti-mom-2 is expressed in the anchor cell from the L3 stage (Figure 3D). Oti-lin-44 is expressed in the tail region and in the L3 stage in P6.p daughters (Figure 3, Figure S3, and Figure S6). Similar to cwn-1 in C. elegans (Harterink et al. 2011; Minor et al. 2013), we found that Oti-lin-44 is additionally expressed in the sex myoblast precursors that are located to the left and right of the anchor cell in the L3 stage (Figure S3). As the sex myoblast expression of Oti-lin-44 differed from the reported uterus/anchor cell pattern in C. elegans using lacZ staining or fluorescent reporters (Inoue et al. 2004), we localized lin-44 by smFISH in C. elegans and saw similar expression in the sex myoblasts (identified by labeling with hlh-8::GFP (Harfe et al. 1998) and P6.px, and none in the uterus and anchor cell (Figure S6). In conclusion, the larval expression patterns of the five Wnt genes were thus similar in O. tipulae and C. elegans.
From the Oti-egl-20 expression pattern and mutant phenotype, the EGL-20 protein is produced from the posterior of the animal and promotes Pn.p cell competence as far as P4.p. P3.p is not competent and does not divide in O. tipulae (Félix and Sternberg 1997; Delattre and Félix 2001), and is thus not affected by Wnt pathway mutations, whereas it is highly sensitive to Wnt pathway modulation in C. elegans (Pénigault and Félix 2011b). However, the difference in phenotype severity between Oti-mig-14 or Oti-mom-5 mutants on one hand, and Oti-egl-20 (including the sy464 allele with a stop codon) on the other hand, suggests that other Wnt signals, perhaps mostly CWN-1 from the posterior as in C. elegans (Gleason et al. 2006), may act jointly to promote Pn.p competence.
Overall, the major differences between C. elegans and O. tipulae for this class of mutants are: (1) Wnt pathway mutations were not found in the first vulva mutant screens in C. elegans; (2) the miscentering of the 1° fate on P5.p is much more penetrant in O. tipulae than in C. elegans (Figure 3, see Discussion); and (3) egl-20/Wnt mutations lead to a low division frequency of P8.p in O. tipulae compared to C. elegans, and a comparable or even weaker effect on P4.p: Oti-egl-20(sy464) and Cel-egl-20(n585) animals show 30 and 1% loss of division of P8.p, compared to 22 and 54% loss of division of P4.p, respectively (Dichtel et al. 2001; Myers and Greenwald 2007). Note that the Oti-egl-20(sy464) allele is a nonsense mutation in the Wnt domain while Cel-egl-20(n585) is a missense mutation, which may alternatively explain differential effects.
The hyperinduced mutations affect plexin and semaphorin genes
Much more unexpected is the identification of the loci resulting in the strongest vulva hyperinduction phenotypes. We had identified two loci, iov-2 and iov-3, with similar hyperinduction defects: by cell lineage and attachment criteria, P4.p and P8.p were transformed to a 2° fate at an intermediate penetrance of ∼50% for P4.p and 80% for P8.p in the strongest, TMP-UV alleles (Dichtel-Danjoy and Félix 2004b). The iov-3 locus turned out to correspond to the Oti-plx-1 gene, coding for a plexin (one small out-of-frame deletion as the TMP-UV allele mf78 and two missense alleles). The iov-2(mf76) mutant shows a 634-bp deletion in the Oti-smp-1 gene, coding for a semaphorin-type ligand of plexin (Figure 4A). Although this latter gene’s identification relies on a single allele at the iov-2 locus, since both the iov-2 and iov-3 loci show similar hyperinduced mutant phenotypes (Dichtel-Danjoy and Félix 2004b), this lesion makes Oti-smp-1 a very likely candidate for the causal mutation for this phenotype. This implicates a new intercellular signaling pathway in vulval cell fate patterning and induction.
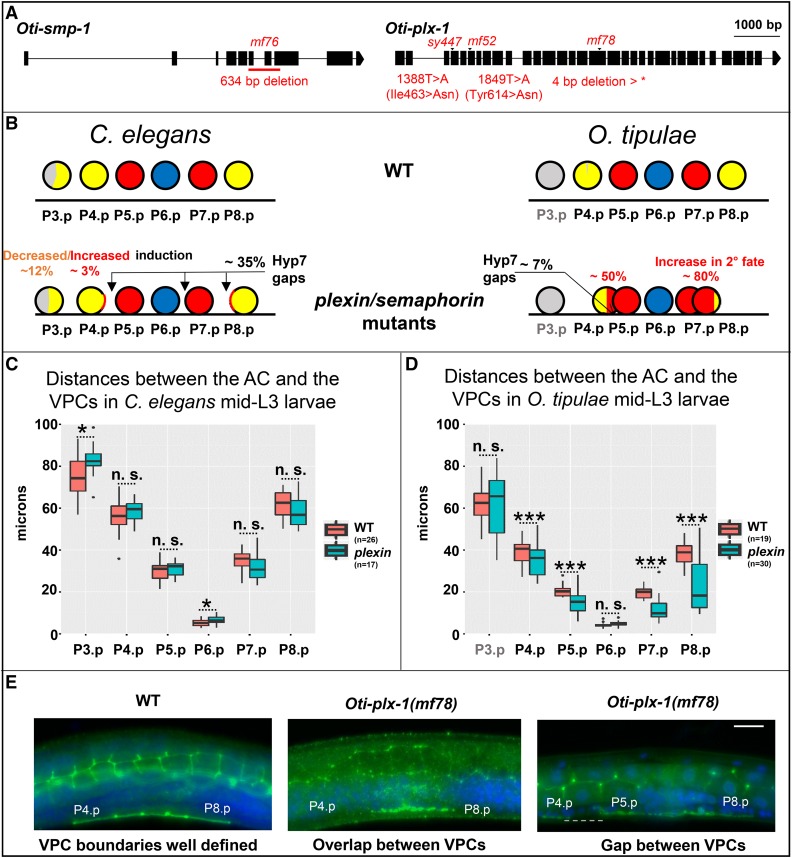
Figure 4 .
O. tipulae plexin/semaphorin mutants present defects in vulval induction and closer VPC cells. (A) Gene models of Oti-smp-1 and Oti-plx-1 with their respective mutations in O. tipulae. (B) Schematic depiction of the phenotypic effects of plexin/semaphorin mutants in C. elegans and O. tipulae on the induction and localization of Pn.p cells. Quantifications can be found in Table S4. In C. elegans, the percentages shown are the highest found along alleles, namely those in plx-1(ev724) mutants. Arrows show the most common localization of intercellular spaces (gaps) between VPCs. Each VPC diagram (circle) is colored according to the frequency of its acquired fate (yellow, red, and blue for the 3°, 2°, and 1° fates, respectively, and gray for undivided). Data from Dichtel-Danjoy and Félix (2004b). (C) Normalized distances between the AC and the VPCs in C. elegans WT and plx-1(ev724) animals at the mid-L3 stage after DU division. Only the distances between the AC and P3.p, and P6.p, are significantly larger in plx-1 mutants compared to WT (Wilcoxon rank sum test, P < 0.05). (D) Normalized distances between the AC and the VPCs in O. tipulae WT and Oti-plx-1(mf78) animals at the mid-L3 stage after DU division. Distances between each of P(4–8).p and the AC, except for P6.p, are all significantly smaller in Oti-plx-1(mf78) mutants relative to WT (Wilcoxon rank sum test, with P-values < 10−4). (E) Immunostaining of cell junctions with MH27 antibody (in green), with DAPI staining in blue. The central panel shows overlapping VPCs, while the right panel shows a rare instance of a gap (dotted line) in Oti-plx-1(mf78) animals. All the images are set to the same scale. Bar, 10 μm. Anterior is to the left in all images, and the ventral side is down. AC, anchor cell; n.s., not significant; VPC, vulval precursor cell; WT, wild-type; *: p<0.05; ***: p<0.001.
The plexin-semaphorin pathway is well known for contact-dependent growth inhibition between neurons, acting in many organisms (Kolodkin et al. 1992; Luo et al. 1993; Winberg et al. 1998). In C. elegans, mutations in smp-2/mab-20, smp-1, and plx-1 (Roy et al. 2000; Fujii et al. 2002; Ginzburg et al. 2002; Dalpé et al. 2004; Pickett et al. 2007; Nukazuka et al. 2008) were first found and most studied for their effect on the displacement of sensory organs (rays) in the male tail. Their impact on vulval formation concerns late morphogenesis events in formation of the vulval rings, events that take place after the three rounds of Pn.p divisions (Dalpé et al. 2005; Liu et al. 2005; Pellegrino et al. 2011). When examined, their effect on vulval induction is very weak (Liu et al. 2005) and mostly in the direction of hypoinduction of the 2° fate, as shown in Figure 4B and Table S4 for plx-1 mutations. The existing plx-1 alleles may not be null, as nc36 and nc37 only affect some long forms from an upstream transcriptional site, and ev724 may lead to a truncated protein with some activity (Dalpé et al. 2004). Therefore, by CRISPR and nonhomologous end-joining repair in the plx-1(ev724) background, we generated two new alleles (mf159 and mf160) corresponding to a second molecular lesion (a 26-bp deletion and an 11-bp insertion, respectively) upstream of ev724 in an exon corresponding to the semaphorin-binding domain. The plx-1(mf159ev724) and plx-1(mf160ev724) combinations are predicted to code only for incomplete and nonfunctional forms of PLX-1, independently of the transcriptional start site; we can conclude that these animals are true molecularly null mutants for plx-1. Defects in vulval induction in these animals were not stronger than those seen in plx-1(ev724) alone, and in this case did not include any hyperinduction (n = 76 and 72 for mf159ev724 and mf160ev724, respectively; Table S4). To rule out that the difference in vulval phenotypes between the two species is due to a difference in paralog usage, we quantified the vulval induction in animals bearing mutations for plx-2, the paralog of plx-1; we did not see any vulval defect in the predicted plx-2(ev773) null mutants (n = 57; Table S4) (Ikegami et al. 2004) nor in plx-2(ev773); plx-1(ev724) double mutants (n = 47; Table S4). These experiments clearly demonstrated that alterations of the plexin-semaphorin pathways in C. elegans do not result in vulval hyperinduction defects that are as strong as those in O. tipulae.
The hyperinduction of P4.p and P8.p in the O. tipulae iov-2/smp-1 and iov-3/plx-1 mutants is a transformation of 3°–2° fate. The ectopically induced cells never adopt a 1° fate; they join the main vulval invagination, and therefore the adult phenotype is a protruding vulva and not additional bumps on the cuticle, as in the C. elegans Multivulva mutants. This contrasts with the C. elegans hyperinduced mutants, which correspond to an excess of Ras pathway signaling, leading to ectopic 1° and 2° fates.
To understand why plexin and semaphorin mutations cause vulval hyperinduction in O. tipulae, we measured cell position at the time of induction, before the formation of the vulval invagination, using nuclear position as a proxy (Grimbert et al. 2016). As in C. elegans plexin and semaphorin mutants, we observed that the VPCs do not form an anteroposterior row as in wild-type animals (Dalpé et al. 2005; Liu et al. 2005; Pellegrino et al. 2011), but instead either overlap left and right of each other, or sometimes show a lack of a junction and a gap between successive cells (Figure 4, B–E). In contrast to C. elegans plx-1(ev724) mutants, gaps are rare in O. tipulae plx-1(mf78) mutants and do not often involve the three central cells. Instead, left–right overlaps occur between P4.p and P5.p, and between P7.p and P8.p. As these overlaps could alter the distance between the anchor cell and the Pn.p cells, we measured these distances and found that they were shorter in the O. tipulae plx-1(mf78) mutant, but not in the C. elegans counterpart plx-1(ev724) (Figure 4, C and D) (both alleles are deletion alleles, creating an early stop codon before the plexin domain and subjected to nonsense-mediated degradation, and thus putatively comparable null alleles). As a consequence, the VPCs tend to be closer to the anchor cell in O. tipulae, likely explaining the excess of 2° fate induction in the first induction wave.
In summary, the identification of these four different mutations points to the plexin-semaphorin signaling pathway being important for the correct induction of the VPCs, due to its effect on VPC positioning.
Discussion
The unsurprising single Vulvaless mutation in O. tipulae
In the initial C. elegans screens for vulval induction defects, most Vulvaless mutations corresponding to induction defects affected the genes lin-2, lin-7, or lin-10 (Horvitz and Sulston 1980; Ferguson and Horvitz 1985; Ferguson et al. 1987). Only rare tissue-specific reduction-of-function alleles were recovered in lin-3 and let-23, coding for EGF and the EGF receptor, respectively. Downstream factors in the EGFR-Ras/MAP kinase cascade were only subsequently obtained by suppressor or enhancer screens (Sternberg and Han 1998).
For lin-3, the first C. elegans allele to be isolated, e1417, turned out to be a base substitution affecting a cis-regulatory E-box (Hwang and Sternberg 2004). The second viable allele, n378, is a substitution in the signal peptide, showing high tissue-specificity for reasons still unknown (Liu et al. 1999). Further lin-3 alleles were obtained in noncomplementation screens or screens for lethal mutants (Ferguson and Horvitz 1985; Liu et al. 1999). In summary, besides the lin-2/lin-7/lin-10 genes, a main target for a Vulvaless mutation appeared to be the cis-regulatory element that activates lin-3 expression in the anchor cell in a tissue-specific manner. Given this, the sole Vulvaless mutation we found in mutagenesis of O. tipulae, iov-1(mf86), is a remarkably predictable hit: a deletion in a noncoding region homologous to that mutated in Cel-lin-3(e1417) (Barkoulas et al. 2016). Both screens requiring that hermaphrodites be viable and fertile, we recovered a similar (orthologous) cis-regulatory mutation in the anchor cell element: random mutagenesis ended up being as targeted as the CRISPR/Cas9 experiment that confirmed the importance of this E-box (Figure 2).
Concerning lin-2, lin-7, or lin-10, we now know that the proteins LIN-2/CASK, LIN-7/Velis, and LIN-1/Mint1 bind to the C-terminus of the LET-23/EGFR receptor and help to localize it to the basolateral membrane facing the anchor cell (Simske et al. 1996; Kaech et al. 1998). Mutations in any of these three loci have so far not been recovered in C. briggsae and P. pacificus, nor here in O. tipulae (where screens were extensive but not saturated), whereas they are frequent in C. elegans (see above). Thus, it is likely that either the loss-of-function in these genes is lethal or it does not affect the vulva. It will be interesting to delete them using reverse genetic methods such as CRISPR/Cas9-mediated genome modification.
A surprise signaling pathway found only in O. tipulae vulva mutant screens
In stark contrast, the identification of the plexin-semaphorin pathway using the hyperinduced mutations in O. tipulae was unpredictable from results in C. elegans, C. briggsae (Seetharaman et al. 2010; Sharanya et al. 2012, 2015), and P. pacificus (Jungblut and Sommer 1998, 2001; Schlager et al. 2006; Tian et al. 2008). In the case of C. elegans, the vulval fate specification errors in plexin/semaphorin mutants are indeed rare and occur at low penetrance, and in directions of both excess and loss of induction. Instead, in O. tipulae, the specification of P4.p or P8.p as a 2° fate is quite penetrant and only excess of induction is observed (Figure 4B and Table S4). The cell positioning defects in the O. tipulae plexin/semaphorin mutants explain that the hyperinduction of vulval fates is gonad-dependent (Dichtel-Danjoy and Félix 2004b). In contrast to this gonad-dependence, C. elegans hyperinduced mutants, such as lin-1, lin-13, lin-15, lin-31, and lin-34(d), retain some vulval induction upon anchor cell ablation (or in lin-3 double mutants) (Ferguson et al. 1987; Han and Sternberg 1990).
What explains the difference between C. elegans and O. tipulae in the effect of mutations in the plexin-semaphorin pathway? In both species, semaphorin and plexin appear to act via contact inhibition of the VPCs while they grow and contact each other (Liu et al. 2005) (Figure 4). We propose that two phenomena are involved in the fate specification difference that are not mutually exclusive. First, the VPCs are, on average, closer to the anchor cell in the early L3 stage in Oti-plx-1 mutants compared to the corresponding C. elegans plx-1 mutants (Figure 4, C and D; Table 2); this likely increases the exposure of P4.p and P8.p to Oti-LIN-3 from the anchor cell, hence the 2° fate. The smaller body size of O. tipulae may also play a role. Second, the 2° fate is in part induced in C. elegans by direct contact between P6.p and other VPCs through transmembrane Delta ligands. In O. tipulae, due to differences in the fate patterning mechanism, we have no evidence of lateral signaling, whereby the 1°-fated cell induces the 2° fate in its neighbors, nor of Notch pathway involvement, except maybe later through Oti-delta expression in P6.p daughters; indeed, P5.p, P6.p, and P7.p do not appear to differ from each other before their division, although this may be due to a lack of adequate markers (Félix and Sternberg 1997). Signaling from the anchor cell at a distance is thus potentially stronger in O. tipulae than in C. elegans.
Table 2. Quantification of large interspaces (gaps) between VPCs in C. elegans and O. tipulae plexin mutants, as determined by MH27 staining.
| Species | Strain | Phenotype | Number of animals |
|---|---|---|---|
| C. elegans | N2 | WT | > 50 |
| C. elegans | ST54: plx-1(nc37) | WT | 37 |
| Gap P7.p–P8.p | 13 | ||
| Gap P6.p–P7.p | 6 | ||
| Gaps P4.p–P5.p and P6.p–P7.p | 1 | ||
| O. tipulae | CEW1 | WT | > 30 |
| O. tipulae | JU108: Oti-plx-1(mf78) | WT | 35 |
| Gap P4.p–P5.p | 2 | ||
| Gaps P4.p–P5.p and P7.p–P8.p | 1 |
“Gap P7.p–P8.p” refers to a gap between the P7.p and P8.p cells. WT, wild-type.
In C. elegans, VPCs are attracted toward the anchor cell in response to LIN-3 signaling, thus creating positive feedback whereby the most induced cell moves closest to the anchor cell (Grimbert et al. 2016). The same feedback may be at stake for the 2° cells, but we never observed an excess of 1°-fated cells in O. tipulae. This correlates with the fact that we do not observe other VPCs overlapping with P6.p nor contacting the anchor cell in the plexin/semaphorin mutants. It is thus possible that lateral inhibition from P6.p to its neighbors takes place in these mutants, preventing the positioning of two VPCs below the anchor cell.
In sum, the vulval hyperinduction of plexin-semaphorin pathway mutants in O. tipulae is a secondary effect of VPC mispositioning, which is an unexpected finding as it does not exist in C. elegans. As the effect of the semaphorin pathway is on cell position, not induction per se, the 2° fate must be induced by another signaling pathway.
Wnt and EGF pathways act jointly in vulval competence and induction
We find that O. tipulae Wnt pathway mutants affect Pn.p competence and induction (2–3° and 3° to F (fusion with hyp7 in the L2 stage) transformations, Figure 3A), and result in centering of the 1° fate on P5.p. The initial genetic screens for C. elegans vulva mutants did not identify the Wnt pathway. The corresponding mutants were found later by specifically screening for mutants that had a variably expressed protruding vulva phenotype (Eisenmann et al. 1998; Eisenmann and Kim 2000). It would be tempting to conclude that there is a difference in Wnt pathway involvement in O. tipulae compared to C. elegans vulva induction. However, we propose that the difference is subtle.
In C. elegans, the Wnt pathway is mostly known to maintain vulval precursor competence to receive the LIN-3 signal in the L2 and L3 stages (Eisenmann et al. 1998). In the absence of Wnts, the Pn.p cells adopt an F fate instead of the 3° fate (one division in the L3 stage before fusion to hyp7) (Gleason et al. 2006). This prevents them from being induced to a vulval fate. In other words, the Wnt signaling pathway establishes competence (F to 3° fate transformation) for the next round of signaling (EGF, which induces 1° and 2° fates). Yet the inductions by Wnt and EGF in C. elegans are partially intermingled. Indeed, the Wnt pathway also participates to the induction of 2° vulval fates vs. the 3° fate (Eisenmann et al. 1998; Gleason et al. 2002; Braendle and Félix 2008; Milloz et al. 2008; Seetharaman et al. 2010). Conversely, the LIN-3/EGF pathway participates in “competence maintenance” (F vs. 3°) (Myers and Greenwald 2007). Thus, both pathways appear to jointly act in C. elegans to promote both competence (a very first induction) and 2° vulval fate induction.
The same holds true in O. tipulae, with quantitative variations in mutant phenotypes. In the Oti-lin-3(mf86) mutant, the 1° fate is abolished while the 2° fate is reduced. The intermediate level of 2° fate may be due to some remaining Oti-lin-3 gene expression (Figure 2C). Alternatively, another signal, such as Wnts, may participate in 2° fate induction. Accordingly, a double mutant between the EGF and Wnt pathways, Oti-mom-5(sy493); lin-3(mf86), abolishes induction, as in C. elegans (Eisenmann et al. 1998; Braendle and Félix 2008) (Figure 3A). Thus, we conclude that despite quantitative differences in mutant penetrance, the joint involvement of the Wnt and EGF pathways in the induction of vulval fates appears similar in C. elegans and O. tipulae.
This joint induction by Wnts and LIN-3 differs from the situation described in an outgroup nematode, P. pacificus (Kiontke et al. 2007). In this species, similar to O. tipulae and unlike C. elegans, the induction of vulval fates by the gonad occurs gradually before and after Pn.p divisions (2° then 1°) (Sigrist and Sommer 1999; Kiontke et al. 2007). However, in contrast to both O. tipulae and C. elegans, no EGF/Ras pathway component appears to be involved and the Wnt pathway is considered to be the only inducer from the gonad (Tian et al. 2008). This induction by the gonad starts before the anchor cell is born (Sigrist and Sommer 1999) and the Wnt ligand secreted by the gonad appears to be Ppa-LIN-44, based on in situ hybridization in the uterine precursor region (Tian et al. 2008) (no Ppa-lin-44 mutant is available). Since we have now shown that the expression of lin-44 orthologs in O. tipulae and C. elegans occurs in the sex myoblasts and not the uterine precursors (Figure 3, Figure S3, and Figure S6), it will be good to clarify the site of expression of Ppa-lin-44 (the sex myoblasts are located on either side of the uterus, and fixation for in situ hybridization followed by alkaline phosphatase reaction may render the distinction between uterus and sex myoblast expression difficult). This is important, as Ppa-LIN-44 can represent the vulval induction signal as proposed only if it is expressed in the gonad precursors ablated in Sigrist and Sommer (1999). Indeed, the only other Wnt ligand expressed in the P. pacificus gonad is Ppa-mom-2, but its expression in the anchor cell appears to start much later than when the induction of 2° fates begins (Sigrist and Sommer 1999; Kiontke et al. 2007; Tian et al. 2008).
Another argument in Tian et al. (2008) regarding the Wnt pathway being the main vulva-inducing pathway in P. pacificus comes from the high penetrance of the Ppa-bar-1/armadillo(0) mutant compared to its C. elegans counterpart (Tian et al. 2008). However, only two β-catenins were found in P. pacificus, as there are no wrm-1 or sys-1 orthologs (Tian et al. 2008). The null mutation in the armadillo homolog gene that is called Ppa-bar-1 was obtained by a targeted reverse genetic approach and is maternal-effect lethal, unlike in C. elegans. In C. elegans, wrm-1; bar-1 double mutants were noted by Green et al. (2008) to be fully Vulvaless. Thus, the strong vulval phenotype of the Ppa-bar-1 mutant could in part be explained by less armadillo gene redundancy in P. pacificus than in C. elegans and O. tipulae. Upstream of these armadillo paralogs, in P. pacificus as in C. elegans, the multiple Wnt ligands and receptors are partially redundant (Gleason et al. 2006; Tian et al. 2008). In C. elegans, triple mutants in Wnts have a strong vulval phenotype, including loss of division characteristic of the 3° fate in C. elegans. Note that there is no equivalent to the 3° fate in P. pacificus; on the anterior side, the noncompetent cells die by apoptosis, and on the posterior side, P8.p is competent early on to replace P(5–7).p then fuses to hyp7 without division after the onset of vulval induction, which occurs earlier than in C. elegans compared to larval molts (Sommer 1997; Sigrist and Sommer 1999; Jungblut and Sommer 2000).
In sum, there is so far no indication that the induction by LIN-3 seen in O. tipulae and C. elegans is present in the outgroup P. pacificus. Further work is needed to substantiate this result and, most interestingly, systematic phylogenetic analysis with more outgroup species is required to polarize the character.
The Wnt pathway is required for correct centering of the vulval pattern
The clearest difference of Wnt pathway mutant phenotypes between C. elegans and O. tipulae lies in the centering of the 1° fate on P5.p, and the likely correlated higher penetrance of the F fate in P7.p. In C. elegans, only a small percentage of Wnt pathway mutant animals display P5.p centering, which has been shown to reflect the posterior displacement of P6.p compared to the anchor cell and higher variance in cell positions (Milloz et al. 2008; Grimbert et al. 2016). In Oti-mom-5 animals, a strong shift in anchor cell position relative to P6.p and P5.p in the L2 stage has also been observed (Louvet-Vallée et al. 2003). Quantitative differences between the various phenotypes in the two species likely correspond to the extent of cell displacement.
The phenotypes of Oti-mom-5/cov-4 and Oti-mig-14/cov-5 mutants closely resemble that of Oti-mig-13/cov-3, the homolog of mammalian low-density lipoprotein receptor-related protein 12 identified by Besnard et al. (2017). These three loci were grouped in the same O. tipulae phenotypic subclass in Louvet-Vallée et al. (2003), corresponding to a loss of VPC competence and miscentering of the 1° fate on P5.p. In C. elegans, mig-13 mutations were first identified as affecting the anterior migration of QR neuroblasts (Sym et al. 1999), a process that is also affected at many steps by Wnt pathway mutations including mom-5 and a noncanonical Wnt pathway for the long-range migration (Rella et al. 2016). mig-13 acts cell autonomously in the migrating QR neuroblast lineage, where its expression is under direct positive regulation by the lin-39 Hox gene (Wang et al. 2013). The MIG-13 protein is located at the plasma membrane of migrating QR neuroblasts, and is required for coronin and actin cytoskeleton polarization (Wang et al. 2013). We previously showed that C. elegans mig-13 mutants show a mild phenotype of VPC competence loss and miscentering of the 1° fate on P5.p (Besnard et al. 2017). The relationship between the Wnt pathway and MIG-13 in vulval development remains unclear, but it is tempting to speculate that, as for QR neuroblasts, they may jointly affect VPC polarity, growth, and movement via the actin cytoskeleton (Wang et al. 2013; Grimbert et al. 2016).
Conclusions and future directions
This study reveals the two faces of genetic screens: necessity and contingency. On the side of necessity, Cel-lin-3(e1417) and Oti-lin-3(mf86) were obtained among the first mutations defective in anchor cell induction of the vulva, both reducing the function of a tissue-specific cis-regulatory element of the inductive signal. This convergent result exemplifies the power and near predictability of genetic screens in targeting mutations in a lethal pathway in a tissue-specific manner. On the side of contingency, the mutations causing hyperinduction of the vulva hit contrasting molecular pathways in C. elegans vs. O. tipulae, contingent on particular features of the genetic architecture and anatomy of each species. For example, in C. elegans, initial discovery of lin-12/Notch involvement relied on rare gain-of-function mutations (Greenwald et al. 1983), and the finding of the Synthetic Multivulva genetic pathway hinged on the lin-15 operon and the chance double hit of the lin-8; lin-9 mutant (Ferguson and Horvitz 1985, 1989; Clark et al. 1994; Huang et al. 1994; Saffer et al. 2011). In O. tipulae, those same pathways could also be important but less easily targeted in vulval screens (which are not yet saturated), while differences in cell positioning may explain the stronger requirement for the plexin-semaphorin signaling pathway.
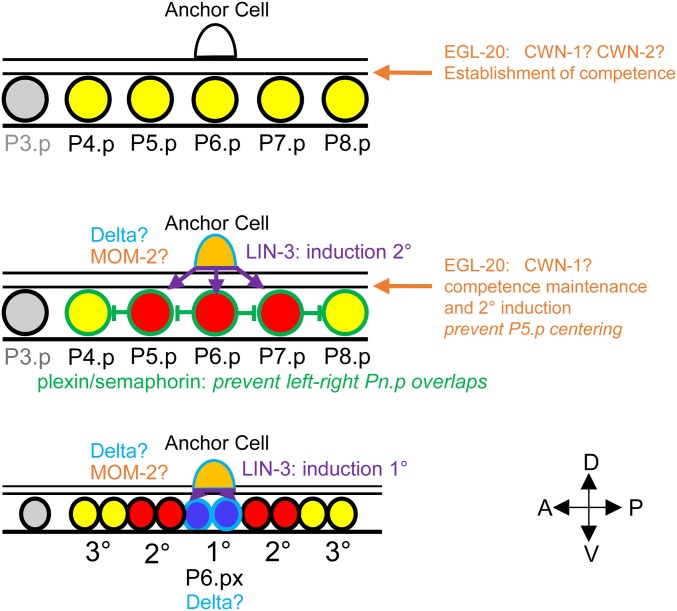
Figure 5 presents our current model of the vulval cell fate patterning mechanism in O. tipulae. Oti-LIN-3 produced by the anchor cell is important for the induction of 2° then of 1° fates. Oti-LIN-3 is thus likely the inductive signal for both steps of induction, as defined in Félix and Sternberg (1997). The 1° fate induction appears to always occur upon contact with the anchor cell, which may represent a requirement for a transmembrane ligand or simply a high concentration of the ligand. Thus, although the cell fate patterning mechanism unraveled through anchor cell ablation is derived in C. elegans compared to many other nematode species (Kiontke et al. 2007), the involvement of LIN-3 in vulval induction appears to be older.
Figure 5.
Expression of signaling molecules and vulval cell fate patterning in O. tipulae. The VPCs are color-coded according to their fate as in previous figures. Their boundary is color-coded according to the signaling molecules that they express (at least as mRNAs): LIN-3 in purple, Wnts in orange, plexin in green, and Delta in light blue. A question mark indicates that the effect of removing this signal is not known. Note that in addition, the sex myoblasts left and right of the AC express lin-44/Wnt. 1°, primary; 2°, secondary; 3°, tertiary; A, anterior; AC, anchor cell; D, dorsal; P, posterior; V, ventral; VPC, vulval precursor cell.
Our findings on the effects of both the Wnt and semaphorin pathways on VPC positioning relative to the anchor cell emphasize the importance of cell positioning in vulval cell fate patterning, since gradients of signaling molecules (EGF and Wnt) are involved. The VPC positioning defect may link the Wnt and MIG-13 pathways to cell polarity, growth, movement, and the actin cytoskeleton (Wang et al. 2013; Grimbert et al. 2016).
Finally, a future direction concerns Notch pathway involvement. It may not be surprising that mutations in this pathway were not found in the screens as they are sterile or lethal in C. elegans. In P. pacificus, the Notch pathway appears relevant to intraspecific evolution (Kienle and Sommer 2013), but no mutant was found or produced by reverse genetics. However, we do not rule out their possible involvement, in light of dsl expression in P6.p daughters and the role of apx-1 in intraspecific evolution in P. pacificus (Kienle and Sommer 2013).
Acknowledgments
We thank Aurélien Richaud for performing a smFISH experiment; Joao Picao Osorio for comments on the manuscript; Michalis Barkoulas for his advice on Oscheius smFISH; Mark Blaxter’s laboratory for their continuous support of nematode genomics as well as for maintaining the Caenorhabditis website (http://www.caenorhabditis.org/); Benjamin Podbilewicz for advice on the immunofluorescence experiments; Sana Dieudonné, Aurélien Richaud, and Clément Dubois for advice and help in the use of the mapping-by-sequencing technique; the Associate Editor Meera Sundaram and the anonymous reviewers for helpful input; and WormBase. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). This work was funded by grants from the Agence Nationale de la Recherche (ANR12-BSV2-0004-01 and ANR10-LABX-54 MEMOLIFE) and Paris Sciences et Lettres (Vuldemorph). We also acknowledge the support of the Bettencourt Schueller Foundation (Coup d’Elan 2011) and the support of the Human Frontier Science Program (RGP030/2016).
Author contributions: M.-A.F., F.B., and A.M.V.-V. designed the experiments. M.-A.F., and F.B. performed crosses, isolated DNA, and analyzed the sequences, with input from A.M.V.-V. F.B., M.-A.F., and A.M.V.-V. identified O. tipulae gene homologs. A.M.V.-V. performed the smFISH, CRISPR, and DIC analyses. A.M.V.-V. and M.-A.F. wrote the manuscript, with input from F.B.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7624559.
Communicating editor: M. Sundaram
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bänziger C., Soldini D., Schutt C., Zipperlen P., Hausmann G., et al. , 2006. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125: 509–522. 10.1016/j.cell.2006.02.049 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., van Zon J. S., Milloz J., van Oudenaarden A., Félix M. A., 2013. Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Dev. Cell 24: 64–75. 10.1016/j.devcel.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Vargas Velazquez A. M., Peluffo A. E., Félix M. A., 2016. Evolution of new cis-regulatory motifs required for cell-specific gene expression in Caenorhabditis. PLoS Genet. 12: e1006278 10.1371/journal.pgen.1006278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F., Koutsovoulos G., Dieudonné S., Blaxter M., Félix M.-A., 2017. Toward universal forward genetics: using a draft genome sequence of the nematode Oscheius tipulae to identify mutations affecting vulva development. Genetics 206: 1747–1761. 10.1534/genetics.117.203521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C., Félix M.-A., 2008. Plasticity and errors of a robust developmental system in different environments. Dev. Cell 15: 714–724. 10.1016/j.devcel.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. G., Lu X., Horvitz H. R., 1994. The C. elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpé G., Zhang L. W., Zheng H., Culotti J. G., 2004. Conversion of cell movement responses to Semaphorin-1 and Plexin-1 from attraction to repulsion by lowered levels of specific RAC GTPases in C. elegans. Development 131: 2073–2088. 10.1242/dev.01063 [DOI] [PubMed] [Google Scholar]
- Dalpé G., Brown L., Culotti J. G., 2005. Vulva morphogenesis involves attraction of plexin 1-expressing primordial vulva cells to semaphorin 1a sequentially expressed at the vulva midline. Development 132: 1387–1400. 10.1242/dev.01694 [DOI] [PubMed] [Google Scholar]
- Delattre M., Félix M.-A., 2001. Polymorphism and evolution of vulval precursor cell lineages within two nematode genera, Caenorhabditis and Oscheius. Curr. Biol. 11: 631–643. 10.1016/S0960-9822(01)00202-0 [DOI] [PubMed] [Google Scholar]
- Dichtel M.-L., Louvet-Vallée S., Viney M. E., Félix M.-A., Sternberg P. W., 2001. Control of vulval cell division number in the nematode Oscheius/Dolichorhabditis sp. CEW1. Genetics 157: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy M.-L., Félix M.-A., 2004a. Phenotypic neighborhood and micro-evolvability. TIG 20: 268–276. 10.1016/j.tig.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Dichtel-Danjoy M.-L., Félix M.-A., 2004b. The two steps of vulval induction in Oscheius tipulae CEW1 recruit common regulators including a MEK kinase. Dev. Biol. 265: 113–126. 10.1016/j.ydbio.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Kim S. K., 2000. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans development. Genetics 156: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Maloof J. N., Simske J. S., Kenyon C., Kim S. K., 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125: 3667–3680. [DOI] [PubMed] [Google Scholar]
- Félix M.-A., 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 17: 103–114. 10.1016/j.cub.2006.12.024 [DOI] [PubMed] [Google Scholar]
- Félix M.-A., 2012. Caenorhabditis elegans vulval cell fate patterning. Phys. Biol. 9: 045001 10.1088/1478-3975/9/4/045001 [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Barkoulas M., 2012. Robustness and flexibility in nematode vulva development. TIG 28: 185–195. 10.1016/j.tig.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Félix M. A., Duveau F., 2012. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10: 59 10.1186/1741-7007-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Sternberg P. W., 1997. Two nested gonadal inductions of the vulva in nematodes. Development 124: 253–259. [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Sternberg P. W., 1998. A gonad-derived survival signal for vulva precursor cells in two nematode species. Curr. Biol. 8: 287–290. 10.1016/S0960-9822(98)70111-3 [DOI] [PubMed] [Google Scholar]
- Félix M.-A., De Ley P., Sommer R. J., Frisse L., Nadler S. A., et al. , 2000a. Evolution of vulva development in the Cephalobina (Nematoda). Dev. Biol. 221: 68–86. 10.1006/dbio.2000.9665 [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Delattre M., Dichtel M.-L., 2000b. Comparative developmental studies using Oscheius/Dolichorhabditis sp. CEW1 (Rhabditidae). Nematology 2: 89–98. 10.1163/156854100508809 [DOI] [Google Scholar]
- Ferguson E., Horvitz H. R., 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1989. The Multivulva phenotype of certain C. elegans mutants results from defects in two functionally-redundant pathways. Genetics 123: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Sternberg P. W., Horvitz H. R., 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326: 259–267 (erratum: Nature 327:82). 10.1038/326259a0 [DOI] [PubMed] [Google Scholar]
- Francis R., Waterston R. H., 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114: 465–479. 10.1083/jcb.114.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Nakao F., Shibata Y., Shioi G., Kodama E., et al. , 2002. Caenorhabditis elegans PlexinA, PLX-1, interacts with transmembrane semaphorins and regulates epidermal morphogenesis. Development 129: 2053–2063. [DOI] [PubMed] [Google Scholar]
- Ginzburg V. E., Roy P. J., Culotti J. G., 2002. Semaphorin 1a and semaphorin 1b are required for correct epidermal cell positioning and adhesion during morphogenesis in C. elegans. Development 129: 2065–2078. [DOI] [PubMed] [Google Scholar]
- Gish W., States D. J., 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3: 266–272. 10.1038/ng0393-266 [DOI] [PubMed] [Google Scholar]
- Gleason J. E., Korswagen H. C., Eisenmann D. M., 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. 10.1101/gad.981602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E., Szyleyko E. A., Eisenmann D. M., 2006. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev. Biol. 298: 442–457. 10.1016/j.ydbio.2006.06.050 [DOI] [PubMed] [Google Scholar]
- Green J. L., Inoue T., Sternberg P. W., 2008. Opposing Wnt pathways orient cell polarity during organogenesis. Cell 134: 646–656. 10.1016/j.cell.2008.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R., 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444. 10.1016/0092-8674(83)90377-X [DOI] [PubMed] [Google Scholar]
- Grimbert S., Tietze K., Barkoulas M., Sternberg P. W., Félix M.-A., et al. , 2016. Anchor cell signaling and vulval precursor cell positioning establish a reproducible spatial context during C. elegans vulval induction. Dev. Biol. 416: 123–135. 10.1016/j.ydbio.2016.05.036 [DOI] [PubMed] [Google Scholar]
- Han M., Sternberg P. W., 1990. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63: 921–931. 10.1016/0092-8674(90)90495-Z [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Vaz Gomes A., Kenyon C., Liu J., Krause M., et al. , 1998. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12: 2623–2635. 10.1101/gad.12.16.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M., Kim D. H., Middelkoop T. C., Doan T. D., van Oudenaarden A., et al. , 2011. Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138: 2915–2924. 10.1242/dev.064733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Tzou P., Sternberg P. W., 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5: 395–411. 10.1091/mbc.5.4.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. J., Sternberg P. W., 2004. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development 131: 143–151. 10.1242/dev.00924 [DOI] [PubMed] [Google Scholar]
- Ikegami R., Zheng H., Ong S. H., Culotti J., 2004. Integration of semaphorin-2A/MAB-20, ephrin-4, and UNC-129 TGF-beta signaling pathways regulates sorting of distinct sensory rays in C. elegans. Dev. Cell 6: 383–395. 10.1016/S1534-5807(04)00057-7 [DOI] [PubMed] [Google Scholar]
- Inoue T., Oz H. S., Wiland D., Gharib S., Deshpande R., et al. , 2004. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell 118: 795–806. 10.1016/j.cell.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Ji, N., and A. van Oudenaarden, 2012 Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos (December 13, 2012), WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org. 10.1895/wormbook.1.153.1 [DOI] [PMC free article] [PubMed]
- Jungblut B., Sommer R. J., 1998. The Pristionchus pacificus mab-5 gene is involved in the regulation of ventral epidermal cell fates. Curr. Biol. 8: 775–778. 10.1016/S0960-9822(98)70301-X [DOI] [PubMed] [Google Scholar]
- Jungblut B., Sommer R. J., 2000. Novel cell-cell interactions during vulva development in Pristionchus pacificus. Development 127: 3295–3303. [DOI] [PubMed] [Google Scholar]
- Jungblut B., Sommer R. J., 2001. The nematode even-skipped homolog vab-7 regulates gonad and vulva position in Pristionchus pacificus. Development 128: 253–261. [DOI] [PubMed] [Google Scholar]
- Kaech S. M., Whitfield C. W., Kim S. K., 1998. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94: 761–771. 10.1016/S0092-8674(00)81735-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W. S., Hill R. J., Clandinin T. R., Sternberg P. W., 1995. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82: 297–307. 10.1016/0092-8674(95)90317-8 [DOI] [PubMed] [Google Scholar]
- Kienle S., Sommer R. J., 2013. Cryptic variation in vulva development by cis-regulatory evolution of a HAIRY-binding site. Nat. Commun. 4: 1714 10.1038/ncomms2711 [DOI] [PubMed] [Google Scholar]
- Kimble J., 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 87: 286–300. 10.1016/0012-1606(81)90152-4 [DOI] [PubMed] [Google Scholar]
- Kiontke K., Barrière A., Kolotuev I., Podbilewicz B., Sommer R. J., et al. , 2007. Trends, stasis and drift in the evolution of nematode vulva development. Curr. Biol. 17: 1925–1937. 10.1016/j.cub.2007.10.061 [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Matthes D. J., O’Connor T. P., Patel N. H., Admon A., et al. , 1992. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron 9: 831–845. 10.1016/0896-6273(92)90237-8 [DOI] [PubMed] [Google Scholar]
- Kolotuev I., Podbilewicz B., 2004. Pristionchus pacificus vulva formation: polarized division, cell migration, cell fusion, and evolution of invagination. Dev. Biol. 266: 322–333. 10.1016/j.ydbio.2003.10.029 [DOI] [PubMed] [Google Scholar]
- Kolotuev I., Podbilewicz B., 2008. Changing of the cell division axes drives vulva evolution in nematodes. Dev. Biol. 313: 142–154. 10.1016/j.ydbio.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tzou P., Hill R. J., Sternberg P. W., 1999. Structural requirements for the tissue-specific and tissue-general functions of the C. elegans epidermal growth factor LIN-3. Genetics 153: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Fujii T., Nukazuka A., Kurokawa R., Suzuki M., et al. , 2005. C. elegans PlexinA PLX-1 mediates a cell contact-dependent stop signal in vulval precursor cells. Dev. Biol. 282: 138–151. 10.1016/j.ydbio.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Louvet-Vallée S., Kolotuev I., Podbilewicz B., Félix M.-A., 2003. Control of vulval competence and centering in the nematode Oscheius sp. 1 CEW1. Genetics 163: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Raible D., Raper J. A., 1993. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 75: 217–227. 10.1016/0092-8674(93)80064-L [DOI] [PubMed] [Google Scholar]
- Milloz J., Duveau F., Nuez I., Félix M.-A., 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 22: 3064–3075. 10.1101/gad.495308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I., Stephen G., Bayer M., Cock P. J., Pritchard L., et al. , 2013. Using tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 14: 193–202. 10.1093/bib/bbs012 [DOI] [PubMed] [Google Scholar]
- Minor P. J., He T. F., Sohn C. H., Asthagiri A. R., Sternberg P. W., 2013. FGF signaling regulates Wnt ligand expression to control vulval cell lineage polarity in C. elegans. Development 140: 3882–3891. 10.1242/dev.095687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers T. R., Greenwald I., 2007. Wnt signal from multiple tissues and lin-3/EGF signal from the gonad maintain vulval precursor cell competence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 20368–20373. 10.1073/pnas.0709989104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukazuka A., Fujisawa H., Inada T., Oda Y., Takagi S., 2008. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2alpha in Caenorhabditis elegans. Genes Dev. 22: 1025–1036. 10.1101/gad.1644008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M. W., Farooqui S., Frohli E., Rehrauer H., Kaeser-Pebernard S., et al. , 2011. LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development 138: 4649–4660. 10.1242/dev.071951 [DOI] [PubMed] [Google Scholar]
- Pénigault J.-B., Félix M.-A., 2011a. Evolution of a system sensitive to stochastic noise: P3.p cell fate in Caenorhabditis. Dev. Biol. 357: 419–427. 10.1016/j.ydbio.2011.05.675 [DOI] [PubMed] [Google Scholar]
- Pénigault J.-B., Félix M.-A., 2011b. High sensitivity of C. elegans vulval precursor cells to the dose of posterior Wnts. Dev. Biol. 357: 428–438. 10.1016/j.ydbio.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Pickett C. L., Breen K. T., Ayer D. E., 2007. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev. Biol. 310: 226–239. 10.1016/j.ydbio.2007.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S., van Oudenaarden A., Tyagi S., 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5: 877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rella L., Fernandes Povoa E. E., Korswagen H. C., 2016. The Caenorhabditis elegans Q neuroblasts: a powerful system to study cell migration at single-cell resolution in vivo. Genesis 54: 198–211. 10.1002/dvg.22931 [DOI] [PubMed] [Google Scholar]
- Rocheleau C. E., Downs W. D., Lin R., Wittman C., Bei Y., et al. , 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90: 707–716. 10.1016/S0092-8674(00)80531-0 [DOI] [PubMed] [Google Scholar]
- Roy P. J., Zheng H., Warren C. E., Culotti J. G., 2000. mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development 127: 755–767. [DOI] [PubMed] [Google Scholar]
- Saffer A. M., Kim D. H., van Oudenaarden A., Horvitz H. R., 2011. The Caenorhabditis elegans synthetic Multivulva genes prevent Ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF. PLoS Genet. 7: e1002418 10.1371/journal.pgen.1002418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schlager B., Roseler W., Zheng M., Gutierrez A., Sommer R. J., 2006. HAIRY-like transcription factors and the evolution of the nematode vulva equivalence group. Curr. Biol. 16: 1386–1394. 10.1016/j.cub.2006.06.058 [DOI] [PubMed] [Google Scholar]
- Seetharaman A., Cumbo P., Bojanala N., Gupta B. P., 2010. Conserved mechanism of Wnt signaling function in the specification of vulval precursor fates in C. elegans and C. briggsae. Dev. Biol. 346: 128–139. 10.1016/j.ydbio.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Sharanya D., Thillainathan B., Marri S., Bojanala N., Taylor J., et al. , 2012. Genetic control of vulval development in Caenorhabditis briggsae. G3 (Bethesda) 2: 1625–1641. 10.1534/g3.112.004598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanya D., Fillis C. J., Kim J., Zitnik E. M., Jr., Ward K. A., et al. , 2015. Mutations in Caenorhabditis briggsae identify new genes important for limiting the response to EGF signaling during vulval development. Evol. Dev. 17: 34–48. 10.1111/ede.12105 [DOI] [PubMed] [Google Scholar]
- Sigrist C. B., Sommer R. J., 1999. Vulva formation in Pristionchus pacificus relies on continuous gonadal induction. Dev. Genes Evol. 209: 451–459. 10.1007/s004270050278 [DOI] [PubMed] [Google Scholar]
- Simske J. S., Kaech S. M., Harp S. A., Kim S. K., 1996. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85: 195–204. 10.1016/S0092-8674(00)81096-X [DOI] [PubMed] [Google Scholar]
- Sommer R. J., 1997. Evolutionary changes of developmental mechanisms in the absence of cell lineage alterations during vulva formation in the Diplogastridae (Nematoda). Development 124: 243–251. [DOI] [PubMed] [Google Scholar]
- Sommer, R. J., 2005 Evolution of development in nematodes related to C. elegans (December 14, 2005), WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org. 10.1895/wormbook.1.46.1 [DOI] [PMC free article] [PubMed]
- Sommer, R. J., 2006 Pristionchus pacificus (August 14, 2006), WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org. 10.1895/wormbook.1.102.1 [DOI] [PMC free article] [PubMed]
- Sommer R. J., Sternberg P. W., 1994. Changes of induction and competence during the evolution of vulva development in nematodes. Science 265: 114–118. 10.1126/science.8016644 [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Sternberg P. W., 1996. Apoptosis and change of competence limit the size of the vulva equivalence group in Pristionchus pacificus: a genetic analysis. Curr. Biol. 6: 52–59. 10.1016/S0960-9822(02)00421-9 [DOI] [PubMed] [Google Scholar]
- Song S., Zhang B., Sun H., Li X., Xiang Y., et al. , 2010. A Wnt-Frz/Ror-Dsh pathway regulates neurite outgrowth in Caenorhabditis elegans. PLoS Genet. 6: e1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P. W., 2005 Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community, http://www.wormbook.org. 10.1895/wormbook.1.6.1 [DOI] [PMC free article] [PubMed]
- Sternberg P. W., Han M., 1998. Genetics of RAS signaling in C. elegans. TIG 14: 466–472. 10.1016/S0168-9525(98)01592-3 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1982. Postembryonic nongonadal cell lineages of the nematode Panagrellus redivivus: description and comparison with those of Caenorhabditis elegans. Dev. Biol. 93: 181–205. 10.1016/0012-1606(82)90251-2 [DOI] [PubMed] [Google Scholar]
- Sulston J., Horvitz H. R., 1977. Postembryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., White J. G., 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78: 577–597. 10.1016/0012-1606(80)90353-X [DOI] [PubMed] [Google Scholar]
- Sym M., Robinson N., Kenyon C., 1999. MIG-13 positions migrating cells along the anteroposterior body axis of C. elegans. Cell 98: 25–36. 10.1016/S0092-8674(00)80603-0 [DOI] [PubMed] [Google Scholar]
- Tian H., Schlager B., Xiao H., Sommer R. J., 2008. Wnt signaling induces vulva development in the nematode Pristionchus pacificus. Curr. Biol. 18: 142–146. 10.1016/j.cub.2007.12.048 [DOI] [PubMed] [Google Scholar]
- True J. R., Haag E. S., 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119. 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- Tuli, M. A., A. Daul, and T. Schedl, 2018 Caenorhabditis nomenclature (August 8, 2018), WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org. 10.1895/wormbook.1.183.1 [DOI] [PMC free article] [PubMed]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhou F., Lv S., Yi P., Zhu Z., et al. , 2013. Transmembrane protein MIG-13 links the Wnt signaling and Hox genes to the cell polarity in neuronal migration. Proc. Natl. Acad. Sci. USA 110: 11175–11180. 10.1073/pnas.1301849110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg M. L., Noordermeer J. N., Tamagnone L., Comoglio P. M., Spriggs M. K., et al. , 1998. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95: 903–916. 10.1016/S0092-8674(00)81715-8 [DOI] [PubMed] [Google Scholar]
- Yang P. T., Lorenowicz M. J., Silhankova M., Coudreuse D. Y., Betist M. C., et al. , 2008. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev. Cell 14: 140–147. 10.1016/j.devcel.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental Tables are available through the Figshare portal (https://doi.org/10.25386/genetics.7624559):
Table S1. List of strains used in this study.
Table S2. Sequences of DNA primers used in this study. Sequencing primers to verify by Sanger sequencing the mutations identified by the mapping-by-sequencing approach, and to identify the molecular lesions in additional alleles.
Table S3. Sequences of smFISH probes used in this study. The fluorophore coupled to each probe is noted at the end of the set name.
Table S4. smFISH quantifications, distance measurements, and vulval cell fates used in this study.
Code for mutant identification is available at https://github.com/fabfabBesnard/Andalusian_Mapping. Sequencing reads used for mapping-by-sequencing can be find at the Sequence Read Archive portal under the bioproject number PRJNA514975.